Abstract
We tested the hypothesis that alterations in the intestinal microbiota are linked with the progression of type 1 diabetes (T1D). Herein, we present results from a study performed in subjects with islet autoimmunity living in the U.S. High-throughput sequencing of bacterial 16S rRNA genes and adjustment for sex, age, autoantibody presence, and HLA indicated that the gut microbiomes of seropositive subjects differed from those of autoantibody-free first-degree relatives (FDRs) in the abundance of four taxa. Furthermore, subjects with autoantibodies, seronegative FDRs, and new-onset patients had different levels of the Firmicutes genera Lactobacillus and Staphylococcus compared with healthy control subjects with no family history of autoimmunity. Further analysis revealed trends toward increased and reduced abundances of the Bacteroidetes genera Bacteroides and Prevotella, respectively, in seropositive subjects with multiple versus one autoantibody. Canonical discriminant analysis suggested that the gut microbiomes of autoantibody-positive individuals and seronegative FDRs clustered together but separate from those of new-onset patients and unrelated healthy control subjects. Finally, no differences in biodiversity were evident in seropositive versus seronegative FDRs. These observations suggest that altered intestinal microbiota may be associated with disease susceptibility.
Introduction
Type 1 diabetes (T1D) is an autoimmune disorder that involves β-cell inflammation and destruction (1). Although the mechanisms that trigger the disease are not yet clear, human and animal studies implicate both genetic and environmental factors in the disease process. The intestinal microbiota plays a key role in the development and function of the immune system (2). Data from human and animal studies have led to the hypothesis that altered gut microbiota (“dysbiosis”) could be associated with mechanisms of metabolic and immune-mediated disorders, such as obesity, celiac disease, type 2 diabetes, and inflammatory bowel disease (3,4).
Dysbiosis has been postulated to be associated with mechanisms of T1D (5–11). The development of T1D in animal models, such as the NOD (6) and RIP-B7.1 (12) mice and the diabetes-prone BioBreeding (13) and LEW1.WR1 (10) rats, is linked with changes in the intestinal microbiome. Human studies performed in subjects at risk for T1D provided evidence for a decline and an increase in the abundance of the bacterial phyla Firmicutes and Bacteroidetes, respectively (14). Others reported the absence of Bifidobacterium species and increased levels of the genus Bacteroides in children with two or more islet autoantibodies (15). A study performed in subjects with T1D further demonstrated an increase in the abundances of Bacteroidetes and Clostridium spp. in addition to a reduction in the genera Lactobacillus and Bifidobacterium (16). Finally, significant alterations in microbial interaction networks with no differences in bacterial diversity, microbial composition, or the level of bacterial genera were observed in seropositive and seronegative children (17).
In this study, we tested the hypothesis that alterations in the human intestinal microbiota are linked with the progression of human T1D. Analysis of 16S bacterial rRNA sequencing data demonstrated that the abundances of four bacterial genera were altered in seropositive subjects compared with seronegative first-degree relatives (FDRs). Furthermore, seropositive subjects and seronegative FDRs as well as new-onset patients have a reduction in the abundance of the Firmicutes genera Lactobacillus or Staphylococcus compared with unrelated healthy control subjects. The data suggest that gastrointestinal tract dysbiosis may be associated with disease progression.
Research Design and Methods
Subject Characteristics
We analyzed the gut microbiome from four subject cohorts with or without evidence of islet autoimmunity residing in the Denver metro area. These studies included 35 subjects with newly diagnosed T1D (up to 6 months from disease diagnosis) recruited during their routine visits to the Barbara Davis Center for Diabetes. Twenty-one individuals with one to four autoantibodies and 32 seronegative FDRs of subjects with islet autoimmunity were recruited from the Type 1 Diabetes TrialNet Natural History Study (Table 1). Only two seronegative subjects had siblings in the seropositive group. Twenty-three individuals without any family history of autoimmunity (unrelated healthy control subjects) were recruited from university employees and children of employees. The presence of antibodies against GAD65, insulin, ICA512, and ZnT8 was determined by the Autoantibody/HLA Service Center at the Barbara Davis Center for Diabetes. Typing for HLA DR3 and DR4 of study subjects was performed by PCR analysis (18). Thirty-two of the 35 new-onset patients (91%) had DR3 and/or DR4 HLA diabetes risk alleles. All autoantibody seropositive subjects (21 of 21) and 29 of 32 seronegative FDRs carried DR3 and/or DR4 (91%). Twelve out of the 23 unrelated healthy subjects had DR3 and/or DR4 (52%). The median HbA1c level in the seropositive and new-onset subject cohorts was 5.0% (range 4.2–5.3) and 7.9% (range 5.5–15), respectively. Subjects who received antibiotic therapy up to 4 weeks prior to sample collection and individuals with known infections or gastrointestinal disorders were not included in the study. One subject in each of the seropositive and new-onset groups and three from the seronegative cohort reported being vegetarians. The study was approved by the institutional review board at the University of Colorado Denver. Fecal samples were collected at the Barbara Davis Center for Diabetes or at the home of the study participants using stool collection tubes and stored at −80°C until use. Samples collected at home were stored at −20°C and delivered to the Barbara Davis Center for Diabetes on ice by study subjects or laboratory personnel.
Table 1.
Cohort characteristics
| Variable | Unrelated healthy control subjects | New-onset patients | Seropositive | Seronegative FDRs |
|---|---|---|---|---|
| (n = 23) | (n = 35) | (n = 21) | (n = 32) | |
| Females/males, n/n | 9/14 | 16/19 | 12/9 | 14/18 |
| Age (years), median (range) | 12 (4–24) | 11 (2–20) | 9 (4–49) | 12 (3–45) |
| HLA, n (%) | ||||
| 3 alone | 5 (22) | 10 (29) | 5 (24) | 11 (34) |
| 4 alone | 3 (13) | 13 (37) | 12 (57) | 8 (25) |
| 3 plus 4 | 4 (17) | 9 (26) | 4 (19) | 10 (31) |
| x/x | 11 (48) | 3 (8) | 0 | 3 (10) |
| Autoantibodies, n (%) | ||||
| Zero | – | 2 (6) | 0 | 32 (100) |
| One | – | 12 (34) | 5 (24) | 0 |
| Two to four | – | 21 (60) | 16 (76) | 0 |
| HbA1c, median (range) | – | 7.9 (5.5–15.0) | 5.0 (4.2–5.3) | – |
| Age at onset (years), median (range) | – | 10 (2–20) | – | – |
| Disease duration (weeks), median (range) | – | 5.1 (0.3–17.3) | – | – |
| Impaired glucose metabolism, n (%) | – | – | 5 (24) | – |
Microbiome Analysis
Bacterial profiles were determined by broad-range amplification and sequence analysis of 16S rRNA genes. In brief, amplicons were generated using primers that target ∼280 base pairs of the V4 variable region of the 16S rRNA gene (19). PCR products were normalized using a SequalPrep kit (Invitrogen, Carlsbad, CA), pooled, lyophilized, purified, and concentrated using a DNA Clean & Concentrator Kit (Zymo, Irvine, CA). The amplicon pool was quantified using Qubit 2.0 Fluorometer (Invitrogen), diluted to 4 nmol/L, and denatured with 0.2 N NaOH at room temperature. The denatured DNA was diluted to 15 pmol/L and spiked with 25% of the Illumina PhiX Control DNA prior to loading the sequencer. Illumina paired-end sequencing was performed on the MiSeq platform with version 2.3.0.8 of the MiSeq Control Software and version 2.3.32 of MiSeq Reporter, using a 600-cycle version 3 reagent kit.
As previously described (9), Illumina MiSeq paired-end sequences were sorted by sample via barcodes in the paired reads with a python script. The sorted paired reads were assembled using phrap (20,21). Pairs that did not assemble were discarded. Assembled sequence ends were trimmed over a moving window of five nucleotides until average quality met or exceeded 20. Trimmed sequences with more than one ambiguity or shorter than 200 nucleotides were discarded. Potential chimeras identified with Uchime (usearch6.0.203_i86linux32) (22) using the Schloss (23) Silva reference sequences were removed from subsequent analyses. Assembled sequences were aligned and classified with SINA (1.2.11) using the 629,124 bacterial sequences in Silva 111 (24) as reference configured to yield the Silva taxonomy. Operational taxonomic units (OTUs) were produced by clustering sequences with identical taxonomic assignments. A median of 131,000 sequences (range 8,100–313,000) were generated per sample, with a median Good coverage score of 99.97%. The software package Explicet (v2.9.4, www.explicet.org) (25) was used for display, analysis (rarefied values for median Good coverage), and figure generation of results.
Statistical Analysis
Patient demographics were described using percentages and medians and ranges across the four groups for the categorical and continuous variables, respectively. Shannon diversity was calculated using rarefaction in Explicet and compared across groups using ANOVA. For ease of interpretation, effective number of species was calculated from Shannon diversity (26,26). These values represent the expected number of taxa in an even community. Wilcoxon rank-based tests were used to compare the relative abundance of each taxa across groups after adjusting for clinical covariates (age, sex, autoantibody presence, and HLA genotype). The relative abundance for each taxa was transformed to ranks, and these transformed variables were used as dependent variables in a linear regression model. If the overall P value of the differences between the groups was significant, then the pairwise differences were calculated using linear contrasts and a false discovery rate adjustment for multiple comparisons was used. An unadjusted P value for the overall test across groups and the subgroup analysis used a 0.05 cutoff and the adjusted P value used for the individual comparisons used a 0.1 cutoff to identify differentially expressed bacterial groups. Patterns in OTU prevalence and/or abundance in seropositive subjects with one (n = 5) versus two to four autoantibodies (n = 16) were analyzed using Wilcoxon rank-based tests without the false discovery rate correction. Canonical discriminant analysis was performed to multivariately assess differences in the bacterial community across the four groups. A small constant (1/total) was added to the counts to eliminate zeros prior to the application of the centered log ratio transformation recommended for multivariate analysis of compositional data (27,28).
Results
Taxa in the Stool From Subjects With and Without Islet Autoimmunity
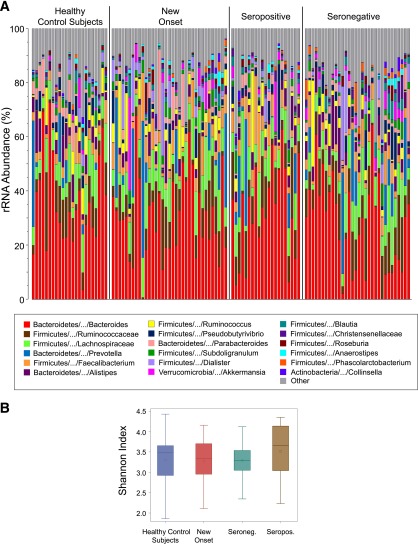
We postulated that alterations in the intestinal microbiome would be involved in the progression of human T1D. To test this possibility, we sequenced the V4 region of 16S rRNA genes (primers 534F–805R [9,10]) from fecal samples of seropositive individuals with one to four autoantibodies (n = 21) compared with seronegative FDRs (n = 32). Comparisons were also made between the gut bacterial content of autoantibody-positive individuals and seronegative FDRs to that of new-onset patients (n = 35) and autoantibody-free healthy control subjects with no family history of autoimmunity (n = 23). A median of 131,000 (range 8,100–313,000) high-quality 16S rRNA gene sequences were obtained for each sample. The data presented in Fig. 1A demonstrate that similar overall gut bacterial profiles were observed among the subject cohorts. Furthermore, comparable Shannon bacterial diversity indices were measured in seropositive versus seronegative FDRs (Fig. 1B) (P = 0.44). These observations suggest that islet autoimmunity is probably not associated with altered bacterial diversity or striking differences in the intestinal microbiota.
Figure 1.
A: Stacked bar chart of median percent counts of OTUs representing bacterial genera with a frequency of ≥1% of total counts in the stool from subjects with and without islet autoimmunity as indicated in the figure. The relative abundances are inferred from 16S rRNA sequence counts in datasets. The x and y axes represent the sample name and percentages of bacterial taxa, respectively. B: The distribution of Shannon indices across groups is displayed using box plots. The area inside the box represents the interquartile range (25th to 75th percentiles), and the median and mean are denoted by a line and a circle, respectively. The whiskers extend 1.5 interquartile range from the box; the observations outside of this range are displayed as points. Seroneg., seronegative; Seropos., seropositive.
Gut Bacterial Communities in Individuals With Islet Autoimmunity
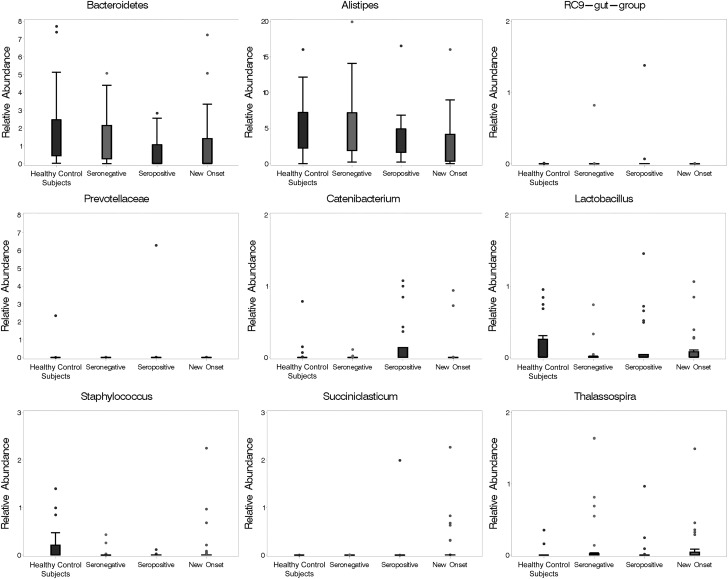
We next compared the abundance of individual bacterial taxa from autoantibody-positive versus autoantibody-free subjects. Similar numbers of bacterial phyla and genera (i.e., OTU richness) were observed in the subject cohorts (data not shown). Wilcoxon rank-based tests with adjustment for covariates (i.e., age, sex, autoantibody presence, and HLA) and multiple comparisons across groups revealed significant differences in the abundance of nine bacterial taxa, of which four were found to be different in seropositive versus seronegative cohorts (Fig. 2, Table 2, and Supplementary Table 1). Parameter estimates further demonstrated that the level of the majority of the taxa with significant differences across groups was not associated with age, sex, or HLA3/4 (Supplementary Table 2). Only the level of a group of bacteria that belong to the Bacteroidetes phylum and could not be classified to lower levels (termed by us Bacteroidetes other) was observed to be associated with age (P = 0.02). The limited sample size of this study did not provide sufficient power to assess the effect of the autoantibody number on the gut bacterial composition. The covariate-adjusted analysis indicated that a significant increase was detectable in the relative median abundance of the Firmicutes genera Catenibacterium and of the Bacteroidetes family and genus Prevotellaceae and RC9 gut group, respectively, in seropositive compared with seronegative FDRs (adjusted P = 0.02 for all) (Fig. 2, Table 2, and Supplementary Table 1). In contrast, a reduction in the abundance of the taxa classified only to Bacteroidetes (Bacteroidetes other) was evident in seropositive versus seronegative FDRs (adjusted P = 0.09).
Figure 2.
Median percent abundance of bacterial communities in subjects with and without islet autoimmunity. The plot displays those taxa that were significantly different across groups after adjustment for covariates (statistical significance and numeric values are shown in Table 2 and Supplementary Table 1, respectively).
Table 2.
Results from Wilcoxon rank-based tests after adjustment for covariates indicating the pairwise comparisons for the genera with a statistically significant difference across the groups
| Taxa | Overall P value | Comparison | F value | P valuea | FDR P value |
|---|---|---|---|---|---|
| Bacteroidetes otherb | 0.006 | NO vs. Ab+ vs. Ab− | 2.42 | 0.094 | 0.154 |
| Ab+ vs. Ab− vs. Co | 4.82 | 0.010 | 0.029 | ||
| Ab+ vs. Co | 9.48 | 0.003 | 0.019 | ||
| Ab− vs. Co | 1.69 | 0.197 | 0.276 | ||
| NO vs. Ab− | 2.86 | 0.094 | 0.154 | ||
| NO vs. Ab+ | 0.26 | 0.614 | 0.682 | ||
| Ab+ vs. Ab− | 4.13 | 0.045 | 0.090 | ||
| Co vs. NO | 8.65 | 0.004 | 0.019 | ||
| Alistipes | 0.013 | NO vs. Ab+ vs. Ab− | 4.52 | 0.013 | 0.034 |
| Ab+ vs. Ab− vs. Co | 1.10 | 0.338 | 0.422 | ||
| Ab+ vs. Co | 1.30 | 0.256 | 0.336 | ||
| Ab− vs. Co | 0.04 | 0.848 | 0.893 | ||
| NO vs. Ab− | 9.04 | 0.003 | 0.019 | ||
| NO vs. Ab+ | 1.73 | 0.191 | 0.273 | ||
| Ab+ vs. Ab− | 2.01 | 0.159 | 0.231 | ||
| Co vs. NO | 7.02 | 0.009 | 0.028 | ||
| RC9 gut group | 0.024 | NO vs. Ab+ vs. Ab− | 3.93 | 0.023 | 0.053 |
| Ab+ vs. Ab− vs. Co | 4.91 | 0.009 | 0.028 | ||
| Ab+ vs. Co | 7.52 | 0.007 | 0.025 | ||
| Ab− vs. Co | 0.03 | 0.864 | 0.893 | ||
| NO vs. Ab− | 0.89 | 0.348 | 0.422 | ||
| NO vs. Ab+ | 3.77 | 0.055 | 0.107 | ||
| Ab+ vs. Ab− | 7.72 | 0.006 | 0.024 | ||
| Co vs. NO | 1.15 | 0.287 | 0.369 | ||
| Prevotellaceae | 0.006 | NO vs. Ab+ vs. Ab− | 5.06 | 0.008 | 0.026 |
| Ab+ vs. Ab− vs. Co | 5.53 | 0.005 | 0.022 | ||
| Ab+ vs. Co | 7.59 | 0.007 | 0.025 | ||
| Ab− vs. Co | 0.01 | 0.912 | 0.912 | ||
| NO vs. Ab− | 4.61 | 0.034 | 0.070 | ||
| NO vs. Ab+ | 1.30 | 0.257 | 0.336 | ||
| Ab+ vs. Ab− | 9.44 | 0.003 | 0.019 | ||
| Co vs. NO | 3.69 | 0.058 | 0.108 | ||
| Catenibacterium | 0.015 | NO vs. Ab+ vs. Ab− | 5.37 | 0.006 | 0.024 |
| Ab+ vs. Ab− vs. Co | 5.13 | 0.007 | 0.025 | ||
| Ab+ vs. Co | 2.59 | 0.111 | 0.172 | ||
| Ab− vs. Co | 2.22 | 0.139 | 0.209 | ||
| NO vs. Ab− | 0.71 | 0.402 | 0.469 | ||
| NO vs. Ab+ | 6.03 | 0.016 | 0.039 | ||
| Ab+ vs. Ab− | 10.23 | 0.002 | 0.017 | ||
| Co vs. NO | 0.55 | 0.462 | 0.533 | ||
| Lactobacillus | <0.001 | NO vs. Ab+ vs. Ab− | 2.32 | 0.104 | 0.164 |
| Ab+ vs. Ab− vs. CO | 8.82 | <0.001 | 0.008 | ||
| Ab+ vs. Co | 8.42 | 0.005 | 0.020 | ||
| Ab− vs. Co | 16.94 | <0.001 | 0.004 | ||
| NO vs. Ab− | 4.63 | 0.034 | 0.070 | ||
| NO vs. Ab+ | 1.04 | 0.310 | 0.393 | ||
| Ab+ vs. Ab− | 0.87 | 0.352 | 0.422 | ||
| Co vs. NO | 4.87 | 0.030 | 0.066 | ||
| Staphylococcus | 0.001 | NO vs. Ab+ vs. Ab− | 1.88 | 0.158 | 0.231 |
| Ab+ vs. Ab− vs. Co | 8.52 | <0.001 | 0.008 | ||
| Ab+ vs. Co | 6.79 | 0.011 | 0.029 | ||
| Ab− vs. Co | 16.79 | <0.001 | 0.004 | ||
| NO vs. Ab− | 3.59 | 0.061 | 0.109 | ||
| NO vs. Ab+ | 0.23 | 0.630 | 0.691 | ||
| Ab+ vs. Ab− | 1.53 | 0.219 | 0.294 | ||
| Co vs. NO | 5.91 | 0.017 | 0.041 | ||
| Succiniclasticum | 0.001 | NO vs. Ab+ vs. Ab− | 6.58 | 0.002 | 0.017 |
| Ab+ vs. Ab− vs. Co | 0.20 | 0.818 | 0.877 | ||
| Ab+ vs. Co | 0.35 | 0.556 | 0.634 | ||
| Ab− vs. Co | 0.01 | 0.907 | 0.912 | ||
| NO vs. Ab− | 11.73 | <0.001 | 0.011 | ||
| NO vs. Ab+ | 6.77 | 0.011 | 0.029 | ||
| Ab+ vs. Ab− | 0.27 | 0.605 | 0.681 | ||
| Co vs. NO | 11.33 | 0.001 | 0.012 | ||
| Thalassospira | 0.008 | NO vs. Ab+ vs. Ab− | 1.61 | 0.204 | 0.283 |
| Ab+ vs. Ab− vs. Co | 4.73 | 0.011 | 0.029 | ||
| Ab+ vs. Co | 1.54 | 0.217 | 0.294 | ||
| Ab− vs. Co | 9.26 | 0.003 | 0.019 | ||
| NO vs. Ab− | 0.03 | 0.873 | 0.893 | ||
| NO vs. Ab+ | 2.34 | 0.129 | 0.197 | ||
| Ab+ vs. Ab− | 2.76 | 0.100 | 0.160 | ||
| Co vs. NO | 8.92 | 0.004 | 0.019 |
Ab−, seronegative; Ab+, seropositive; CO, unrelated control; FDR, false discovery rate; NO, new onset.
aSignificant differences are in bold font.
bA group of bacteria that belong to the Bacteroidetes phylum and could not be classified to lower taxa levels.
We further analyzed the gut microbiomes of seropositive and seronegative subjects compared with those of new-onset patients and healthy control subjects (Fig. 2, Table 2, and Supplementary Table 1). Differences in the bacterial level between these cohorts were noted in genera that belong to the phylum Firmicutes. A reduction and an increase were observed in the relative abundance of Succiniclasticum in new-onset subjects compared with seropositive and seronegative subjects, respectively (adjusted P = 0.01 and P = 0.03, respectively). A decrease of Catenibacterium was observed in new-onset subjects compared with autoantibody-positive subjects (adjusted P = 0.04). An increase in the abundance of the Bacteroidetes genera Alistipes was detected in seronegative FDRs versus new-onset patients (adjusted P = 0.02). A decrease in the abundance of the Bacteroidetes family Prevotellaceae and the Firmicutes genera Lactobacillus and Succiniclasticum was seen in seronegative FDRs versus new-onset patients (adjusted P = 0.07 for Prevotellaceae and Lactobacillus and P = 0.01 for Succiniclasticum). Spearman rank correlation coefficients indicated that the relative abundance of all taxa found to be significantly different in new-onset subjects versus seropositive or seronegative FDRs was not associated with the HbA1c level, age of disease onset, or disease duration (data not shown). The level of the Firmicutes genera Lactobacillus and Staphylococcus was elevated in unrelated healthy control subjects compared with seropositive individuals (adjusted P = 0.02 and P = 0.03, respectively), whereas the level of the Bacteroidetes family and genus Prevotellaceae and RC9 gut group, respectively, was elevated in seropositive individuals compared with unrelated healthy control subjects (adjusted P = 0.03 for both). The abundances of Lactobacillus and Staphylococcus were also higher in the unrelated healthy cohort compared with the new-onset and seropositive cohorts (adjusted P = 0.07 and P = 0.04, respectively).
Because of the wide age range of subjects in the seropositive and seronegative FDR groups (Table 1), we assessed whether the differences observed in the gut bacterial composition may be related to age. To this end, we reanalyzed the intestinal microbiome data after excluding those subjects older than 18 years of age (n = 16 for unrelated healthy control subjects, 34 for new onset, 17 for seropositive, and 21 for seronegative). We found that the abundances of six out of nine bacterial taxa found to be different between cohorts (Fig. 2, Table 2, and Supplementary Table 1) were also different when comparing cohorts of individuals younger than 18 years (data not shown). Similar to cohorts of subjects from all age-groups (Fig. 2, Table 2, and Supplementary Table 1), the median abundances of Staphylococcus and Lactobacillus were diminished in genetically susceptible individuals under the age of 18 years with or without autoimmunity compared with unrelated healthy control subjects, but these differences did not reach statistical significance. Furthermore, unlike that observed in groups comprising both children and adults, the abundance of the bacterial genus Alistipes was similar among individuals under the age of 18 years. Finally, the abundance of the Bacteroidetes genus Barnesiella was found to be significantly different among children and adolescents (data not shown). A similar analysis in subjects older than 18 years was not performed due to a limited sample number in each cohort.
Collectively, these findings may suggest that alterations in the abundances of specific bacterial groups can be observed in the intestine of genetically susceptible individuals prior to and after disease onset and some of these changes may be age related.
Gut Microbiome in Subjects at Low Versus High Risk for Disease Development
Next, we sought to identify trends in the gut microbiota of subjects at high risk versus low risk for T1D. To do so, we analyzed the intestinal microbiome of subjects with two to four autoantibodies (n = 16) versus one autoantibody (n = 5) using less stringent criteria, without adjustment for multiple comparisons, for this preliminary subanalysis. The data shown in Table 3 demonstrate a 6- and 12-fold increase in the abundances of the genera Bacteroides and Akkermansia, respectively, in subjects with two to four versus one autoantibody (unadjusted P = 0.01 and P = 0.04, respectively). In contrast, a considerable 260-fold reduction was observed in the abundance of Prevotella in subjects with multiple versus one autoantibody (unadjusted P = 0.01). Finally, reduced abundances, albeit to a lesser degree than seen for Prevotella, were observed in the level of the genera Butyricimonas, Coprococcus, and Butyrivibrio (unadjusted P = 0.03 for Butyricimonas and Coprococcus and P < 0.01 for Butyrivibrio). These findings suggest that seropositivity with more than one autoantibody may be linked with alterations in the abundance of bacteria that belong to the Bacteroidetes and Firmicutes phyla.
Table 3.
Median percent abundances of bacterial taxa in subjects with one versus multiple autoantibodies
| Taxaa | One autoantibody (n = 5) | Two to four autoantibodies (n = 16) | Unadjusted P valueb |
|---|---|---|---|
| Bacteria/Bacteroidetes/Bacteroidia/Bacteroidales/Bacteroidaceae/Bacteroides | 6.37 (5.67–6.96) | 35.62 (13.20–49.29) | 0.01 |
| Bacteria/Bacteroidetes/Bacteroidia/Bacteroidales/Prevotellaceae/Prevotella | 12.91 (9.62–26.77) | 0.05 (0.01–1.82) | 0.01 |
| Bacteria/Bacteroidetes/Bacteroidia/Bacteroidales/Porphyromonadaceae/Butyricimonas | 0.53 (0.28–0.54) | 0.23 (0.00–0.33) | 0.03 |
| Bacteria/Firmicutes/Clostridia/Clostridiales/Lachnospiraceae/Coprococcus | 1.37 (0.62–2.16) | 0.26 (0.09–1.02) | 0.03 |
| Bacteria/Firmicutes/Clostridia/Clostridiales/Lachnospiraceae/Butyrivibrio | 0.08 (0.04–0.44) | 0.00 (0.00–0.00) | <0.01 |
| Bacteria/Verrucomicrobia/Verrucomicrobiae/Verrucomicrobiales/Verrucomicrobiaceae/Akkermansia | 0.06 (0.02–0.08) | 0.74 (0.15–4.46) | 0.04 |
Data are median relative abundance of 16S rRNA sequences for bacterial clade. Shown in parentheses are the ranges of the bacterial abundance.
aPhylogeny of bacterial genera, assigned by 16S rRNA sequencing analysis.
bResults from Wilcoxon rank-based test.
Canonical Discriminant Analysis
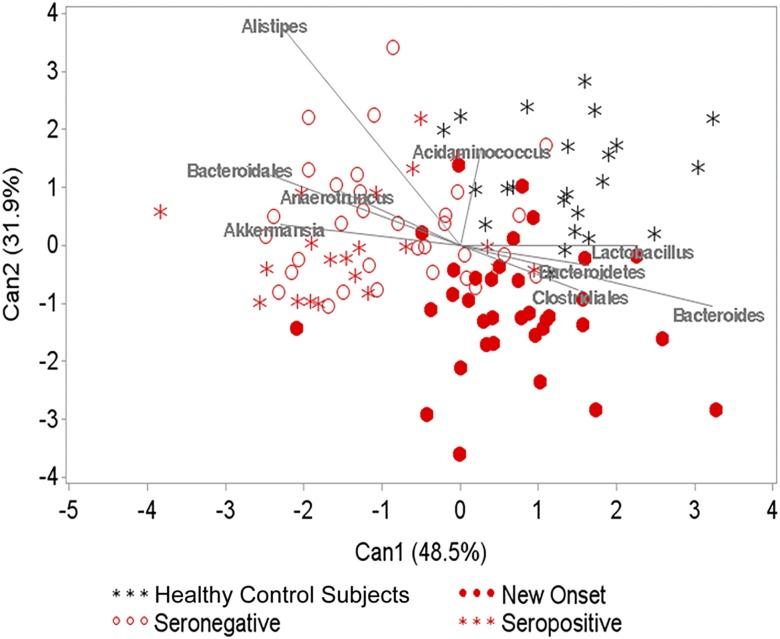
We used canonical discriminant analysis to identify constituents of the gut microbiota that multivariately differentiate across the groups. In this analysis, taxa with relative abundance <1% were combined into a single rare category. Healthy control subjects and new-onset patients formed fairly well-separated clusters, whereas seronegative FDRs and seropositive subjects clustered together but separate from the other two groups (Fig. 3 and Supplementary Tables 3 and 4). The majority of the separation was due to the first component (P = 0.02 for the first component and P = 0.15 for the first two). The first component separated the new-onset from the remaining groups, and the second component separated the healthy control subjects and new-onset cluster from the seropositive and seronegative FDRs. These data raise the hypothesis that the gut microbiomes of seropositive subjects and seronegative FDRs are similar to each other but distinct from those of new-onset patients and unrelated healthy individuals. The data may also imply that the gut microbiota of new-onset subjects is different from that of unrelated healthy individuals.
Figure 3.
Plot from canonical discriminant analysis used to discriminate between seropositive and seronegative subjects. The weights attributed to the taxa that contributed the most to the components are displayed as vectors in the plot.
Discussion
The intestinal microbiota plays a pivotal role in maintaining immune homeostasis in the gut and periphery, protecting against microbial infections and promoting the development and functionality of the immune system (29). A shift in the compositional structure of the gut microbiome could potentially lead to the disruption of the normal interplay between the microbiota and the host, resulting in adverse effects on health (30). Indeed, recent human and animal studies have linked alterations in the gut microbiota to both proinflammatory and metabolic disorders (29). The composition of the gut microbiota is influenced by environmental factors, such as diet, exposure to microbes, geography, and cultural differences (reviewed in Yatsunenko et al. [31]). Herein, we addressed the potential link between compositional changes in the intestinal microbiota and disease development. We present for the first time microbiome data from at-risk and new-onset subjects living in the U.S. We made four key observations. First, no clear bacterial signature of predominant OTUs was detected in individuals with islet autoimmunity prior to and after disease onset (seropositive and newly diagnosed subjects). Second, the abundances of four bacterial genera were altered in seropositive subjects with one or more autoantibodies compared with seronegative FDRs. Third, the gut microbiota of healthy control subjects with no family history of autoimmunity had increased abundances of Lactobacillus spp. and Staphylococcus spp. versus new-onset patients and seropositive and seronegative FDRs. Last, canonical discriminant analysis suggested that the intestinal microbiomes of seropositive subjects and seronegative FDR cohorts are similar to each other but distinct from those of new-onset patients and unrelated healthy control subjects and that the microbiota of new-onset patients is different than that of unrelated healthy subjects. On the basis of these data, we hypothesize that alterations in the intestinal microbiome may be linked with diabetes susceptibility and T1D onset.
How alterations in the abundances of the bacterial phylum Bacteroidetes (Bacteroidetes other) and the Bacteroidetes genus and family RC9 gut group and Prevotellaceae, respectively, that we observed in seropositive subjects compared with seronegative FDRs is not yet clear. Changes in the level of the RC9 gut group were observed by us in diabetes-susceptible Toll-like receptor (TLR)9-deficient versus diabetes-resistant mice (11). The altered abundances of the bacterial genus and family Catenibacterium and Prevotellaceae, respectively, in seropositive versus negative individuals are reminiscent of observations made in subjects with HIV. In patients with HIV, the fecal microbiota contained higher levels of Catenibacterium and Prevotellaceae (32). It remains to be seen whether the increase in these microbes in subjects with islet autoimmunity may be caused at least in part by proinflammatory responses, as was recently suggested for individuals with HIV (32).
We observed a reduction in the relative abundances of Staphylococci and Lactobacilli in the new-onset, seropositive, and seronegative FDR groups versus the unrelated healthy control group. The data could raise the hypothesis that subjects in the former groups have relatively limited ability to regulate proinflammatory responses. Indeed, potentially probiotic members of Lactobacilli (33) have been associated with beneficial effects on proinflammatory disorders (34). Support for the possibility that Lactobacilli could potentially downmodulate inflammation is provided by recent data that dendritic cells cocultured with species of Lactobacilli induce polarization of regulatory T cells (35,36). As to Staphylococcus, it is an important inhabitant of the human skin and the intestine (37). Its beneficial effect on the gut microbiome could potentially be associated with promoting the growth of anaerobic bacteria, including species of the genera Bifidobacterium, Clostridium, and Bacteroides, bacterial groups that promote maturation of the neonatal gut (33,38). Adding to the complexity of a possible role of Staphylococcus in T1D is the fact that these bacteria were also associated with the ability to promote proinflammatory responses (39). Thus, future studies are required to elucidate whether and how changes in the abundance of Staphylococci and Lactobacilli are linked with disease progression.
The mechanisms leading to alterations in the intestinal microbiota or the timing of these changes in genetically susceptible individuals are not yet known. Our recent animal studies suggested that dysbiosis may develop as a result of microbial infections (10,11). Another potential mechanism could be associated, at least in part, with changes in immunity (32). We have recently shown that monocytes and dendritic cells from autoantibody-positive and new-onset subjects have enhanced TLR-induced interleukin-1β responses (12). That the immune system can reshape the gut microbiome with subsequent health changes is based on recent mouse studies. For example, mice with aberrant expression of TLR5, interleukin-22, or the inflammasome have altered gut bacteria associated with colitis (40–42) and metabolic syndrome (43). Furthermore, Wen et al. (6) and our studies (11) demonstrated that diabetes-susceptible mice deficient in various TLR pathways have altered intestinal microbiota. The possibility that the altered gut bacterial composition observed in seropositive subjects and autoantibody-free FDRs compared with new-onset patients and unrelated healthy individuals is linked with differences in the expression of high-risk diabetes HLA alleles is not supported by our data, as we did not find significant association between bacterial abundances and the expression HLA3 and/or HLA4 versus other HLA types. Previous studies in mice (44) and rats (45) have demonstrated that MHC expression is one of the factors that governs the gut bacterial composition. It remains to be determined whether, like mice and rats, the gut bacterial composition in humans is influenced by the HLA profile.
Whether the altered gut microbiome observed in new-onset patients is associated with disease onset is unknown, since it could be linked at least in part with inflammation observed in patients after disease onset (46). Our data do not support the possibility that a shift in the gut microbiome in this cohort is linked with levels of HbA1c, age at onset, or disease duration (data not shown). It could be that alterations in the gut microbiota of new-onset patients are a result of changes in diet that often ensue after disease onset (reviewed in Maslowski and Mackay [47]).
We found a pattern toward increased abundances of the Bacteroidetes genus Bacteroides and a reduction in the Bacteroidetes genus Prevotella and the phylum Firmicutes in subjects with multiple autoantibodies versus one autoantibody. These data are consistent with two Finnish studies showing that seropositive individuals with multiple autoantibodies have altered abundance of the Bacteroides genus (48). An increase in Bacteroides has been linked with the “Western diet” characterized by content high in protein and fat and low in plant fiber, whereas an increase in Prevotella has been linked to a diet rich in plant fibers (49). Prevotella is highly prevalent in African children with a diet rich in grains (50), and taxa from both Prevotella and Firmicutes can digest plant polysaccharides (49,51) and promote the production of short-chain acids known for their anti-inflammatory properties (52). Through similar mechanisms, a Western diet may also be involved in the global rise in the incidence of T1D observed in recent decades (47,53). Our data further demonstrate a trend toward a reduction in the level of Butyricimonas and Coprococcus and an increase in Akkermansia in subjects with multiple autoantibodies compared with one autoantibody. How these changes may be involved in disease progression is unclear. The abundance of the Firmicutes genus Coprococcus was shown to be elevated in Crohn disease and decreased in HIV individuals (54), whereas taxa that belong to Butyricimonas, Butyrivibrio, and Akkermansia were linked with the synthesis of short-chain fatty acids (55–57).
Our observations are somewhat different than data from previous reports that demonstrated increased microbial diversity in patients with diabetes (15) or increased abundances of the Bacteroidaceae family and the Bacteroides genus in seropositive individuals compared with autoantibody-negative control subjects (48). There could be various reasons for these seemingly disparate observations, as the gut microbiome is influenced by multiple environmental factors, such as diet, exposure to microbes, geography, climate, and cultural differences that exist between different countries and communities (reviewed in Yatsunenko et al. [31]). Finally, the use of different data analysis approaches in previous studies compared with those used by us could also have led to a different outcome.
This study has a number of potential limitations. First, our results must be confirmed in larger, multicenter subject cohorts to assess their validity and generalizability. The relatively small sample size limited our ability to detect significant differences in the microbiota of individuals with multiple autoantibodies versus one autoantibody. Second, due to the cross-sectional design of the study, we were unable to determine whether alterations in the microbiome within an individual were associated with disease progression. Despite these limitations, our cohorts were very well characterized, and the approach used enabled us to analyze for the first time the gut microbiome of genetically susceptible individuals with and without islet autoimmunity versus the general population.
In summary, our data raise the hypothesis that alterations in the structural composition of the intestinal microbiota are associated with T1D progression. Identifying bacteria and immune pathways associated with early diabetes may lead to a new class of immunotherapies to modulate the gut microbiota and prevent islet destruction.
Supplementary Material
Article Information
Funding. This study was supported by JDRF grant 17-2011-655 (to D.Z.) and National Institutes of Health grant HG-005964 (to D.N.F.).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. A.K.A. recruited study participants. N.H. was involved in sample storage and performed HLA typing. P.A.G. contributed to subject recruitment. D.I. produced the DNA library and performed the sequencing. C.E.R. and B.D.W. performed data and bioinformatic analyses. D.N.F. designed and oversaw all aspects of the sequencing experiments and data analysis, researched the data, and participated in writing the manuscript. D.Z. oversaw the project and wrote the manuscript. D.Z. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db14-1847/-/DC1.
References
- 1.Gianani R, Eisenbarth GS. The stages of type 1A diabetes: 2005. Immunol Rev 2005;204:232–249 [DOI] [PubMed] [Google Scholar]
- 2.Chervonsky A. Innate receptors and microbes in induction of autoimmunity. Curr Opin Immunol 2009;21:641–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frank DN, Zhu W, Sartor RB, Li E. Investigating the biological and clinical significance of human dysbioses. Trends Microbiol 2011;19:427–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peterson DA, Frank DN, Pace NR, Gordon JI. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe 2008;3:417–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunne JL, Triplett EW, Gevers D, et al. The intestinal microbiome in type 1 diabetes. Clin Exp Immunol 2014;177:30–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wen L, Ley RE, Volchkov PY, et al. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature 2008;455:1109–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brugman S, Klatter FA, Visser JT, et al. Antibiotic treatment partially protects against type 1 diabetes in the Bio-Breeding diabetes-prone rat. Is the gut flora involved in the development of type 1 diabetes? Diabetologia 2006;49:2105–2108 [DOI] [PubMed] [Google Scholar]
- 8.Roesch LF, Lorca GL, Casella G, et al. Culture-independent identification of gut bacteria correlated with the onset of diabetes in a rat model. ISME J 2009;3:536–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Markle JGM, Frank DN, Mortin-Toth S, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 2013;339:1084–1088 [DOI] [PubMed] [Google Scholar]
- 10.Hara N, Alkanani AK, Ir D, et al. Prevention of virus-induced type 1 diabetes with antibiotic therapy. J Immunol 2012;189:3805–3814 [DOI] [PubMed] [Google Scholar]
- 11.Alkanani AK, Hara N, Lien E, et al. Induction of diabetes in the RIP-B7.1 mouse model is critically dependent on TLR3 and MyD88 pathways and is associated with alterations in the intestinal microbiome. Diabetes 2014;63:619–631 [DOI] [PubMed] [Google Scholar]
- 12.Alkanani AK, Rewers M, Dong F, Waugh K, Gottlieb PA, Zipris D. Dysregulated Toll-like receptor-induced interleukin-1β and interleukin-6 responses in subjects at risk for the development of type 1 diabetes. Diabetes 2012;61:2525–2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lau K, Benitez P, Ardissone A, et al. Inhibition of type 1 diabetes correlated to a Lactobacillus johnsonii N6.2-mediated Th17 bias. J Immunol 2011;186:3538–3546 [DOI] [PubMed] [Google Scholar]
- 14.Giongo A, Gano KA, Crabb DB, et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J 2011;5:82–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Goffau MC, Fuentes S, van den Bogert B, et al. Aberrant gut microbiota composition at the onset of type 1 diabetes in young children. Diabetologia 2014;57:1569–1577 [DOI] [PubMed] [Google Scholar]
- 16.Murri M, Leiva I, Gomez-Zumaquero JM, et al. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-control study. BMC Med 2013;11:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Endesfelder D, zu Castell W, Ardissone A, et al. Compromised gut microbiota networks in children with anti-islet cell autoimmunity. Diabetes 2014;63:2006–2014 [DOI] [PubMed] [Google Scholar]
- 18.Steck AK, Armstrong TK, Babu SR, Eisenbarth GS; Type 1 Diabetes Genetics Consortium . Stepwise or linear decrease in penetrance of type 1 diabetes with lower-risk HLA genotypes over the past 40 years. Diabetes 2011;60:1045–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caporaso JG, Lauber CL, Walters WA, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 2011;108(Suppl. 1):4516–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 1998;8:186–194 [PubMed] [Google Scholar]
- 21.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 1998;8:175–185 [DOI] [PubMed] [Google Scholar]
- 22.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011;27:2194–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schloss PD, Westcott SL. Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl Environ Microbiol 2011;77:3219–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 2013;41:D590–D596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robertson CE, Harris JK, Wagner BD, et al. Explicet: graphical user interface software for metadata-driven management, analysis and visualization of microbiome data. Bioinformatics 2013;29:3100–3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jost L. Entropy and diversity. Oikos 2006;113:363–375 [Google Scholar]
- 27.Aitchinson J. The Statistical Analysis of Compositional Data. Monographs on Statistics and Applied Probability. London, Chapman & Hall, 1986 [Google Scholar]
- 28.Filzmoser P, Hron K, Reimann C. Univariate statistical analysis of environmental (compositional) data: problems and possibilities. Sci Total Environ 2009;407:6100–6108 [DOI] [PubMed] [Google Scholar]
- 29.Brenchley JM, Douek DC. Microbial translocation across the GI tract. Annu Rev Immunol 2012;30:149–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang DW, Park JG, Ilhan ZE, et al. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS ONE 2013;8:e68322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature 2012;486:222–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lozupone CA, Li M, Campbell TB, et al. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe 2013;14:329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Penders J, Thijs C, Vink C, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 2006;118:511–521 [DOI] [PubMed] [Google Scholar]
- 34.Hammer GE, Ma A. Molecular control of steady-state dendritic cell maturation and immune homeostasis. Annu Rev Immunol 2013;31:743–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bron PA, van Baarlen P, Kleerebezem M. Emerging molecular insights into the interaction between probiotics and the host intestinal mucosa. Nat Rev Microbiol 2012;10:66–78 [DOI] [PubMed] [Google Scholar]
- 36.Smits HH, Engering A, van der Kleij D, et al. Selective probiotic bacteria induce IL-10-producing regulatory T cells in vitro by modulating dendritic cell function through dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin. J Allergy Clin Immunol 2005;115:1260–1267 [DOI] [PubMed] [Google Scholar]
- 37.Sánchez E, Ribes-Koninckx C, Calabuig M, Sanz Y. Intestinal Staphylococcus spp. and virulent features associated with coeliac disease. J Clin Pathol 2012;65:830–834 [DOI] [PubMed] [Google Scholar]
- 38.Marques TM, Wall R, Ross RP, Fitzgerald GF, Ryan CA, Stanton C. Programming infant gut microbiota: influence of dietary and environmental factors. Curr Opin Biotechnol 2010;21:149–156 [DOI] [PubMed] [Google Scholar]
- 39.Edwards LA, O’Neill C, Furman MA, et al. Enterotoxin-producing staphylococci cause intestinal inflammation by a combination of direct epithelial cytopathy and superantigen-mediated T-cell activation. Inflamm Bowel Dis 2012;18:624–640 [DOI] [PubMed] [Google Scholar]
- 40.Carvalho FA, Koren O, Goodrich JK, et al. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe 2012;12:139–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elinav E, Strowig T, Kau AL, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis (Abstract). Cell 2011;145:745–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zenewicz LA, Yin X, Wang G, et al. IL-22 deficiency alters colonic microbiota to be transmissible and colitogenic. J Immunol 2013;190:5306–5312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vijay-Kumar M, Aitken JD, Carvalho FA, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 2010;328:228–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toivanen P, Vaahtovuo J, Eerola E. Influence of major histocompatibility complex on bacterial composition of fecal flora. Infect Immun 2001;69:2372–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin P, Bach M, Asquith M, et al. HLA-B27 and human β2-microglobulin affect the gut microbiota of transgenic rats. PLoS ONE 2014;9:e105684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Devaraj S, Dasu MR, Rockwood J, Winter W, Griffen SC, Jialal I. Increased toll-like receptor (TLR) 2 and TLR4 expression in monocytes from patients with type 1 diabetes: further evidence of a proinflammatory state. J Clin Endocrinol Metab 2008;93:578–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maslowski KM, Mackay CR. Diet, gut microbiota and immune responses. Nat Immunol 2011;12:5–9 [DOI] [PubMed] [Google Scholar]
- 48.de Goffau MC, Luopajärvi K, Knip M, et al. Fecal microbiota composition differs between children with β-cell autoimmunity and those without. Diabetes 2013;62:1238–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011;334:105–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A 2010;107:14691–14696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holmes E, Li JV, Marchesi JR, Nicholson JK. Gut microbiota composition and activity in relation to host metabolic phenotype and disease risk. Cell Metab 2012;16:559–564 [DOI] [PubMed] [Google Scholar]
- 52.Smith PM, Howitt MR, Panikov N, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013;341:569–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brown CT, Davis-Richardson AG, Giongo A, et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS ONE 2011;6:e25792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sands SA, Tsau S, Yankee TM, Parker BL, Ericsson AC, LeVine SM. The effect of omeprazole on the development of experimental autoimmune encephalomyelitis in C57BL/6J and SJL/J mice. BMC Res Notes 2014;7:605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Belzer C, de Vos WM. Microbes inside—from diversity to function: the case of Akkermansia. ISME J 2012;6:1449–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sakamoto M, Takagaki A, Matsumoto K, Kato Y, Goto K, Benno Y. Butyricimonas synergistica gen. nov., sp. nov. and Butyricimonas virosa sp. nov., butyric acid-producing bacteria in the family ‘Porphyromonadaceae’ isolated from rat faeces. Int J Syst Evol Microbiol 2009;59:1748–1753 [DOI] [PubMed] [Google Scholar]
- 57.Lee WJ, Hase K. Gut microbiota-generated metabolites in animal health and disease. Nat Chem Biol 2014;10:416–424 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.