Abstract
Previously it was reported that Alzheimer’s disease (AD) patients have reduced amyloid (Aβ1–42) and elevated total tau (t-tau) and phosphorylated tau (p-tau181p) in the cerebro-spinal fluid (CSF), suggesting that these same measures could be used to detect early AD pathology in healthy elderly (CN) and mild cognitive impairment (MCI). In this study, we tested the hypothesis that there would be an association among rates of regional brain atrophy, the CSF biomarkers Aβ1–42, t-tau, and p-tau181p and ApoE ε4 status, and that the pattern of this association would be diagnosis specific. Our findings primarily showed that lower CSF Aβ1–42 and higher tau concentrations were associated with increased rates of regional brain tissue loss and the patterns varied across the clinical groups. Taken together, these findings demonstrate that CSF biomarker concentrations are associated with the characteristic patterns of structural brain changes in CN and MCI that resemble to a large extent the pathology seen in AD. Therefore, the finding of faster progression of brain atrophy in the presence of lower Aβ1–42 levels and higher p-tau levels supports the hypothesis that CSF Aβ1–42 and tau are measures of early AD pathology. Moreover, the relationship among CSF biomarkers, ApoE ε4 status, and brain atrophy rates are regionally varying, supporting the view that the genetic predisposition of the brain to amyloid and tau mediated pathology is regional and disease stage specific.
Keywords: MRI, Alzheimer’s disease, cerebrospinal fluid, biomarkers, cortical thickness, atrophy, ApoE
INTRODUCTION
There is an increasing body of evidence from in vivo imaging and post mortem studies indicating that Alzheimer’s disease (AD) is associated with a sequence of pathophysiological events that can occur over a long period (approximately 20-years) before clinical symptoms become apparent [1]. A slow disease progression provides potentially a window for early interventions to reduce or even stop progression of AD. Histopathological studies showed that the hallmarks of the disease, Aβ-rich amyloid plaques and neurofibrillary tangles formed by abnormal tau, precede neuron loss in presymptomatic AD patients [1]. Substantial accumulations of plaques and tangles in the brain can also be found in non-demented subjects with mild cognitive impairment (MCI), individuals at an increased risk of developing AD or other dementias [2–4]. Consistent with histopathological findings, cerebrospinal fluid (CSF) chemistry studies have pointed to alterations in CSF Aβ (in particular Aβ1–42), total tau (t-tau) and phosphorylated tau (p-tau181p) concentrations preceding clinical symptoms of AD [5]. In general, studies found that increased CSF t-tau and p-tau181p were associated with neuronal and axonal damage, whereas reduced CSF Aβ1–42, the form of Aβ that most readily fibrillizes and deposits earliest in plaques, has been implicated to reflect higher amyloid plaque burden in the brain [6, 7]. However, the CSF measures are not easily interpretable because their origins are not exclusively brain derived and they provide no information about the regional spread of brain damage. Despite this, there is considerable agreement that measuring CSF Aβ1–42, t-tau, and p-tau181p improves the diagnostic accuracy for AD [8].
Independent of biomarker studies, numerous structural MRI studies have shown a characteristic pattern of brain atrophy in AD and a similar pattern in MCI, affecting primarily regions in the parietotemporal lobe, including the hippocampus, which plays a central role in memory formation [9–22]. In addition, an increasing number of longitudinal MRI studies show that both AD and MCI are also associated with a regional pattern of increased rates of brain tissue loss compared to normal aging [23–29]. With the emerging findings of CSF biomarker and structural imaging alterations in AD, there is considerable interest in utilizing the CSF and MRI measures together to improve detection of early signs of AD, as well as, in unraveling relationships between CSF Aβ1–42, t-tau, and p-tau181p and MRI measures of regional brain alterations. Recently it has been shown that the combination of CSF biomarkers and atrophy rates can provide better prediction of AD than either source of data alone [30–32]. However, whether relationships between brain atrophy rates and CSF biomarkers help further to improve predictions has not fully been explored.
Moreover, the role of the apolipoprotein E allele ε4 (ApoE ε4) gene, a major risk factor for AD, ought to be considered for a comprehensive evaluation. Presence of ApoE ε4 is related to abnormal CSF biomarker concentrations [33, 34], as well as, to higher rates of brain atrophy [35–39]. The relationships among all three factors, CSF biomarkers, ApoE ε4, and rates of regional brain atrophy, might therefore provide important information about the vulnerability of the brain to AD. Our overall goal in this study was therefore to unravel the relationships among all three factors: brain atrophy rates, CSFbiomarker concentrations, and presence of ApoE ε4. Toward the goal of identifying an AD biomarker, it will be important to fully understand the relationship between CSF biomarker concentrations and brain degeneration, such as neuron loss, which is thought to underlie the clinical symptoms in AD [4, 5]. While CSF biomarkers relate to cumulative AD pathology in the brain as peripheral measures, MRI as an external tool elucidates the distribution of the AD related neurodegeneration (i.e., brain atrophy in terms of tissue loss and ventricular enlargement). However, relatively few MRI studies so far have reported correlations between CSF biomarkers and the pattern of brain atrophy or the rate of atrophy progression [37, 40–46]. Specifically, in healthy elderly individuals, it has been shown that low CSF levels of Aβ1–42 correlate with ventricular expansion and volumetric reductions in widespread brain areas [45]. In individuals with progressive MCI, low CSF Aβ1–42 concentration and high concentrations of CSF p-tau181p and t-tau are associated with higher subsequent rates of hippocampal atrophy [37, 40–42]. In AD patients, elevated CSF p-tau181p concentrations were associated with higher subsequent rates of hippocampal atrophy and medial temporal atrophy [40–43], while low CSF Aβ1–42 concentrations exhibited larger rates of medial temporal atrophy [43]. However, the majority of previous MRI studies in this context focused on hippocampal and temporal lobe atrophy and ventricular expansion in MCI and AD patients, while relatively little is known about relations between the CSF biomarker concentrations and atrophy rates of other regions throughout the brain. In addition, variations in these relationships across the spectrum of cognitive impairments have not been comprehensively studied for regions across the brain.
Our main goal in this study was to test the hypothesis that relations between CSF biomarkers (i.e., Aβ1–42, t- tau, and p-tau181p concentrations) and rates of regional brain atrophy not only vary across brain regions but also across the cognitive spectrum, including healthy elderly individuals (CN), individuals with MCI, and AD patients. In particular we tested that (1) low Aβ1–42 and high t-tau and p-tau181p concentrations were associated with smaller absolute cortical thickness including parieto-temporal and prefrontal cortical regions in CN, MCI, and AD, (2) low Aβ1–42 and high t-tau and p-tau181p concentrations were associated with increased rates of regional brain atrophy including parieto-temporal, especially medial temporal, precuneus, and posterior cingulate cortical regions in CN, MCI, and AD, and (3) the patterns of association were group-specific. In addition, we tested whether abnormal CSF biomarker concentrations and ApoE ε4 status separately or together were associated with higher rates of brain atrophy.
COHORT AND METHODS
We examined the baseline cortical thickness and the rate of change in cortical thickness across the brain in CN, individuals with MCI, and AD patients. Structural magnetic resonance imaging (MRI) brain scans at multiple time points (four time point scans – baseline, 6, 12, and 24 months – for CN and AD subjects and five time point scans – baseline, 6, 12, 18, and 24 months – for individuals with MCI) were acquired at multiple Alzheimer’s Disease Neuroimaging Initiative (ADNI) sites using 1.5 Tesla MRI scanners. Using FreeSurfer longitudinal processing framework, local cortical thickness throughout the entire cortex was automatically measured at each time point. In each diagnostic group separately, generalized linear mixed effect models followed by pair-wise maximum likelihood tests were performed to test:1) if baseline CSF biomarker concentrations predict absolute local thickness at baseline;2) if baseline CSF biomarker concentrations modulate the rates of brain atrophy (i.e., the rate of change in cortical thickness); and 3) if CSF biomarkers and ApoE ε4 modulate the rates of brain atrophy jointly or independently, after accounting for variations in age, sex, and education. Finally, we tested if the observed modulation effects of baseline CSF biomarkers on rates of atrophy differ among groups. The methodological details are explained herein.
Participants
The participants in this study were recruited through the ADNI, a longitudinal, multicenter study launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies, and non-profit organizations, as a $60 million, 5-year public–private partnership to define biomarkers of early Alzheimer’s disease for clinical trials (http://www.adni-info.org). The Principal Investigator of this initiative is Michael W. Weiner, MD of the Veteran Affairs Medical Center and University of California in San Francisco.
Briefly, inclusion criteria for the CN group were Mini-Mental State Examination (MMSE) scores between 24 and 30, a Clinical Dementia Rating -Sum of Boxes (CDR-SB) score of 0, and lack of depression, MCI, or dementia. Inclusion criteria for the MCI group followed the Peterson criteria [47] for amnestic MCI, which required a subjective memory complaint, objective memory loss measured by education-adjusted Wechsler Memory Scale-Revised Logical Memory II scores, a CDR-SB of 0.5, absence of significant impairment in other cognitive domains, preserved activities of daily living, and an absence of dementia. AD participants met the National Institute for Neurological and Communicative Disorders and Stroke-Alzheimer’s Disease and Related Disorder Association (NINDS/ADRDA) criteria for probable AD, had an MMSE between 18 and 26, and a CDR-SB of 0.5 to 1.0. Exclusion criteria included history of structural brain lesions or head trauma, significant neurological disease other than incipient AD, and use of psychotropic medications that could affect memory. The full details of the inclusion and exclusion criteria for the ADNI can be found at ht//www.adni-info.org. Written consent was obtained from all subjects participating in the study according to the Declaration of Helsinki (Br Med J 1991;302 :1194), and the study was approved by the institutional review board at each participating site.
The population in this study included ADNI subjects with valid test result for all three CSF biomarkers and successful longitudinal FreeSurfer processing of MR images from at least two time points. Overall, the study population was comprised of 77 CN, 119 MCI, and 53 AD subjects. Details of CSF biomarker concentration measurement and longitudinal structural MR image processing are described in the following sections. The demographic details of each group are given in Table 1.
Table 1.
Demographic features of study groups
| CN | MCI | AD | |
|---|---|---|---|
| N (baseline) | 77 | 119 | 54 |
| N follow-up (6, 12, 18, 24 months) | 77, 76, 0, 63 | 119, 109, 96, 77 | 54, 53, 0, 31 |
| Baseline age (years) | 75 ± 5.0 | 74 ± 7.6 | 74 ± 8.0 |
| Gender (F/M) | 40/37 | 46/73 | 25/29 |
| Baseline CSF Aβ1–42 (pg/mL) | 203 ± 51.8 | 165 ± 57.5 | 142 ± 39.7 |
| Baseline CSF t-tau (pg/mL) | 70 ± 29.5 | 101 ± 50.0 | 128 ± 53.1 |
| Baseline CSF p-tau181p (pg/mL) | 26 ± 15.0 | 35 ± 16.5 | 42 ± 16.8 |
| ApoE ε4 status (3/4 – 4/4 carriers) | 18 – 0 | 50 – 13 | 25 – 11 |
| Education (years) | 15.7± 2.9 | 15.7 ± 3.1 | 15.0 ± 2.8 |
| Baseline CDR | 0.0± 0.0 | 0.5 ± 0.0 | 0.72 ± 0.25 |
| Baseline MMSE | 29.1± 1.3 | 26.9 ± 2.7 | 23.5 ± 4.7 |
Structural MRI acquisition
The participants underwent a standardized 1.5 Tesla MRI protocol (http://www.loni.ucla.edu/ADNI/Research/Cores/index.shtml), which included two T1-weighted MRI scans using a sagittal volumetric magnetization prepared rapid gradient echo (MP-RAGE) sequence with the following acquisition parameters: echo time (TE) of 4 ms, repetition time (TR) of 9 ms, flip angle of 8°, acquisition matrix size of 256×256×166 in the x-, y-and z-dimensions with a nominal voxel size of 0.94×0.94×1.2mm3. Only one of the MPRAGE sets was used for analysis. The ADNI MRI quality control center at the Mayo Clinic selected the MP-RAGE image with higher quality and corrected for system-specific image artifacts, as described in [48].
CSF biomarker concentrations
CSF samples were obtained from 53% of ADNI participants, while the rest did not undergo lumbar puncture. The demographics of ADNI subjects with CSF samples are comparable with that in the full ADNI patient population (http://www.adni-info.org).
A small sample of CSF from the lower spine of each subject was collected at baseline by lumbar puncture in the morning after an overnight fast. Lumbar puncture was performed with a 20- or 24-gauge spinal needle as described in the ADNI procedures manual (ht//www.adni-info.org). In brief, CSF was collected into collection tubes provided to each site, then transferred into polypropylene transfer tubes followed by freezing on dry ice within 1 hour after collection, and shipped overnight to the ADNI Biomarker Core laboratory at the University of Pennsylvania Medical Center on dry ice. 0.5 mL aliquots were prepared from these samples after thawing for 1 hour at room temperature and gentle mixing. The aliquots were stored in bar code–labeled polypropylene vials at −80°C. Aβ1–42, t-tau, and p-tau181p were measured in each aliquots using the multiplex xMAP Luminex platform (Luminex Corp, Austin, TX) with Innogenetics (INNO-BIA AlzBio3; Ghent, Belgium; for research use–only reagents) immunoassay kit-based reagents. Full details of this combination of immunoassay reagents and analytical platform are provided elsewhere [49]. The ADNI baseline CSF samples were analyzed over a 14-day period and included test–retest analyses of 29 of the samples that further substantiated the analytical performance (r2 values for comparison of initial test result with retest result of 0.98, 0.90, and 0.85 for t-tau, Aβ1–42, and p-tau181p, respectively for 29 randomly selected samples). Full details of ADNI baseline CSF biomarker measurements are provided elsewhere [7].
FreeSurfer longitudinal MR image processing
Automated cortical thickness measures were performed with FreeSurfer software package, version 4.4 (http://surfer.nmr.mgh.harvard.edu/fswiki). To reduce the confounding effect of intra-subject morphological variability, each subject’s longitudinal data series was processed by FreeSurfer longitudinal workflow. The longitudinal workflow was designed to estimate brain morphometry measurements that were unbiased with respect to any time point. Instead of using information from a specific time point as a prior for other time points, a template image volume was created as an unbiased prior for all time points.
FreeSurfer longitudinal workflow consists of four stages: (1) processing of all time points individually with the cross-sectional workflow; (2) creation of a probabilistic template unbiased toward time points from all time points’ cross-sectional data; (3) processing of unbiased template with the cross-sectional workflow; and finally (4) re-processing of each time point with the longitudinal workflow, which uses the unbiased template results as initial guess for the segmentation and surface reconstruction. For a full description of the FreeSurfer processing steps, see [50, 51], and for a full description of the longitudinal workflow, see http://surfer.nmr.mgh.harvard.edu/fswiki/LongitudinalProcessing.
Vertex-based cortical thickness measurements were obtained as the distance between the reconstructed surface representations of the gray matter/white matter and white matter/CSF tissue interfaces [52]. Each cortical surface was spatially normalized to a template cortical surface using a non-rigid high-dimensional spherical averaging method to align cortical folding patterns. Subject cortical thickness maps were mapped onto the template surface based on this spatial normalization and then smoothed by a surface-based Gaussian blurring kernel with a standard deviation of 10 mm to remove noise-induced variations in the measurements.
The surface reconstruction results were visually examined for anatomical accuracy. Although the FreeSurfer software package allows for manual editing to correct registration and segmentation errors, given the large number of subjects in ADNI data set, only the data with accurate results from fully automated processing were used in the subsequent analysis in the interest of a practical total processing time and avoidance of reader bias. 74% of the MR images passed this quality control, 3% of the images failed the quality control completely, and remaining 23% of the images got partial pass on the quality control. Details of the quality control procedure are posted online at http://www.loni.ucla.edu/twiki/pub/ADNI/ADNIPostProc/UCS-FFreeSurferMethodsSummary.pdf.
According to our FreeSurfer quality control protocol, segmentation and pial surface estimates were checked globally and regionally in the coronal view for regions of overestimation/underestimation, inconsistency with the structural boundaries, or segmention regions not accurately reflecting the underlying anatomy. The quality control procedure accounted only for gross errors; a gross error for these purposes was defined as an area of over/underestimation that was larger than the cursor, and which occurred on two or more slices. This was to account for inevitable small errors in segmentation such as those which may have been the result of partial voluming. Orientations other than the coronal view were used to confirm possible pial border and segmentation gross errors. Subjects with complete segmentation failure or gross errors throughout all brain regions were rated as complete failure and the ones with gross errors in one or more specific brain regions (i.e., temporal lobe regions, superior regions, occipital regions, and insula) were given partial pass rating. All the subjects with passing quality control rating were included in analyses presented in this work.
Statistical analyses
For each subject, variations in cortical thickness were modeled as a function of time starting with the baseline scan (time-point zero) in intervals of subsequent MRI scans in units of years. We employed a general linear mixed effects (GLME) model for analysis of the longitudinal data in which the response variable (i.e., cortical thickness) was regressed against the explanatory variables including time, baseline CSF biomarker concentration (i.e., Aβ1–42, p-tau181p, or t-tau), and the interaction between time and baseline CSF biomarker concentration to estimate the fixed effects in the group, separately from the random effects such as within subject variations in both baseline and longitudinal measures. This concept was used to test the primary hypothesis that variations in CSF biomarker concentrations modulate rates of brain atrophy (i.e., cortical thinning). The fixed effect model was formulated as follows:
Here, Vij represents the size of a brain structure (cortical thickness) from subject i at time point j. Accordingly, Tij indicates the time point of the individual MRI scan, Bi0 represents individual CSF biomarker concentrations at baseline and εij is the mixed effects error. Our goal was to test the significance of the coefficient βYears:CSFbio in explaining structural variations (i.e., the moderator) relative to the coefficients β0, βYears, and βCSFbio and independent of random variations in brain structures at baseline and over time. For a significant interaction to occur, CSF biomarker concentration must modulate the relationship between time and the response variable (i.e., local cortical thickness). To determine if the addition of an interaction term (βYears:CSFbio) between rates of brain atrophy and CSF biomarkers in the model significantly improves the explanatory power of regional variations in cortical thickness as a function of biomarkers, we compared pair-wise GLME models (i.e., with and without the βYears:CSFbio term), fitted by maximum likelihood (ML) via F-tests. These tests were performed separately for each group (i.e., CN, MCI, and AD). Similarly, to determine the significance of CSF biomarker effects on cortical thickness at baseline, the additive term βCSFbio was assessed by pair-wise comparisons of GLME models with and without βCSFbio term, followed by maximum likelihood (ML) via F-tests. Each CSF biomarker was centered on its population mean to reduce colinearity.
To assess if the effect of CSF biomarkers on rates of regional brain atrophy differ across groups (CN, MCI, and AD), we resampled the random effects residual of the fits by 100-fold bootstrap and evaluated differences in distributions by analysis of variance.
Finally, we tested the extent to which ApoE ε4 status contributes to higher brain atrophy rates independent of CSF biomarker concentrations (ApoE ε4 status + CSF biomarker) or via a synergistic interaction with the biomarkers (ApoE ε4 status *CSF biomarker). Again, pair-wise ML tests were performed between models with and without the interaction term (i.e. ApoE ε4 × CSF biomarker) to determine the contribution of the interaction.
Age, gender, and education were included as covariates in each regression model described above. The GLME models and the corresponding pair-wise ML F-tests were evaluated at each surface vertex independently. All statistical analyses were computed using R (the R Project for Statistical Computing; www.r-project.org). To control for false positive findings given the large number of comparisons per brain map, we used the concept of a false discovery rate (FDR) at the level q = 0.05 [53]. For testing the a-priori hypotheses on associations between baseline CSF biomarker levels and the absolute cortical thickness as well as the rates of regional cortical atrophy, which comprised a limited number of planned tests, we used a per comparison error rate of q = 0.05 for each test to determine the probability that any one contrast, after passing FDR, is found by chance.
RESULTS
Effects of baseline CSF biomarker concentrations on absolute cortical thickness
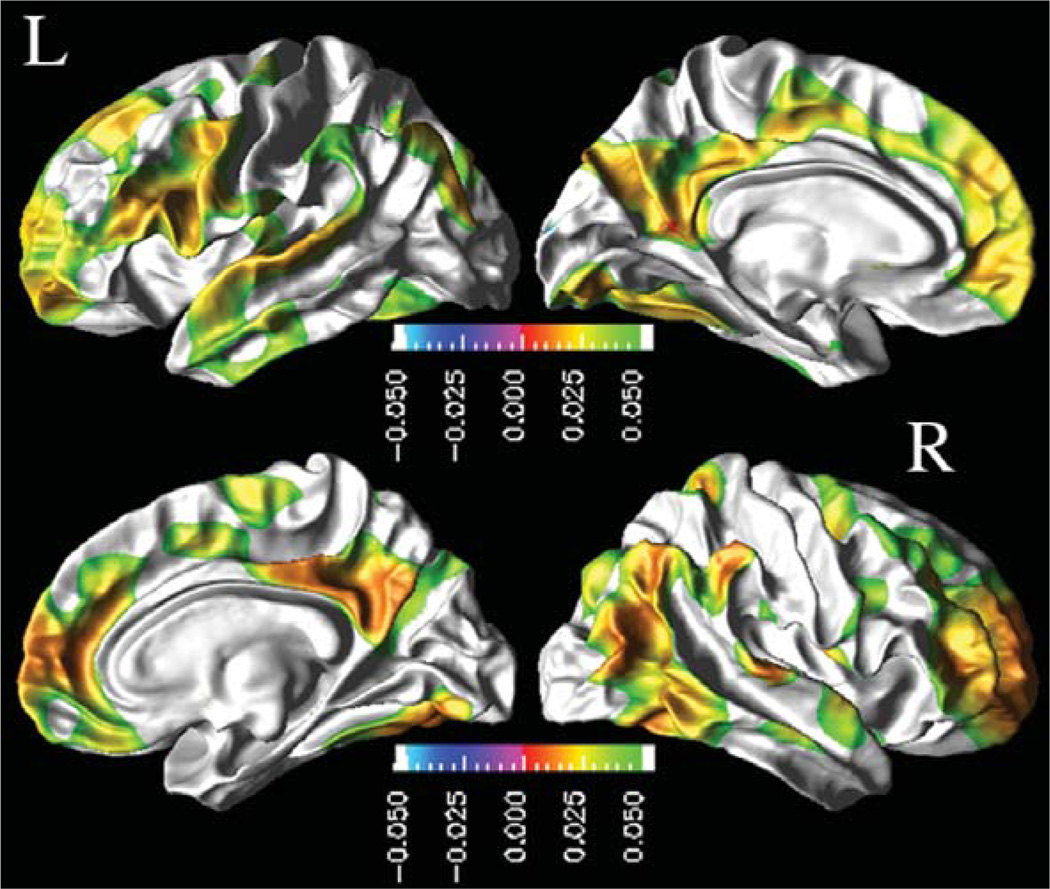
Next, we report effects of CSF biomarker concentrations on absolute cortical thickness after accounting for variations in age, gender, and education across subjects. Figure 1 depicts the regional distribution of CSF Aβ1–42 effects on absolute cortical thickness for CN based on the ML F-tests. In CN, lower baseline CSF Aβ1–42 was associated with a thinner cortex in the bilateral frontal pole, rostral-middle frontal, supramarginal, inferior parietal, middle temporal, inferior temporal, posterior cingulate, precuneus, and fusiform cortices, in the left superior frontal, pars opercularis, pars triangularis, and superior temporal cortices, and in the right superior parietal, medial orbito-frontal, and paracentral lobule cortices. In MCI and AD, no statistically significant association between the baseline CSF biomarker concentrations and the absolute cortical thickness measures were observed.
Fig. 1.
Association between baseline CSF Aβ1–42 concentrations and absolute cortical thickness in CN group: FDR corrected p-value maps at 0.05 level from pair-wise ML F-tests. Hot color (green to red) indicates an association between lower baseline CSF Aβ1–42 and thinner cortex.
Neither t-tau nor p-tau181p showed significant association with absolute cortical thickness in CN, MCI, and AD groups.
Effects of baseline CSF biomarkers concentrations on the rates of regional cortical atrophy
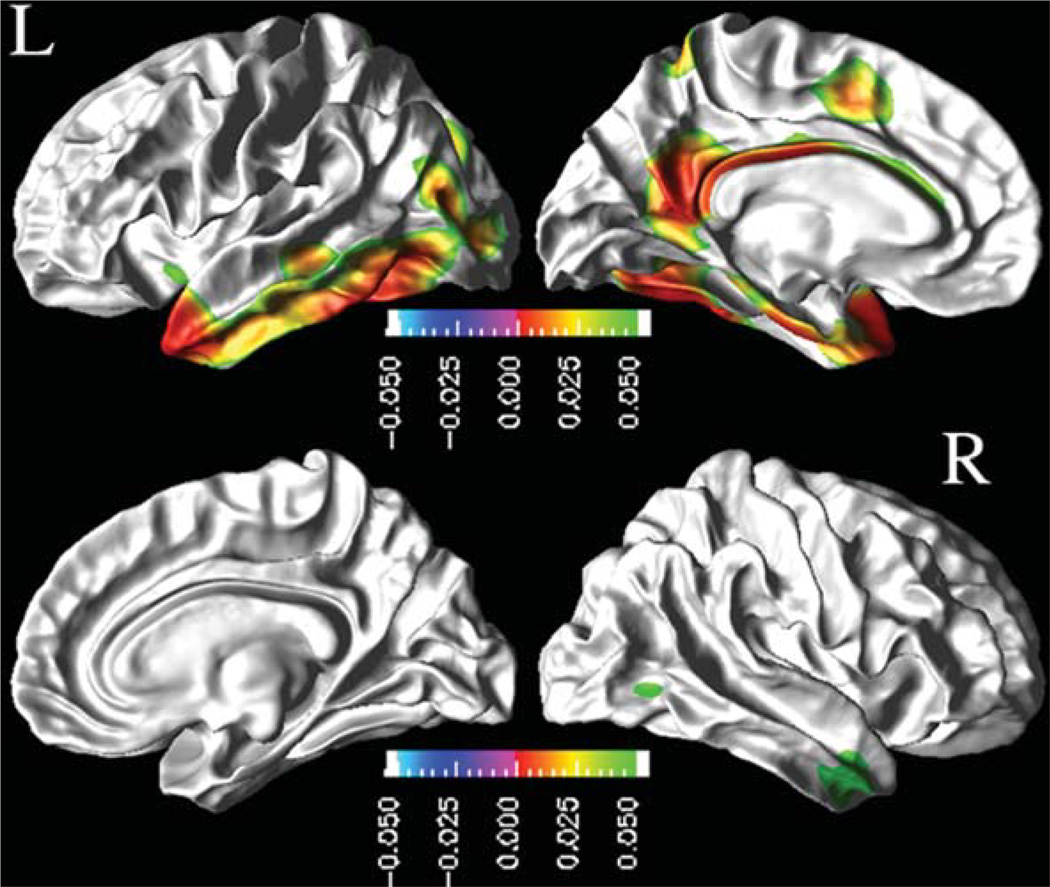
Similarly, we report next the modulation effects of CSF biomarkers on the rates of cortical thinning after accounting for variations in age, gender, and education across subjects. Figure 2 depicts the regional distribution of a CSF Aβ1–42 modulation effects on the rates of cortical thinning for MCI based on the ML F-tests. In MCI, lower concentrations of CSF Aβ1–42 were associated with increased rates of cortical thinning throughout the cortex. The effects were statistically significant (FDR corrected; p < 0.05) in the bilateral inferior temporal and middle temporal cortices and in the left temporal pole, inferior parietal, paracentral lobule, cingulate, isthmus cingulate, precuneus, entorhinal, and fusiform cortices.
Fig. 2.
Effects of baseline CSF Aβ1–42 concentrations on the rate of cortical atrophy in MCI group: FDR corrected p-value map at 0.05 level from pair-wise ML F-tests. Hot color (green to red) indicates an association between lower concentrations of CSF Aβ1–42 and increased rates of cortical thinning.
Neither CN nor AD patients showed significant modulation effects of baseline CSF biomarkers on the cortical atrophy rates after correcting for multiple comparison.
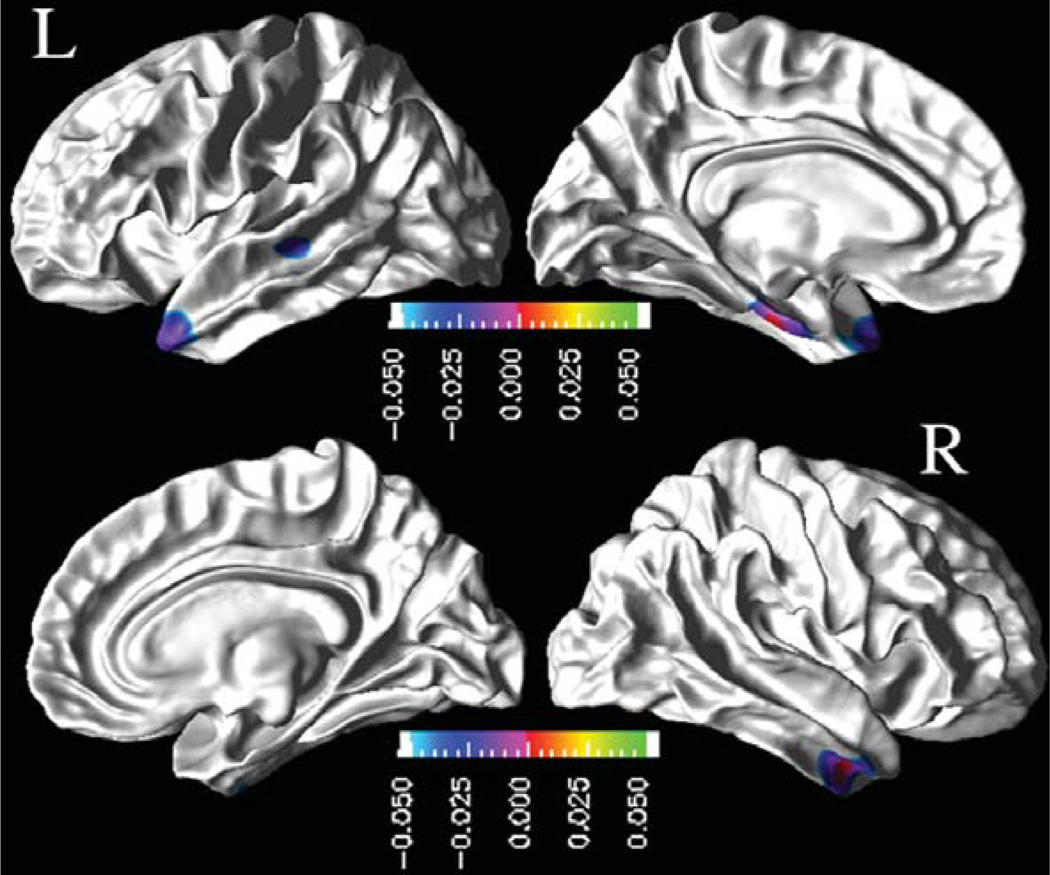
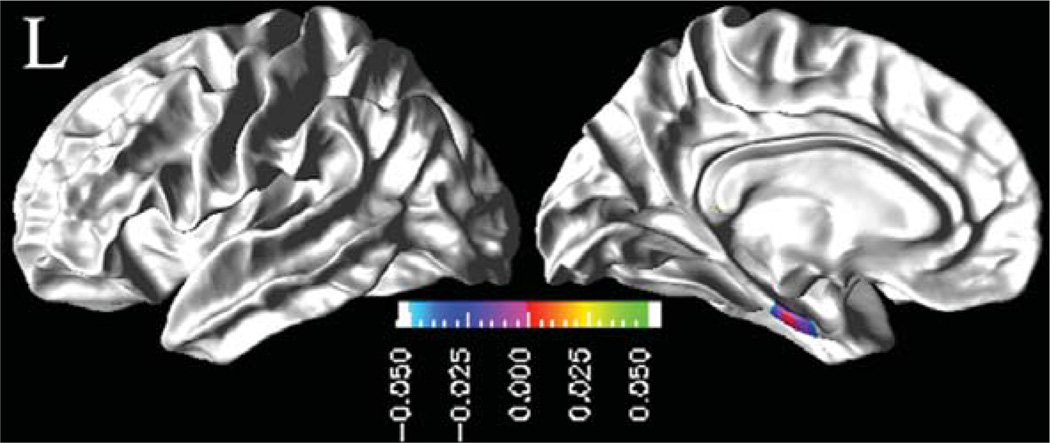
In Fig. 3 are shown the regional distribution of CSF p-tau181p effects on the rates of cortical thinning for MCI. The results indicate that higher baseline CSF p-tau181p concentrations in MCI were associated with higher rates of cortical thinning, significantly in the left temporal pole, superior temporal sulcus, and entorhinal gyrus regions, and in the right inferior and middle temporal cortices. Similarly, higher baseline CSF t-tau concentrations were associated with higher cortical atrophy rates in the left entorhinal gyrus in MCI patients, as shown in Fig. 4.
Fig. 3.
Effects of baseline CSF p-tau181p concentrations on the rates of cortical atrophy in MCI group: FDR corrected p-value mapat 0.05 level from pair-wise ML F-tests. Cold color (blue to purple) indicates an association between higher baseline CSF p-tau181p concentrations and higher rates of cortical thinning.
Fig. 4.
Effects of baseline CSF t-tau concentrations on rate of cortical atrophy in MCI group: FDR corrected p-value map at 0.05 level from pair-wise ML F-tests. Cold color (blue to purple) indicates an association between higher baseline CSF t -tau concentrations and higher rates of cortical thinning.
Group differences in CSF biomarker effects on rates of cortical thinning
By bootstrapping the random effects residuals of the GLME fits with time and baseline CSF biomarker concentration interaction term, we tested if the estimates of the association between biomarkers and atrophy rates significantly differ across populations. Based on pairwise group comparison, we found that differences in the estimations between the groups were all significant (p = 0.05 level). This implies that the relationship among CSF biomarkers and brain atrophy rates was not only regionally specific but also varied across the clinical groups.
ApoE ε4 analyses
In our data set, presence of ApoE ε4 alleles correlated significantly with the CSF Aβ1–42 concentrations (r = −0.50, p<10−5 in CN, r = −0.49, p<10−8 in MCI, and r = −0.53, p<10−5 in AD), after controlling for age. A partial correlation between presence of ApoE ε4 alleles and p-tau181p and t-tau after controlling for age was significant only in MCI group (r = 0.34 with p<10−3 and r = 0.39 with p<10−6, respectively).
Only in individuals with MCI, lower CSF Aβ1–42 and ApoE ε4 together were associated with higher rates of cortical thinning in temporoparietal cortex, including precuneus and posterior cingulate. CSF tau and ApoE ε4 together were associated with higher rates of cortical thinning in the entorhinal cortex, precuneus and temporal pole. In contrast, lower Aβ1–42 alone explained higher rates of cortical thinning in left entorhinal, fusiform, inferior temporal, temporal pole, and parahippocampal cortices without a significant contribution from ApoE ε4. Similarly, higher CSF tau explained higher rates of cortical thinning in entorhinal cortex without a significant contribution from ApoE ε4.
In no case did an interaction between ApoE ε4 status and CSF biomarker concentration (i.e. ApoE ε4 status × CSF biomarker) approach significance in predicting rate of cortical thinning.
DISCUSSION
We have three major findings: (1) In controls, an association between CSF Aβ1–42 and baseline cortical thickness was observed prominently in regions that generally appear affected in AD. (2) In MCI subjects, lower CSF Aβ1–42 and higher p-tau181p and t-tau concentrations were associated with higher rates of brain atrophy in regions of the temporal and parietal cortices implicated in AD pathology. (3) The relationship among CSF biomarkers, ApoE ε4 status and brain atrophy rates was regionally specific and varied across the clinical groups.
Our finding in controls, demonstrating that low baseline CSF Aβ1–42 biomarker concentration is associated with thinner cortex predominantly in the inferior temporal, parietal, frontal, precuneus, and posterior cingulate cortices, provides evidence for a link between variations in peripheral CSF chemistry and regional brain size. Moreover, the result suggests that the link between CSF markers and regional brain size is already established in absence of any apparent clinical symptoms of cognitive deficits. A previous study in cognitively normal elderly also found an association between low levels of CSF Aβ1–42 and smaller whole-brain volume [44]. It has been shown that in healthy elderly individuals, reduction in CSF Aβ1–42 is a predictor of cognitive decline and development of AD [54–56]; therefore, the association between low baseline CSF Aβ1–42 concentration and thin cortex in controls could reflect preclinical AD pathology. However, pathological conditions other than AD might also contribute to the relationship between low CSF Aβ1–42 concentration and cortical thinning.
In individuals with MCI, baseline cortical thickness measures had no significant association with CSF biomarker concentrations. This finding implies a dissociation between CSF biomarkers and their effects on the brain in individuals with MCI. The finding in MCI, showing that variations in CSF biomarker concentrations are associated with a characteristic pattern of altered rates of regional brain atrophy, similar to the pattern seen in AD, further supports the view that these relations reflect brain alterations presymptomatic to AD and could be useful for staging disease severity and assessing disease progression. Specifically, lower CSF Aβ1–42 and increased CSF p-tau181p and t-tau concentrations in MCI were associated with higher atrophy rates involving primarily inferior and medial temporal, parietal, precuneus, and posterior cingulate cortices. Structural MRI studies in AD consistently revealed a pattern of neuroanatomic abnormalities that predominantly involves structures in the medial temporal cortex (i.e., hippocampus and the entorhinal cortex [9–14, 19, 23, 28, 29, 40]) where the early pathological changes are seen, then gradually extends to temporoparietal cortical areas [15–18, 24] as severity of AD progresses [15, 17, 25–27, 57]. Our finding that lower CSF Aβ1–42 and higher CSF p-tau181p and t-tau concentrations were associated with higher atrophy rates of the temporal horn and inferior temporal lobe regions points to a selective vulnerability of these regions to AD pathology, consistent with histopathological findings. The finding that lower CSF Aβ1–42 is associated with a characteristic pattern of brain atrophy in MCI that resembles the atrophy pattern seen in AD is encouraging for the use of CSF Aβ1–42 as an early indicator of AD. Most importantly, elucidating the detrimental relationship between CSF biomarkers and rates of brain atrophy is of great interest to detect AD pathology in early stage, which is fundamental for an accurate early diagnosis of the disease, development of new treatment interventions, and evaluation of clinical trials in AD. The synergistic relationship between CSF biomarker and neurodegeneration patterns are of clinical interest as they may not only improve monitoring AD progression and evaluation of new AD therapies but also aid enrichment of clinical trial cohorts by identifying specific subsets of patients with MCI especially at high risk of developing AD [58–60]. Such a custom tailored cohort selection is desirable since drugs with disease-arresting effects have better efficacy in the preclinical and early phase of the disease before the synaptic and neuronal loss become widespread [61].
The spatial extent of baseline CSF Aβ1–42 modulation effects on brain atrophy observed in individuals with MCI is consistent with previous volumetric studies on patterns of increased atrophy rate in AD patients compared to elderly healthy controls [62]. In addition, a prior autopsy study on AD patients [63] reported neuritic plaques distributed throughout the cortex with the highest densities in the temporal and occipital lobes, while relatively lower plaque densities were found in the parietal lobe. This is consistent with our CSF Aβ1–42 modulation effect findings in MCI. Compared to the prior autopsy study in AD patients [63], the spatial extents of CSF p-tau181p and t-tau modulation effects are consistent with the neurofibrillary tangle distributions in AD pathology. In particular, cortical regions surrounding entorhinal cortex were reported among the most severely affected areas by neurofibrillary tangles. However, since we do not know how many of the MCI subjects in this study will ultimately develop AD, we cannot determine the predictive value of CSF biomarker concentrations for AD. Another observation in the MCI group was the left hemisphere dominance of the CSF biomarkers’ atrophy modulation effects. This observation is consistent with the asymmetric loss of GM (i.e., the left hemisphere atrophies faster than the right hemisphere) in AD [64] and yet again supporting the hypothesis that CSF Aβ1–42, p-tau181p, and t-tau are measures of early AD pathology.
A surprising finding in AD patients was that abnormal CSF biomarker levels were not significantly associated with rates of cortical thinning. Similarly, abnormal CSF biomarker levels in controls were also not significantly associated with rates of cortical thinning. It is also plausible that the finding in AD is the result of a complex relationship between advanced AD pathology in most cortical regions and peripheral biomarker concentration, while the finding in controls might reflect a threshold effect of minimum biomarker concentration on cortical thinning [45]. More studies are warranted to further investigate these issues.
Compared to CN and MCI groups, CSF biomarker concentrations have opposite modulation effects on rates of brain tissue volume change in AD. Specifically, we observed decreased rates of atrophy in the right posterior cingulate and precuneus cortices in the presence of higher concentrations of baseline CSF t-tau (relative to the CSF t-tau concentration distribution in AD group). Although not statistically significant, decelerative effects of CSF biomarker concentrations on rates of cortical atrophy were observed in AD. One explanation of this finding could be the disease-stage specific effects of CSF biomarkers on brain atrophy. AD subjects with lower CSF Aβ1–42 and higher CSF t-tau concentrations (relative to the biomarker concentration distributions in AD group) probably have advanced AD pathology where they reach a plateau in their rate of brain atrophy, which appears as a slower progression of the brain atrophy.
An interesting finding was that relations between the rate of brain atrophy and CSF biomarker concentrations varied across the CSF Aβ1–42, p-tau181p, and t-tau. This is not unexpected because it is known that amyloid plaques and tau containing tangles are distributed discordantly in brain at early stages of the disease. Specifically, the accumulation of amyloid plaques occurs in cortical regions whereas tangles appear in subcortical structures, predominantly involving the hippocampus [1, 63, 65]. Our findings reflect this pattern to some extent.
Finally, the finding of regional variations among CSF biomarkers, ApoE ε4 status, and brain atrophy rate relationships support the view that the genetic predisposition of the brain to amyloid and tau mediated pathology is region and disease stage specific. Interestingly, the most prominent region associated with CSF biomarker regardless of ApoE ε4 status in MCI included the entorhinal cortex, which is thought to be affected early by AD. Moreover, the effect of ApoE ε4 status on the relationships could be dose dependent [33, 34, 66].
Our findings in this study are largely consistent with several similar studies on relationships between CSF biomarkers and brain alterations in MCI and AD [40–42]. Specifically, increased concentrations of CSF p-tau181p are associated with higher subsequent rates of hippocampal atrophy in the progressive MCI and AD patients [40, 41] [42], of medial temporal atrophy in AD patients [43], of the temporal and parietal atrophy in MCI [46], and of right posterior ventricular horn expansion [67]. In contrast, low CSF Aβ1–42 concentration exhibited an association with increased rate of left hippocampal atrophy in the progressive MCI patients [42, 46], of the medial temporal atrophy in AD [43], of the temporal and parietal atrophy in MCI [46], and of ventricular expansion [67]. Elevated CSF t-tau concentrations are associated with higher rates of hippocampal atrophy in stable MCI patients [42]. In controls, it has been shown that low CSF Aβ1–42 concentration correlates with ventricular expansion and volumetric reductions in widespread brain areas, including inferior temporal, inferior parietal, frontal, posterior cingulate, precuneus, caudate, and amygdala regions [45]. Fjell et al. reported generally larger effects of CSF biomarkers on brain tissue change than what we found on the same cohort [45, 46]. However, several methodological differences between our study and that by Fjell et al. complicate direct comparisons. For example, whereas Fjell et al. aimed to evaluate potentially accelerated rates between the first and second scan intervals while accepting mixed effects on rates, we aimed to separate random from fixed effects on rates in order to boost sensitivity while ignoring the possibility of accelerated rates. Since each approach has its estimation bias, the different findings are difficult to interpret.
The majority of previous MRI studies, except [45, 46], in this context focused on the hippocampal and temporal lobe atrophy and ventricular enlargement, while our approach was generalized by assessing various other brain regions. Based on this, we discovered that the association between CSF biomarkers and structural changes are regionally differential. Although this observation is not entirely surprising, given that the pathological processes of plaque and tangle formation, which CSF Aβ1–42 and p-tau181p and t-tau indirectly represent, respectively. The finding of faster progression of brain atrophy in presence of lower baseline concentrations of Aβ1–42 and higher concentrations of p-tau181p/t-tau in MCI together with the similarities between the MCI pattern of CSF biomarker atrophy modulation effects and distribution of tangles and plaques in AD support the hypothesis that CSF biomarkers are measures of early AD pathology. MCI pattern of relations between rate of brain atrophy and CSF biomarker concentrations should be further explored to identify possible pre-symptomatic AD pathology. This finding also suggests a strategy for the potential use of biomarkers in clinical trials. CSF biomarker cut-offs to select fastest progressing cohorts could greatly improve the power of AD prevention trials on healthy elderly and MCI.
Several limitations of our study ought to be mentioned. First, MCI and AD subjects were diagnosed clinically; therefore other pathologies may have contributed to their symptoms and the relationships between CSF biomarkers and brain alterations may be unrelated to AD pathology. Another limitation is that CSF biomarkers, especially CSF Aβ1–42, have been shown to be saturated and may not accurately reflect severity of brain amyloid deposition or plaque density in the later stages of the disease [8, 68]. Therefore, structural brain changes may still occur secondarily to ongoing amyloid deposition or plaque accumulation. Restriction to linear, time-invariant brain atrophy rates is a technical limitation of our study. This is likely a gross simplification because the loss of brain tissue may be compounding and furthermore neurodegeneration in AD may be a dynamic process, which varies during disease progression. Therefore, models with nonlinear atrophy rate characteristics might lead to different results; however, such models are not always robust, given the limited number of serial MRI measurements and they also require careful validation. Finally, another technical limitation is that our study included fewer CN and AD subjects than MCI subjects despite expectations that power to detect atrophy will be higher for MCI than in CN and AD because of higher annual atrophy rates in MCI. Therefore, comparisons between CN and MCI as well as AD and MCI could be biased toward lower sensitivity to detect a change in CN and AD.
In summary, our findings demonstrate that alterations in CSF Aβ1–42, p-tau181p, and t-tau are each associated with characteristic patterns of structural brain changes (cross-sectionally or longitudinally) in CN and MCI that resembles to a large extent the pattern seen in AD pathology. Specifically, the finding of faster progression of brain atrophy in individuals with MCI in the presence of lower baseline CSF Aβ1–42 and higher CSF tau levels supports the view that these CSF biomarkers reflect AD brain pathology. Since the CSF Aβ1–42 and tau levels were also associated with a systematic pattern of regional brain atrophy rates that resembled the pattern known in AD, our findings further support the view that CSF Aβ1–42 and tau reflect brain damage due to AD pathology. Overall, the findings imply that CSF Aβ1–42 and tau taken together with MRI measures of rates of brain atrophy progression are promising candidates as biomarkers for early detection of AD.
ACKNOWLEDGMENTS
This work is funded by the National Institutes of Health (NIH), National Institute of Biomedical Imaging and Bioengineering (NIBIB) [T32 EB001631-05].
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott, AstraZeneca AB, Bayer Schering Pharma AG, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan Corporation, Genentech, GE Healthcare, Glax-oSmithKline, Innogenetics, Johnson and Johnson, Eli Lilly and Co., Medpace, Inc., Merck and Co., Inc., Novartis AG, Pfizer Inc, F. Hoffman-La Roche, Schering-Plough, Synarc, Inc., and Wyeth, as well as non-profit partners the Alzheimer’s Association and Alzheimer’s Drug Discovery Foundation, with participation from the U.S. Food and Drug Administration. Private sector contributions to ADNI are facilitated by the Foundation for the National Institutes of Health (http://www.fnih.org/). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129, K01 AG030514, and the Dana Foundation.
Dr. Schuff received honorary from the Michael J Fox foundation, the British Research Council and Elsevier Publishing company; receives research support from M.J. Fox foundation, Department of Defense (WX), P41 RR023953 (Coinvestigator); P50AG23501 (Coninvestigator).
Dr. Aisen has served as a consultant to Pfizer, Merck, and Novartis.
Dr. Petersen serves as a consultant to Elan Pharmaceuticals, Wyeth Pharmaceuticals, and GE Healthcare; receives royalties from publishing Mild Cognitive Impairment (Oxford University Press, 2003); and receives research support from the NIA [AG 06786 (PI) and AG 16574 (PI)].
Dr. Weiner serves on scientific advisory boards for Bayer Schering Pharma, Eli Lilly, Nestle, CoMentis, Neurochem, Eisai, Avid, Aegis, Genentech, Allergan, Lippincott, Bristol Meyers Squibb, Forest, Pfizer, McKinsey, Mitsubishi, and Novartis. He has received non–industry-supported funding for travel; serves on the editorial board of Alzheimer’s and Dementia; received honoraria from the Rotman Research Institute and BOLT International; receives research support from Merck & Co, Avid, NIH [U01AG024904 (PI), P41 RR023953 (PI), R01AG10897 (PI), P01AG19724 (Coinvestigator), P50AG23501(Coinvestigator), R24 RR021992 (Coinvestigator), R01 NS031966 (Coinvestigator), and P01AG012435 (Coinvestigator)], the Department of Defense [DAMD17-01-1-0764 (PI)], and the Veterans Administration [MIRECC VISN 21 (Core PI)]; and holds stock in Synarc and Elan Pharmaceuticals.
Footnotes
Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (www.loni.ucla.edu ADNI). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. Complete listing of ADNI investigators is available at www.loni.ucla.edu/ADNI/Collaboration/ADNIManuscript Citations.pdf.
DISCLOSURE STATEMENT
Dr. Tosun, Ms. Truran-Sacrey, Dr. Shaw, and Dr. Trojanowski report no disclosures.
REFERENCES
- 1.Price JL, Morris JC. Tangles and plaques in nonde-mented aging and ldquopreclinicalrdquo Alzheimer’s disease. Ann Neurol. 1999;45:358–368. doi: 10.1002/1531-8249(199903)45:3<358::aid-ana12>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 2.Aizenstein HJ, Nebes RD, Saxton JA, Price JC, Mathis CA, Tsopelas ND, Ziolko SK, James JA, Snitz BE, Houck PR, Bi W, Cohen AD, Lopresti BJ, DeKosky ST, Halligan EM, Klunk WE. Frequent amyloid deposition without significant cognitive impairment among the elderly. Arch Neurol. 2008;65:1509–1517. doi: 10.1001/archneur.65.11.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mintun MA, LaRossa GN, Sheline YI, Dence CS, Lee SY, Mach RH, Klunk WE, Mathis CA, DeKosky ST, Morris JC. [11C]PIB in a nondemented population: Potential antecedent marker of Alzheimer disease. Neurology. 2006;67:446–452. doi: 10.1212/01.wnl.0000228230.26044.a4. [DOI] [PubMed] [Google Scholar]
- 4.Jack CR, Jr, Lowe VJ, Weigand SD, Wiste HJ, Senjem ML, Knopman DS, Shiung MM, Gunter JL, Boeve BF, Kemp BJ, Weiner M, Petersen RC The Alzheimer’s Disease Neuroimaging Initiative. Serial PIB and MRI in normal, mild cognitive impairment and Alzheimer’s disease: implications for sequence of pathological events in Alzheimer’s disease. Brain. 2009;132:1355–1365. doi: 10.1093/brain/awp062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fjell AM, Walhovd KB, Amlien I, Bjornerud A, Reinvang I, Gjerstad L, Cappelen T, Willoch F, Due-Tonnessen P, Grambaite R, Skinningsrud A, Stenset V, Fladby T. Morphometric changes in the episodic memory network and tau pathologic features correlate with memory performance in patients with mild cognitive impairment. AJNR Am J Neuroradiol. 2008;29:1183–1189. doi: 10.3174/ajnr.A1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark CM, Xie S, Chittams J, Ewbank D, Peskind E, Galasko D, Morris JC, McKeel DW, Jr, Farlow M, Weitlauf SL, Quinn J, Kaye J, Knopman D, Arai H, Doody RS, DeCarli C, Leight S, Lee VM-Y, Trojanowski JQ. Cerebrospinal fluid tau and β-amyloid: how well do these biomarkers reflect autopsy-confirmed dementia diagnoses? Arch Neurol. 2003;60:1696–1702. doi: 10.1001/archneur.60.12.1696. [DOI] [PubMed] [Google Scholar]
- 7.Shaw LM, Vanderstichele H, Knapik-Czajka M, Clark CM, Aisen PS, Petersen RC, Blennow K, Soares H, Simon A, Lewczuk P, Dean R, Siemers E, Potter W, Lee VM-Y, Trojanowski JQ Initiative AsDN. Cerebrospinal fluid biomarker signature in Alzheimer’s disease neuroimaging initiative subjects. Ann Neurol. 2009;65:403–413. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andreasen N, Minthon L, Vanmechelen E, Vanderstichele H, Davidsson P, Winblad B, Blennow K. Cerebrospinal fluid tau and A[beta]42 as predictors of development of Alzheimer’s disease in patients with mild cognitive impairment. Neurosci Lett. 1999;273:5–8. doi: 10.1016/s0304-3940(99)00617-5. [DOI] [PubMed] [Google Scholar]
- 9.Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, Parikshak N, Hua X, Toga AW, Jack CR, Schuff N, Weiner MW, Thompson PM. Automated 3D mapping of hippocampal atrophy and its clinical correlates in 400 subjects with Alzheimer’s disease, mild cognitive impairment, and elderly controls. Hum Brain Mapp. 2009;30:2766–2788. doi: 10.1002/hbm.20708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, Parikshak N, Toga AW, Jack CR, Jr, Schuff N, Weiner MW, Thompson PM. Automated mapping of hippocampal atrophy in 1-year repeat MRI data from 490 subjects with Alzheimer’s disease, mild cognitive impairment, and elderly controls. NeuroImage. 2009;45:S3–S15. doi: 10.1016/j.neuroimage.2008.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.deToledo-Morrell L, Stoub TR, Bulgakova M, Wilson RS, Bennett DA, Leurgans S, Wuu J, Turner DA. MRI-derived entorhinal volume is a good predictor of conversion from MCI to AD. Neurobiol Aging. 2004;25:1197–1203. doi: 10.1016/j.neurobiolaging.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 12.Schroeter ML, Stein T, Maslowski N, Neumann J. Neural correlates of Alzheimer’s disease and mild cognitive impairment: a systematic and quantitative meta-analysis involving 1351 patients. NeuroImage. 2009;47:1196–1206. doi: 10.1016/j.neuroimage.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, Parikshak N, Hua X, Toga AW, Jack CR, Jr, Weiner MW, Thompson PM. Validation of a fully automated 3D hippocampal segmentation method using subjects with Alzheimer’s disease mild cognitive impairment, and elderly controls. NeuroImage. 2008;43:59–68. doi: 10.1016/j.neuroimage.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson PM, Hayashi KM, de Zubicaray GI, Janke AL, Rose SE, Semple J, Hong MS, Herman DH, Gravano D, Doddrell DM, Toga AW. Mapping hippocampal and ventricular change in Alzheimer disease. NeuroImage. 2004;22:1754–1766. doi: 10.1016/j.neuroimage.2004.03.040. [DOI] [PubMed] [Google Scholar]
- 15.Whitwell JL, Shiung MM, Przybelski SA, Weigand SD, Knopman DS, Boeve BF, Petersen RC, Jack CR., Jr MRI patterns of atrophy associated with progression to AD in amnestic mild cognitive impairment. Neurology. 2008;70:512–520. doi: 10.1212/01.wnl.0000280575.77437.a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hua X, Leow AD, Lee S, Klunder AD, Toga AW, Lepore N, Chou Y-Y, Brun C, Chiang M-C, Barysheva M, Jack CR, Jr, Bernstein MA, Britson PJ, Ward CP, Whitwell JL, Borowski B, Fleisher AS, Fox NC, Boyes RG, Barnes J, Harvey D, Kornak J, Schuff N, Boreta L, Alexander GE, Weiner MW, Thompson PM The Alzheimer’s Disease Neuroimaging I. 3D characterization of brain atrophy in Alzheimer’s disease and mild cognitive impairment using tensor-based morphometry. NeuroImage. 2008;41:19–34. doi: 10.1016/j.neuroimage.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whitwell JL, Przybelski SA, Weigand SD, Knopman DS, Boeve BF, Petersen RC, Jack CR., Jr 3D maps from multiple MRI illustrate changing atrophy patterns as subjects progress from mild cognitive impairment to Alzheimer’s disease. Brain. 2007;130:1777–1786. doi: 10.1093/brain/awm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chetelat Ga, Baron J-C. Early diagnosis of alzheimer’s disease: contribution of structural neuroimaging. NeuroImage. 2003;18:525–541. doi: 10.1016/s1053-8119(02)00026-5. [DOI] [PubMed] [Google Scholar]
- 19.Du AT, Schuff N, Amend D, Laakso MP, Hsu YY, Jagust WJ, Yaffe K, Kramer JH, Reed B, Norman D, Chui HC, Weiner MW. Magnetic resonance imaging of the entorhinal cortex and hippocampus in mild cognitive impairment and Alzheimer’s disease. J Neurol Neurosurg Psychiatry. 2001;71:441–447. doi: 10.1136/jnnp.71.4.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du AT, Schuff N, Laakso MP, Zhu XP, Jagust WJ, Yaffe K, Kramer JH, Miller BL, Reed BR, Norman D, Chui HC, Weiner MW. Effects of subcortical ischemic vascular dementia and AD on entorhinal cortex and hippocampus. Neurology. 2002;58:1635–1641. doi: 10.1212/wnl.58.11.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kramer JH, Schuff N, Reed BR, Mungas D, Du A-T, Rosen HJ, Jagust WJ, Miller BL, Weiner MW, Chui HC. Hippocampal volume and retention in Alzheimer’s disease. J Int Neuropsychol Soc. 2004;10:639–643. doi: 10.1017/S1355617704104050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duarte A, Hayasaka S, Du A, Schuff N, Jahng G-H, Kramer J, Miller B, Weiner M. Volumetric correlates of memory and executive function in normal elderly, mild cognitive impairment and Alzheimer’s disease. Neurosci Lett. 2006;406:60–65. doi: 10.1016/j.neulet.2006.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stoub TR, Bulgakova M, Leurgans S, Bennett DA, Fleischman D, Turner DA, deToledo-Morrell L. MRI predictors of risk of incident Alzheimer disease :a longitudinal study. Neurology. 2005;64:1520–1524. doi: 10.1212/01.WNL.0000160089.43264.1A. [DOI] [PubMed] [Google Scholar]
- 24.Desikan RS, Fischl B, Cabral HJ, Kemper TL, Guttmann CRG, Blacker D, Hyman BT, Albert MS, Killiany RJ. MRI measures of temporoparietal regions show differential rates of atrophy during prodromal AD. Neurology. 2008;71:819–825. doi: 10.1212/01.wnl.0000320055.57329.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jack CR, Jr, Shiung MM, Gunter JL, O’Brien PC, Weigand SD, Knopman DS, Boeve BF, Ivnik RJ, Smith GE, Cha RH, Tangalos EG, Petersen RC. Comparison of different MRI brain atrophy rate measures with clinical disease progression in AD. Neurology. 2004;62:591–600. doi: 10.1212/01.wnl.0000110315.26026.ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jack CR, Jr, Shiung MM, Weigand SD, O’Brien PC, Gunter JL, Boeve BF, Knopman DS, Smith GE, Ivnik RJ, Tangalos EG, Petersen RC. Brain atrophy rates predict subsequent clinical conversion in normal elderly and amnestic MCI. Neurology. 2005;65:1227–1231. doi: 10.1212/01.wnl.0000180958.22678.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jack CR, Jr, Weigand SD, Shiung MM, Przybelski SA, O’Brien PC, Gunter JL, Knopman DS, Boeve BF, Smith GE, Petersen RC. Atrophy rates accelerate in amnestic mild cognitive impairment. Neurology. 2008;70:1740–1752. doi: 10.1212/01.wnl.0000281688.77598.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du AT, Schuff N, Kramer JH, Ganzer S, Zhu XP, Jagust WJ, Miller BL, Reed BR, Mungas D, Yaffe K, Chui HC, Weiner MW. Higher atrophy rate of entorhinal cortex than hippocampus in AD. Neurology. 2004;62:422–427. doi: 10.1212/01.wnl.0000106462.72282.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Du AT, Schuff N, Zhu XP, Jagust WJ, Miller BL, Reed BR, Kramer JH, Mungas D, Yaffe K, Chui HC, Weiner MW. Atrophy rates of entorhinal cortex in AD and normal aging. Neurology. 2003;60:481–486. doi: 10.1212/01.wnl.0000044400.11317.ec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brys M, Glodzik L, Mosconi L, Switalski R, De Santi S, Pirraglia E, Rich K, Kim BC, Mehta P, Zinkowski R, Pratico D, Wallin A, Zetterberg H, Tsui WH, Rusinek H, Blennow K, de Leon MJ. Magnetic resonance imaging improves cerebrospinal fluid biomarkers in the early detection of Alzheimer’s disease. J Alzheimers Dis. 2009;16:351–362. doi: 10.3233/JAD-2009-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vemuri P, Wiste HJ, Weigand SD, Shaw LM, Trojanowski JQ, Weiner MW, Knopman DS, Petersen RC, Jack CR, Jr On behalf of the Alzheimer’s Disease Neuroimaging, Initiative. MRI and CSF biomarkers in normal, MCI, and AD subjects: predicting future clinical change. Neurology. 2009;73:294–301. doi: 10.1212/WNL.0b013e3181af79fb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vemuri P, Wiste HJ, Weigand SD, Shaw LM, Trojanowski JQ, Weiner MW, Knopman DS, Petersen RC, Jack CR, Jr On behalf of the Alzheimer’s Disease Neuroimaging, Initiative. MRI and CSF biomarkers in normal, MCI, and AD subjects: diagnostic discrimination and cognitive correlations. Neurology. 2009;73:287–293. doi: 10.1212/WNL.0b013e3181af79e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glodzik-Sobanska L, Pirraglia E, Brys M, de Santi S, Mosconi L, Rich KE, Switalski R, Louis LS, Sadowski MJ, Martiniuk F, Mehta P, Pratico D, Zinkowski RP, Blennow K, de Leon MJ. The effects of normal aging and ApoE genotype on the levels of CSF biomarkers for Alzheimer’s disease. Neurobiol Aging. 2009;30:672–681. doi: 10.1016/j.neurobiolaging.2007.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sunderland T, Mirza N, Putnam KT, Linker G, Bhupali D, Durham R, Soares H, Kimmel L, Friedman D, Bergeson J, Csako G, Levy JA, Bartko JJ, Cohen RM. Cerebrospinal fluid β-amyloid1-42 and tau in control subjects at risk for Alzheimer’s disease: the effect of APOE ε4 allele. Biol Psychiatry. 2004;56:670–676. doi: 10.1016/j.biopsych.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 35.Basso M, Gelernter J, Yang J, MacAvoy MG, Varma P, Bronen RA, van Dyck CH. Apolipoprotein E epsilon4 is associated with atrophy of the amygdala in Alzheimer’s disease. Neurobiol Aging. 2006;27:1416–1424. doi: 10.1016/j.neurobiolaging.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 36.Sluimer JD, Vrenken H, Blankenstein MA, Fox NC, Scheltens P, Barkhof F, van der Flier WM. Whole-brain atrophy rate in Alzheimer disease: identifying fast progressors. Neurology. 2008;70:1836–1841. doi: 10.1212/01.wnl.0000311446.61861.e3. [DOI] [PubMed] [Google Scholar]
- 37.Schuff N, Woerner N, Boreta L, Kornfield T, Shaw LM, Trojanowski JQ, Thompson PM, Jack CR, Jr, Weiner MW Alzheimer’s Disease Neuroimaging, Initiative. MRI of hippocampal volume loss in early Alzheimer’s disease in relation to ApoE genotype and biomarkers. Brain. 2009;132:1067–1077. doi: 10.1093/brain/awp007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Potkin SG, Guffanti G, Lakatos A, Turner JA, Kruggel F, Fallon JH, Saykin AJ, Orro A, Lupoli S, Salvi E, Weiner M, Macciardi F For the Alzheimer’s Disease Neuroimaging I. Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for Alzheimer’s disease. PLoS ONE. 2009;4:e6501. doi: 10.1371/journal.pone.0006501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fleisher A, Grundman M, Jack CR, Jr, Petersen RC, Taylor C, Kim HT, Schiller DHB, Bagwell V, Sencakova D, Weiner MF, DeCarli C, DeKosky ST, van Dyck CH, Thal LJ For the Alzheimer’s Disease Cooperative, Study. Sex, apolipoprotein E ε4 status, and hippocampal volume in mild cognitive impairment. Arch Neurol. 2005;62:953–957. doi: 10.1001/archneur.62.6.953. [DOI] [PubMed] [Google Scholar]
- 40.Hampel H, Burger K, Pruessner JC, Zinkowski R, DeBernardis J, Kerkman D, Leinsinger G, Evans AC, Davies P, Moller H-J, Teipel SJ. Correlation of cerebrospinal fluid levels of tau protein phosphorylated at threonine 231 with rates of hippocampal atrophy in Alzheimer disease. Arch Neurol. 2005;62:770–773. doi: 10.1001/archneur.62.5.770. [DOI] [PubMed] [Google Scholar]
- 41.Henneman WJP, Vrenken H, Barnes J, Sluimer IC, Verwey NA, Blankenstein MA, Klein M, Fox NC, Scheltens P, Barkhof F, van der Flier WM. Baseline CSFp-tau levels independently predict progression of hippocampal atrophy in Alzheimer disease. Neurology. 2009;73:935–940. doi: 10.1212/WNL.0b013e3181b879ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herukka S-K, Pennanen C, Soininen H, Pirttilä T. CSF Aβ42, tau and phosphorylated tau correlate with medial temporal lobe atrophy. J Alzheimers Dis. 2008;14:51–57. doi: 10.3233/jad-2008-14105. [DOI] [PubMed] [Google Scholar]
- 43.Leow AD, Yanovsky I, Parikshak N, Hua X, Lee S, Toga AW, Jack CR, Jr, Bernstein MA, Britson PJ, Gunter JL, Ward CP, Borowski B, Shaw LM, Trojanowski JQ, Fleisher AS, Harvey D, Kornak J, Schuff N, Alexander GE, Weiner MW, Thompson PM. Alzheimer’s Disease Neuroimaging Initiative: A one-year follow up study using tensor-based morphometry correlating degenerative rates, biomarkers and cognition. NeuroImage. 2009;45:645–655. doi: 10.1016/j.neuroimage.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fagan AM, Head D, Shah AR, Marcus D, Mintun M, Morris JC, Holtzman DM. Decreased cerebrospinal fluid A beta(42) correlates with brain atrophy in cognitively normal elderly. Ann Neurol. 2009;65:176–183. doi: 10.1002/ana.21559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fjell AM, Walhovd KB, Fennema-Notestine C, McEvoy LK, Hagler DJ, Holland D, Blennow K, Brewer JB, Dale AM The Alzheimer’s Disease Neuroimaging, Initiative. Brain atrophy in healthy aging is related to CSF levels of Aβ1–42. Cereb Cortex. 2010;279:2069–2079. doi: 10.1093/cercor/bhp279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fjell AM, Walhovd KB, Fennema-Notestine C, McEvoy LK, Hagler DJ, Holland D, Brewer JB, Dale AM For the Alzheimer’s Disease Neuroimaging, Initiative. CSF biomarkers in prediction of cerebral and clinical change in mild cognitive impairment and Alzheimer’s disease. J. Neurosci. 2010;30:2088–2101. doi: 10.1523/JNEUROSCI.3785-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petersen RC, Smith GE, Waring SC, Ivnik RJ, Tangalos EG, Kokmen E. Mild cognitive impairment: clinical characterization and outcome. Arch Neurol. 1999;56:303–308. doi: 10.1001/archneur.56.3.303. [DOI] [PubMed] [Google Scholar]
- 48.Jack CR, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, Whitwell JL, Ward C, Dale AM, Felmlee JP, Gunter JL, Hill DLG, Killiany R, Schuff N, Fox-Bosetti S, Lin C, Studholme C, DeCarli CS, Krueger G, Ward HA, Metzger GJ, Scott KT, Mallozzi R, Blezek D, Levy J, Debbins JP, Fleisher AS, Albert M, Green R, Bartzokis G, Glover G, Mugler J, Weiner MW, Study A. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Resonance Imag. 2008;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olsson A, Vanderstichele H, Andreasen N, De Meyer G, Wallin A, Holmberg B, Rosengren L, Vanmechelen E, Blennow K. Simultaneous measurement of {beta}-amyloid(1–42), total tau, and phosphorylated tau (Thr181) in cerebrospinal fluid by the xMAP technology. Clin Chem. 2005;51:336–345. doi: 10.1373/clinchem.2004.039347. [DOI] [PubMed] [Google Scholar]
- 50.Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- 51.Fischl B, van der Kouwe A, Destrieux C, Halgren E, Segonne F, Salat DH, Busa E, Seidman LJ, Goldstein J, Kennedy D, Caviness V, Makris N, Rosen B, Dale AM. Automatically parcellating the human cerebral cortex. Cereb Cortex. 2004;14:11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- 52.Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci U S A. 2000;97:11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 54.Stomrud E, Hansson O, Blennow K, Minthon L, Londos E. Cerebrospinal fluid biomarkers predict decline in subjective cognitive function over 3 years in healthy elderly. Dement Geriatr Cogn Disord. 2007;24:118–124. doi: 10.1159/000105017. [DOI] [PubMed] [Google Scholar]
- 55.Skoog I, Davidsson P, Aevarsson ì, Vanderstichele H, Vanmechelen E, Blennow K. Cerebrospinal Fluid β-Amyloid 42 Is Reduced before the Onset of Sporadic Dementia: A Population-Based Study in 85-Year-Olds. Dement Geriatr Cogn Disord. 2003;15:169–176. doi: 10.1159/000068478. [DOI] [PubMed] [Google Scholar]
- 56.Leinonen V, Koivisto AM, Savolainen S, Rummukainen J, Tamminen JN, Tillgren T, Vainikka S, Pyykkä OT, Mälsä J, Fraunberg M, Pirttilä T, Jääskeläinen JE, Soininen H, Rinne J, Alafuzoff I. Amyloid and tau proteins in cortical brain biopsy and Alzheimer’s disease. Ann Neurol. 2010;68:446–453. doi: 10.1002/ana.22100. [DOI] [PubMed] [Google Scholar]
- 57.DeCarli C, Frisoni GB, Clark CM, Harvey D, Grundman M, Petersen RC, Thal LJ, Jin S, Jack CR, Jr, Scheltens P For the Alzheimer’s Disease Cooperative Study, Group. Qualitative Estimates of Medial Temporal Atrophy as a Predictor of Progression From Mild Cognitive Impairment to Dementia. Arch Neurol. 2007;64:108–115. doi: 10.1001/archneur.64.1.108. [DOI] [PubMed] [Google Scholar]
- 58.Blennow K, Hampel H. CSF markers for incipient Alzheimer’s disease. Lancet Neurol. 2003;2:605–613. doi: 10.1016/s1474-4422(03)00530-1. [DOI] [PubMed] [Google Scholar]
- 59.Hampel H, Teipel SJ, Fuchsberger T, Andreasen N, Wiltfang J, Otto M, Shen Y, Dodel R, Du Y, Farlow M, Moller HJ, Blennow K, Buerger K. Value of CSF β-amyloid1-42 and tau as predictors of Alzheimer’s disease in patients with mild cognitive impairment. Mol Psychiatry. 2003;9:705–710. doi: 10.1038/sj.mp.4001473. [DOI] [PubMed] [Google Scholar]
- 60.John HG. Incorporating biomarkers into clinical drug trials in Alzheimer’s disease. J Alzheimers Dis. 2001;3:287–292. doi: 10.3233/jad-2001-3303. [DOI] [PubMed] [Google Scholar]
- 61.Shaw LM, Korecka M, Clark CM, Lee VMY, Trojanowski JQ. Biomarkers of neurodegeneration for diagnosis and monitoring therapeutics. Nat Rev Drug Discov. 2007;6:295–303. doi: 10.1038/nrd2176. [DOI] [PubMed] [Google Scholar]
- 62.Scahill RI, Schott JM, Stevens JM, Rossor MN, Fox NC. Mapping the evolution of regional atrophy in Alzheimer’s disease: Unbiased analysis of fluid-registered serial MRI. Proc Natl Acad Sci U S A. 2002;99:4703–4707. doi: 10.1073/pnas.052587399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arnold SE, Hyman BT, Flory J, Damasio AR, Van Hoesen GW. The topographical and neuroanatomical distribution of neurofibrillary tangles and neuritic plaques in the cerebral cortex of patients with Alzheimer’s disease. Cereb Cortex. 1991;1:103–116. doi: 10.1093/cercor/1.1.103. [DOI] [PubMed] [Google Scholar]
- 64.Thompson PM, Hayashi KM, de Zubicaray G, Janke AL, Rose SE, Semple J, Herman D, Hong MS, Dittmer SS, Doddrell DM, Toga AW. Dynamics of Gray Matter Loss in Alzheimer’s Disease. J. Neurosci. 2003;23:994–1005. doi: 10.1523/JNEUROSCI.23-03-00994.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 66.Andersson C, Blennow K, Johansson SE, Almkvist O, Engfeldt P, Lindau M, Eriksdotter-Jonhagen M. Differential CSF Biomarker Levels in APOE- e4-Positive and -Negative Patients with Memory Impairment. Dement Geriatr Cogn Disord. 2007;23:87–95. doi: 10.1159/000097354. [DOI] [PubMed] [Google Scholar]
- 67.Chou Y-Y, Avedissian LeporβÈN, Madsen C, Parikshak SK, Hua N, Shaw X, Trojanowski LM, Weiner JQ, Toga MW, Thompson AWPM. Mapping correlations between ventricular expansion and CSF amyloid and tau biomarkers in 240 subjects with Alzheimer’s disease, mild cognitive impairment and elderly controls. NeuroImage. 2009;46:394–410. doi: 10.1016/j.neuroimage.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stefani A, Martorana A, Bernardini S, Panella M, Mercati F, Orlacchio A, Pierantozzi M. CSF markers in Alzheimer disease patients are not related to the different degree of cognitive impairment. J Neurol Sci. 2006;251:124–128. doi: 10.1016/j.jns.2006.09.014. [DOI] [PubMed] [Google Scholar]