Abstract
Phenotypic changes induced by extracellular vesicles have been implicated in mesenchymal stromal cell–promoted recovery of AKI. MicroRNAs are potential candidates for cell reprogramming toward a proregenerative phenotype. The aim of this study was to evaluate whether microRNA deregulation inhibits the regenerative potential of mesenchymal stromal cells and derived extracellular vesicles in a model of glycerol-induced AKI in severe combined immunodeficient mice. We generated mesenchymal stromal cells depleted of Drosha to alter microRNA expression. Drosha-knockdown cells produced extracellular vesicles that did not differ from those of wild-type cells in quantity, surface molecule expression, and internalization within renal tubular epithelial cells. However, these vesicles showed global downregulation of microRNAs. Whereas wild-type mesenchymal stromal cells and derived vesicles administered intravenously induced morphologic and functional recovery in AKI, the Drosha-knockdown counterparts were ineffective. RNA sequencing analysis showed that kidney genes deregulated after injury were restored by treatment with mesenchymal stromal cells and derived vesicles but not with Drosha-knockdown cells and vesicles. Gene ontology analysis showed in AKI an association of downregulated genes with fatty acid metabolism and upregulated genes with inflammation, matrix-receptor interaction, and cell adhesion molecules. These alterations reverted after treatment with wild-type mesenchymal stromal cells and extracellular vesicles but not after treatment with the Drosha-knockdown counterparts. In conclusion, microRNA depletion in mesenchymal stromal cells and extracellular vesicles significantly reduced their intrinsic regenerative potential in AKI, suggesting a critical role of microRNAs in recovery after AKI.
Keywords: acute renal failure, gene expression, stem cell
Several studies showed that mesenchymal stromal cells (MSCs) may promote recovery from AKI. The mechanism has been ascribed mainly to a paracrine action of MSC-derived mediators.1 Bi et al.2 showed that the conditioned medium of MSCs mimics the therapeutic effects of the cells. We found that extracellular vesicles (EVs) released from MSCs contribute to their regenerative properties.3 The regenerative potential of MSC-derived EVs has been described in several models of AKI and chronic kidney injury.4,5
The mechanism of action of vesicles has been attributed to the transfer of information between cells.6 After internalization, EVs may deliver their proteins, lipids, or nucleic acid content to recipient cells, changing their fate.7–9 EVs released by MSCs were shown to express proteins involved in MSC self-renewal and differentiation capability.10 Moreover, analysis of the nucleic acid composition showed the presence of MSC-specific mRNA and mature microRNAs (miRNAs).3,11 Recently, extracellular RNAs have been implicated as both promising biomarkers for a variety of pathologies12,13 and mediators of several biologic processes.14 EVs represent a mechanism for the delivery of the nucleic acids in a degrading enzyme–protective way.15,16
miRNAs17 are a class of small noncoding RNAs that is abundant in the cell and functions as endogenous mediators of RNA silencing. miRNAs regulate protein expression by blocking both the mRNA transcription and the protein translation.18 Two crucial enzymes play a relevant role in the regulation of miRNA maturation inside the cells: the RNase III Drosha and Dicer enzymes. The first one acts in the nucleus, where it cleaves the inactive pri-miRNA into the precursor miRNA, a double–stranded hairpin structure of about 70 nucleotides.19 The precursor miRNA is then exported from the nucleus to the cytoplasm to finally generate a functional mature miRNA by Dicer cleavage.20 EV-mediated transfer of functional miRNAs has been shown to occur in several contexts.21–23
Whether EV-mediated transfer of miRNAs is implicated in the healing properties of MSC-derived vesicles in AKI has not yet been investigated. To address this issue, we evaluated the effect of the global suppression of miRNA biogenesis24 by generation of Drosha-knockdown MSCs to be used as EV donors. The choice of Drosha as a target enzyme was on the basis of the knowledge that Drosha activity is restricted to the generation of miRNAs.25–27 Here, we reported that depletion of Drosha in MSCs generated EVs with a deregulation of their miRNA content and altered the intrinsic regenerative potential of MSCs and derived EVs. These data support the relevance of miRNAs shuttled by EVs in the healing properties of MSCs.
Results
Generation of Drosha-Knockdown MSCs
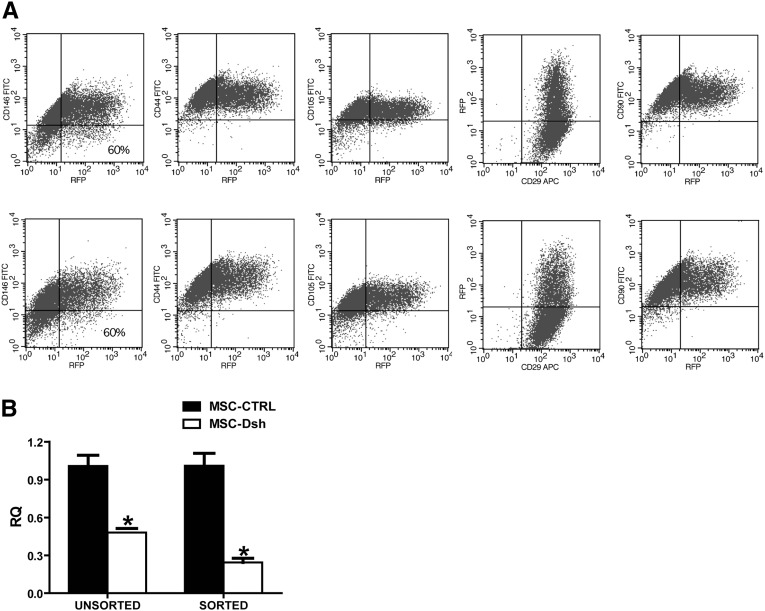
miRNA reduction was obtained by MSC transduction with a lentiviral, tetracycline-inducible vector containing a short hairpin RNA (shRNA) targeting Drosha (MSC-Dsh) and the red fluorescence protein (RFP). Transduced MSCs maintained classic mesenchymal markers on their surface (Figure 1A) as detected by FACS analysis and conserved their ability to differentiate in chondrocytes, adipocytes, and osteocytes (data not shown). Knockdown of Drosha was assessed by quantitative real-time (qRT)-PCR on Dsh-transduced MSCs cultured with doxycycline compared with control-transduced (MSC-CTRL) cells. In cell population from three separate donors, we observed a 2-fold reduction (P<0.01) of Drosha transcript level (Figure 1B) in MSC-Dsh with respect to MSC-CTRL. This reduction was enhanced in sorted RFP-positive MSC-Dsh (Figure 1B) (P<0.01). In all experiments, sorted MSC-CTRL and MSC-Dsh were used.
Figure 1.
MSC-Dsh maintained the classical MSC surface markers. (A) FACS analysis of transduced MSC-CTRL (upper panel) and MSC-Dsh (lower panel). Efficiency of transduction was measured as the percentage of RFP-positive cells. (B) Comparison of Drosha transcript expression in unsorted or RFP-sorted MSC-CTRL and MSC-Dsh measured by qRT-PCR. GAPDH transcript was used to normalize RNA input. Three different lines were tested. Data are expressed as relative quantification levels (RQ) (means±SDs) of three different experiments. *P<0.05. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Characterization of EVs Released from MSC-Dsh
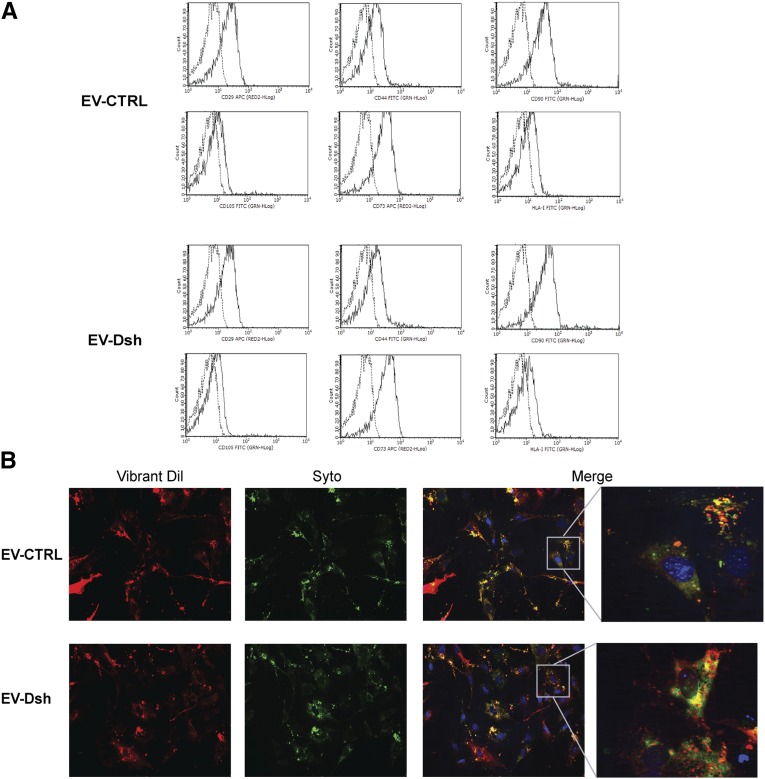
EVs from MSC-Dsh (EV-Dsh) showed a size distribution comparable with the one released by MSC-CTRL (EV-CTRL), with mean diameters of 166±16 and 152±23 nm, respectively (detected by the Nanosight instrument). Cytofluorimetric analyses of EV-CTRL and EV-Dsh showed the expression by EVs of antigens characteristic of the cells of origin, such as CD29, CD44, CD105, CD73, CD90, and HLA class I (Figure 2A). EV-CTRL and EV-Dsh (3×109 parts per milliliter) were equally incorporated by murine tubular epithelial cells (mTECs) as observed by confocal microscopy after 24 hours of incubation (Figure 2B). SYTO-RNA carried by EVs was transferred to target cells (Figure 2B).
Figure 2.
Drosha knockdown does not promote changes in EV surface markers and up-take by mTEC. (A) Representative FACS analyses of the expression of classic MSC markers (CD29, CD44, CD105, CD73, CD90, and HLA class I) by EV-CTRL and EV-Dsh. Dotted lines indicate the isotype controls. Three different EV preparations were tested with similar results. (B) Representative micrograph of incorporation of EV-CTRL and EV-Dsh by mTECs after 24 hours of incubation. EVs were collected from MSCs double stained with Vybrant Dil (red) and SYTO-RNA (which labels the RNA; green). Three experiments were performed with similar results. Original magnification, ×200; enlarged image magnification, ×630.
miRNA Profile in EV-Dsh
miRNA screening and comparison in EV-Dsh and EV-CTRL were evaluated by qRT-PCR. Using a cutoff <35 Ct value in miRNA expression, we found 161 miRNAs detected in all the EV-CTRL samples (n=4). Comparing the miRNA content between EV-CTRL and EV-Dsh, a global tendency to downregulation of miRNAs in EV-Dsh with respect to EV-CTRL was observed (55%±15% of all of the expressed miRNAs). The intersection of the list of miRNAs downregulated in different EV-Dsh with respect to EV-CTRL identified 49 miRNAs that are commonly downregulated in EV-Dsh (Table 1) (P<0.05).
Table 1.
miRNAs downregulated in EV-Dsh with respect to EV-CTRL
| Target Name | Rq Mean | SD | t Test |
|---|---|---|---|
| hsa-miR-215 | 0.04 | 0.04 | 2.27E-05 |
| hsa-miR-28–3p | 0.28 | 0.08 | 0.000415 |
| hsa-miR-376a | 0.43 | 0.09 | 0.001068 |
| hsa-miR-24 | 0.67 | 0.06 | 0.001268 |
| hsa-miR-1227 | 0.23 | 0.13 | 0.001392 |
| hsa-miR-30e-3p | 0.53 | 0.09 | 0.001867 |
| hsa-miR-411 | 0.45 | 0.11 | 0.002006 |
| mmu-miR-129–3p | 0.13 | 0.18 | 0.002474 |
| hsa-miR-486 | 0.22 | 0.17 | 0.00274 |
| hsa-miR-218 | 0.33 | 0.15 | 0.002831 |
| hsa-miR-889 | 0.22 | 0.17 | 0.002908 |
| hsa-miR-1243 | 0.19 | 0.19 | 0.003278 |
| hsa-miR-345 | 0.38 | 0.15 | 0.003789 |
| mmu-miR-140 | 0.50 | 0.13 | 0.004866 |
| hsa-miR-186 | 0.34 | 0.19 | 0.005884 |
| hsa-miR-382 | 0.22 | 0.22 | 0.00613 |
| hsa-miR-146b | 0.55 | 0.13 | 0.006271 |
| hsa-miR-202 | 0.18 | 0.24 | 0.006639 |
| hsa-miR-10a | 0.28 | 0.22 | 0.007002 |
| hsa-miR-323–3p | 0.27 | 0.22 | 0.007399 |
| hsa-miR-214 | 0.49 | 0.16 | 0.007832 |
| hsa-miR-744 | 0.45 | 0.18 | 0.008342 |
| mmu-miR-134 | 0.39 | 0.20 | 0.008937 |
| hsa-miR-199a | 0.27 | 0.25 | 0.00985 |
| hsa-let-7b | 0.60 | 0.15 | 0.01334 |
| hsa-miR-483–5p | 0.28 | 0.28 | 0.014658 |
| hsa-miR-106a | 0.50 | 0.20 | 0.015468 |
| hsa-miR-590–5p | 0.30 | 0.29 | 0.017129 |
| hsa-miR-376c | 0.57 | 0.18 | 0.017469 |
| hsa-miR-133a | 0.47 | 0.22 | 0.018088 |
| hsa-miR-423–5p | 0.22 | 0.19 | 0.018506 |
| hsa-miR-425–5p | 0.46 | 0.24 | 0.020084 |
| hsa-miR-29a | 0.67 | 0.15 | 0.022468 |
| hsa-miR-574–3p | 0.57 | 0.20 | 0.024011 |
| hsa-let-7d | 0.59 | 0.19 | 0.024324 |
| hsa-miR-150 | 0.64 | 0.18 | 0.026056 |
| hsa-miR-19b | 0.62 | 0.19 | 0.029332 |
| hsa-miR-33a# | 0.24 | 0.40 | 0.031748 |
| hsa-miR-1208 | 0.54 | 0.24 | 0.032548 |
| hsa-miR-191 | 0.63 | 0.20 | 0.033122 |
| hsa-miR-203 | 0.58 | 0.23 | 0.035758 |
| hsa-miR-200c | 0.40 | 0.34 | 0.03695 |
| hsa-miR-181a | 0.55 | 0.25 | 0.038543 |
| hsa-miR-487b | 0.43 | 0.32 | 0.03889 |
| hsa-miR-17 | 0.63 | 0.21 | 0.04191 |
| hsa-miR-532 | 0.54 | 0.27 | 0.04398 |
| hsa-miR-148a | 0.34 | 0.25 | 0.044801 |
| hsa-miR-328 | 0.48 | 0.31 | 0.044856 |
| hsa-miR-29c | 0.56 | 0.28 | 0.049784 |
The relative expression (RQ) of miRNAs in EV-Dsh was defined as FC evaluated as 2−ΔΔCt with respect to the reference sample (EV-CTRL) as described in Concise Methods (n=4 in each group; P<0.05).
Impaired Protective Effect of MSC-Dsh in a Model of Glycerol-Induced AKI
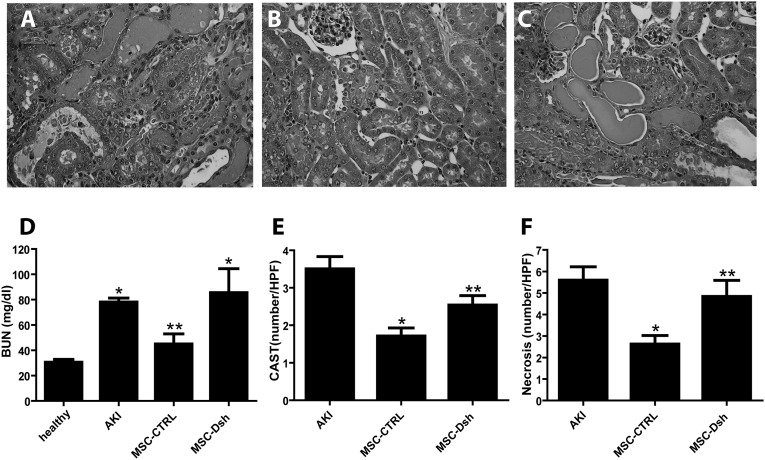
At day 3 after AKI induction, 75,000 MSC-CTRL or MSC-Dsh cells were administered intravenously. Mice were then euthanized after 5 days from the induction of AKI. The lesions observed in AKI mice included tubular hyaline casts, vacuolization, and presence of necrosis of proximal and distal tubular epithelium (Figure 3A). Morphologic damage correlated with the significant rise in BUN (Figure 3D). Injection of MSC-CTRL resulted in a decrease of tubular lesions and evidence of renal tubular repair with respect to AKI mice treated with vehicle alone (Figure 3B), which parallel the reduction of BUN levels (Figure 3D). On the contrary, injection of MSC-Dsh did not significantly improve morphologic (Figure 3C) or functional injury (Figure 3D). The quantitative evaluation of casts (Figure 3E) and tubular necrosis (Figure 3F) showed a significant reduction in MSC-CTRL– but not MSC-Dsh–treated mice.
Figure 3.
Injection of MSC-Dsh does not protect from AKI injury. (A–C) Representative micrographs of renal histology of (A) untreated AKI mice injected with PBS and treated AKI mice injected with (B) MSC-CTRL or (C) MSC-Dsh cells (75,000 cells) and euthanized at day 5 after glycerol administration. Original magnification, ×400. (D) BUN (milligrams per deciliter) levels measured in healthy, untreated AKI, and AKI mice treated with MSC-CTRL or MSC-Dsh 5 days after glycerol administration. ANOVA with Newman–Keuls multicomparison test. *P<0.05 AKI and MSC-Dsh versus healthy; **P<0.05 MSC-CTRL versus MSC-Dsh. (E and F) Morphometric evaluation of (E) hyalin casts and (F) tubular necrosis in untreated AKI and AKI mice treated with MSC-CTRL or MSC-Dsh 5 days after glycerol administration. Results are expressed as means±SEMs (eight mice per group). ANOVA with Newman–Keuls multicomparison test. *P<0.05 MSC-CTRL versus AKI; **P<0.05 MSC-Dsh versus MSC-CTRL. CAST, hyalin casts; HPF, high-power field.
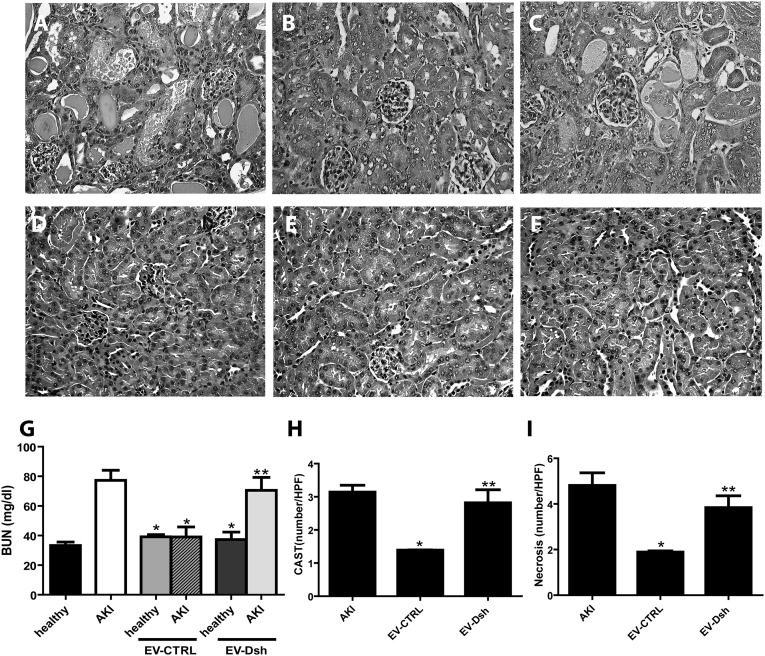
We also compared the effect of EV-Dsh with that of EV-CTRL. For this purpose, we injected 2.2×108 particles per mouse EV-CTRL and EV-Dsh at day 3 after AKI induction. In mice treated with EV-CTRL but not EV-Dsh, the tubular lesions at day 5 were significantly less severe than those in untreated AKI mice (Figure 4, A–C). In control healthy mice injected with EV-CTRL and EV-Dsh, the histologic analysis and BUN measurement showed no negative effects associated with the EV-CTRL and EV-Dsh injection (Figure 4, D–G) in the absence of AKI damage. EV-Dsh injection in AKI mice did not inhibit the rise of BUN (Figure 4G). Also, the counts of hyaline casts and tubular necrosis showed the inefficiency of EV-Dsh with respect to the EV-CTRL (Figure 4, H and I).
Figure 4.
EV-Dsh injection does not induce recovery from AKI damage. (A–C) Representative micrographs of renal histology of (A) untreated AKI mice and AKI mice treated with (B) EV-CTRL or (C) EV-Dsh and euthanized at day 5 after glycerol administration. Original magnification, ×400. (D–F) Representative micrographs of renal histology of (D) untreated healthy and healthy mice injected with (E) EV-CTRL or (F) EV-Dsh and euthanized at day 2 after EV administration. (G) BUN (milligrams per deciliter) levels measured in healthy, untreated AKI, and AKI mice treated with EV-CTRL or EV-Dsh (5 healthy mice per group and 16 AKI mice per group). ANOVA with Newman–Keuls multicomparison test. *P<0.05 versus AKI; **P<0.05 AKI EV-Dsh versus AKI EV-CTRL. (H and I) Morphometric evaluation of (H) hyalin casts and (I) tubular necrosis in untreated AKI and AKI-treated mice injected with EV-CTRL or EV-Dsh 5 days after glycerol administration. Results are expressed as means±SEMs; ANOVA with Newman–Keuls multicomparison test. *P<0.05 EV-CTRL versus AKI; **P<0.05 EV-Dsh versus EV-CTRL. CAST, hyalin casts; HPF, high-power field.
MSC-CTRL but Not MSC-Dsh Induced a Proregenerative Gene Signature in the Kidney
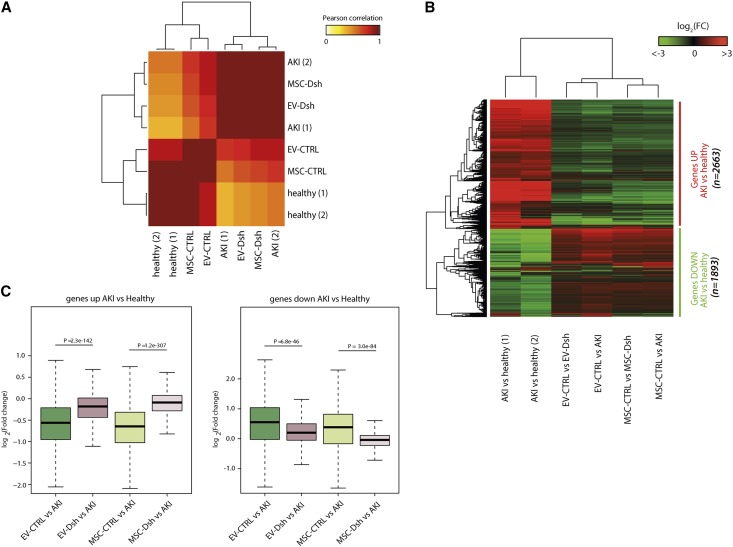
To detect the molecular changes that occurred in the kidneys of treated animals, we performed RNA-Seq analysis (Gene Expression Omnibus accession number GSE59958). We observed a strong correlation between MSC-CTRL– and EV-CTRL–treated mice (Pearson correlation coefficient [PCC]=0.96). Interestingly, comparison of MSC-CTRL and EV-CTRL treatment with the AKI condition showed a lower degree of correlation (PCC=0.72 and PCC=0.74, respectively), indicating that both treatments partially reverted the molecular changes occurring in the kidney after glycerol-induced damage. This was confirmed by the observation that healthy mice significantly correlated with MSC-CTRL and EV-CTRL samples (PCC=0.966 and PCC=0.858, respectively) (Figure 5A). On the contrary, the transcriptome profile of MSC-Dsh– and EV-Dsh–treated mice showed high correlation with the untreated AKI mice (PCC=0.997 and PCC=0.988, respectively) and low or no correlation with healthy samples (PCC=0.486 and PCC=0.384, respectively), indicating no healing properties associated with Drosha-knockdown MSC or EV-Dsh treatments (Figure 5A).
Figure 5.
MSC-Dsh treatment induced in the kidney a different gene expression signature with respect to MSC-CTRL. (A) Pearson correlation of the global transcriptome profile of kidneys of healthy, untreated AKI, and AKI treated with MSC-CTRL, MSC-Dsh, EV-CTRL, and EV-Dsh mice. EV-CTRL and MSC-CTRL clustered into two major groups, supporting an overlap in the genes regulated by both cells and EVs in AKI. Moreover, healthy mice significantly correlated with MSC-CTRL and EV-CTRL samples. On the contrary, the transcriptomic profile of MSC-Dsh– and EV-Dsh–treated mice showed a high correlation with the untreated AKI mice. (B) Heat map of the distribution of AKI-deregulated genes in MSC-CTRL, MSC-Dsh, EV-CTRL, and EV-Dsh samples (n=2663 upregulated AKI versus healthy; n=1893 downregulated AKI versus healthy). (C) Median distribution of the FCs in MSC-CTRL, MSC-Dsh, EV-CTRL, and EV-Dsh with respect to AKI samples showing statistically different global distribution of all genes in two groups. FC, fold change.
Consistently, differential expression analysis of all genes deregulated in AKI versus healthy kidney samples (n=2663, upregulated; n=1893, downregulated) showed an extremely similar pattern (Figure 5B). The EV-CTRL and MSC-CTRL were clustered into two major groups, supporting a relevant overlap in the genes regulated by both cells and EVs during AKI repair (Figure 5B). Interestingly, MSC-Dsh and EV-Dsh kidneys showed a global gene expression pattern very similar to that of AKI (Figure 5B). These results suggested that MSCs depleted of Dsh were unable to activate the proregenerative transcriptomic program in injured kidneys. In fact, genes altered in AKI versus healthy condition showed a significant reversion to physiologic levels with EV-CTRL and MSC-CTRL treatments compared with EV-Dsh and MSC-Dsh treatments (EV-CTRL versus EV-Dsh, P<0.001 for both upregulated and downregulated genes; MSC-CTRL versus MSC-Dsh, P<0.001 for both upregulated and downregulated genes) (Figure 5C).
Gene Ontology Analyses
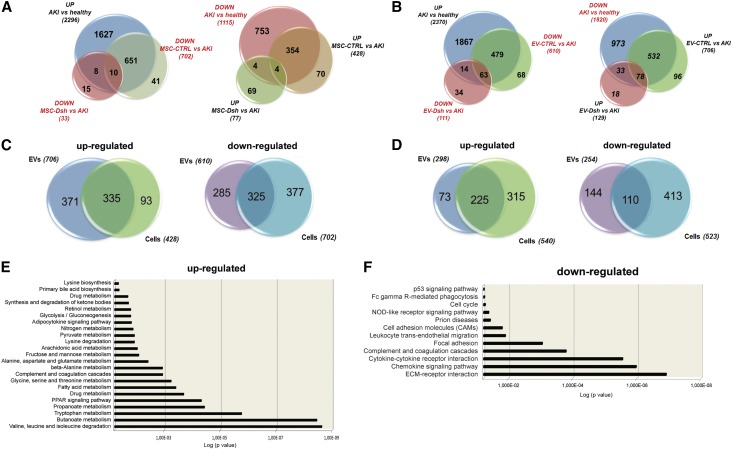
Focusing on genes with a significant fold change (FC) among all of the treatments (FC≥|1|), we detected a relevant group of transcripts significantly reverted after MSC-CTRL (n=702 and n=428 downregulated and upregulated with respect to AKI, respectively) (Figure 6A) and EV-CTRL (n=610 and n=706 downregulated and upregulated with respect to AKI, respectively) stimulation (Figure 6B). The same modulation was not observed for the MSC-Dsh and EV-Dsh treatments (n=33 and n=77 for MSC-Dsh and n=111 and n=129 for EV-Dsh upregulated and downregulated with respect to AKI, respectively) (Figure 6, A and B).
Figure 6.
MSC and EV-CTRL modulated common genes in target cells associated with kidney recovery. (A and B) Venn diagrams showing the number of genes upregulated or downregulated (FC≥|1|) in the kidneys from AKI mice treated with (A) MSC-CTRL and MSC-Dsh or (B) EV-CTRL and EV-Dsh with respect to untreated AKI. (C and D) Venn diagrams showing the number of genes commonly upregulated or downregulated in (C) the kidneys from AKI mice treated with MSC-CTRL or EV-CTRL with respect to untreated AKI and (D) AKI treated with MSC-Dsh or EV-Dsh. (E and F) GO analysis reveals that common modulated genes in AKI mice treated with MSC-CTRL or EV-CTRL with respect to untreated AKI are enriched for (E) genes associated with metabolic pathways, complement, and coagulation cascades response (for the upregulated genes) and (F) genes associated with response to inflammation, ECM-receptor interaction, cell adhesion molecules, and cell cycle (for the downregulated genes). GO, gene ontology; ECM, extracellular matrix.
Gene ontology (GO) analysis was performed on a subset of genes that showed comparable high modulation in both MSC and EV treatments. This yielded final subsets of 335 and 325 upregulated and downregulated genes, respectively, common to MSC-CTRL and EV-CTRL treatments with respect to AKI (Figure 6C) or 225 and 110 upregulated and downregulated genes, respectively, common to MSC-CTRL and EV-CTRL treatments with respect to MSC-Dsh and EV-Dsh (Figure 6D). The GO enrichment analysis revealed that common upregulated transcripts in EV-CTRL and MSC-CTRL with respect to AKI were enriched for genes associated with the regulation of metabolic pathways (valine, leucine, and isoleucine degradation; Tryptophan and Butanoate metabolism; and peroxisome proliferator-activated receptors signaling pathway: P<0.001 for all the groups) as well as genes involved in the complement and coagulation cascades (Figure 6E) (P<0.001). The most representative GO of the downregulated genes was associated with response to inflammation, extracellular matrix (ECM)-receptor interaction, cell adhesion molecules, and cell cycle (Figure 6F) (P<0.001, P<0.001, P=0.02, and P=0.04, respectively). Cross-match of the common genes modulated after MSC-Dsh or EV-Dsh treatment with respect to AKI showed no significant enrichment of these pathways (data not shown). Genes comodulated in EV-CTRL and MSC-CTRL with respect to MSC-Dsh or EV-Dsh were associated with ECM remodeling and focal adhesion for the downregulated transcripts and metabolic regulation for the upregulated transcripts (Supplemental Table 1).
miRNA:mRNA Data Integration
To identify potential mRNA targets for vesicle-carried miRNAs, we first clustered the miRNAs expressed in EV-CTRL (n=161) into families according to their seed sequence and scanned the 3′ untranslated region (3′-UTR) of AKI-expressed genes for perfect seed-match occurrences (6–8 nucleotides). Moreover, to account for potential cooperative action of different miRNAs, we restricted our research to those genes targeted by at least two expressed miRNA families. Our analysis produced a list of 16 significantly enriched miRNA families (P<0.05, hypergeometric distribution) associated in 15 pairs with targets that show significant downregulation [log2(FC)≤−1] in EV-CTRL versus AKI mice (Supplemental Table 2). For eight of these miRNA families, at least one representative miRNA was differentially expressed in EV-Dsh, supporting their involvement in the EV healing process. In particular, we found a significant enrichment for miR-483–5p, miR-191, miR-28–3p, miR-423–5p, miR-744, miR-129–3p, miR-24, and miR-148a families. The union of the targets for 15 pairs of miRNA families identified 209 genes potentially modulated by these miRNAs, of which 165 (77.8%) genes were not reverted in EV-Dsh treatment with respect to AKI. GO enrichment analysis of these targets showed the over-representation of processes associated with ECM-receptor interaction and focal adhesion (P<0.001 and P=0.003, respectively) and Wnt and p53 pathways (P=0.02 and P=0.04, respectively) (Supplemental Table 3).
Effect of Drosha Knockdown on the Expression of Markers of Kidney Injury
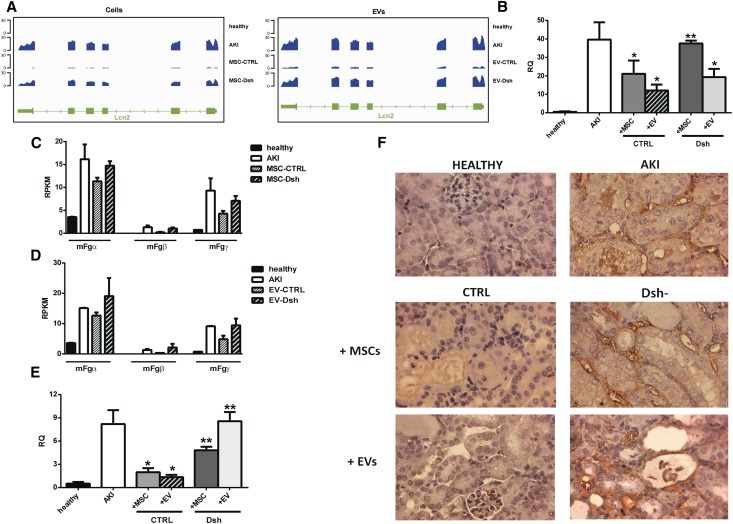
Kidneys from untreated AKI mice showed elevated levels of transcripts coding for markers of tubular damage, such as lipocalin 2 (Lcn2)28 and fibrinogen (Fg) subunits,29 as detected by RNA-Seq analysis. In healthy mice, no expression of Lcn2 was observed. The treatment with MSC-CTRL and EV-CTRL leads to a significant decrease of Lcn2 (Figure 7A), whereas MSC-Dsh– or EV-Dsh–injected mice maintained elevated levels of the Lcn2 transcript (Figure 7A). Lcn2 downregulation after both MSC-CTRL and EV-CTRL treatments was confirmed by qRT-PCR (n=6 mice per group) (Figure 7B). MSC-Dsh– and EV-Dsh–injected animals maintained elevated levels of Lcn2 (Figure 7B).
Figure 7.
MSC and EV-Dsh injected mice maintained elevated levels of AKI markers. (A) Representative genomic occupancy profile of the Lcn2 gene among healthy, untreated AKI, MSC-treated AKI (left panel), or EV-treated AKI (right panel). (B) qRT-PCR confirmation of Lcn2 modulation in mice after MSC and EV treatments with respect to untreated AKI mice (n=6 mice per group). Healthy mice are used as negative controls. (C and D) Expression levels of the three Fg subunits detected by RNA-Seq analysis among healthy, untreated AKI, (C) MSC-treated AKI, or (D) EV-treated AKI mice. (E) qRT-PCR confirmation of Fg-β expression in mice after MSC and EV treatments with respect to untreated AKI mice (n=6 mice per group). In healthy mice, no or minimal expression of all of the Fg subunits was observed. (F) Immunohistochemistry analysis of Fg-β among healthy, untreated AKI, MSC-treated, and EV-treated animals. Tubular staining was not observed in kidneys from healthy mice and was markedly reduced in mice treated with MSC-CTRL and EV-CTRL. On the contrary, AKI untreated, MSC-Dsh–treated, and EV-Dsh–treated animals showed a strong peritubular staining and some cytoplasmic tubular staining. For qRT-PCR analysis, data are expressed as means ± SDs; ANOVA with Newman–Keuls multicomparison test. *P<0.05 versus AKI; **P<0.05 versus CTRL.
We also evaluated the levels of Fg as another important marker of AKI damage.29 AKI induced the upregulation of all of the Fg subunits (subunits-α, -β, and -γ) (Figure 7C). MSC-CTRL– or EV-CTRL–treated mice showed significant downregulation of Fg-β and less relevant reductions of α- and γ-subunits. MSC-Dsh– or EV-Dsh–injected mice maintained elevated levels of all of the Fg subunits (Figure 7, C and D). In healthy mice, no or minimal expression of all of the Fg subunits was observed. qRT-PCR of the whole kidney confirmed minimal expression of Fg-β in control kidneys (Figure 7E). A robust increase of Fg-β was observed after 5 days in AKI untreated mice as well as mice treated with MSC-Dsh and EV-Dsh. MSC-CTRL and EV-CTRL reverted the Fg-β expression, showing transcript expression comparable with that of healthy mice (Figure 7E). Healthy mice injected with EV-CTRL and EV-Dsh showed levels of expression of Lcn2 and Fg-β comparable with those of healthy untreated animals (data not shown). These data collectively indicated that injured renal tubules can initiate local Fg synthesis.29 Immunohistochemistry analysis of Fg-β in untreated AKI mice showed a strong peritubular staining for Fg and some positivity in the tubular cell cytoplasm (Figure 7F). Tubular staining was not observed in kidneys from healthy mice and was markedly reduced in mice treated with MSC-CTRL and EV-CTRL but not MSC-Dsh and EV-Dsh (Figure 7F).
Discussion
The results of this study showed that downregulation of miRNAs in human MSCs reduced the intrinsic kidney proregenerative properties of these cells and their derived EVs. This effect was associated with changes in miRNA composition of cells and EVs. Previous studies showed that EVs derived from human MSCs were able to favor recovery of AKI by activation of antiapoptotic and proregenerative programs in tubular epithelial cells.3,30–32 Transfer of mRNAs from MSCs to tubular epithelial cells was observed both in vitro and in vivo.3 The mRNA transfer, which was also observed in other models,33,34 was followed by their transient transduction into proteins. We recently showed an EV-mediated transfer of miRNAs from MSCs to tubular epithelial cells in an in vitro model of renal tubular cell injury induced by ATP depletion.35 miRNA modulation in renal tubular cells was associated with functional recovery and phenotypic changes.36 In this study, we evaluated whether EV-shuttled miRNAs play a critical role in the healing properties of MSCs in an in vivo model of AKI. We observed that knockdown of Drosha in MSCs globally reduced miRNA content in EVs. Moreover, MSC-Dsh and EV-Dsh exhibited a significantly reduced proregenerative with respect to wild-type MSCs and EVs. The lack of EV-Dsh efficacy was not dependent on reduced internalization or expression of surface molecules, but rather, it was dependent on the molecular content. The efficacy of wild-type MSCs and EVs in AKI recovery was observed at not only functional and morphologic levels but also, at molecular level. Transcriptome analysis of kidneys from AKI mice showed the alteration of about 4556 genes with respect to healthy mice. The genes downregulated by AKI were associated with metabolism, such as valine, leucine, and isoleucine degradation, Tryptophan and Butanoate, and fatty acid metabolism. Different reports documented the importance of mitochondrial fatty acid oxidation in the cytoprotection of proximal tubule cells during the development of AKI.36 Wild-type MSCs and EVs upregulated genes acting in metabolic pathways associated with fatty acid oxidation, glycolysis, gluconeogenesis, and ketone bodies generation. These processes have been involved in the regulation of energy metabolism of damaged tubular epithelial cells by MSC treatment.37
Moreover, AKI damage was associated with the upregulation of genes involved in inflammation, ECM-receptor interaction, cell adhesion molecules, and cell cycle with respect to healthy controls. Treatment with wild-type MSCs and EVs restored a normal transcriptome pattern. In contrast, Dsh-knockdown cells and EVs were unable to inhibit the molecular alteration observed in AKI, and the RNA-Seq analysis showed a superimposable pattern between AKI and Dsh-treated mice. Recent studies correlated Lcn2 and Fg-β expression with AKI development in mice.28,29 Overexpression of these markers of renal injury was reduced by wild-type MSCs and EVs but not MSC-Dsh and EV-Dsh. Inefficiency of Drosha-knockdown cells and EVs in inducing renal repair was observed at not only morphologic level but also, functional levels as indicated by the persistence of elevated BUN levels. In an attempt to identify the possible healing miRNAs shuttled by EVs, we performed bioinformatic mRNA:miRNA data analysis. Here, we showed a statistically significant correlation of 16 miRNA families (associated in 15 pairs) with targets that show significant downregulation in EV-CTRL–treated animals with respect to AKI. Of them, eight miRNA families were significantly reduced in EV-Dsh (miR-483–5p, miR-191, miR-28–3p, miR-423–5p, miR-744, miR-129–3p, miR-24, and miR-148a families). Union of genes of the 15 pairs of miRNAs, identified 209 genes potentially modulated by these miRNAs. GO enrichment analysis of these targets showed the over-representation of processes associated with the first stages of kidney damage and repair by stem cell administration, such as the cell adhesion molecules and MAPK signaling.38 Interestingly, the Wnt signaling pathway,39 ECM remodeling,40 and the p53 pathway41 were also present among the significant biologic processes identified (data not shown). These processes are usually associated with relevant stages in the transition from acute to chronic injury, such as vascular failure, interstitial fibrosis, and glomerulosclerosis development.38–41 By selecting in this set, only genes not reverted in EV-Dsh treatment with respect to AKI, 165 genes resulted as the possible miRNA targets. GO enrichment analysis of these genes showed that EV-Dsh–treated mice overexpressed genes associated with processes related with the progression of kidney damage, such as the Wnt pathway, p53 signaling, ECM remodeling, and focal adhesion processes, like the untreated AKI. These results represent an initial step for additional biologic validations of miRNAs shuttled by EVs and involved in the healing pathways associated with MSC treatment of AKI.
In conclusion, the results of this study indicate that miRNAs play a critical role in the regenerative potential of MSCs. The global downregulation of miRNAs obtained by Drosha knockdown in MSCs abrogated the ability to promote AKI recovery by both cells and EVs.
Concise Methods
Culture of Bone Marrow MSCs
Bone marrow cells were obtained by Lonza (Basel, Switzerland) and cultured as previously described.3 Cells were seeded at a density of 10,000 cells/cm2 and used within the seven passage. Cytofluorimetric analysis was performed as described,3 and the following mAbs conjugated with Allophycocyanin (APC) or FITC were used: anti-CD146, anti-CD44, anti-CD105, anti-CD29, and anti-CD90. Mouse IgG isotypic controls were from Miltenyi (Bergisch Gladbach, Germany). All of the cell preparations at different passages of culture expressed the typical MSC markers: CD105, CD73, CD44, CD90, CD166, and CD146. They also expressed HLA class I. The adipogenic and osteogenic differentiation abilities of MSC were determined as previously described.3
Transduction of MSCs with Lentiviral Particles
Lentiviral particles were produced using the third generation core packing plasmids provided by Calautti (Department of Molecular Biotechnology and Life Sciences, Torino, Italy). Virus particles were released by 293T cells after overnight collection, filtered through a 0.45-mm filter, and concentrated by ultracentrifugation as previously described. For the construct targeting Drosha, approximately 105 passage 0 hMSCs were transduced42 with particles containing the pTRIPZ lentiviral–inducible vector shRNAmiR targeting Drosha (clone V2THS_71783; Open Biosystem) with a single infection. Transduction efficiency was determined by measuring the percentage of RFP-positive cells using different doses of lentivirus. Wild-type cells were transduced with a pTRIPZ lentiviral–inducible vector scramble shRNAmiR. After transduction, cells were grown out to 70% confluence in complete medium containing doxycycline (1 μg/ml). RFP-positive cells were selected using a MoFlo Cell Sorter (Dakocytomation, Copenhagen, Denmark). Sorted RFP+ cells were characterized by incubation with FITC- or APC-conjugated antibodies; 10,000 cells were analyzed at each experimental point. Doxycycline was added to cultured cells every time that they reached the semiconfluent stage, and the medium was changed after 72 hours during the duration of the experiment to avoid fluctuation in the Drosha transcript level. Drosha expression was evaluated by qRT-PCR using the following primers: F1 5-CATGCACCAGATTCTCCTGTA-3 and R1 5-GTCTCCTGCATAACTCAACTG-3 as previously described.43
Isolation of EVs
EVs were obtained from supernatants of MSCs cultured in RPMI deprived of FCS and supplemented with 0.5% BSA (Sigma-Aldrich) in the presence of doxycycline. The viability of cells incubated overnight without serum was >98% as detected by trypan blue exclusion. No apoptotic cells were detected by terminal deoxynucleotidyl transferase–mediated digoxigenin-deoxyuridine nick-end labeling assay. Supernatants of MSCs after removal of cell debris and apoptotic bodies were purified by ultracentrifugation for 1 hour at 4°C at 100,000×g (Beckman Coulter Optima L-90K ultracentrifuge; Beckman Coulter, Fullerton, CA) as previously described.44
EV dimension and profile were analyzed by Nanosight LM10 (NanoSight Ltd., Minton Park, United Kingdom), and the protein content was quantified by Bradford (Bio-Rad, Hercules, CA). Endotoxin contamination of EVs was excluded by Limulus testing according to the manufacturer’s instruction (Charles River Laboratories, Wilmington, MA), and EVs were stored at −80°C. To trace EVs by fluorescence microscopy, MSCs were labeled with the SYTO-RNA Select green fluorescent cell stain (Molecular Probes; Life Technology) that specifically stains RNA and a lipophilic membrane stain that diffuses laterally to stain the entire cell (Life Technology).44 EVs obtained from labeled cells were washed by ultracentrifugation as described below.
EV Characterization
EVs were characterized by cytofluorimetric analysis using FITC- or APC-conjugated antibodies against CD73, CD44, CD105, CD90, CD29, and HLA class I. FITC or APC mouse nonimmune isotypic IgGs (Miltenyi) were used as controls. Briefly, EVs (1.5×108 particles) were incubated for 15 minutes at 4°C with antibodies, diluted in 300 microL and immediately acquired as previously described.45 FACS analysis was acquired using the Guava easyCyte Flow Cytometer (EMD Millipore, Billerica, MA) and analyzed with InCyte software.45
Isolation and Culture of mTECs
Kidneys were obtained from healthy female C57 mice. mTECs were isolated, cultured, characterized for expression of tubular markers, and negative for endothelial and glomerular markers as previously described.3 To determine the incorporation efficacy of EV-Dsh with respect to EV-CTRL by mTECs, we incubated EVs (3×109 parts per milliliter) double labeled with SYTO-RNASelect and Vybrant Dil (Molecular Probes)44 with mTECs for 24 hours. The uptake of EVs was analyzed confocal microscopy (Zeiss LSM 5 Pascal; Carl Zeiss, Oberkochen, Germany).
RNA Isolation and Screening
Total RNA was isolated from cells and EVs using the mirVana RNA isolation kit (Applied Biosystems) according to the manufacturer’s protocol. For the extraction of the RNA from kidneys, Trizol reagent was used. RNA was then quantified spectrophotometrically (ND-1000; Nanodrop, Wilmington DE), and the RNA quality was assessed by capillary electrophoresis on an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) using the Total Eukaryotic Nano and Small RNA kits.
RNA-Seq
For RNA-Seq library preparation, approximately 2 μg total RNA from mouse kidney was subjected to poly(A) selection, and libraries were prepared using the TruSeq RNA Sample Prep Kit (Illumina) following the manufacturer’s instructions. Sequencing was performed on the Illumina HiScanSQ platform. Reads were mapped to the Mus musculus mm9 reference assembly using TopHat v2.0.10,46 and RPKMs (reads per kilobase per million mapped reads) for transcripts were calculated with Cufflinks v2.1.1.47 Transcripts with RPKM≥1 were further considered for differential expression analysis. Genes with log2(FC)≥1 and log2(FC)≤−1 were considered as upregulated or downregulated, respectively. GO analysis was performed using DAVID Bioinformatics Resource.48
RNA-Seq data have been deposited in the Gene Expression Omnibus (accession number GSE59958; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE59958).
RNA and miRNA Expression Analyses
RNA expression confirmation was conducted by qRT-PCR. First–strand cDNA preparation and qRT-PCR experiments were performed as previously described.35 qRT-PCR was performed using a 96-well StepOne Real Time System (Applied Biosystems). FC in mRNA expression was calculated as 2−ΔΔCt using mGAPDH gene as the normalizer. All of the sequence-specific oligonucleotide primers were purchased from MWG-Biotech AG (Ebersberg, Germany; www.mwg-biotech.com).
To analyze the miRNA content in MSC-CTRL, EV-CTRL, MSC-Dsh, and EV-Dsh, the Applied Biosystems TaqManH MicroRNA Assay Human Panel Early Access Kit (Life Technology) was used to profile 754 mature miRNAs by sequential steps of reverse transcription (Megaplex RT Pools; Life Technology) using an Applied Biosystems 7900H qRT-PCR instrument. Raw Ct values, automatic baseline, and threshold were calculated using the SDS software, version 2.3. Comparison of miRNA expression was conducted using the Expression Suite software (Life Technology). FC (Rq) in miRNA expression in EV-Dsh with respect to EV-CTRL was calculated as 2−ΔΔCt using the RNU-48 as normalizer. Rq was measured for each sample and obtained comparing eight EV samples (n=4 in each group). Statistical significance was set at P<0.05 and measured by paired t tests. Only statistically reduced miRNAs in all samples tested would be considered for additional studies.
miRNA Target Sites Enrichment Analyses
For miRNA target sites analysis, miRNAs expressed in EV-CTRL were clustered in families according to their seed sequence (nucleotides 2–7). For each downregulated gene with a log2(FC) of at least −1 in EV-CTRL versus AKI mice, we extracted the 3′-UTR sequence and performed a screening for perfect targets (6–8mers) for each miRNA family. For genes with multiple isoforms, only the isoform with the longest 3′-UTR was used. To account for potential miRNA cooperatively, we searched for targets enriched for seed-match sequences of at least two expressed miRNA families. The expected number of targets (used as background) for each pair of miRNA families was calculated on the whole Refseq annotation (21,164 genes). Statistical significance was assessed using a hypergeometric test.
SCID Mice Model of AKI
Studies were approved by the Ethics Committee of Turin University and conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. A model of rhabdomyolisis-induced AKI was performed in male SCID mice as previously described.3 On day 3 after glycerol administration, mice received an intravenous injection into the tail vein of 75,000 MSC-CTRL or MSC-Dsh cells and EV-CTRL or EV-Dsh in 150 μl saline or saline alone. The number of particles injected (2.2×108 particles per mouse) corresponds to an average of the amount of EVs released overnight by 75,000 MSCs (average of 2,900 EVs per cell), which showed a therapeutic effect on this AKI model.3 Mice were killed on day 5 (n=8 per group) after glycerol administration, and blood samples for BUN determination and kidney specimens were collected. For gene expression, small pieces of the kidney cortical regions were stored in RNA later solution (Applied Biosystems) at −20°C until required.
Statistical Analyses
Data were analyzed using the GraphPad Prism 6.0 Demo program. Statistical analyses were conducted using one-way ANOVA with Newman–Keuls or Dunnett post-tests and t tests where appropriate. Statistical significance was established at P<0.05.
Disclosures
F.C. and C.T. (Fresenius Medical Care) are employed by a commercial company and contributed to the study as researchers. C.T. and G.C. are named inventors in related patents.
Supplementary Material
Acknowledgments
We thank Federica Antico and Dr. Claudia Cavallari for technical help.
Research reported in this publication was supported by European Community Marie Curie Actions IAPP2013_EV, Stem Injury Award 612224, and National Center for Advancing Translational Sciences of the National Institutes of Health Award UH2-TR000880.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
See related editorial, “Exosomes to the Rescue,” on pages 2303–2304.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2014070710/-/DCSupplemental.
References
- 1.Humphreys BD, Valerius MT, Kobayashi A, Mugford JW, Soeung S, Duffield JS, McMahon AP, Bonventre JV: Intrinsic epithelial cells repair the kidney after injury. Cell Stem Cell 2: 284–291, 2008 [DOI] [PubMed] [Google Scholar]
- 2.Bi B, Schmitt R, Israilova M, Nishio H, Cantley LG: Stromal cells protect against acute tubular injury via an endocrine effect. J Am Soc Nephrol 18: 2486–2496, 2007 [DOI] [PubMed] [Google Scholar]
- 3.Bruno S, Grange C, Deregibus MC, Calogero RA, Saviozzi S, Collino F, Morando L, Busca A, Falda M, Bussolati B, Tetta C, Camussi G: Mesenchymal stem cell-derived microvesicles protect against acute tubular injury. J Am Soc Nephrol 20: 1053–1067, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruno S, Grange C, Collino F, Deregibus MC, Cantaluppi V, Biancone L, Tetta C, Camussi G: Microvesicles derived from mesenchymal stem cells enhance survival in a lethal model of acute kidney injury. PLoS ONE 7: e33115, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He J, Wang Y, Sun S, Yu M, Wang C, Pei X, Zhu B, Wu J, Zhao W: Bone marrow stem cells-derived microvesicles protect against renal injury in the mouse remnant kidney model. Nephrology (Carlton) 17: 493–500, 2012 [DOI] [PubMed] [Google Scholar]
- 6.Akers JC, Gonda D, Kim R, Carter BS, Chen CC: Biogenesis of extracellular vesicles (EV): Exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J Neurooncol 113: 1–11, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee Y, El Andaloussi S, Wood MJ: Exosomes and microvesicles: Extracellular vesicles for genetic information transfer and gene therapy. Hum Mol Genet 21: R125–R134, 2012 [DOI] [PubMed] [Google Scholar]
- 8.Ratajczak J, Miekus K, Kucia M, Zhang J, Reca R, Dvorak P, Ratajczak MZ: Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: Evidence for horizontal transfer of mRNA and protein delivery. Leukemia 20: 847–856, 2006 [DOI] [PubMed] [Google Scholar]
- 9.Aliotta JM, Pereira M, Amaral A, Sorokina A, Igbinoba Z, Hasslinger A, El-Bizri R, Rounds SI, Quesenberry PJ, Klinger JR: Induction of pulmonary hypertensive changes by extracellular vesicles from monocrotaline-treated mice. Cardiovasc Res 100: 354–362, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim HS, Choi DY, Yun SJ, Choi SM, Kang JW, Jung JW, Hwang D, Kim KP, Kim DW: Proteomic analysis of microvesicles derived from human mesenchymal stem cells. J Proteome Res 11: 839–849, 2012 [DOI] [PubMed] [Google Scholar]
- 11.Collino F, Deregibus MC, Bruno S, Sterpone L, Aghemo G, Viltono L, Tetta C, Camussi G: Microvesicles derived from adult human bone marrow and tissue specific mesenchymal stem cells shuttle selected pattern of miRNAs. PLoS ONE 5: e11803, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maluf DG, Dumur CI, Suh JL, Scian MJ, King AL, Cathro H, Lee JK, Gehrau RC, Brayman KL, Gallon L, Mas VR: The urine microRNA profile may help monitor post-transplant renal graft function. Kidney Int 85: 439–449, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M: microRNAs in cancer management. Lancet Oncol 13: e249–e258, 2012 [DOI] [PubMed] [Google Scholar]
- 14.Blelloch R, Gutkind JS: Epigenetics, noncoding RNAs, and cell signaling—crossroads in the regulation of cell fate decisions. Curr Opin Cell Biol 25: 149–151, 2013 [DOI] [PubMed] [Google Scholar]
- 15.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M: Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A 108: 5003–5008, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quesenberry PJ, Goldberg LR, Aliotta JM, Dooner MS, Pereira MG, Wen S, Camussi G: Cellular phenotype and extracellular vesicles: Basic and clinical considerations. Stem Cells Dev 23: 1429–1436, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cech TR, Steitz JA: The noncoding RNA revolution-trashing old rules to forge new ones. Cell 157: 77–94, 2014 [DOI] [PubMed] [Google Scholar]
- 18.Bartel DP: MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN: The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev 18: 3016–3027, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leisegang MS, Martin R, Ramírez AS, Bohnsack MT: Exportin t and Exportin 5: tRNA and miRNA biogenesis - and beyond. Biol Chem 393: 599–604, 2012 [DOI] [PubMed] [Google Scholar]
- 21.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO: Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9: 654–659, 2007 [DOI] [PubMed] [Google Scholar]
- 22.Yuan A, Farber EL, Rapoport AL, Tejada D, Deniskin R, Akhmedov NB, Farber DB: Transfer of microRNAs by embryonic stem cell microvesicles. PLoS ONE 4: e4722, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xin H, Li Y, Buller B, Katakowski M, Zhang Y, Wang X, Shang X, Zhang ZG, Chopp M: Exosome-mediated transfer of miR-133b from multipotent mesenchymal stromal cells to neural cells contributes to neurite outgrowth. Stem Cells 30: 1556–1564, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jinek M, Doudna JA: A three-dimensional view of the molecular machinery of RNA interference. Nature 457: 405–412, 2009 [DOI] [PubMed] [Google Scholar]
- 25.Merritt WM, Bar-Eli M, Sood AK: The dicey role of Dicer: Implications for RNAi therapy. Cancer Res 70: 2571–2574, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Volk N, Shomron N: Versatility of MicroRNA biogenesis. PLoS ONE 6: e19391, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie M, Steitz JA: Versatile microRNA biogenesis in animals and their viruses. RNA Biol 11: 673–681, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kashiwagi E, Tonomura Y, Kondo C, Masuno K, Fujisawa K, Tsuchiya N, Matsushima S, Torii M, Takasu N, Izawa T, Kuwamura M, Yamate J: Involvement of neutrophil gelatinase-associated lipocalin and osteopontin in renal tubular regeneration and interstitial fibrosis after cisplatin-induced renal failure. Exp Toxicol Pathol 66: 301–311, 2014 [DOI] [PubMed] [Google Scholar]
- 29.Sörensen-Zender I, Rong S, Susnik N, Lange J, Gueler F, Degen JL, Melk A, Haller H, Schmitt R: Role of fibrinogen in acute ischemic kidney injury. Am J Physiol Renal Physiol 305: F777–F785, 2013 [DOI] [PubMed] [Google Scholar]
- 30.Qi S, Wu D: Bone marrow-derived mesenchymal stem cells protect against cisplatin-induced acute kidney injury in rats by inhibiting cell apoptosis. Int J Mol Med 32: 1262–1272, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim JH, Park DJ, Yun JC, Jung MH, Yeo HD, Kim HJ, Kim DW, Yang JI, Lee GW, Jeong SH, Roh GS, Chang SH: Human adipose tissue-derived mesenchymal stem cells protect kidneys from cisplatin nephrotoxicity in rats. Am J Physiol Renal Physiol 302: F1141–F1150, 2012 [DOI] [PubMed] [Google Scholar]
- 32.Zhou Y, Xu H, Xu W, Wang B, Wu H, Tao Y, Zhang B, Wang M, Mao F, Yan Y, Gao S, Gu H, Zhu W, Qian H: Exosomes released by human umbilical cord mesenchymal stem cells protect against cisplatin-induced renal oxidative stress and apoptosis in vivo and in vitro. Stem Cell Res Ther 4: 34, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Del Tatto M, Ng T, Aliotta JM, Colvin GA, Dooner MS, Berz D, Dooner GJ, Papa EF, Hixson DC, Ramratnam B, Aswad BI, Sears EH, Reagan J, Quesenberry PJ: Marrow cell genetic phenotype change induced by human lung cancer cells. Exp Hematol 39: 1072–1080, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomasoni S, Longaretti L, Rota C, Morigi M, Conti S, Gotti E, Capelli C, Introna M, Remuzzi G, Benigni A: Transfer of growth factor receptor mRNA via exosomes unravels the regenerative effect of mesenchymal stem cells. Stem Cells Dev 22: 772–780, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindoso RS, Collino F, Bruno S, Araujo DS, Sant’Anna JF, Tetta C, Provero P, Quesenberry PJ, Vieyra A, Einicker-Lamas M, Camussi G: Extracellular vesicles released from mesenchymal stromal cells modulate miRNA in renal tubular cells and inhibit ATP depletion injury. Stem Cells Dev 23: 1809–1819, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Negishi K, Noiri E, Sugaya T, Li S, Megyesi J, Nagothu K, Portilla D: A role of liver fatty acid-binding protein in cisplatin-induced acute renal failure. Kidney Int 72: 348–358, 2007 [DOI] [PubMed] [Google Scholar]
- 37.da Costa MR, Pizzatti L, Lindoso RS, Sant’Anna JF, DuRocher B, Abdelhay E, Vieyra A: Mechanisms of kidney repair by human mesenchymal stromal cells after ischemia: A comprehensive view using label-free MS(E). Proteomics 14: 1480–1493, 2014 [DOI] [PubMed] [Google Scholar]
- 38.Fang Y, Tian X, Bai S, Fan J, Hou W, Tong H, Li D: Autologous transplantation of adipose-derived mesenchymal stem cells ameliorates streptozotocin-induced diabetic nephropathy in rats by inhibiting oxidative stress, pro-inflammatory cytokines and the p38 MAPK signaling pathway. Int J Mol Med 30: 85–92, 2012 [DOI] [PubMed] [Google Scholar]
- 39.Guo Y, Xiao L, Sun L, Liu F: Wnt/beta-catenin signaling: A promising new target for fibrosis diseases. Physiol Res 61: 337–346, 2012 [DOI] [PubMed] [Google Scholar]
- 40.Gonçalves JG, de Bragança AC, Canale D, Shimizu MH, Sanches TR, Moysés RM, Andrade L, Seguro AC, Volpini RA: Vitamin D deficiency aggravates chronic kidney disease progression after ischemic acute kidney injury. PLoS ONE 9: e107228, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ying Y, Kim J, Westphal SN, Long KE, Padanilam BJ: Targeted deletion of p53 in the proximal tubule prevents ischemic renal injury [published online ahead of print May 22, 2014]. J Am Soc Nephrol .doi:ASN.2013121270 [DOI] [PMC free article] [PubMed]
- 42.Oskowitz AZ, Penfornis P, Tucker A, Prockop DJ, Pochampally R: Drosha regulates hMSCs cell cycle progression through a miRNA independent mechanism. Int J Biochem Cell Biol 43: 1563–1572, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aagaard L, Amarzguioui M, Sun G, Santos LC, Ehsani A, Prydz H, Rossi JJ: A facile lentiviral vector system for expression of doxycycline-inducible shRNAs: Knockdown of the pre-miRNA processing enzyme Drosha. Mol Ther 15: 938–945, 2007 [DOI] [PubMed] [Google Scholar]
- 44.Bruno S, Collino F, Deregibus MC, Grange C, Tetta C, Camussi G: Microvesicles derived from human bone marrow mesenchymal stem cells inhibit tumor growth. Stem Cells Dev 22: 758–771, 2013 [DOI] [PubMed] [Google Scholar]
- 45.Grange C, Tapparo M, Bruno S, Chatterjee D, Quesenberry PJ, Tetta C, Camussi G: Biodistribution of mesenchymal stem cell-derived extracellular vesicles in a model of acute kidney injury monitored by optical imaging. Int J Mol Med 33: 1055–1063, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL: TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L: Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31: 46–53, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang W, Sherman BT, Lempicki RA: Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57, 2009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.