Abstract
The three-dimensional profile method expresses the three-dimensional structure of a protein as a table, the profile, which represents the local environment of each residue. The score of an amino acid sequence, aligned with the three-dimensional profile, reflects its compatibility with the profiled structure. In the original implementation, each local environment was characterized by its polarity, the area buried of its side chain, and its secondary structure. Here we describe a modified three-dimensional profile algorithm that characterizes the local environment in terms of the statistical preferences of the profiled residue for neighbors of specific residue types, main-chain conformations, or secondary structure. Combined profiles of the original and the three new types were tested on beta/alpha-barrel protein structures. The method identified the following enzymes of unknown three-dimensional structure as probable beta/alpha-barrels, all of which catalyze reactions in the biosynthesis of aromatic amino acids: anthranilate phosphoribosyltransferase (trpD), glutamine amidotransferase (trpG), and phosphoribosylformimino-5-aminoimidazole carboxamide ribotide isomerase (hisA).
Full text
PDF
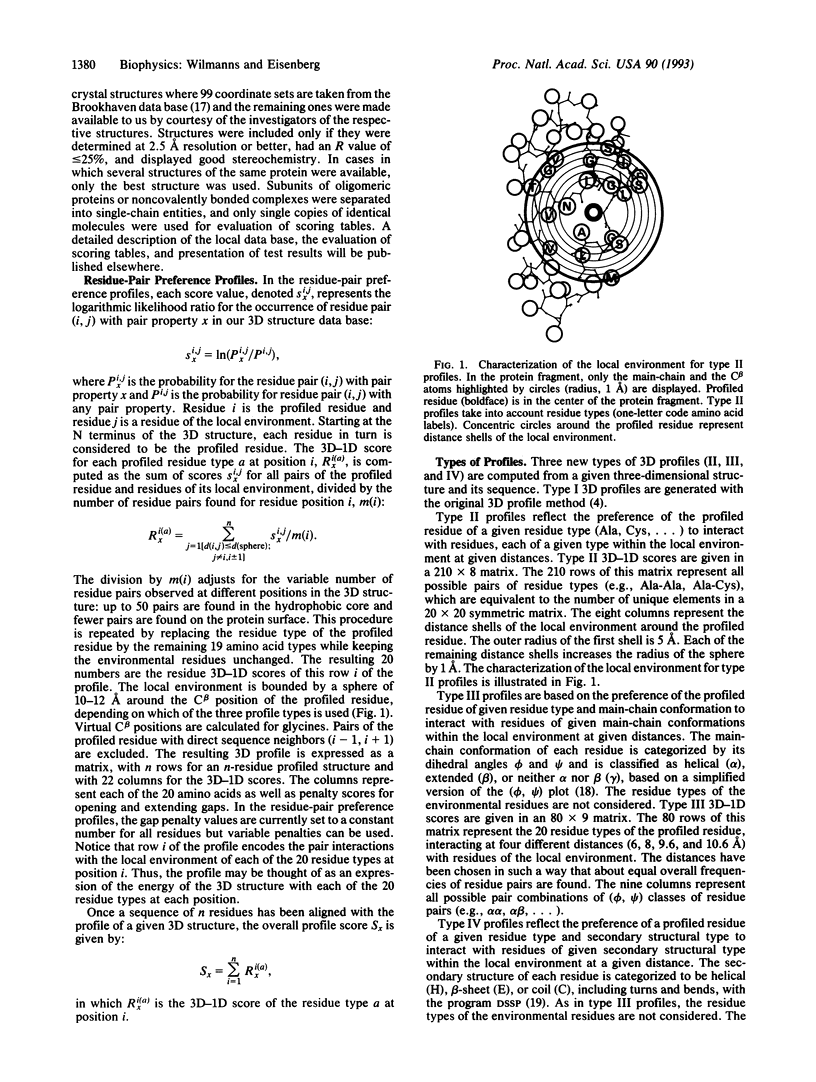



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bowie J. U., Lüthy R., Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional structure. Science. 1991 Jul 12;253(5016):164–170. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- Casari G., Sippl M. J. Structure-derived hydrophobic potential. Hydrophobic potential derived from X-ray structures of globular proteins is able to identify native folds. J Mol Biol. 1992 Apr 5;224(3):725–732. doi: 10.1016/0022-2836(92)90556-y. [DOI] [PubMed] [Google Scholar]
- Crawford I. P., Niermann T., Kirschner K. Prediction of secondary structure by evolutionary comparison: application to the alpha subunit of tryptophan synthase. Proteins. 1987;2(2):118–129. doi: 10.1002/prot.340020206. [DOI] [PubMed] [Google Scholar]
- Curmi P. M., Cascio D., Sweet R. M., Eisenberg D., Schreuder H. Crystal structure of the unactivated form of ribulose-1,5-bisphosphate carboxylase/oxygenase from tobacco refined at 2.0-A resolution. J Biol Chem. 1992 Aug 25;267(24):16980–16989. [PubMed] [Google Scholar]
- Farber G. K., Petsko G. A. The evolution of alpha/beta barrel enzymes. Trends Biochem Sci. 1990 Jun;15(6):228–234. doi: 10.1016/0968-0004(90)90035-a. [DOI] [PubMed] [Google Scholar]
- Godzik A., Kolinski A., Skolnick J. Topology fingerprint approach to the inverse protein folding problem. J Mol Biol. 1992 Sep 5;227(1):227–238. doi: 10.1016/0022-2836(92)90693-e. [DOI] [PubMed] [Google Scholar]
- Gregoret L. M., Cohen F. E. Novel method for the rapid evaluation of packing in protein structures. J Mol Biol. 1990 Feb 20;211(4):959–974. doi: 10.1016/0022-2836(90)90086-2. [DOI] [PubMed] [Google Scholar]
- Gribskov M., Lüthy R., Eisenberg D. Profile analysis. Methods Enzymol. 1990;183:146–159. doi: 10.1016/0076-6879(90)83011-w. [DOI] [PubMed] [Google Scholar]
- Gribskov M., McLachlan A. D., Eisenberg D. Profile analysis: detection of distantly related proteins. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4355–4358. doi: 10.1073/pnas.84.13.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendlich M., Lackner P., Weitckus S., Floeckner H., Froschauer R., Gottsbacher K., Casari G., Sippl M. J. Identification of native protein folds amongst a large number of incorrect models. The calculation of low energy conformations from potentials of mean force. J Mol Biol. 1990 Nov 5;216(1):167–180. doi: 10.1016/S0022-2836(05)80068-3. [DOI] [PubMed] [Google Scholar]
- Hinds D. A., Levitt M. A lattice model for protein structure prediction at low resolution. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):2536–2540. doi: 10.1073/pnas.89.7.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobohm U., Scharf M., Schneider R., Sander C. Selection of representative protein data sets. Protein Sci. 1992 Mar;1(3):409–417. doi: 10.1002/pro.5560010313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde C. C., Ahmed S. A., Padlan E. A., Miles E. W., Davies D. R. Three-dimensional structure of the tryptophan synthase alpha 2 beta 2 multienzyme complex from Salmonella typhimurium. J Biol Chem. 1988 Nov 25;263(33):17857–17871. [PubMed] [Google Scholar]
- Jones D. T., Taylor W. R., Thornton J. M. A new approach to protein fold recognition. Nature. 1992 Jul 2;358(6381):86–89. doi: 10.1038/358086a0. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983 Dec;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Knight S., Andersson I., Brändén C. I. Crystallographic analysis of ribulose 1,5-bisphosphate carboxylase from spinach at 2.4 A resolution. Subunit interactions and active site. J Mol Biol. 1990 Sep 5;215(1):113–160. doi: 10.1016/S0022-2836(05)80100-7. [DOI] [PubMed] [Google Scholar]
- Lasters I., Wodak S. J., Alard P., van Cutsem E. Structural principles of parallel beta-barrels in proteins. Proc Natl Acad Sci U S A. 1988 May;85(10):3338–3342. doi: 10.1073/pnas.85.10.3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist Y. Refined structure of spinach glycolate oxidase at 2 A resolution. J Mol Biol. 1989 Sep 5;209(1):151–166. doi: 10.1016/0022-2836(89)90178-2. [DOI] [PubMed] [Google Scholar]
- Lolis E., Petsko G. A. Crystallographic analysis of the complex between triosephosphate isomerase and 2-phosphoglycolate at 2.5-A resolution: implications for catalysis. Biochemistry. 1990 Jul 17;29(28):6619–6625. doi: 10.1021/bi00480a010. [DOI] [PubMed] [Google Scholar]
- Ozenberger B. A., Brickman T. J., McIntosh M. A. Nucleotide sequence of Escherichia coli isochorismate synthetase gene entC and evolutionary relationship of isochorismate synthetase and other chorismate-utilizing enzymes. J Bacteriol. 1989 Feb;171(2):775–783. doi: 10.1128/jb.171.2.775-783.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooman M. J., Kocher J. P., Wodak S. J. Prediction of protein backbone conformation based on seven structure assignments. Influence of local interactions. J Mol Biol. 1991 Oct 5;221(3):961–979. doi: 10.1016/0022-2836(91)80186-x. [DOI] [PubMed] [Google Scholar]
- Sippl M. J. Calculation of conformational ensembles from potentials of mean force. An approach to the knowledge-based prediction of local structures in globular proteins. J Mol Biol. 1990 Jun 20;213(4):859–883. doi: 10.1016/s0022-2836(05)80269-4. [DOI] [PubMed] [Google Scholar]
- Tanaka S., Scheraga H. A. Medium- and long-range interaction parameters between amino acids for predicting three-dimensional structures of proteins. Macromolecules. 1976 Nov-Dec;9(6):945–950. doi: 10.1021/ma60054a013. [DOI] [PubMed] [Google Scholar]
- Wierenga R. K., Noble M. E., Vriend G., Nauche S., Hol W. G. Refined 1.83 A structure of trypanosomal triosephosphate isomerase crystallized in the presence of 2.4 M-ammonium sulphate. A comparison with the structure of the trypanosomal triosephosphate isomerase-glycerol-3-phosphate complex. J Mol Biol. 1991 Aug 20;220(4):995–1015. doi: 10.1016/0022-2836(91)90368-g. [DOI] [PubMed] [Google Scholar]
- Wilmanns M., Hyde C. C., Davies D. R., Kirschner K., Jansonius J. N. Structural conservation in parallel beta/alpha-barrel enzymes that catalyze three sequential reactions in the pathway of tryptophan biosynthesis. Biochemistry. 1991 Sep 24;30(38):9161–9169. doi: 10.1021/bi00102a006. [DOI] [PubMed] [Google Scholar]
- Wilmanns M., Priestle J. P., Niermann T., Jansonius J. N. Three-dimensional structure of the bifunctional enzyme phosphoribosylanthranilate isomerase: indoleglycerolphosphate synthase from Escherichia coli refined at 2.0 A resolution. J Mol Biol. 1992 Jan 20;223(2):477–507. doi: 10.1016/0022-2836(92)90665-7. [DOI] [PubMed] [Google Scholar]
- Xia Z. X., Mathews F. S. Molecular structure of flavocytochrome b2 at 2.4 A resolution. J Mol Biol. 1990 Apr 20;212(4):837–863. doi: 10.1016/0022-2836(90)90240-M. [DOI] [PubMed] [Google Scholar]




