Abstract
The prebiotic fructooligosaccharides (FOS) content of yacon makes this root an attractive alternative for the supplementation of a variety of food products. The preservation of yacon by fermentation has been proposed as an alternative to increase the probiotic content of the root concomitantly with its shelf life. Thus the fermented yacon could have significant functional content. The objective of this research was to characterize the biochemistry and microbiology of spontaneous yacon fermentation and define the viability of the proposed process. The biochemical analysis of spontaneous heterolactic fermentation of yacon showed a progressive drop in pH with increased lactic and acetic acids, and the production of mannitol during fermentation. The microbial ecology of yacon fermentation was investigated using culture-dependent and culture-independent methods. Bacterial cell counts revealed a dominance of lactic acid bacteria (LAB) over yeasts, which were also present during fermentation. Results showed that the heterofermentative LAB were primarily Leuconostoc species, which dominated the fermentation. The fermentation of yacon by Leuconostoc spp. is thus presented as a viable method to achieve long term preservation of this root.
Keywords: fermented vegetables, lactic acid bacteria, heterolactic fermentation
1. Introduction
Fermentation of fruits and vegetables is one of the oldest technologies known to man. It results in the improvement of quality and safety of foods, due to the microbial biosynthesis of vitamins and essential amino acids, and the preservation of ascorbic acid, phenolic compounds and pigments (Galvez et al., 2007). Foods to be fermented carry a microbial load dependent on the soil, water, air and type of produce, with reported values of 103–105 CFU/g total aerobes (Rodríguez et al., 2009). The type of microorganisms and their development are determining factors of the course of the fermentation and the quality of the final product (Rodríguez et al., 2009). Typically, vegetable fermentations rely on spontaneous growth of indigenous lactic acid bacteria (LAB). Relevant determinants of the course of vegetable fermentations include temperature, initial pH, nutrients, and salt (NaCl) concentration in the cover brine solutions (Fleming et al., 1995). The concentration of NaCl used in vegetable fermentations ranges from 2% to 10% or more, depending on the type of products manufactured. NaCl favors the growth of LAB which produce lactic acid consequently dropping the medium pH and inhibiting the proliferation of competing undesirable bacteria. LAB are widely used in the food industry in the production of enzymes and metabolites, nutraceuticals, as vehicles of vaccines, as starter cultures in controlled fermentations, and as probiotics (Rodríguez et al., 2009). Although a large number of LAB starters are routinely used in dairy, meat and bakery fermentation, only a few cultures have been characterized and used at commercial scale for vegetable fermentations and none have been applied to yacon fermentation.
Yacon (Smallanthus sonchifolius Poepp.Endl) is an Andean root with known medicinal properties. The root has a sweet taste and crisp texture and is consumed raw, boiled, baked or as juice. Yacon is now considered a functional food due to its fructooligosaccharides (FOS) and polyphenol contents (Flores et al., 2003; Genta et al., 2009; Ojansivu et al., 2010). Many yacon-based new products have been developed during the last 10 years, such as flour, syrup, jam, chips, and pickles among others (Reina et al., 2008; Ribeiro, 2008). The yacon, rich in FOS, reducing sugars, polyphenols, amino acids and minerals, has a considerable potential to make value-added products. Although, previous studies have shown that LAB are involved in yacon fermentation, the specific species involved are still unknown (Reina et al., 2009). As in many other vegetable fermentations (Ballesteros et al., 1999), the microbial diversity of yacon fermentation is significantly influenced by the amount of salt added. While yeasts predominate in salt-free yacon fermentation, heterofermentative and homofermentative LAB predominate in fermentations with 2–4% and more than 5% salt, respectively (Reina et al., 2009).
The fermentation of yacon is expected to preserve its nutritional value long term. Moreover, a non-alcoholic fermentation can impart an acceptable flavor and quality, while ensuring the safety of the good for human consumption. A fermentation lead by heterofermentative LAB has the potential to add value to the processed good, naturally rich in nutrients, by producing prebiotic exopolyssacharides from the indigenous FOS. Several heterofermentative LAB are able to produce mannitol and exopolysaccharides (EPS) from fructose/glucose mixtures. Even though these compounds are not produced industrially by fermentation, the prospect of producing them using a food grade LAB is promising (Saha and Racine, 2011). The production of mannitol and EPS in food by LAB could result in the manufacture of food products with an added nutritional value and use as functional foods. The objective of this study was to characterize the microbial diversity of yacon spontaneous fermentation in the presence of 2% salt, using culture-dependent and independent methods.
2. Material and methods
2.1. Yacon
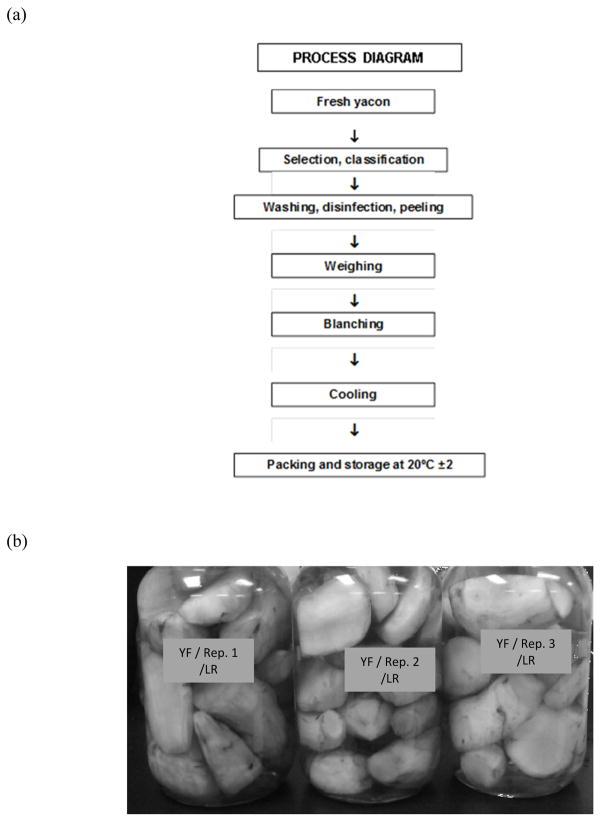
Yacon was obtained from a commercial grower in California, USA. Roots with no physical damage and equal consistency to the touch were selected, peeled (Butler and Rivera, 2004) and processed as described by Reina et al. (2009), with some modifications. Briefly, samples were blanched at 95 ºC for 3 min and immediately cooled in the same amount (V/W) of 4% cover brine (NaCl) at 4 ºC, so that the salt would equilibrate at 2% (Figure 1). The blanching step was introduced to reduce the number of indigenous yeasts and inactivate polyphenoloxidase (Narai-Kanayama et al., 2007). Samples were then vacuum-packed in sterile glass containers and kept at 20 ± 2 ºC until use. Each experiment consisted of 3 replicates. Two independent lots of yacon were tested.
Figure 1.
Flow chart to prepare pickled yacon for fermentation (a) and yacon packed with cover brine (b).
2.2. Microbiological Analyses
Cover brine samples were serially diluted in saline solution and plated on plate count agar (PCA, Difco Laboratories, Detroit, MI), violet-red bile agar (VRBG, Difco Laboratories) supplemented with 1% glucose, Lactobacilli de Man Rogosa and Sharpe agar (MRS, Difco Laboratories) supplemented with 0.5 mg/L of L-Cys (Sigma-Aldrich, St. Louis, MO), and yeast extract malt agar (YMA, Difco Laboratories) containing 250 mg/liter chlortetracycline and 250 mg/liter chloramphenicol (Sigma Aldrich) to enumerate the total aerobic microbiota, Enterobacteriaceae, LAB, and yeasts and molds, respectively. MRS plates were incubated in anaerobic jars at 30 and 37 ºC for 48–72 h. YMA and PCA plates were incubated aerobically at 30 ºC; while VRBG plates were incubated at 37 ºC. Plating and plate counting were performed using a spiral plater (model 4000; Spiral Biotech, Norwood, MA) with an automated colony counter (Protos Plus; Microbiology International, Frederick, MD). Colonies were randomly selected from MRS agar plates for sampling days 0, 2, 7, 15, and 30 and streaked on the same medium to obtain pure cultures. Frozen stocks of the pure cultures were prepared in MRS broth containing 20% glycerol (Sigma-Aldrich) and stored at −80 ºC.
2.3. Total Genomic DNA Extraction
A propidium monoazide (PMA, Biotium, Inc., Hayword, CA) treatment was applied to the homogenized and filtered yacon samples as described by Pan and Breidt (2007) prior to DNA extraction. Total genomic DNA was extracted using the PowerSoil DNA isolation kit (MO BIO Laboratories Inc., Carlsbad, CA) following the manufacturer’s instructions. DNA from isolated colonies growing on MRS was extracted using the InstaGene Matrix DNA extraction kit (Bio-Rad, Hercules, CA) following the manufacturer’s instructions.
2.4. 16S rDNA sequencing
Partial 16S rDNA sequencing was performed using the total genomic DNA extracted from complex samples or the bacterial isolates chromosomal DNA. The PCR mix contained 2X master mix (Bio-Rad), chromosomal DNA (5–10 ng), and 0.6 μM of primers RBUP (5-AGAGTTTGATCCTGGCTCAG-3’) and 1492r (5’–GGTTACCTTGTTACGACTT-3’) (Wilson et al., 1990). The PCR cycle consisted of 4 min at 95 ºC followed by 30 cycles of 30 s at 95 ºC, 30 s at 61 ºC, and 30 s at 72 ºC, with a final extension step of 7 min at 72 ºC and stored at 4 ºC until used. PCR products were purified using the Qiagen PCR purification kit (Qiagen, Valencia, CA) and sequenced by Eton Bioscience Inc. (Durham, NC). The sequences obtained were subjected to the basic local alignment search tool (BLAST) (Altschul et al., 1990) using the 16S ribosomal RNA sequence database to determine the strains identity.
2.5 Total DNA analysis by PCR-NMR
Total genomic DNA extracted as described above was delivered to AthoGen (Carlsbad, CA) for a broad range bacterial rDNA analysis using their Ibis Technology for microbial screening.
2.6. 16S rDNA bacterial amplicon sequencing using Ion Torrent PGM
For amplicon library preparation, the V1–V2 hypervariable region of the 16S rDNA was amplified from total bacterial DNA isolated from each sample using the forward primer composed of the Ion Torrent adapter A (5’-CCATCTCATCCCTGCGTGTCTCCGACTCAG -3’), a 10bp IonXpress ™ barcode, unique to each sample (Life Technologies, Grand Island, NY) and the universal bacterial primer 8F (5’-AGAGTTTGATCCTGGCTCAG-3’). The reverse primer consisted of the Ion Torrent trP1 adapter (5’-CCTCTCTATGGGCAGTCGGTGAT -3’) (Life Technologies, Grand Island, NY) followed by the reverse bacterial primer 338R (5′-GCTGCCTCCCGTAGGAGT-3′). PCR reactions contained 5–10 ng of DNA template, 2.5 units of HotStar Hi-fidelity DNA polymerase (Qiagen, Valencia, CA), 1x HotStar Hi-Fidelity PCR buffer containing dNTPs, and 0.6 μM of each primer. Reaction conditions consisted of an initial denaturation for 5 min at 94 ºC followed by 35 cycles of denaturing at 94 ºC for 60 s, annealing at 57 ºC for 60 s, and extension at 72 ºC for 60 s, and a final extension of 72 ºC for 10 min. Negative controls, not containing template, were amplified for all barcode-primer sets. The PCR products of 411 bp in size for eash sample were confirmed and gel purified using E-Gel Electrophoresis System (Invitrogen, Life Technologies division). DNA concentrations were quantified using QuantiT™ PicoGreen ® dsDNA Reagent (Molecular Probes, Life Technologies division) and mixed at equimolar concentrations. All kits were used according to the respective manufacturers’ instructions. The prepared amplicon library pool was submitted to the Microbiome Core Facility (UNC, Chapel Hill, NC) for sequencing on the Ion Torrent PGM instrument. Initial data analysis, base pair calling and trimming of each sequence was performed on the Ion Torrent browser to yield high quality reads.
2.7. Amplicon high-throughput sequencing data analysis
Raw Ion Torrent fastq files were demultiplexed, quality-filtered, and analyzed using QIIME (Caporaso et al., 2010). The 400-bp reads were truncated at any site if more than three sequential bases receiving a quality score of <20, and any read containing ambiguous base calls or barcode/primer errors were discarded, as were truncated reads. Operational Taxonomic Units (OTUs) were assigned using the QIIME implementation of UCLUST (Edgar, 2010), with a threshold of 97% pairwise identity, and representative sequences from each OTU selected for taxonomy assignment. Chimera detection and removal was carried out using ChimeraSlayer. Beta diversity estimates were calculated within QIIME using weighted and unweighted Unifrac distances (Lozupone and Knight, 2005) between samples at a depth of 989 sequences per sample. From these estimates, principal coordinates were computed to compress dimensionality into two- and three-dimensional principal coordinate analysis plots. QIIME was also used to calculate alpha diversity on rarefied OTU tables to assess sampling depth coverage using observed species, Shannon and phylogenetic diversity (PD) metrics.
2.8. Chemical Analyses
pH measurements were done using an Acumet® Research 25 pH meter (Fischer Scientific, CA, USA) equipped with a gel-filled pencil-thin pH combination electrode (Fischer Scientific, PA, USA). pH measurements were done from the aseptically collected cover brine samples or the extract of the homogenized fresh or fermented yacon samples. Yacon samples (10g) were homogenized at maximum speed for 1 min (Laboratory Blender, Stomacher 400, Tekmar, Cincinnati, OH) using filtered bags. The filtrate was used for pH measurements.
Brine samples (2 mL) were withdrawn aseptically at selected time points during the fermentations for analysis by high-performance liquid chromatography (HPLC). Organic acids, sugars, and ethanol concentrations were measured with a Thermo Separation Products HPLC (ThermoQuest Inc., San Jose, CA) system consisting of a P1000 pump, an SCM100 solvent degasser, an AS3000 autosampler, and a UV6000 diode array detector (ThermoQuest). An HPX-87H column, 300 by 7.8 mm (Bio-Rad) was used with a differential refractometer (Waters model 410 Millipore, Milford, MA) and a UV detector (UV6000LP, Thermo Separation Products, San Jose, CA) for detection of the analytes. Operating conditions of the system included a sample tray at 6 ºC, a column at 65 ºC and 0.03 N H2SO4 eluent at a flow rate of 0.8 mL/min. The UV6000 detector was set to 210 nm at a rate of 1 Hz for data collection and 2 Hz was used for refractive index data. ChromQuest version 4.1 chromatography software was used to control the system and analyze the data. The pH was measured with an Accumet AR25 pH meter (Fisher, Atlanta, GA).
3. Results and Discussion
3.1. Culture-dependent analysis of the microbial diversity of yacon fermentation
Previous studies have shown the effect of different salt concentrations on the microbial diversity of yacon fermentation (Reina et al., 2009). Yacon fermentation is dominated by yeasts, reaching populations of 2x106 CFU/g if sodium chloride free cover brine is applied. A yacon fermentation with 2% NaCl at equilibrium, is dominated by LAB (109 CFU/g) followed by yeasts that reached a population of 101–2 CFU/g. The metabolic activity of LAB drop the pH of the yacon to 3.7, which is well below the 4.6 pH required by the safety guidelines. Fermentation occurs rapidly (24 h), and the resulting product is stable. These observations coincide with results reported from the fermentation of other vegetables, in which 2% salt concentration favors the growth of LAB such as Leuconostoc and Lactococcus.
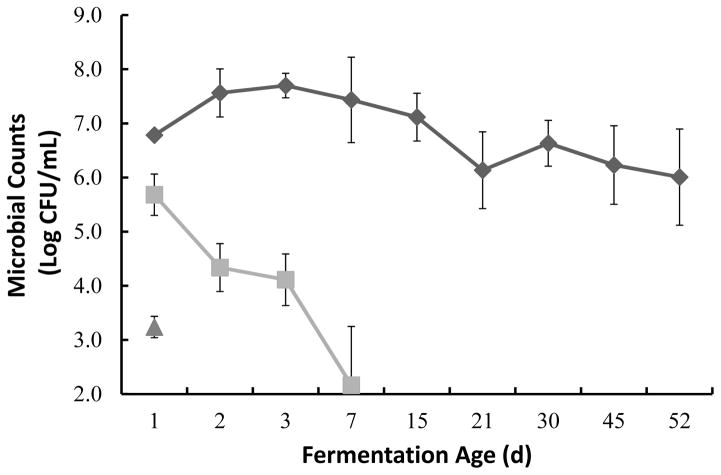
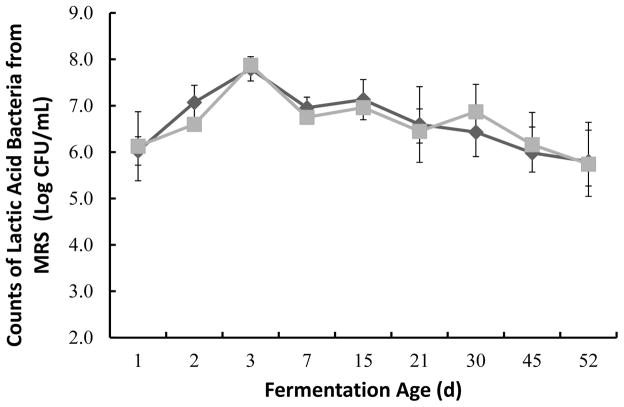
In this study the fresh yacon indigenous microbiota was found to be primarily composed of aerobic mesophilic bacteria (6.27 ±0.3 Log CFU/mL), Enterobacteriaceae (5.04 ± 0.6 Log CFU/mL) and some yeasts (3.24 ±0.28 Log CFU/mL). LAB composed the majority of the mesophilic population with initial counts of 5.11 ± 1 Log CFU/mL. The total mesophilic bacteria, primarily composed of lactic acid bacteria, increased with time, reaching close to 8 log CFU/mL after 3 d of fermentation (Figure 2 and 3). The total number of Enterobacteriaceae decreased after 48 h of fermentation reaching numbers below the limits of detection as the pH decreased (Figure 2 and 5). Although, LAB counts were slightly higher when the plates were incubated at 30 ºC as compared to 37 ºC the differences in counts were not significant.
Figure 2.
Microbial counts during yacon fermentation. Total aerobic counts from PCA (◆), counts for Enterobacteriaceae from VRBG (■) and counts for yeasts from YMA (▲) represent the average of triplicate samples with corresponding standard errors. Counts from YMA were below the detection levels after day 1. Minimum detection level was 2.4 Log CFU/mL. Fermentation age axis is not presented to scale.
Figure 3.
Lactic acid bacteria counts during yacon fermentation. Counts obtained from MRS agar plates incubated at 30 °C (◆) and 37 °C (■) under anaerobic conditions, represent the average of triplicate samples with corresponding standard errors. Fermentation age axis was not drawn to scale.
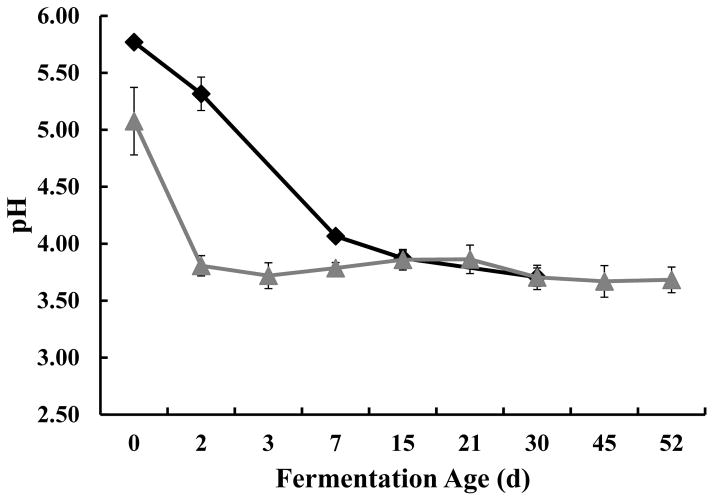
Figure 5.
Changes of pH during yacon fermentation. Numbers presented for the cover brine samples (
 ) are the averages of triplicate from two independent replicates. Numbers presented for the brined yacon samples (◆) represent triplicates of one lot of yacon with corresponding standard deviations. The fermentation age axis is not drawn to scale.
) are the averages of triplicate from two independent replicates. Numbers presented for the brined yacon samples (◆) represent triplicates of one lot of yacon with corresponding standard deviations. The fermentation age axis is not drawn to scale.
Yeasts were initially present in low numbers (Figure 2) and decreased below the limit of detection by the second day of the fermentation. An amplicon for the 18S rDNA using total eukaryotic DNA isolated from the yacon fermentation samples could not be obtained (data not shown). Together these data suggest that the blanching step and supplementation of the cover brine with 4% salt were sufficient to prevent the prevalence of yeasts in the process. Molds were not detected in the fresh yacon samples.
Colonies of LAB (39 total) from representative MRS plates at different points of fermentation were selected for further study. Table 2 shows the colonies identification using partial sequencing of the 16S rDNA. Three Leuconostoc spp. dominated the fermentation, mesenteroides, pseudomesenteroides and citreum. Staphylococcus warneri was detected in the fresh yacon only.
Table 2.
Culture independent detection of predominant bacteria using PCR-NMR as reported by AthoGen.
| Fermentation Age (d) | Quality Score | Amplicon Identifications |
|---|---|---|
| 1 | 0.99 | Leuconostoc mesenteroides |
| 2 | 0.99 | Leuconostoc mesenteroides |
| 7 | 0.99 | Leuconostoc gasicomitatum/kimchii/pseudomesenteroides/sp. |
| 15 | 0.99 | Leuconostoc mesenteroides |
| 0.97 | Enterobacter sp. |
3.2. Culture-independent analysis of the microbial diversity of yacon fermentation
Total genomic DNA was isolated from samples treated with PMA to eliminate free DNA derived from dead cells. Analysis of total genomic DNA samples from days 1, 2, 7 and 15 using AthoGen technology for bacterial identification suggested that Leuconsotoc mesenteroides was the dominant bacterium in yacon fermentation during the first two weeks (Table 2).
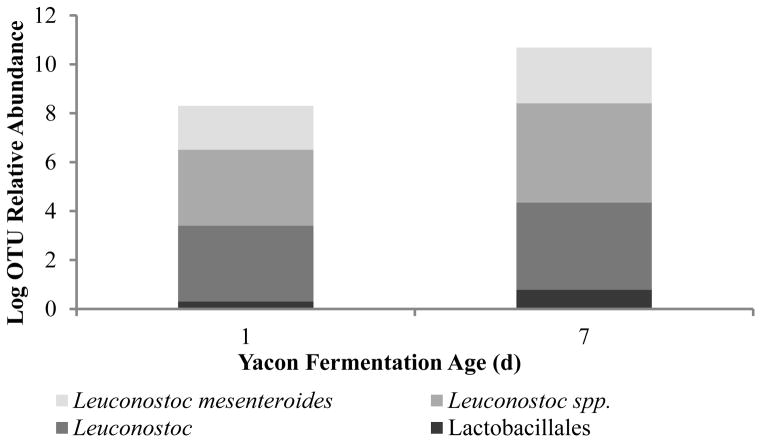
To further characterize the dominant bacterial groups at the beginning and at peak of a fermentation, we conducted a microbiome analysis of samples. Figure 4 presents the results of sequencing analysis using the Ion Torrent platform, which are in agreement with the data from the culture-based method and the PCR-NMR approach. Amplicon sequencing of the 16S ribosomal gene confirmed that the dominant bacterial community in yacon fermentations are Leuconostoc species, including L. mesenteroides. This analysis additionally showed that the community of Lactobacillales is dominated by Leuconostoc species, presumably due to the low percentage of salt added in the cover brine (2%). Species of Leuconostoc are commonly found in fruits and vegetables and they play important roles in the fermentation of dairy products, vegetables and meats given their ability to ferment sugars (sucrose, glucose, fructose) producing lactic acid, glucans, fructans, and mannitol (Groben et al., 2001; Kim et al., 2002; Patra et al., 2011).
Figure 4.
Distribution of bacterial species detected by amplicon sequencing of the 16S ribosomal gene at the beginning and peak of a yacon fermentation.
3.3. Biochemical characteristics of yacon fermentation
The fresh yacon had an initial pH of 5.77 ± 0.3, which decreased gradually with time to reach a final pH of 3.71 ± 0.11 after 7 d of fermentation (Figure 5). This reduction of pH coincided with the growth of LAB during the fermentation (Figure 2). Cover brine and yacon pH reached an equilibrium after 15 d of fermentation (figure 5). The use of small pre-cut pieces of the fruit may allow for a faster equilibration, which would likely induce a quicker suppression of the undesired enterobacteriaceae population. Thus the use of small pre-cut pieces of fresh yacon should be considered as the best choice for the manufacture of a safer fermented good.
To assess the overall metabolic activity of the microbial community, the concentrations of sucrose, glucose, fructose, and organic acids were measured by HPLC (Table 3). The results suggest that yacon is a rich source of glucose, and fructose, and that these sugars were utilized as the carbon sources for the production of acetic and lactic acids in equimolar concentrations, and mannitol and ethanol. Malic acid was present in the fresh yacon to 1.30 ± 0.57 mM, but was not utilized during the fermentation (data not shown). Propionic and butyric acids, and glycerol were not detected. The detected metabolites, in particular mannitol, represent those typically produced by heterofermentative L. mesenteroides in natural fermentations (Saha and Racine, 2011). The equimolar concentrations of acetic and lactic acids detected confirms that a heterofermentative LAB dominated the fermentation. Relatively high fructose concentrations were detected in some samples, suggesting the production of this sugar from the acid hydrolysis of FOS during the fermentation (Narai-Kanayama et al., 2007; Reina et al., 2009). FOS are composed of linear chains of fructose units and often terminate in a glucose unit, ranging from 2–70 monomers. They function as prebiotic by stimulating the growth of probiotics in the intestine. Acidic pH affect the concentration of prebiotics in the final product by hydrolysis to smaller chains easier utilized by bacteria. Longer chains may still be present in the final product as well. However it has been demonstrated that acidic conditions as occurring in fermented food products had little influence on prebiotic activity of FOS containing between 2 and 70 monomers, which are stable even at pH as low as 3 (Huebener, et al. 2008) as indicated by a bacterial mass assay. Further studies will evaluate FOS metabolism during yacon fermentation.
Table 3.
Biochemical changes during yacon spontaneous fermentation. Values presented are the average of triplicate samples.
| Fermentation Age (days) | Components Detected by HPLC Analysis (mM) | |||||
|---|---|---|---|---|---|---|
| Glucose | Fructose | Lactic Acid | Acetic Acid | Mannitol | Ethanol | |
| 0 | 81.6 ± 8 | 278.4 ± 28 | ND | ND | ND | ND |
| 2 | 19.1 ± 4 | 102.1 ± 10 | 2.7 ± 0.8 | 8.5 ± 0.7 | 20.1 ± 3 | ND |
| 7 | 6.3 ± 0.7 | ND | 27.6 ± 4 | 31.1 ± 4 | 72.4 ± 2 | 22.6 ± 0.5 |
| 15 | N/A¥ | ND | 36.2 ± 7 | 39.9 ± 6 | 77.1 ± 2 | 25.9 ± 3 |
| 30 | N/A | ND | 41.5 ± 8 | 45.1 ± 7 | 82.8 ± 2 | 31.7 ± 0.2 |
ND: not detected
Data not available
To our knowledge this is the first study showing the identification of LAB in yacon fermentation. Our results show the prevalence of heterofermtative LAB, mainly Leuconostoc in the indigenous yacon fermentation with 2% NaCl. Among the Leuconostoc spp. identified, one species, mesenteroides, dominated across the three techniques used for microbial identification. Utilization of a starter culture of L. mesenteroides or a combination of Leuconostoc spp. and of small pre-cut pieces of yacon may be advantageous in yacon fermentation with 2% salt to accelerate acid production and the equilibration of the system, respectively. Such strategy should result in the quicker reduction of the undesired and indigenous Enterobacteriacea and minimized their proliferation.
Table 1.
Identity of cultivated bacteria from yacon fermentation as determined by the partial sequencing of the 16S rDNA
| Fermentation Age (d) | Best BLAST Hits | GeneBank Accession No. for Representative Sequences | No. of Colonies Identified |
|---|---|---|---|
| 0 (fresh) | Staphylococcus warneri | KF661311 | 2 |
| 2 | Leuconostoc mesenteroides | KF661314 | 6 |
| 7 |
Leuconostoc mesenteroides
Leuconostoc pseudomesenteroides |
KF661315 KF661313 |
6 1 |
| 15 |
Leuconostoc mesenteroides
Leuconostoc pseudomesenteroides |
KF661302, KF661308 KF661309 |
7 1 |
| 30 |
Leuconostoc mesenteroides
Leuconostoc citreum |
KF661319, KF661320 KF661310 |
12 1 |
Highlights.
Lactic acid bacteria predominate in yacon fermentation with 2% salt.
Sugars from yacon are microbiologically converted to organic acids and mannitol.
Leuconostoc spp. lead yacon fermentation with 2% salt.
Yacon can be preserved long term by fermentation with 2% salt.
Acknowledgments
The author thanks Ms. Sandra Parker and Ms. Janet Hayes at the USDA-ARS, SAA Food Science Research Unit located in Raleigh, NC for secretarial support and technical assistance, respectively. We also thank Nevermore Farm in Arbuckle, CA, USA for supplying yacon. This research was partially supported (10%) by the National Institutes of Health grant number P30 DK34987, and a Simmons award to M. Andrea Azcarate-Peril. A Postdoctoral Fellowship from the Spanish Government (MECD) fully funded Dr. Eduardo Medina.
Footnotes
Mention of a trademark or proprietary product does not constitute a guarantee or warranty of the product by the U. S. Department of Agriculture or North Carolina Agricultural Research Service, nor does it imply approval to the exclusion of other products that may be suitable.
References
- Altschul SF, Gish W, Miller W, Meyers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2.) in the GenBank ( Benson DA, Boguski MS, Lipman DJ, Ostell J. GenBank. Nucleic Acids Research. 1997;25:1–6. doi: 10.1093/nar/25.1.1.
- Ballesteros C, Palop L, Sanchez I. Influence on sodium chloride concentration on the control lactic acid fermentation of “Almagro” eggplants. International Journal of Food Microbiology. 1999;53:13–20. doi: 10.1016/s0168-1605(99)00130-0. [DOI] [PubMed] [Google Scholar]
- Butler G, Rivera D. Project Report. CIP; Lima-Peru: 2004. Innovations in peeling technology for yacon. [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. Qiime allows analysis of high-throughput community sequence data. Nature Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Fleming H, McDonald L, McFeeters R, Thompson R, Humphries E. Fermentation of cucumber without sodium chloride. Journal of Food Science. 1995;60:312–315. 319. [Google Scholar]
- Flores H, Walker T, Guimaraes R, Harsh P, Vivanco J. Andean root and tuber crops: Underground Rainbow. HortScience. 2003;38(2):161–167. [Google Scholar]
- Galvez A, Abrioul H, Lopez R, Omar N. Bacteriocin-based strategies for food biopreservation. International Journal of Food Microbiology. 2007;120:51–70. doi: 10.1016/j.ijfoodmicro.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Genta S, Cabrera W, Habib N, Pons J, Carillo IM, Grau A, Sánchez S. Yacon syrup: Beneficial effects on obesity and insulin resistance in humans. Clinical Nutrition. 2009;28(2):182–87. doi: 10.1016/j.clnu.2009.01.013. [DOI] [PubMed] [Google Scholar]
- Grobben GJ, Peters SWPG, Wouter Wisselink H, Weusthuis RA, Hoefnagel MHN, Hugenholtz J, Eggink G. Spontaneous formation of a mannitol-producing variant of Leuconostoc pseudomesenteroides grown in the presence of fructose. Applied Environmental Microbiology. 2001;67(6):2867–2870. doi: 10.1128/AEM.67.6.2867-2870.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebner J, Wehlinga RL, Parkhurstb A, Hutkins RW. Effect of processing conditions on the prebiotic activity of commercial prebiotics. International Dairy Journal. 2008;(18):287–293. [Google Scholar]
- Kim CY, Lee JH, Kim BH, Yoo SK, Seo ES, Cho KS, Day DF, Kim D. Production of mannitol using Leuconostoc mesenteroides NRRL B-1149. Biotechnology and Bioprocess Engineering. 2002;7:234–236. [Google Scholar]
- Lacumin L, Comi G, Cantoni C, Cocolin L. Ecology and dynamics of coagulase-negative cocci isolated from sausages. Systematic and Applied Microbiology. 2006;29(6):480–486. doi: 10.1016/j.syapm.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Lozupone CA, Knight R. UniFrac: A new phylogenetic method for comparing microbial communities. Applied and Environmental Microbiology. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narai-Kanayama A, Tokita N, Aso K. Dependence of fructooligosacharide content on activity of fructooligosachardie-metabolizing enzymes in yacón (Smallanthus sonchifolius) tuberous roots during storage. Journal of Food Science. 2007;72(6):S381–S387. doi: 10.1111/j.1750-3841.2007.00422.x. [DOI] [PubMed] [Google Scholar]
- Ojansivu I, Ferreira C, Salminen S. Yacon, a new source of prebiotic oligosaccharides with history of safe use. Trends in Food Science and Technology. 2010;22:40–46. [Google Scholar]
- Pan Y, Breidt F. Enumeration of Viable Listeria monocytogenes Cells by real-time PCR with propidium monoazide and ethidium monoazide in the presence of dead cells. Applied and Environmental Microbiology. 2007;73(24):8028–8031. doi: 10.1128/AEM.01198-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra F, Tomar S, Rajput Y, Singh R. Characterization of mannitol producing strains of Leuconostoc species. World Journal of Microbiology and Biotechnology. 2011;27(4):933–939. [Google Scholar]
- Reina LD, Guija E, Reategui O, Parodi C, Encizo J. Yacon preservation by natural fermentation. Cientifica. 2009;6(3):221–231. [Google Scholar]
- Reina LD, Guija E, Reategui O, Parodi C, Encizo J, Pita D. Pickled yacon as a functional food. Cientifica. 2008;5:201–203. [Google Scholar]
- Ribeiro J. Dissertation (Master in Food Science) Chapter 3. Federal University of Lavras; Lavras, MG: 2008. Chemical and biochemical studies of in natura and processed yacon and influence of its consumption in the glycemic levels and fecal lipids of rats; pp. 107–132. [Google Scholar]
- Rodríguez H, Curiel JA, Landete JM, de las Rivas B, López de Felipe F, Gómez-Cordovés C, Mancheño JM, Muñoz R. Food phenolics and lactic acid bacteria. International Journal of Food Microbiology. 2009;132 (2–3):79–90. doi: 10.1016/j.ijfoodmicro.2009.03.025. [DOI] [PubMed] [Google Scholar]
- Saha BC, Racine FM. Biotechnology production of mannitol and its applications. Applied Microbiology and Biotechnology. 2011;89(4):879–91. doi: 10.1007/s00253-010-2979-3. [DOI] [PubMed] [Google Scholar]
- Wilson KH, Blitchington RB, Greene RC. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. Journal of Clinical Microbiology. 1990;28:942–1946. doi: 10.1128/jcm.28.9.1942-1946.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]