Abstract
Primary hyperoxaluria (PH) is a rare autosomal recessive disease characterized by oxalate accumulation in the kidneys and other organs. Three loci have been identified: AGXT (PH1), GRHPR (PH2), and HOGA1 (PH3). Here, we compared genotype to phenotype in 355 patients in the Rare Kidney Stone Consortium PH registry and calculated prevalence using publicly available whole-exome data. PH1 (68.4% of families) was the most severe PH type, whereas PH3 (11.0% of families) showed the slowest decline in renal function but the earliest symptoms. A group of patients with disease progression similar to that of PH3, but for whom no mutation was detected (11.3% of families), suggested further genetic heterogeneity. We confirmed that the AGXT p.G170R mistargeting allele resulted in a milder PH1 phenotype; however, other potential AGXT mistargeting alleles caused more severe (fully penetrant) disease. We identified the first PH3 patient with ESRD; a homozygote for two linked, novel missense mutations. Population analysis suggested that PH is an order of magnitude more common than determined from clinical cohorts (prevalence, approximately 1:58,000; carrier frequency, approximately 1:70). We estimated PH to be approximately three times less prevalent among African Americans than among European Americans because of a limited number of common European origin alleles. PH3 was predicted to be as prevalent as PH1 and twice as common as PH2, indicating that PH3 (and PH2) cases are underdiagnosed and/or incompletely penetrant. These results highlight a role for molecular analyses in PH diagnostics and prognostics and suggest that wider analysis of the idiopathic stone-forming population may be beneficial.
Keywords: genetic renal disease, kidney stones, molecular genetics
The primary hyperoxalurias (PHs) are rare autosomal recessive inborn errors of hepatic glyoxylate metabolism characterized by oxalate overproduction and elevated excretion.1–3 Calcium oxalate oversaturation leads to recurrent urolithiasis and/or nephrocalcinosis, with reduced renal elimination due to renal damage resulting in oxalate deposition in all tissues (systemic oxalosis).1,2,4,5
The estimated PH prevalence is <3:1,000,000, but in approximately 20%–50% of cases severe renal insufficiency, or recurrent disease after transplantation, occurs before diagnosis.3,6–8 Underdiagnosis probably occurs because of the phenotypic heterogeneity of PH, ranging from infantile nephrocalcinosis with kidney failure to only occasional stone formation (similar to idiopathic stone disease), and unfamiliarity with this rare monogenic disorder.3,9 Three genetic forms of PH have been defined: PH1–3, associated with mutations to AGXT, GRHPR, and HOGA1, respectively.10–12
PH1, caused by deficiency of the liver-specific peroxisomal enzyme alanine-glyoxylate aminotransferase (AGT), is the most severe PH form, accounting for about 80% of genetically characterized patients.1,3,13 It can manifest as infantile oxalosis, resulting in early death,14,15 but a more typical course is recurrent urolithiasis with progressive nephrocalcinosis and ESRD by 20–30 years of age.5,16–18 A total of 178 AGXT mutations have been described; the three most common, p.G170R, c.33dupC, and p.I244T, account for approximately 30%, 11%, and 6% of AGXT mutant alleles, respectively; p.I244T is common in Spanish/North African populations.19–24
AGXT p.G170R is associated with mistargeting of the AGT homodimer to mitochondria and the associated unmasking of the p.P11L mitochondrial targeting sequence (MTS) of the “minor” p.P11L/p.I340M haplotype.25–29 Patients with p.G170R have milder renal disease and respond to pyridoxine treatment, a cofactor that reduces enzyme mistargeting.25,27,28,30–32 The mutations p.G41R, p.F152I and p.I244T also unmask the MTS if present on the minor allele, and anecdotal evidence suggests that patients with p.F152I benefit from pyridoxine treatment.29,30,32,33 Detecting genotype-phenotype correlations beyond the minor allele requiring (MiR) variants is complicated by the allelic heterogeneity and marked phenotypic variability, both intrafamilial and among unrelated patients with the same allelic combination, suggesting environmental and modifier gene roles.5,25
PH2 is generally less severe than PH1 but with a similar age at first symptoms.1,2,4 It is caused by deficiency of glyoxylate reductase/hydroxypyruvate reductase (GR/HPR), and accounts for about 10% of genetically characterized PH cases.1,34,35 To date, 28 different mutations have been described, with c.103delG and c.403_404+2delAAGT accounting for 37% and 18% of mutant alleles, respectively; c.403_404+2delAAGT is predominantly a mutation found in Asians.4,22,35
PH3 is the least severe form, with good preservation of kidney function in most patients. The typical presentation is recurrent urolithiasis and marked hypercalciuria in the first decade, but less active stone formation later.1,2,36,37 HOGA1 encodes the liver-specific mitochondrial enzyme 4-hydroxy-2-oxogluterate aldolase (HOGA), and mutations cause hydroxy-2-oxogluterate aldolase build-up, inhibiting GR/HPR function.11,38 PH3 accounts for approximately 10% of genetically characterized cases, with some carriers found to be idiopathic stone formers, suggesting sensitivity to haploinsufficiency.1,39 There are 19 described mutations with c.700+5G>T accounting for about 50% of all HOGA1 alleles; p.E315del is found predominantly in Ashkenazi Jews.22,36,37,39 No allelic correlations have been established for PH2 or PH3.
Here, we evaluated genotype-phenotype correlations at the genic and allelic level using a large collection of PH patients from the Rare Kidney Stone Consortium Primary Hyperoxaluria registry (RKSC PH registry). In addition, we used publicly available whole-exome sequencing data to provide population-based estimates of PH prevalence. Overall, these studies provide new insight into PH phenotypes and the significance of particular alleles. In addition, they indicate a carrier rate far higher than estimated from clinical populations, suggesting a significant level of underdiagnosis and/or incomplete penetrance/variable expressivity.
Results
Genic and Allelic Analysis of the RKSC PH Registry Population
We mutation-screened the three known PH loci in 301 PH families (355 patients) using Sanger sequencing. Of these, 68.4% (206 pedigrees, 247 patients) were PH1, 9.3% (28 pedigrees, 35 patients) were PH2, and 11.0% (33 pedigrees, 38 patients) were PH3. Two mutant alleles were detected in each case. The remaining 11.3% (34 pedigrees, 35 patients) had a clinical phenotype consistent with PH according to the RKSC PH registry entry criteria (Concise Methods), yet no mutation was detected (NMD) in the known genes.
In total, we identified 121 different mutations within this cohort (83 AGXT, 21 GRHPR, and 17 HOGA1), of which 36 have not been previously described in the Human Gene Mutation Database (HGMD 2013.3 Professional) or the PH Mutation Database22 (PHMD; 18 AGXT, 13 GRHPR, and 5 HOGA1) (Supplemental Figure 1, Table 1). One of the novel mutations, GRHPR c.[-4G>A, -3C>T], is upstream of the transcription start codon generating a frameshifted novel start site with a stronger Kozak consensus (Supplemental Figure 2A).
Table 1.
Novel pathogenic variants identified within the RKSC PH registry
| Gene/Exon/IVS | cDNAa | Protein | In silico Analysis Scoreb | Second Allelec | Pedigree No.c |
|---|---|---|---|---|---|
| AGXT | |||||
| 1 | c.34_35dupAA | p.K12fs | – | c.34_35dupAA | 393–01 |
| 1 | c.126dupG | p.L43fs | – | c.126dupG | 287–01 |
| 2 | c.188G>A | p.G63D | C65/Affect Protein/Prob Damaging; 7/7 | p.G63D | 400–01 |
| 2 | c.292G>C | p.D98H | C0/Affect Protein/Prob Damaging; 6/7 | p.D98H | 401–01 |
| 2 | c.299_307dupTCCTGGTTG | p.V102_G103insVLV | – | p.S158L | 115–01 |
| 3 | c.402C>G | p.Y134* | – | p.Y134* | 419–01 |
| 4 | c.469delGd | p.E157fs | – | p.G170R | 157–01 |
| 5 | c.560C>A | p.S187Y | C15/Affect Protein/Prob Damaging; 6/7 | p.G170R | 369–01 |
| 5 | c.560_561insGGT | p.S187_L188insG | – | c.560_561insGGT | 69–01 |
| IVS5 | c.595+1G>T | p.G200fs | Donor splice site eliminated | c.595+1G>T | 104–01 |
| 8 | c.779A>G | p.Y260C | C45/Affect Protein/Prob Damaging; 5/7 | p.Y260C | 117–01 |
| 8 | c.781C>G | p.H261D | C65/Affect Protein/Prob Damaging; 7/7 | c.33dupC | 171–01 |
| 8 | c.815T>C | p.L272P | C65/Affect Protein/Prob Damaging; 7/7 | p.L272P | 462–01 |
| 8 | c.832delC | p.A277fs | – | p.S158* | 218–01 |
| 10 | c.949C>T | p.R317W | C65/Affect Protein/Prob Damaging; 7/7 | p.G190R | 155–01 |
| 10 | c.973delG | p.A325fs | p.I279T | 182–01 | |
| 11 | c.1078C>T | p.R360W | C65/Affect Protein/Prob Damaging; 7/7 | c.33dupC | 153–01 |
| 11 | c.1084G>Ad | p.G362S | C55/Affect Protein/Prob Damaging; 7/7 | c.847–3C>G | 43–01 |
| GRHPR | |||||
| 1 | c.[-4G>A, -3C>T] | p.M1ext-4 | WT (0.67) to Mut (0.71)e | c.103delG | 376–02 |
| 3 | c.271delG | p.D91fs | – | p.S209F | 380–02 |
| 3/IVS3 | c.287_287+4delGGTAAinsCCC | p.R96fs | Donor splice site eliminated | c.287_287+4delGGTAAinsCCC | 432–02 |
| 4 | c.337G>T | p.E113* | – | p.R302P | 28–02 |
| IVS4 | c.404+5G>A | p.N135fs | WT (0.99) to Mut (0.34)f | c.103delG, c.103delG, p.N135* | 34–02, 356–02, 147–02 |
| 5 | c.454dupA | p.T152fs | – | c.103delG, c.103delG | 184–02, 191–02 |
| 7 | c.626C>T | p.S209F | C65/Affect Protein/Prob Damaging; 7/7 | c.271delG, c.769dupG | 380–02, 78–02 |
| 7 | c.694delC | p.Q232fs | – | c.694delC | 333–02 |
| IVS7 | c.734+1G>A | p.R246fs | Donor splice site eliminated | p.A297T | 213–02 |
| 8 | c.769dupG | p.Q256fs | – | p.S209F | 78–02 |
| 9 | c.889G>A | p.A297T | C55/Affect Protein/Prob Damaging; 7/7 | c.734+1G>A | 213–02 |
| 9 | c.890_891dupCC | p.T298fs | – | p.E113K | 135–02 |
| 9 | c.905G>C | p.R302P | C65/Affect Protein/Prob Damaging; 7/7 | p.E113* | 28–02 |
| HOGA1 | |||||
| 2 | c.227G>A | p.G76D | C65/Affect Protein/Prob Damaging; 7/7 | p.M292T | 282–03 |
| 2 | c.308A>Tg | p.N103Ig | C55/Affect Protein/Benign; 1/7 | c.700+5G>T | 362–03 |
| 4 | c.533T>Ch | p.L178Ph | C65/Affect Protein/Prob Damaging; 7/7 | p.P179T | 457–03 |
| 4 | c.535C>Ah | p.P179Th | C35/Affect Protein/Possibly Damaging; 7/7 | p.L178P | 457–03 |
| 7 | c.973G>A | p.G325S | C55/Affect Protein/Prob Damaging; 7/7 | c.700+5G>T | 193–03 |
Nucleotide numbering based on NM_000030.2 (AGXT), NM_012203.1 (GRHPR), and NM_138413.3 (HOGA1).
In silico evaluation of novel missense mutations: AlignGVGD/SIFT/PolyPhen-2 and identity with multiple sequence alignment (human, bull, mouse, rat, dog, chicken, and zebrafish).
Second allele is listed in respect to pedigree order.
These variants were also identified as novel AGXT alleles in the recent publication from Mandrile et al.47
Kozak mutation was scored using NetStart 1.0; higher score more likely acts as transcription initiation codon.
Novel splicing was assessed using BDGP; higher score more likely acts as splice site.
Detail of this substitution shown in Supplemental Figure 2C.
Detail of this double substitution shown in Supplemental Figure 2B.
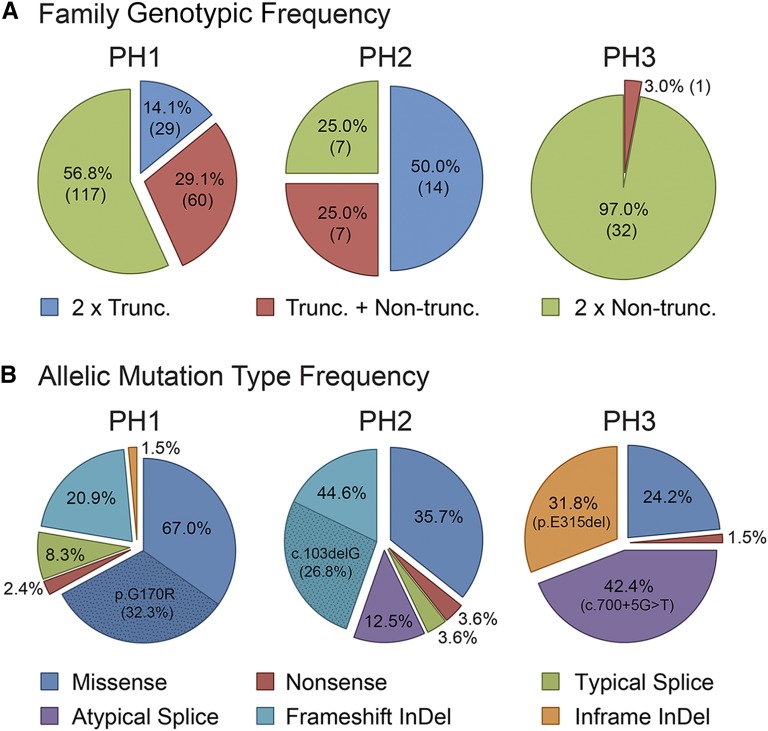
The genotypic and allelic breakdown varied considerably between the different genes (Figure 1). For instance, 50.0% of PH2 pedigrees contained two truncating mutations, an allelic combination that accounted for only the minority of PH1 genotypes (14.1%) and was absent in PH3 (Figure 1A). GRHPR was more prone to frameshifting InDels (insertion, duplication, deletion, or insertion+deletion), accounting for 44.6% of all mutant alleles, whereas most AGXT mutations were missense (67.0%) (Figure 1B). These enrichments were driven by common mutations; the AGXT mistargeting change p.G170R was found in 50.5% of PH1 families and GRHPR c.103delG in 35.7% of PH2 families. HOGA1 showed considerably less allelic heterogeneity with the two most common alleles (c.700+5G>T and p.E315del), accounting for 74.2% of the total (Figure 1B).
Figure 1.
Genotypic and allelic breakdown varies considerably between the different PH types in the mutation resolved 267 pedigrees. (A) Genotypic breakdown by PH type showing patients with two truncating alleles (nonsense, splice, and frameshifting InDels [insertion, duplication, deletion, or insertion+deletion]), two nontruncating alleles (missense and inframe InDels), or a truncating plus a nontruncating allele. Number in parentheses represents number of families with that genotype. (B) Allelic analysis by PH type of different mutation types. Common alleles of each PH type plus their frequencies are also highlighted. AGXT p.G170R was found homozygously (hom) in 29 families and heterozygously (het) in 75 families, GRHPR c.103delG (hom, 5 families; het, 5 families), HOGA1 c.700+5G>T (hom, 8 families; het, 12 families), HOGA1 p.E315del (hom, 7 families; het, 7 families).
Genotype-Phenotype Comparisons between PH Types
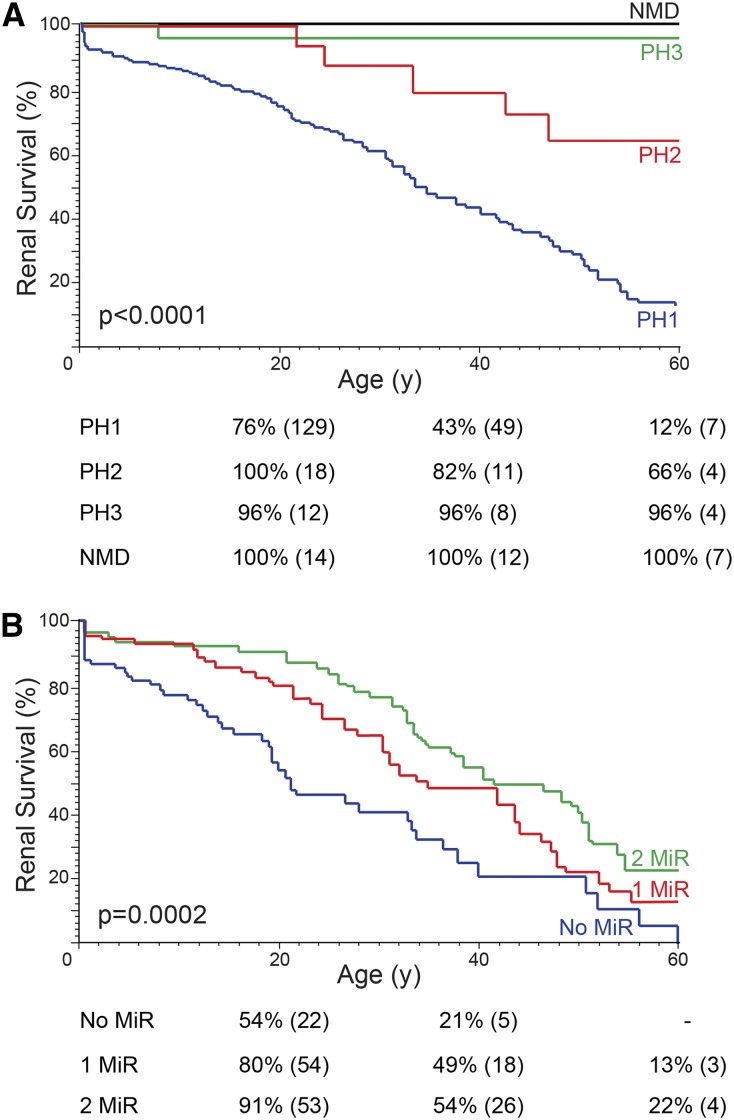
Within our cohort, the median (25th, 75th percentiles) age at PH symptom onset was 5.2 years (1.7, 15 years) and age at last contact was 21 years (9, 43 years). To date, 22 of 355 patients have died, and 139 developed ESRD. Renal survival analysis showed that PH1 patients were more likely to reach ESRD and at an earlier age, followed by PH2 and then PH3 (Figure 2A).1,2 No NMD patient experienced ESRD by age 60 years (Figure 2A), although two had later renal failure. Patients with PH1 had a higher incidence of nephrocalcinosis and higher urine oxalate, lower calcium, and lower citrate levels (Table 2). Patients with PH3 had earlier onset of symptoms than did those with PH1 or PH2, but had lower urine oxalate excretion and, in general, more slowly progressive disease. The NMD patients were the latest to show symptoms and had oxalate levels similar to those in patients with PH3. Elevated glycolate or l-glycerate levels indicated PH1 or PH2, respectively (Table 2).4,5
Figure 2.
Renal Survival plots showing (A) poorer renal survival for PH1 patients followed by PH2, and (B) better renal survival for PH1 patients with two MiR alleles. (A) Kaplan-Meier renal survival plot of the PH1, PH2, PH3, and NMD cohorts. (B) Kaplan-Meier renal survival plot of PH1 patients categorized as having two MiR alleles (homozygous, or compound heterozygous; AGXT p.G41R, p.F152I, p.G170R, p.I244T, 89 patients), one MiR allele in combination with a non-MiR allele (97 patients), or two alleles of which neither was a MiR allele (no MiR, 81 patients). Tables below Kaplan-Meier plots show survival estimates with number of patients at risk in parentheses.
Table 2.
Clinical presentation of RKSC PH patients based on PH type
| Variable | PH1 (n=247) | PH2 (n=35) | PH3 (n=38) | NMD (n=35) | P Value, Adjusted |
|---|---|---|---|---|---|
| Age at symptoms | 0.01 | ||||
| Median (y) | 5.2 | 7.4 | 2.6 | 11.5 | |
| Q1, Q3 (y) | (2.0, 14.0) | (1.7, 15.8) | (0.9, 5.5) | (3.9, 24.1) | |
| Minimum, maximum (y) | (0.1, 53.0) | (0.6, 42.0) | (0.3, 31.0) | (0.1, 69.7) | |
| Patients (n) | 213 | 31 | 33 | 29 | |
| Nephrocalcinosis, n (%) | <0.0001 | ||||
| Yes | 49 (30.6) | 5 (16.7) | 1 (2.7) | 5 (16.7) | |
| No | 111 (69.4) | 25 (83.3) | 36 (97.3) | 25 (83.3) | |
| Urine chemistry | |||||
| Ox24 (mmol/1.73 m2) (normal < 0.46)a | <0.0001 | ||||
| Median | 1.8 | 1.7 | 1.1 | 1.2 | |
| Q1, Q3 | (1.2, 2.7) | (1.1, 2.2) | (0.9, 1.4) | (1.0, 1.5) | |
| Patients (n) | 121 | 23 | 34 | 23 | |
| Ca24 (mg/1.73m2) (normal 100–300)a | <0.0001 | ||||
| Median | 56.8 | 91.5 | 91.4 | 135.9 | |
| Q1, Q3 | (34.1, 91.2) | (55.8, 134.2) | (63.5, 153.0) | (79.9, 236.8) | |
| Patients (n) | 82 | 19 | 26 | 20 | |
| Cit24 (mg/1.73 m2) (normal 320–1240)a | <0.0001 | ||||
| Median | 278.5 | 722.9 | 676.9 | 371.0 | |
| Q1, Q3 | (108.2, 488.3) | (245.2, 1285.0) | (416.3, 812.9) | (289.3, 954.5) | |
| Patients (n) | 74 | 15 | 27 | 20 | |
| Glycolate (mg/g creatinine) (normal 0–78)b | <0.0001 | ||||
| Median | 102.2 | 28.0 | 18.0 | 33.0 | |
| Q1, Q3 | (46.5, 218.0) | (22.0, 41.0) | (8.0, 42.0) | (14.0, 54.0) | |
| Patients (n) | 96 | 22 | 23 | 22 | |
| l-glycerate (mg/g creatinine) (normal 0–8)b | <0.0001 | ||||
| Median | 3.0 | 785.0 | 1.0 | 7.0 | |
| Q1, Q3 | (2.0, 9.0) | (271.0, 1467.0) | (0.0, 6.0) | (2.0, 11.0) | |
| Patients (n) | 91 | 19 | 21 | 21 |
Q1, Q3, first and third quartiles; Ox24, 24-hour oxalate; Ca24, 24-hour calcium; Cit24, 24-hour citrate.
Normal values are for adult patients. Values in table have been adjusted for body surface area to permit comparison of adult and pediatric patients.52,53
Normal values may be higher in children from birth to 5 years of age.
Allelic Associations with Phenotype in PH1
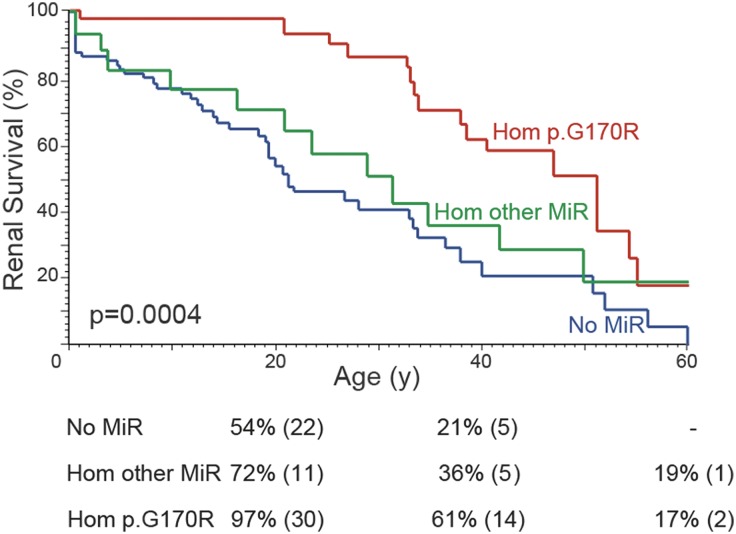
To compare AGXT genotype with PH1 phenotype, we characterized patients by their combination of mutation types, defined as truncating, nontruncating, or MiR (p.G41R, p.F152I, p.G170R, p.I244T), with the hypothesis that nontruncating/MiR mutations might still have some residual function. Patients with two MiR mutations had later onset of symptoms, progressed more slowly to ESRD, and had lower oxalate and glycolate levels than patients with any other genotypes (Figure 2B, Supplemental Table 1, Table 3). These results are consistent with previous findings for p.G170R25 and highlight the incomplete penetrance/variable expressivity of these alleles. No significant difference in phenotype was noted between patients with two truncating or two nontruncating mutations when MiR mutations were excluded (Supplemental Figure 3, Supplemental Table 2), suggesting that non-MiR, nontruncating alleles are usually fully inactivating. However, we did notice a renal survival benefit among those heterozygous for a MiR allele compared with those having no MiR allele, as previously reported for p.G170R (Figure 2B).25 This milder phenotype was also evident by lower urine glycolate levels but not by other urine chemistries, the age at disease onset, or the incidence of nephrocalcinosis (Table 3). To evaluate whether all MiR alleles behaved similarly with regard to phenotype, we separated p.G170R from the other MiR alleles (p.G41R, p.F152I, p.I244T). Kaplan-Meier renal survival curves revealed that only p.G170R homozygotes had milder disease, not patients homozygous for the other MiR alleles (Figure 3). The survival advantage for p.G170R persisted, although significance decreased, even after adjustment for treatment with pyridoxine before renal failure (Figure 3); this occurred in 53% of p.G170R homozygotes, 26% of other MiR homozygotes, and 38% of patients without a MiR allele. The proportion of patients treated did not significantly differ between the groups (P=0.14). Therefore, although all MiR alleles unmask the functional MTS, their penetrance significantly differs.29
Table 3.
Evaluation of PH1 severity based on number of MiR variants per genotype
| Variable | Hom MiR (n=69) | Het MiR (n=97) | No MiR (n=81) | P Value, Adjusted |
|---|---|---|---|---|
| Age at symptoms | 0.006 | |||
| Median (y) | 10.6 | 4.7 | 4.3 | |
| Q1, Q3 (y) | (2.4, 22.0) | (1.7, 12.4) | (0.7, 8.4) | |
| Minimum, maximum (y) | (0.5, 49.8) | (0.1, 53.0) | (0.1, 41.5) | |
| Patients (n) | 59 | 86 | 68 | |
| Nephrocalcinosis, n (%) | 0.08 | |||
| Yes | 7 (18.4) | 21 (30.4) | 21 (39.6) | |
| No | 31 (81.6) | 48 (69.6) | 32 (60.4) | |
| Urine chemistry | ||||
| Ox24 (mmol/1.73 m2) (normal < 0.46)a | <0.001 | |||
| Median | 1.1 | 2.0 | 2.3 | |
| Q1, Q3 | (0.7, 1.7) | (1.3, 2.6) | (1.7, 2.9) | |
| Patients (n) | 26 | 63 | 32 | |
| Ca24 (mg/1.73m2) (normal 100–300)a | 0.10 | |||
| Median | 79.5 | 54.1 | 54.9 | |
| Q1, Q3 | (29.0, 157.5) | (35.8, 78.6) | (32.7, 69.2) | |
| Patients (n) | 19 | 42 | 21 | |
| Cit24 (mg/1.73 m2) (normal 320–1240)a | 0.71 | |||
| Median | 402.5 | 275.7 | 281.7 | |
| Q1, Q3 | (125.4, 584.7) | (112.7, 436.2) | (87.9, 471.6) | |
| Patients (n) | 16 | 36 | 22 | |
| Glycolate (mg/g creatinine) (normal 0–78)b | 0.002 | |||
| Median | 45.0 | 96.0 | 182.0 | |
| Q1, Q3 | (19.0, 143.0) | (48.0, 182.0) | (89.0, 310.0) | |
| Patients (n) | 23 | 47 | 26 |
Hom, homozygous; Het, heterozygous; Q1, Q3, first and third quartiles; Ox24, 24-hour oxalate; Ca24, 24-hour calcium; Cit24, 24-hour citrate.
Normal values are for adult patients. Values in table have been adjusted for body surface area to permit comparison of adult and pediatric patients.52,53
Normal values may be higher in children from birth to 5 years of age.
Figure 3.
Renal survival plots showing that only homozygous p.G170R patients but not homozygotes of other MiR genotypes had a renal survival advantage. Kaplan-Meier renal survival plot plus survival estimate table for PH1 patients homozygous for p.G170R (34 patients), homozygous or compound heterozygous for another MiR allele (p.G41R, p.F152I, p.I244T, 19 patients), or having no MiR allele (81 patients). Pairwise comparisons show greater renal survival in patients homozygous for AGXT p.G170R but not the other MiR alleles compared with patients without a MiR allele (Hom p.G170R versus no MiR; P<0.0001 [P<0.001], Hom other MiR versus no MiR P=0.27 [P=0.35], Hom p.G170R versus Hom other MiR P=0.04 [P=0.05]). P values in square brackets are adjusted for the effect of pyridoxine treatment, reducing the overall significance but upholding the survival advantage of AGXT p.G170R homozygotes.
Allelic Associations with Phenotype in PH2 and PH3
To evaluate genotype-phenotype correlations in our PH2 cohort, patients were grouped according to mutation type (two truncating, two nontruncating, or a truncating plus a nontruncating allele). However, no significant associations were observed with age at symptoms or ESRD, nephrocalcinosis, and urine chemistry (Supplemental Table 3).
Patients with PH3 were grouped by the common mutations (homozygous for c.700+5G>T or p.E315del; heterozygous, compound heterozygous for c.700+5G>T/p.E315del, or lacking these mutations). As for PH2, no significant genotype-phenotype associations in our relatively small PH3 cohort were apparent (Supplemental Table 4). Interestingly, one PH3 patient developed ESRD at 8 years. This patient was homozygous for a pair of closely linked, nonconservative missense variants predicted to be highly pathogenic, rather than the common HOGA1 alleles (Supplemental Figure 2B, Table 1). This patient was symptomatic at age 18 months, with multiple calcium oxalate stones that were surgically removed; CKD was first noted at age 6 years, at least partly attributed to obstructive uropathy.
Disease Variability among Patients with the Same Genotype and within Families
Intrafamilial variable disease expression and phenotypic heterogeneity among patients with the same allelic combinations have been observed in PH1.5,25 In the current cohort, five PH1 families had differences of >20 years between siblings regarding the timing of ESRD onset, and considerable phenotypic variability was present between patients homozygous for specific AGXT alleles (Supplemental Table 5A). Within the PH2 and PH3 cohorts, homozygotes for common mutations also displayed considerable variability of disease expression (Supplemental Table 5A). Within one PH2 family homozygous for GRHPR c.103delG, one of three affected siblings reached ESRD at age 21 years, whereas the other two manifested with only occasional stone disease, with eGFRs of 40 and 85 ml/min per 1.73 m2 at 56 and 53 years, respectively.
Estimation of PH Prevalence Using Public Whole-Exome Sequencing Data
From the National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project (ESP) data, we extracted allelic counts of 22 known PH mutations (12 AGXT, 5 GRHPR, and 5 HOGA1) found within the population of 4300 European Americans (EA) and 2203 African Americans (AA) (Supplemental Figure 1, Supplemental Table 6).40 Further, we included all frameshifting or nonsense changes and scored all rare, nonsynonymous variants not previously identified as PH mutations using in silico tools, and thus identified 20 additional likely pathogenic variants (4 AGXT, 6 GRHPR, and 10 HOGA1) (Supplemental Figure 1, Supplemental Table 7).
With use of the Hardy-Weinberg equation, an overall PH carrier frequency (CF) of 1:71 with a predicted prevalence of 1:58,243 was calculated from the 22 known mutant alleles (Supplemental Table 6, Table 4). Including the predicted pathogenic alleles increased the CF and prevalence to 1:58 and 1:38,630, respectively (Supplemental Tables 7 and 8). PH was predicted to be approximately 60% less prevalent in AAs than in EAs (EA: CF, 1:61, prevalence, 1:40,276; AA: CF, 1:104, prevalence, 1:100,380) (Table 4). This difference was largely driven by the common EA allele HOGA1 c.700+5G>T (CF, 1:165; AA: CF, 1:1102) (Figure 4, Supplemental Table 6). The prevalence of PH1 was similar in EAs and AAs, even though AGXT p.G170R (EA: CF, 1:429) was unique to the EA population, but was balanced by AGXT p.R289H, the most prevalent AA allele, which accounted for approximately 33% of all AA mutant alleles (CF, 1:289; EA: CF, 1:1956) (Figure 4, Supplemental Table 6). Interestingly, the AGXT “minor” allele variants p.P11L/p.I340M were 15 times less prevalent in AAs than EAs (EA: CF, 1:3, prevalence, 1:15; AA: CF, 1:8, prevalence, 1:236) (Supplemental Table 6). As with PH1, PH2 prevalence was also similar in EAs versus AAs as GRHPR c.103delG was relatively common in both populations (EA: CF, 1:375; AA: CF, 1:426) (Figure 4, Supplemental Table 6).
Table 4.
Carrier frequency and prevalence based on PH mutant alleles found in the NHLBI ESP
| Variable | EA Patients | AA Patients | All Patients (EA + AA) |
|---|---|---|---|
| PH1+PH2+PH3 | |||
| Allele count (Alt/Ref) | 70/8495 | 21/4361 | 91/12,856 |
| Mutant allele frequency (%) | 0.824 | 0.482 | 0.708 |
| Prevalence (1 in)a | 40,276 | 100,380 | 58,243 |
| CF (1 in)a | 61 | 104 | 71 |
| PH1 (AGXT) | |||
| Allele count (Alt/Ref) | 22/8502 | 11/4359 | 33/12,861 |
| Mutant allele frequency (%) | 0.259 | 0.252 | 0.257 |
| Prevalence (1 in) | 149,347 | 157,032 | 151,887 |
| CF (1 in) | 194 | 199 | 195 |
| PH2 (GRHPR) | |||
| Allele count (Alt/Ref) | 15/8459 | 8/4348 | 23/12,807 |
| Mutant allele frequency (%) | 0.177 | 0.184 | 0.180 |
| Prevalence (1 in) | 318,021 | 295,392 | 310,055 |
| CF (1 in) | 282 | 272 | 279 |
| PH3 (HOGA1) | |||
| Allele count (Alt/Ref) | 33/8524 | 2/4377 | 35/12,901 |
| Mutant allele frequency (%) | 0.387 | 0.046 | 0.271 |
| Prevalence (1 in) | 66,720 | 4,789,532 | 135,866 |
| CF (1 in) | 130 | 1095 | 185 |
Prevalence was determined by combining the individual prevalence of all PH types with bigenic PH cases counting as carriers.
Figure 4.
Carrier frequencies of common PH alleles found in the NHLBI ESP account for significant racial differences. Depiction of the most common PH alleles separated by EA and AA CF. AGXT p.G170R (CF in EAs, 0.23%) and p.R289H (CF in AAs, 0.35%) account for 45.1% and 68.8% of the total PH1 CF, respectively. Similarly, GRHPR c.103delG accounts for 75.3% and 63.4% of the total EA or AA PH2 CF, respectively, and HOGA1 c.700+5G>T accounts for 78.4% and 100% of the total EA or AA PH3 CF, respectively.
When separated by PH type, PH1 and PH3 had a similar CF (1:195 versus 1:185) and prevalence (1:151,887 versus 1:135,866) (Table 4), whereas in our patient screen PH1 was six times more common (68.4% of families versus 11.0%). These data suggest a distinct underdiagnosis and/or lack of penetrance of PH3. The frequency of PH3 was predominantly driven by HOGA1 c.700+5G>T, which had an approximately 2.8 times greater CF (1 in 232) than AGXT p.G170R (1 in 649) in the ESP data, yet they accounted for 5.2% and 24.9% of all PH alleles, respectively, within our patient cohort. On the basis of these calculations, PH2 was predicted to be about 2.2 times less prevalent than PH1 or PH3 (CF, 1:279; prevalence, 1:310,055) (Table 4), but in our disease cohort its prevalence was similar to that of PH3 and approximately seven times rarer than PH1.
Discussion
Here we present genotype-phenotype correlations in one of the largest fully genotyped PH cohorts to date and for the first time calculate PH prevalence based on large population data. Because of the population size, we have significantly increased the number of unique PH mutations (AGXT by 10.1%, GRHPR by 46.4%, and HOGA1 by 26.3%) (Supplemental Figure 1). This analysis highlights the allelic heterogeneity of these disorders, although the population importance of a few common alleles is also reinforced (Figure 1). While elevated urinary glycolate,5 l-glycerate,4 or HOGA38 levels can often provide strong evidence regarding the PH gene involved (Table 2), there is significant overlap between PH types and normal individuals. Thus, we emphasize here the value of molecular analyses as a gold standard method to diagnose PH. As sequencing costs fall and new methods are adopted, it is increasingly practical to implement a protocol that fully screens the coding regions of all genes, not just commonly inherited variants,5 and thoroughly evaluates all detected variants.
Our genic prevalence in mutation resolved families—77.1% PH1, 10.5% PH2, 12.4% PH3—coincides with previous estimates (PH1, 80%; PH2, 10%; PH3, 10%) and highlights PH1 as the most common form, while PH3 is now at least as common as PH2.1–3 Interestingly, 11.3% of our clinically defined cohort (34 pedigrees) had no mutation detected in the known PH genes. Given the phenotype in these families (see below) and the reliability of molecular diagnostics (with no single PH mutations detected in our NMD group), further genetic heterogeneity in PH seems likely. Although our NMD cases met the RKSC PH registry criteria (Concise Methods), first symptoms occurred 4–9 years later than for genetically defined PH (Table 2), urine oxalate excretion was lower than PH1 and PH2 (but similar to PH3), and ESRD was rare, although there was considerable interfamilial variability (Supplemental Table 5B). These data indicate that rigorous clinical/molecular assessment is required to identify a population suitable for further gene discovery efforts in PH.
Our PH1 cohort displayed many characteristics reported in smaller cohorts; two AGXT variants accounted for 47.8% of PH1 mutant alleles (p.G170R [32.3%] and c.33dupC [15.5%]), and most mutations were missense, even after exclusion of common alleles (Figure 1).6,25,41–43 PH1 was the most severe PH type (Figure 2, Table 2), with a median age of first symptoms (5.2 years) and a cumulative renal survival (76%, 43%, and 12% at 20 years, 40 years, and 60 years, respectively) similar to that reported in the literature.1,2,5,18,25 Further, although elevated urinary excretion of glycolate suggested PH1, the range (6.0–1183.0 mg/g creatinine) overlapped with other PH types and NMD patients (minimum, maximum urine glycolate mg/g creatinine: PH2, 0.0, 105.0; PH3, 0.0, 116.0; NMD, 1.0, 168.0) (Table 2), indicating that glycolate analysis alone cannot unequivocally identify this PH form.5 Patients homozygous for one of the four MiR alleles29 had milder PH1 (Figure 2, Table 3), as suggested for p.G170R,25 but separating p.G170R from the other MiR alleles highlighted that later onset of ESRD was associated only with p.G170R (Figure 3); renal survival of the other MiR patients was similar to that of patients with truncating or non-MiR nontruncating alleles. These results emphasize the incompletely penetrant nature/variable expressivity of p.G170R and suggest a different pathomechanism for other MiR alleles, such as monomer aggregation or active site disruption, as indicated for p.G41R.29,44 This is important in considering pyridoxine therapy for patients with other MiR alleles because this cofactor is thought to stabilize the monomer/dimer. Nevertheless, anecdotal evidence suggests pyridoxine response in a variety of AGXT mutation patients (including p.F152I or two truncating alleles), indicating that alternative therapeutic benefits of pyridoxine may exist (e.g., influence on gene expression and/or enzyme activity).30,32,45–47
We found no difference in phenotype between patients with two non-MiR nontruncating versus two truncating alleles, suggesting that all these AGXT alleles are fully inactivating (Supplemental Figure 3, Supplemental Table 2). This contrasts with a recent publication that reported better renal survival of patients with two non-p.G170R nontruncating variants versus two truncating variants (16.9 years versus 9.9 years).47 Although it is possible we did not observe this correlation because of our smaller cohort size (247 patients in the RKSC PH registry versus 410 patients in the OxalEurope Consortium), and the significance of pairwise comparisons was not provided.47 The OxalEurope investigators also analyzed allelic effects between specific missense variants,47 but given the observed intrafamilial/intragenotype phenotypic heterogeneity (Supplemental Table 5A)5,25,47 due to genetic/environmental modifier effects, caution is required with interpretation in these small populations. More detailed allelic analysis of PH1 will be possible as population sizes increase and through pooled analysis of available cohorts.
In our cohort, PH2 was similar to PH1 in terms of age at symptoms and oxalate levels; however, progression to ESRD was significantly slower (Figure 2, Table 2).2,4 Elevated urinary l-glycerate levels separated PH2 cases from PH1 and PH3 cases, but some overlap with the genetically unresolved group was seen (minimum, maximum urine l-glycerate mg/g creatinine: PH1, 0.0, 129.0; PH2, 151.0, 4355.0; PH3, 0.0, 40.0; NMD, 0.0, 152.0).4 Similarly, dramatic intrafamilial/intragenotype phenotypic heterogeneity was observed (Supplemental Table 5A), suggesting significant genetic/environmental modifier effects in PH2. No significant associations of phenotype with mutation type were detected in patients with PH2 (Supplemental Table 3), although our cohort was probably too small to identify subtle differences in disease severity. Interestingly, most GRHPR mutant alleles were frameshifting, largely driven by the common c.103delG variant.
In our cohort, no PH3 family had two truncating alleles (Figure 1), with only one such patient described.36 Phenotypic analysis of patients with varying or no detected HOGA levels will prove interesting and provide insight into whether most HOGA1 alleles are incompletely or fully penetrant, consistent with structural analyses.48 Interestingly, some HOGA1 carriers present with mild hyperoxaluria or idiopathic urinary stone disease39 (unlike other forms of PH), and the first reported patient with PH3 to reach ESRD was homozygous for two novel, linked missense variants (Supplemental Figure 2); suggesting that more penetrant mutations may have a stronger phenotypic effect. However, the ESRD in this single patient with PH3 may have, at least in part, been aggravated by renal injury from recurrent stone removal surgeries. Paradoxically, patients with PH3 present with symptoms at the earliest age but subsequently have relatively mild disease (Figure 2, Table 2), suggesting decreased sensitivity to HOGA levels with aging. The lack of a detected genotype-phenotype correlation between the two most common HOGA1 alleles (c.700+5G>T and p.E315del) may again be due to the small populations and other genetic/environmental modifier effects (Supplemental Figure 5A).
The most important finding from this study is that the overall carrier frequency (approximately 1:70) and the inferred prevalence (approximately 1:58,000) calculated from population data are close to an order of magnitude greater than estimates from clinical studies. Because these new estimates come from counting known mutant alleles and are largely driven by a few common alleles, this study is unlikely to overestimate the carrier frequency. The estimates of PH1 prevalence are about twice the previous estimates, but the difference is much more notable for PH3, which has an observed population carrier frequency higher than PH1 but is 6-fold less common in currently identified clinical populations. PH2 has a population carrier frequency half that of PH1 but is more than 7-fold less common in clinical populations. It is notable that PH is determined to be about 2.5-fold less prevalent in AAs than EAs, a difference mainly driven by the HOGA1 c.700+5G>T mutation (Figure 4). Interestingly, AGXT p.G170R is exclusively found in EAs, and the AGXT MTS variants (p.P11L, p.I340M, minor haplotype) are much rarer in AAs than EAs (Figure 4, Supplemental Table 6).49 This finding suggests that mutations predicted to mistarget AGT might generally remain asymptomatic in AAs because presence of the minor haplotype is required for the MTS to be active.29 Interestingly, PH1 prevalence is nonetheless similar between EAs and AAs because of the most common AA mutation, AGXT p.R289H (reported in the PHMD22). This variant is significantly more common than any PH allele (excluding EA: HOGA1 c.700+5G>T) (Figure 4) and scores poorly,40 calling into question its true pathogenic nature. Removal of this allele would decrease PH1 prevalence by 48% (CF, 1:270; prevalence, 1:291,256) and overall PH prevalence by 18% (CF, 1:79; prevalence, 1:71,333, Supplemental Table 9). Further, PH1 prevalence in AAs would decrease by 87% (CF, 1:549; prevalence, 1:1,203,908) and PH prevalence overall within the AA population by 56% (CF, 1:156; prevalence, 1:225,971). Consequently, PH would be about 82% less prevalent in AAs than EAs.
The difference between the expected and observed prevalence, especially for PH3 and to a lesser extent for PH2, is likely due to underdiagnosis of these diseases, which have overall milder phenotypes and are much less likely than PH1 to result in ESRD. In addition, relatively mild disease in a subset of PH1 patients (especially p.G170R homozygotes) and a wide variability in age at first symptoms (Table 2), may account for the 2-fold underdiagnosis of PH1. Further, misdiagnosis of PH for other renal diseases, as illustrated by homozygosity for an AGXT mutation in a patient with nephronophthisis,50 may also play a role in underestimation of PH prevalence. Therefore, PH genotype analysis of idiopathic stone disease populations and other patients with PH phenocopies but no known mutations are likely to be informative for undiagnosed PH cases. Indeed, within a stone clinic setting the diagnosis of PH should be carefully considered among patients with unexplained hyperoxaluria and/or CKD, according to the published diagnostic algorithm.51 This consists of an initial comprehensive analysis of urine oxalate and chemistries plus plasma oxalate, followed by molecular testing of the PH genes in suspected cases.
Concise Methods
RKSC PH Registry
The relevant institutional review boards and ethics committees approved the study, and all participants gave informed consent. The RKSC PH registry is a secure web-based registry that enables international contributions.3 Patients within the registry met at least one of the following criteria: (1) Liver biopsy confirmed specific enzyme deficiency of AGT, GR/HPR, or HOGA, (2) molecular diagnostic testing of AGXT, GRHPR, and HOGA1 identified two mutant alleles in one of the genes, (3) urinary oxalate excretion was >0.8 mmol/1.73 m2 per day in the absence of an identifiable secondary cause of hyperoxaluria, and/or (4) ESRD had developed, along with (a) predialysis plasma oxalate>60 μmol/L and renal biopsy confirming extensive oxalate deposition or (b) systemic oxalosis (retinal oxalate deposits; oxalate deposits in bone marrow, skin, or other tissue; nephrocalcinosis; or calcium oxalate nephrolithiasis). As of June 2014, 422 patients from 37 countries had been enrolled in the RKSC PH registry, of whom 355 were screened for mutations in the known PH genes.
DNA Collection and Sanger Mutation Screening
Patients enrolled in the RKSC PH registry who consented to molecular testing had blood drawn for DNA isolation. Sanger sequencing was performed of all coding exons±20 bp (AGXT, NM_000030.2; GRHPR, NM_012203.1; HOGA1, NM_138413.3) using M13-tailed primers (Beckman Coulter). Primer sequences can be obtained by request. All Sanger chromatograms were analyzed using Mutation Surveyor, version 4.06 (Softgenetics), and identified variants were categorized as nonsense, missense, frameshifting, or inframe InDels; typical splice (±2 bp from exon boundary); or atypical splice (>±2 bp from exon boundary). A total of 115 families (approximately 38%) were screened for the coding regions of all three genes simultaneously, while the remainder were screened sequentially until two pathogenic alleles were identified (first AGXT, second GRHPR, third HOGA1). None of the NMD cases had a single pathogenic allele in any of the three genes, and none of the patients with PH1, 2, or 3 had a third pathogenic allele in the same or another PH gene.
Evaluation of Novel PH Alleles
Amino acid substitutions not previously reported in HGMD or PHMD22 were evaluated using the in silico prediction programs AlignGVGD (http://agvgd.iarc.fr/index.php), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/), and SIFT (http://sift.jcvi.org). The novel GRHPR atypical splice site mutation (c.404+5G>A) was evaluated using the Berkeley Drosophila Genome Project (http://www.fruitfly.org) with the DNA sequence of the altered ±2 exons as input. The novel GRHPR Kozak sequence mutation (c.[−4G>A, −3C>T]) was assessed using NetStart 1.0 (http://www.cbs.dtu.dk/services/NetStart/) with the full mRNA+5′ untranslated region as input. Additionally, for amino acid substitutions, multiple sequence alignments using AGT, GR/HPR, and HOGA protein orthologs of bull (Bos taurus), mouse (Mus musculus), rat (Rattus norvegicus), dog (Canis lupus), chicken (Gallus gallus), and zebrafish (Danio rerio) were used to evaluate evolutionary conservation. Amino acid substitutions were scored as pathogenic if two of three in silico tools predicted them as damaging, the change was evolutionarily conserved, and/or the variant segregated appropriately.
Statistical Analyses
Phenotype analyses used the first available 24-hour urine (oxalate, calcium, citrate, glycolate, l-glycerate) and first renal imaging (nephrocalcinosis). More than 80% of these tests were done from 1 year before to 3 years after PH diagnosis. Comparisons of genotypic groups with continuous and nominal phenotypic factors at disease presentation were done using the Kruskal–Wallis rank test and chi-squared tests, respectively. Mixed-effects models (urine chemistry ranks) and generalized estimating equations (nephrocalcinosis) were also used to test genotypic effects while adjusting for possible pedigree effects. Time from birth to development of renal failure was estimated using Kaplan–Meier curves, with comparisons based on the log-rank test and censoring subjects without renal failure at last follow-up. The Cox model with robust variance estimator was used to test associations of phenotype with time to ESRD, with adjustment for potential within-pedigree correlations. The adjustment for the effect of pyridoxine treatment was performed using a time-dependent covariate in the Cox model. All statistical tests were two sided, with an α level of 0.05; only P values adjusted for potential within-pedigree effects as described above were reported.
Prevalence Calculations
The data of the NHLBI ESP40 were downloaded from http://evs.gs.washington.edu/EVS/ in VCF file format and analyzed using SNP & Variation Suit, version 7 (GoldenHelix). Variants within AGXT, GRHPR, and HOGA1 that were known as PH mutations (HGMD, PHMD, RKSC PH registry) were annotated (Supplemental Figure 1, Supplemental Table 6). All other variants with an MAF <0.01% were scored for potential pathogenicity using the criteria described above (Supplemental Figure 1, Supplemental Table 7). The only PH mutation reported as homozygous in ESP was GRHPR c.103delG. However, the variant is located in a poly-G stretch, which often results in misalignment; hence, genotype and allele counts were adjusted to represent this homozygous call as heterozygous. Using alternate and total allele counts, prevalence (1/p2) and CFs [1/(1−p2−q2)=1/(2pq)] were calculated according to Hardy-Weinberg equilibrium for each PH type using the sum of all alternate PH1, 2, or 3 alleles (p, known [Table 4] or known+scored as pathogenic [Supplemental Table 8]) and all WT alleles (1−p=q). Calculations for PH overall were done by adding the p2 values of each PH type, with bigenic PH individuals counted as carriers.
Disclosures
None.
Supplementary Material
Acknowledgments
We thank all of the patients and families who have participated in the RKSC PH registry as well as the many physicians who collected the detailed clinical records (see below and Supplemental Material for the list of RKSC coordinators and participation/contribution sites, respectively). Further, we thank the study coordinators who collected clinical data and biological samples, as well as Ramila A. Mehta for her statistical advice.
The RKSC is a part of the National Institutes of Health Rare Diseases Clinical Research Network. Funding and/or programmatic support for this project has been provided by U54-DK083908 from the National Institute of Diabetes and Digestive and Kidney Diseases, National Center for Advancing Translational Sciences, Mayo Hyperoxaluria Center, and Mayo Foundation. This work was further supported by the Oxalosis and Hyperoxaluria Foundation and the American Society of Nephrology Ben J. Lipps Research Fellowship (K.H.).
This work was partially presented as two abstracts at the Annual Meeting of the American Society of Nephrology, 2013 (Atlanta, GA), and the 11th International Primary Hyperoxaluria Workshop, 2014 (Chicago, IL).
RKSC Coordinators and Coordination Sites: Craig Langman (Children's Memorial Hospital, Chicago), Lawrence Copelvitch (Children's Hospital of Philadelphia), Yaacov Frishberg (Shaare Zedek Medical Center, Jerusalem, Israel), Vidar Edvardsson (Landspitali University Hospital, Reykjavik, Iceland), David Goldfarb (New York University), Dean Assimos (University of Alabama, Birmingham), and Todd Lowther (Wake Forest University School of Medicine, Winston-Salem).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2014070698/-/DCSupplemental.
References
- 1.Hoppe B: An update on primary hyperoxaluria. Nat Rev Nephrol 8: 467–475, 2012 [DOI] [PubMed] [Google Scholar]
- 2.Cochat P, Rumsby G: Primary hyperoxaluria. N Engl J Med 369: 649–658, 2013 [DOI] [PubMed] [Google Scholar]
- 3.Lieske JC, Monico CG, Holmes WS, Bergstralh EJ, Slezak JM, Rohlinger AL, Olson JB, Milliner DS: International registry for primary hyperoxaluria. Am J Nephrol 25: 290–296, 2005 [DOI] [PubMed] [Google Scholar]
- 4.Rumsby G: Primary hyperoxaluria type 2. In: GeneReviews(R), edited by Pagon RA, Adam MP, Bird TD, Dolan CR, Fong CT, Smith RJH, Stephens K, Seattle, WA, University of Washington, 1993, Updated 2011 [Google Scholar]
- 5.Coulter-Mackie MB, White CT, Hurley RM, Chew BH, Lange D: Primary hyperoxaluria type 1. In: GeneReviews(R), edited by Pagon RA, Adam MP, Bird TD, Dolan CR, Fong CT, Smith RJH, Stephens K, Seattle, WA, University of Washington, 1993, Updated 2011 [Google Scholar]
- 6.Pirulli D, Marangella M, Amoroso A: Primary hyperoxaluria: Genotype-phenotype correlation. J Nephrol 16: 297–309, 2003 [PubMed] [Google Scholar]
- 7.Harambat J, van Stralen KJ, Espinosa L, Groothoff JW, Hulton SA, Cerkauskiene R, Schaefer F, Verrina E, Jager KJ, Cochat P, European Society for Pediatric Nephrology/European Renal Association-European Dialysis and Transplant Association (ESPN/ERA-EDTA) Registry : Characteristics and outcomes of children with primary oxalosis requiring renal replacement therapy. Clin J Am Soc Nephrol 7: 458–465, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Hoeven SM, van Woerden CS, Groothoff JW: Primary hyperoxaluria type 1, a too often missed diagnosis and potentially treatable cause of end-stage renal disease in adults: Results of the Dutch cohort. Nephrol Dial Transplant 27: 3855–3862, 2012 [DOI] [PubMed] [Google Scholar]
- 9.Danpure CJ, Jennings PR, Fryer P, Purdue PE, Allsop J: Primary hyperoxaluria type 1: Genotypic and phenotypic heterogeneity. J Inherit Metab Dis 17: 487–499, 1994 [DOI] [PubMed] [Google Scholar]
- 10.Cramer SD, Ferree PM, Lin K, Milliner DS, Holmes RP: The gene encoding hydroxypyruvate reductase (GRHPR) is mutated in patients with primary hyperoxaluria type II. Hum Mol Genet 8: 2063–2069, 1999 [DOI] [PubMed] [Google Scholar]
- 11.Belostotsky R, Seboun E, Idelson GH, Milliner DS, Becker-Cohen R, Rinat C, Monico CG, Feinstein S, Ben-Shalom E, Magen D, Weissman I, Charon C, Frishberg Y: Mutations in DHDPSL are responsible for primary hyperoxaluria type III. Am J Hum Genet 87: 392–399, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Purdue PE, Lumb MJ, Fox M, Griffo G, Hamon-Benais C, Povey S, Danpure CJ: Characterization and chromosomal mapping of a genomic clone encoding human alanine:glyoxylate aminotransferase. Genomics 10: 34–42, 1991 [DOI] [PubMed] [Google Scholar]
- 13.Danpure CJ, Jennings PR: Peroxisomal alanine:glyoxylate aminotransferase deficiency in primary hyperoxaluria type I. FEBS Lett 201: 20–24, 1986 [DOI] [PubMed] [Google Scholar]
- 14.Millan MT, Berquist WE, So SK, Sarwal MM, Wayman KI, Cox KL, Filler G, Salvatierra O, Jr, Esquivel CO: One hundred percent patient and kidney allograft survival with simultaneous liver and kidney transplantation in infants with primary hyperoxaluria: A single-center experience. Transplantation 76: 1458–1463, 2003 [DOI] [PubMed] [Google Scholar]
- 15.Cochat P: Primary hyperoxaluria type 1. Kidney Int 55: 2533–2547, 1999 [DOI] [PubMed] [Google Scholar]
- 16.Hoppe B: Evidence of true genotype-phenotype correlation in primary hyperoxaluria type 1. Kidney Int 77: 383–385, 2010 [DOI] [PubMed] [Google Scholar]
- 17.Cochat P, Liutkus A, Fargue S, Basmaison O, Ranchin B, Rolland MO: Primary hyperoxaluria type 1: Still challenging! Pediatr Nephrol 21: 1075–1081, 2006 [DOI] [PubMed] [Google Scholar]
- 18.Hoppe B, Beck BB, Milliner DS: The primary hyperoxalurias. Kidney Int 75: 1264–1271, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danpure CJ: Molecular aetiology of primary hyperoxaluria type 1. Nephron, Exp Nephrol 98: e39–e44, 2004 [DOI] [PubMed] [Google Scholar]
- 20.Danpure CJ, Lumb MJ, Birdsey GM, Zhang X: Alanine:glyoxylate aminotransferase peroxisome-to-mitochondrion mistargeting in human hereditary kidney stone disease. Biochim Biophys Acta 1647: 70–75, 2003 [DOI] [PubMed] [Google Scholar]
- 21.Williams EL, Acquaviva C, Amoroso A, Chevalier F, Coulter-Mackie M, Monico CG, Giachino D, Owen T, Robbiano A, Salido E, Waterham H, Rumsby G: Primary hyperoxaluria type 1: Update and additional mutation analysis of the AGXT gene. Hum Mutat 30: 910–917, 2009 [DOI] [PubMed] [Google Scholar]
- 22.Rumsby G: Primary Hyperoxaluria Mutation Database. 2013. https://www.uclh.nhs.uk/OurServices/ServiceA-Z/PATH/PATHBIOMED/CBIO/Pages/Phmdatabase.aspx. Accessed October 12, 2014
- 23.Benhaj Mbarek I, Abroug S, Omezzine A, Zellama D, Achour A, Harbi A, Bouslama A: Selected AGXT gene mutations analysis provides a genetic diagnosis in 28% of Tunisian patients with primary hyperoxaluria. BMC Nephrol 12: 25, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santana A, Salido E, Torres A, Shapiro LJ: Primary hyperoxaluria type 1 in the Canary Islands: A conformational disease due to I244T mutation in the P11L-containing alanine:glyoxylate aminotransferase. Proc Natl Acad Sci U S A 100: 7277–7282, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harambat J, Fargue S, Acquaviva C, Gagnadoux MF, Janssen F, Liutkus A, Mourani C, Macher MA, Abramowicz D, Legendre C, Durrbach A, Tsimaratos M, Nivet H, Girardin E, Schott AM, Rolland MO, Cochat P: Genotype-phenotype correlation in primary hyperoxaluria type 1: The p.Gly170Arg AGXT mutation is associated with a better outcome. Kidney Int 77: 443–449, 2010 [DOI] [PubMed] [Google Scholar]
- 26.Danpure CJ, Cooper PJ, Wise PJ, Jennings PR: An enzyme trafficking defect in two patients with primary hyperoxaluria type 1: Peroxisomal alanine/glyoxylate aminotransferase rerouted to mitochondria. J Cell Biol 108: 1345–1352, 1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cellini B, Lorenzetto A, Montioli R, Oppici E, Voltattorni CB: Human liver peroxisomal alanine:glyoxylate aminotransferase: Different stability under chemical stress of the major allele, the minor allele, and its pathogenic G170R variant. Biochimie 92: 1801–1811, 2010 [DOI] [PubMed] [Google Scholar]
- 28.Pey AL, Salido E, Sanchez-Ruiz JM: Role of low native state kinetic stability and interaction of partially unfolded states with molecular chaperones in the mitochondrial protein mistargeting associated with primary hyperoxaluria. Amino Acids 41: 1233–1245, 2011 [DOI] [PubMed] [Google Scholar]
- 29.Fargue S, Lewin J, Rumsby G, Danpure CJ: Four of the most common mutations in primary hyperoxaluria type 1 unmask the cryptic mitochondrial targeting sequence of alanine:glyoxylate aminotransferase encoded by the polymorphic minor allele. J Biol Chem 288: 2475–2484, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monico CG, Olson JB, Milliner DS: Implications of genotype and enzyme phenotype in pyridoxine response of patients with type I primary hyperoxaluria. Am J Nephrol 25: 183–188, 2005 [DOI] [PubMed] [Google Scholar]
- 31.Monico CG, Rossetti S, Olson JB, Milliner DS: Pyridoxine effect in type I primary hyperoxaluria is associated with the most common mutant allele. Kidney Int 67: 1704–1709, 2005 [DOI] [PubMed] [Google Scholar]
- 32.van Woerden CS, Groothoff JW, Wijburg FA, Annink C, Wanders RJ, Waterham HR: Clinical implications of mutation analysis in primary hyperoxaluria type 1. Kidney Int 66: 746–752, 2004 [DOI] [PubMed] [Google Scholar]
- 33.Cellini B, Montioli R, Paiardini A, Lorenzetto A, Voltattorni CB: Molecular insight into the synergism between the minor allele of human liver peroxisomal alanine:glyoxylate aminotransferase and the F152I mutation. J Biol Chem 284: 8349–8358, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giafi CF, Rumsby G: Kinetic analysis and tissue distribution of human D-glycerate dehydrogenase/glyoxylate reductase and its relevance to the diagnosis of primary hyperoxaluria type 2. Ann Clin Biochem 35: 104–109, 1998 [DOI] [PubMed] [Google Scholar]
- 35.Cregeen DP, Williams EL, Hulton S, Rumsby G: Molecular analysis of the glyoxylate reductase (GRHPR) gene and description of mutations underlying primary hyperoxaluria type 2. Hum Mutat 22: 497, 2003 [DOI] [PubMed] [Google Scholar]
- 36.Williams EL, Bockenhauer D, van’t Hoff WG, Johri N, Laing C, Sinha MD, Unwin R, Viljoen A, Rumsby G: The enzyme 4-hydroxy-2-oxoglutarate aldolase is deficient in primary hyperoxaluria type 3. Nephrol Dial Transplant 27: 3191–3195, 2012 [DOI] [PubMed] [Google Scholar]
- 37.Beck BB, Baasner A, Buescher A, Habbig S, Reintjes N, Kemper MJ, Sikora P, Mache C, Pohl M, Stahl M, Toenshoff B, Pape L, Fehrenbach H, Jacob DE, Grohe B, Wolf MT, Nürnberg G, Yigit G, Salido EC, Hoppe B: Novel findings in patients with primary hyperoxaluria type III and implications for advanced molecular testing strategies. Eur J Hum Genet 21: 162–172, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riedel TJ, Knight J, Murray MS, Milliner DS, Holmes RP, Lowther WT: 4-Hydroxy-2-oxoglutarate aldolase inactivity in primary hyperoxaluria type 3 and glyoxylate reductase inhibition. Biochim Biophys Acta 1822: 1544–1552, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monico CG, Rossetti S, Belostotsky R, Cogal AG, Herges RM, Seide BM, Olson JB, Bergstrahl EJ, Williams HJ, Haley WE, Frishberg Y, Milliner DS: Primary hyperoxaluria type III gene HOGA1 (formerly DHDPSL) as a possible risk factor for idiopathic calcium oxalate urolithiasis. Clin J Am Soc Nephrol 6: 2289–2295, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Exome Variant Server: NHLBI GO Exome Sequencing Project (ESP). http://evs.gs.washington.edu/EVS/. Accessed October 20, 2014
- 41.Amoroso A, Pirulli D, Florian F, Puzzer D, Boniotto M, Crovella S, Zezlina S, Spanò A, Mazzola G, Savoldi S, Ferrettini C, Berutti S, Petrarulo M, Marangella M: AGXT gene mutations and their influence on clinical heterogeneity of type 1 primary hyperoxaluria. J Am Soc Nephrol 12: 2072–2079, 2001 [DOI] [PubMed] [Google Scholar]
- 42.Takayama T, Nagata M, Ichiyama A, Ozono S: Primary hyperoxaluria type 1 in Japan. Am J Nephrol 25: 297–302, 2005 [DOI] [PubMed] [Google Scholar]
- 43.Coulter-Mackie MB, Rumsby G: Genetic heterogeneity in primary hyperoxaluria type 1: Impact on diagnosis. Mol Genet Metab 83: 38–46, 2004 [DOI] [PubMed] [Google Scholar]
- 44.Cellini B, Montioli R, Paiardini A, Lorenzetto A, Maset F, Bellini T, Oppici E, Voltattorni CB: Molecular defects of the glycine 41 variants of alanine glyoxylate aminotransferase associated with primary hyperoxaluria type I. Proc Natl Acad Sci U S A 107: 2896–2901, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fargue S, Rumsby G, Danpure CJ: Multiple mechanisms of action of pyridoxine in primary hyperoxaluria type 1. Biochim Biophys Acta 1832: 1776–1783, 2013 [DOI] [PubMed] [Google Scholar]
- 46.Hoyer-Kuhn H, Kohbrok S, Volland R, Franklin J, Hero B, Beck BB, Hoppe B: Vitamin B6 in primary hyperoxaluria I: First prospective trial after 40 years of practice. Clin J Am Soc Nephrol 9: 468–477, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mandrile G, van Woerden CS, Berchialla P, Beck BB, Acquaviva Bourdain C, Hulton SA, Rumsby G: Data from a large European study indicate that the outcome of primary hyperoxaluria type 1 correlates with the AGXT mutation type. Kidney Int 86: 1197–1204, 2014 [DOI] [PubMed] [Google Scholar]
- 48.Riedel TJ, Johnson LC, Knight J, Hantgan RR, Holmes RP, Lowther WT: Structural and biochemical studies of human 4-hydroxy-2-oxoglutarate aldolase: Implications for hydroxyproline metabolism in primary hyperoxaluria. PLoS ONE 6: e26021, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caldwell EF, Mayor LR, Thomas MG, Danpure CJ: Diet and the frequency of the alanine:glyoxylate aminotransferase Pro11Leu polymorphism in different human populations. Hum Genet 115: 504–509, 2004 [DOI] [PubMed] [Google Scholar]
- 50.Gee HY, Otto EA, Hurd TW, Ashraf S, Chaki M, Cluckey A, Vega-Warner V, Saisawat P, Diaz KA, Fang H, Kohl S, Allen SJ, Airik R, Zhou W, Ramaswami G, Janssen S, Fu C, Innis JL, Weber S, Vester U, Davis EE, Katsanis N, Fathy HM, Jeck N, Klaus G, Nayir A, Rahim KA, Al Attrach I, Al Hassoun I, Ozturk S, Drozdz D, Helmchen U, O’Toole JF, Attanasio M, Lewis RA, Nürnberg G, Nürnberg P, Washburn J, MacDonald J, Innis JW, Levy S, Hildebrandt F: Whole-exome resequencing distinguishes cystic kidney diseases from phenocopies in renal ciliopathies. Kidney Int 85: 880–887, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edvardsson VO, Goldfarb DS, Lieske JC, Beara-Lasic L, Anglani F, Milliner DS, Palsson R: Hereditary causes of kidney stones and chronic kidney disease. Pediatr Nephrol 28: 1923–1942, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gibbs DA, Watts RW: The variation of urinary oxalate excretion with age. J Lab Clin Med 73: 901–908, 1969 [PubMed] [Google Scholar]
- 53.Stapenhorst L, Sassen R, Beck B, Laube N, Hesse A, Hoppe B: Hypocitraturia as a risk factor for nephrocalcinosis after kidney transplantation. Pediatr Nephrol 20: 652–656, 2005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.