Abstract
Background
Findings from family studies and recent genome-wide association studies have indicated overlap in the risk genes between schizophrenia and bipolar disorder (BD). After finding a linkage between the ST8SIA2 (ST8 alpha-N-acetyl-neuraminide alpha-2, 8-sicalyltransferase 2 gene) locus (15q26) and mixed families with schizophrenia and BD, several studies have reported a significant association between this gene and schizophrenia or BD. We investigated the genetic association between ST8SIA2 and both schizophrenia and BD in the Korean population.
Methods
A total of 582 patients with schizophrenia, 339 patients with BD, and 502 healthy controls were included. Thirty-one tag single nucleotide polymorphisms (SNPs) across the ST8SIA2 region and three other SNPs showing significant associations in previous studies were genotyped. The associations were evaluated by logistic regression analysis using additive, dominant, and recessive genetic models.
Results
Fourteen of 34 SNPs showed a nominally significant association (p < 0.05) with at least one diagnostic group. These association trends were strongest for the schizophrenia and combined schizophrenia and bipolar I disorder (BD-I) groups. The strongest association was observed in rs11637898 for schizophrenia (p = 0.0033) and BD-I (p = 0.0050) under the dominant model. The association between rs11637898 and the combined schizophrenia and BD-I group (p = 0.0006, under the dominant model) remained significant after correcting for multiple testing.
Discussion
We identified a possible role of ST8SIA2 in the common susceptibility of schizophrenia and BD-I. However, no association trend was observed for bipolar II disorder. Further efforts are needed to identify a specific phenotype associated with this gene crossing the current diagnostic categories.
Introduction
Diagnoses of major psychiatric illnesses are based on phenomenological characteristics, resulting in an overlap in symptoms and treatment between diagnostic categories, as well as an overlap in genetic and biological risk factors [1]. Since the Kraepelinian dichotomy was proposed, schizophrenia and bipolar disorder (BD) have been classified as two distinct clinical entities based on their symptom patterns and course of illness [2]. However, common clinical features, such as psychotic symptoms, emotional dysregulation, and cognitive impairment have been described across the two disorders [3–6]. Findings from genetic studies also suggest shared familial risk and genetic susceptibilities between schizophrenia and BD [1, 7, 8].
Chromosome locus 15q26 has been identified as a candidate region for both schizophrenia and BD in a whole-genome linkage study of eastern Quebec families with combined schizophrenia and BD [9]. This locus has also been reported as a susceptibility locus for BD in a genome-wide linkage analysis in Australian multi-generational families with BD and a broad spectrum of clinical diagnoses, including major depressive disorder and schizoaffective disorder-manic type [10]. ST8SIA2 (ST8 alpha-N-acetyl-neuraminide alpha-2, 8-sicalyltransferase 2 gene) is one of the candidate genes for psychiatric illnesses mapping to this region. In a recent genome-wide association study (GWAS) of Chinese patients with BD, the most strongly associated single nucleotide polymorphisms (SNPs) were close to ST8SIA2 with the lowest p-value of 4.87 × 10−7 [11].
Targeted gene studies for ST8SIA2 have been performed in patients with schizophrenia. Three SNPs (rs3759916, rs3759915, and rs3759914) located in the promoter region show a significant association with schizophrenia in the Japanese (rs3759916 and rs3759914) [12] and Chinese (rs3759915) [13] populations. rs3759916 also shows a sex-specific association with schizophrenia in female Spanish patients with schizophrenia [14]. A BD association study was performed in the Australian population [15] as a fine mapping study of their previous linkage finding [10] in families with BD. In that study, a number of SNPs (rs4586379, rs2035645, rs4777974, rs11637898, rs11074070, rs3858917, rs3784735, and rs2168351) showed a nominally significant association with bipolar spectrum disorders, and a specific risk haplotype was identified. The authors also observed an over-representation of this risk haplotype in an Australian schizophrenia case-control cohort. The same group identified two variants (rs11074064 and rs722645) with putative functional effects that were nominally associated with BD more recently [16].
ST8SIA2 encodes polysialyl transferase, which is involved in the biosynthesis of polysialic acid (polySia) that spatiotemporally modifies neural cell adhesion molecule (NCAM) [17]. The polySia-NCAM complex is predominantly found in the embryonic brain, and its expression in the adult brain is highly restricted to areas where neural plasticity is ongoing, such as the hippocampus, subventricular zone, thalamus, prefrontal cortex, and amygdala [18]. The polySia-NCAM complex is thought to regulate neuronal processes, such as neuronal migration [19] and synaptogenesis [20]; thus, anomalous expression of polySia impairs spatial learning and memory [21–23]. Since growing evidence suggests that a neurodevelopmental deficit is important in the pathophysiological mechanism of schizophrenia and BD [24, 25], ST8SIA2 could be a plausible functional candidate gene.
Our group performed a genome-wide linkage scan of quantitative traits targeting symptom dimensions in multiplex schizophrenia families [26]. The 15q26 locus harboring ST8SIA2 showed a linkage signal that attained the genome-wide empirical threshold for suggestive linkage with the “non-paranoid delusion factor”. Interestingly, lifetime symptoms for this factor include grandiose, religious, and erotic delusions that are frequently observed in patients with schizophrenia or BD [26]. Based on these findings and previous studies of other populations indicating that ST8SIA2 is a candidate gene for both schizophrenia and BD, the current study investigated the genetic association between ST8SIA2 and schizophrenia and BD in the Korean population. We analyzed tag SNPs covering the whole gene locus and applied genotype-based analyses.
Materials and Methods
Study Subjects
Patients who met the DSM-IV [27] diagnostic criteria for schizophrenia (n = 582) and BD (n = 339), including bipolar I disorder (BD-I) (n = 180) and bipolar II disorder (BD-II) (n = 159) were recruited from the inpatient unit and outpatient clinic of Samsung Medical Center and Seoul National University Bundang Hospital.
The healthy subjects (n = 502) consisted of volunteers from the community who were free of any history of clinically significant psychiatric symptoms. Written informed consent was obtained from all subjects after a complete explanation of the study. This study was approved by the institutional review boards of Samsung Medical Center and Seoul National University Bundang Hospital.
SNP Selection
We used the Korean Hapmap database (http://www.khapmap.org) to select ST8SIA2 tag SNPs. Thirty-one tag SNPs were chosen by the program Tagger within Haploview v4.0 (http://www.broad.mit.edu/mpg/haploview) using pairwise tagging of SNPs with an r2 > 0.8 and minor allele frequency (MAF) > 0.05 [28, 29]. An additional three SNPs (rs3759916, rs3759915, and rs3759914), which previously showed a significant association with schizophrenia [12, 13] in other Asian populations, were included in the analysis. The selected SNPs and their genomic or intragenic location, allele types, and minor allele frequencies are summarized in Table 1. All SNPs were intronic, except rs3759916, rs3759915, and rs3759914 on the 5′-untranslated region (UTR) and rs2290492 and rs17600420 on the 3′-UTR. These SNPs spanned the entire ST8SIA2 gene with an average inter-SNP distance of 2.2 kb (range, 56 bp–12.7 kb).
Table 1. Characteristics of the 34 ST8SIA2 tag SNPs.
| SNP | Genomic location | Intragenic location | Alleles | MAF a | SNP | Genomic location | Intragenic location | Alleles | MAF a |
|---|---|---|---|---|---|---|---|---|---|
| rs3759916 | Chr15:90737173 | 5’-UTR | A>G | 0.36 | rs3784732 | Chr15:90787919 | intron 4 | T>C | 0.03 |
| rs3759915 | Chr15:90737392 | 5’-UTR | G>C | 0.48 | rs3784731 | Chr15:90788082 | intron 4 | A>T | 0.29 |
| rs3759914 | Chr15:90737448 | 5’-UTR | A>G | 0.35 | rs1455777 | Chr15:90789744 | intron 5 | T>C | 0.29 |
| rs4777969 | Chr15:90741567 | intron 1 | G>A | 0.37 | rs7166344 | Chr15:90791345 | intron 5 | G>A | 0.30 |
| rs8025225 | Chr15:90741903 | intron 1 | T>C | 0.39 | rs1487982 | Chr15:90795915 | intron 5 | T>C | 0.14 |
| rs2124359 | Chr15:90743092 | intron 1 | G>C | 0.49 | rs1352323 | Chr15:90796644 | intron 5 | C>T | 0.34 |
| rs1487984 | Chr15:90746120 | intron 1 | C>A | 0.11 | rs3784729 | Chr15:90797010 | intron 5 | C>T | 0.30 |
| rs11074067 | Chr15:90747301 | intron 1 | G>C | 0.43 | rs10775256 | Chr15:90798158 | intron 5 | A>G | 0.36 |
| rs881770 | Chr15:90750812 | intron 1 | G>A | 0.49 | rs897463 | Chr15:90801444 | intron 5 | T>A | 0.09 |
| rs4777973 | Chr15:90751886 | intron 1 | A>G | 0.34 | rs11852344 | Chr15:90802524 | intron 5 | A>G | 0.49 |
| rs4777974 | Chr15:90753293 | intron 1 | A>G | 0.11 | rs4777715 | Chr15:90803084 | intron 5 | A>G | 0.39 |
| rs7176813 | Chr15:90753636 | intron 1 | A>G | 0.40 | rs4777988 | Chr15:90803447 | intron 5 | A>G | 0.43 |
| rs11637898 | Chr15:90753853 | intron 1 | A>G | 0.25 | rs3784723 | Chr15:90806762 | intron 5 | T>C | 0.36 |
| rs4777980 | Chr15:90766580 | intron 1 | A>G | 0.28 | rs4777989 | Chr15:90807743 | intron 5 | A>G | 0.46 |
| rs3784737 | Chr15:90768555 | intron 1 | A>G | 0.29 | rs7168443 | Chr15:90808052 | intron 5 | C>T | 0.09 |
| rs8037133 | Chr15:90775460 | intron 2 | A>G | 0.29 | rs2290492 | Chr15:90808977 | 3’-UTR | C>T | 0.11 |
| rs2168351 | Chr15:90784725 | intron 4 | C>T | 0.33 | rs17600420 | Chr15:90812138 | 3’-UTR | A>G | 0.40 |
SNP, single nucleotide polymorphism; MAF, minor allele frequency
a Minor allele frequency based on the control group data
DNA Extraction and Genotyping
Whole blood was collected from each individual into EDTA tubes, and genomic DNA was isolated from peripheral blood leukocytes using the Wizard Genomic DNA Purification kit according to the manufacturer’s instructions (Promega, Madison, WI, USA). MassARRAY Assay Design, version 3.0 software (Sequenom Inc, San Diego, CA, USA) generated three multiplex reactions: 12 SNPs (plex 1), 12 SNPs (plex 2), and 10 SNPs (plex 3). Multiplex SNP genotyping was performed by primer extension and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry using the iPLEX Gold technology from Sequenom. SNP assays were designed using Sequenom MassARRAY Assay Design ver. 3.0 software (primer information is available upon request). The polymerase chain reaction was performed according to the standard iPLEX methodology. Spectra were analyzed by MassARRAY Typer ver. 3.4 software (Sequenom). Quality control was performed to exclude individual SNPs or samples with genotype call rates < 95% and SNP assays with poor-quality spectra/cluster plots.
To confirm the reliability of iPLEX SNP genotyping method, five selected SNPs (rs4777973, rs3784731, rs11637898, rs1455777, and rs897463) for 48 random samples were regenotyped by sequencing reaction using ABI PRISM BigDye Terminator v 3.1 Cycle Sequencing Kits (Applied Biosystems, CA, USA) The concordance rate of genotype data between sequencing and iPLEX SNP genotyping was 99.2%
Statistical Analyses
We used the Kruskal–Wallis test or the chi-square test to reveal differences in demographic variables among the patient and control groups. The Hardy–Weinberg equilibrium was checked with Fisher’s exact test for the genetics analysis, and no significant deviation was observed in any of the SNPs (S1 Table). Genotype-wise association was evaluated by logistic regression analysis with age and sex as covariates. Additive, dominant, and recessive genetic models were considered based on the minor allele of each SNP. The inheritance model with the least Akaike Information Criterion [30] was accepted as the best fitting model. We controlled the experiment-wise type I error using the Bonferroni correction. Thirty-four SNPs were analyzed; thus, a p-value = 0.0015 (0.05/34) was the adjusted level of significance. All statistical analyses were done with snpStats ver. 1.18.0 in R ver. 3.0.2 (http://www.bioconductor.org) [31].
Results
The demographic and clinical characteristics of the subjects are presented in Table 2. More males were present in the schizophrenia group than in the BD-I and control groups, and the BD-II group had more females than in the other groups. The patients with BD-II were older than those in the other groups, and subjects in the control group were younger than the patients.
Table 2. Demographic characteristics of the subjects.
| Demographic characteristics | Control (N = 502) | Schizophrenia (N = 582) | BD-I (N = 180) | BD-II (N = 159) | Statistics a | |
|---|---|---|---|---|---|---|
| Age, mean(SD) | 31.6 (7.9) | 33.9 (9.5) | 33.9 (10.1) | 37.4 (11.4) | F = 16.6 | P<0.001 |
| Sex (Male, %) | 43.8 | 53.6 | 38.3 | 25.2 | χ2 = 46.2 | P<0.001 |
BD-I, bipolar I disorder; BD-II, bipolar II disorder
a After post-hoc analysis, BD-II group was older than the other groups and the control group was younger than the other groups. More males were in the schizophrenia group than in the BD-1 and control groups, and the BD-II group had more females than the other groups.
All SNPs passed quality control with MAF > 0.01, and the missing data rate for each SNP was < 2%. Therefore, all of the genotyped SNPs (n = 34) were included in the statistical analysis.
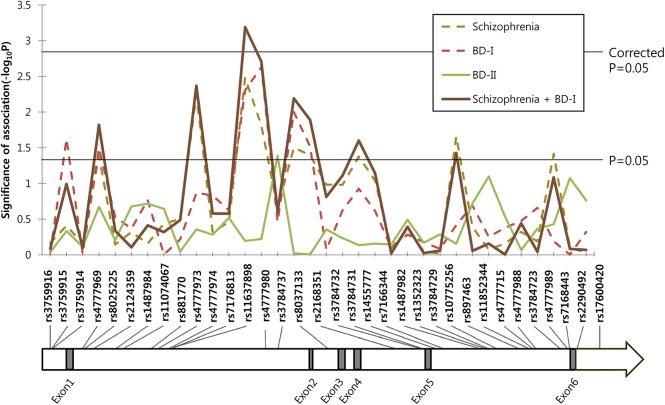
The overall SNP-disease association plot is shown in Fig 1, and the level of significance is shown as –log p-value. Schizophrenia and BD-I showed similar association trends throughout the region, and the association was stronger when schizophrenia and BD-I were analyzed together. But BD-II showed a different pattern with few association signals.
Fig 1. Association pattern between ST8SIA2 tag single nucleotide polymorphisms (SNPs) and schizophrenia and bipolar disorder under the dominant model.
–Log of p-values are represented on the y-axis with relative location of the SNPs in the gene on the x-axis. Two horizontal lines indicate nominal and corrected p-values of 0.05.
The detailed genotypic association analysis results using logistic regression after controlling for the confounding effects of sex and age are summarized in S1 Table. Table 3 presents the results of the SNPs that showed an association with at least one diagnostic category with nominal significance. Statistical values are described only for the best-fit model. We observed nominally significant associations between schizophrenia and 10 SNPs under the additive model (rs4777969, rs4777973, rs11637898, rs4777980, rs8037133, rs2168351, rs3784731, rs1455777, rs897463, and rs7168443). These SNPs also showed associations under the dominant model, except rs3784731. rs2168351, rs3784731, and rs1455777 were also associated with schizophrenia under the recessive model. BD-I showed nominally significant associations with rs3759915, rs4777969, rs11637898, rs4777980, rs8037133, and rs2168351 under the dominant and additive models, and with rs8025225 under the recessive model. Nominally significant associations were observed between BD-II and rs3784737 (under the dominant model) and rs10775256 (under the recessive model). However, none of these associations remained significant after adjusting for multiple testing with Bonferroni’s correction.
Table 3. SNPs associated with either schizophrenia or bipolar disorder with nominally significant p-values (<0.05).
| Comparison with control group (N = 502) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Schizophrenia (N = 582) | BD-I (N = 180) | BD-II (N = 159) | Schizophrenia + BD-I (N = 762) | |||||||||||||
| SNP | P a | OR | CI | Best-fit model b | P a | OR | CI | Best-fit model b | P a | OR | CI | Best-fit model b | P a | OR | CI | Best-fit model b |
| rs3759915 | 0.245 | 0.843 | 0.632–1.124 | Recessive | 0.024* | 1.609 | 1.065–2.430 | Dominant | 0.317 | 0.790 | 0.498–1.253 | Recessive | 0.102 | 1.236 | 0.959–1.594 | Dominant |
| rs4777969 | 0.027* | 0.812 | 0.675–0.977 | Additive | 0.032* | 0.682 | 0.480–0.968 | Dominant | 0.213 | 0.785 | 0.537–1.149 | Dominant | 0.014* | 0.806 | 0.678–0.957 | Additive |
| rs8025225 | 0.412 | 0.870 | 0.623–1.215 | Recessive | 0.024* | 0.519 | 0.294–0.916 | Recessive | 0.621 | 1.102 | 0.749–1.623 | Dominant | 0.131 | 0.782 | 0.569–1.076 | Recessive |
| rs4777973 | 0.006* | 1.420 | 1.107–1.820 | Dominant | 0.135 | 1.311 | 0.919–1.871 | Dominant | 0.439 | 1.162 | 0.794–1.701 | Dominant | 0.004* | 1.405 | 1.113–1.774 | Dominant |
| rs11637898 | 0.003* | 1.443 | 1.130–1.843 | Dominant | 0.005* | 1.641 | 1.162–2.319 | Dominant | 0.484 | 0.770 | 0.370–1.601 | Recessive | 0.0006** | 1.492 | 1.186–1.878 | Dominant |
| rs4777980 | 0.015* | 1.261 | 1.047–1.520 | Additive | 0.002* | 1.729 | 1.215–2.459 | Dominant | 0.134 | 0.550 | 0.251–1.203 | Recessive | 0.002* | 1.440 | 1.143–1.814 | Dominant |
| rs3784737 | 0.220 | 1.165 | 0.913–1.485 | Dominant | 0.343 | 1.182 | 0.837–1.670 | Dominant | 0.041* | 1.485 | 1.017–2.170 | Dominant | 0.249 | 1.144 | 0.910–1.437 | Dominant |
| rs8037133 | 0.026* | 1.232 | 1.025–1.481 | Additive | 0.010* | 1.591 | 1.117–2.264 | Dominant | 0.394 | 0.744 | 0.378–1.467 | Recessive | 0.006* | 1.379 | 1.095–1.739 | Dominant |
| rs2168351 | 0.011* | 1.260 | 1.054–1.507 | Additive | 0.030* | 1.483 | 1.038–2.120 | Dominant | 0.959 | 0.985 | 0.553–1.755 | Recessive | 0.007* | 1.263 | 1.065–1.497 | Additive |
| rs3784731 | 0.021* | 1.656 | 1.080–2.540 | Recessive | 0.238 | 1.231 | 0.871–1.739 | Dominant | 0.583 | 0.900 | 0.619–1.310 | Dominant | 0.024* | 1.227 | 1.027–1.466 | Additive |
| rs1455777 | 0.014* | 1.266 | 1.048–1.529 | Additive | 0.119 | 1.318 | 0.932–1.865 | Dominant | 0.731 | 0.936 | 0.643–1.363 | Dominant | 0.011* | 1.261 | 1.055–1.508 | Additive |
| rs10775256 | 0.178 | 0.775 | 0.535–1.123 | Recessive | 0.213 | 0.708 | 0.410–1.220 | Recessive | 0.039* | 0.514 | 0.274–0.966 | Recessive | 0.125 | 0.763 | 0.539–1.078 | Recessive |
| rs897463 | 0.011* | 0.653 | 0.471–0.907 | Additive | 0.259 | 0.766 | 0.482–1.217 | Recessive | 0.490 | 0.464 | 0.052–4.102 | Recessive | 0.016* | 0.688 | 0.507–0.932 | Additive |
| rs7168443 | 0.029* | 0.702 | 0.511–0.965 | Additive | 0.431 | 0.837 | 0.538–1.303 | Recessive | 0.316 | 0.780 | 0.480–1.267 | Additive | 0.048* | 0.742 | 0.553–0.997 | Additive |
BD-I, bipolar I disorder; BD-II, bipolar II disorder; OR, odds ratio; CI, confidence interval
* p < 0.05
** corrected p < 0.05
a Nominal p-value by logistic regression with age and sex covariates.
b The inheritance model with the least Akaike Information Criterion was accepted as the best fitting model.
As the schizophrenia and BD-I groups showed similar association trends in the ST8SIA2 region (Fig 1), and a previous linkage study defined these two disorders as a common locus phenotype for this region [9], we combined the two groups and applied the same association analysis. Ten SNPs revealed nominally significant associations with lower p-values compared to those of separate analyses for schizophrenia or BD-I. The association with rs11637898 reached the corrected level of significance under the dominant (p = 0.0006) and additive models (p = 0.0011). The minor allele of most of the SNPs showing significant associations was a risk allele for the corresponding illness, except rs4777969, rs8025225, rs897463, and rs7168443 for which the minor allele was a protective allele.
Discussion
ST8SIA2 is located on chromosome 15q25-26 and has been considered a positional and functional candidate gene for schizophrenia and BD, and significant associations have been reported in several different populations [12–15]. Genetic association or linkage with ST8SIA2 has also been observed in patients with major depressive disorder [32, 33] and autism spectrum disorder [34]. These findings suggest that this gene has a role in the occurrence of major psychiatric disorders, which have varying degrees of neurodevelopmental defects.
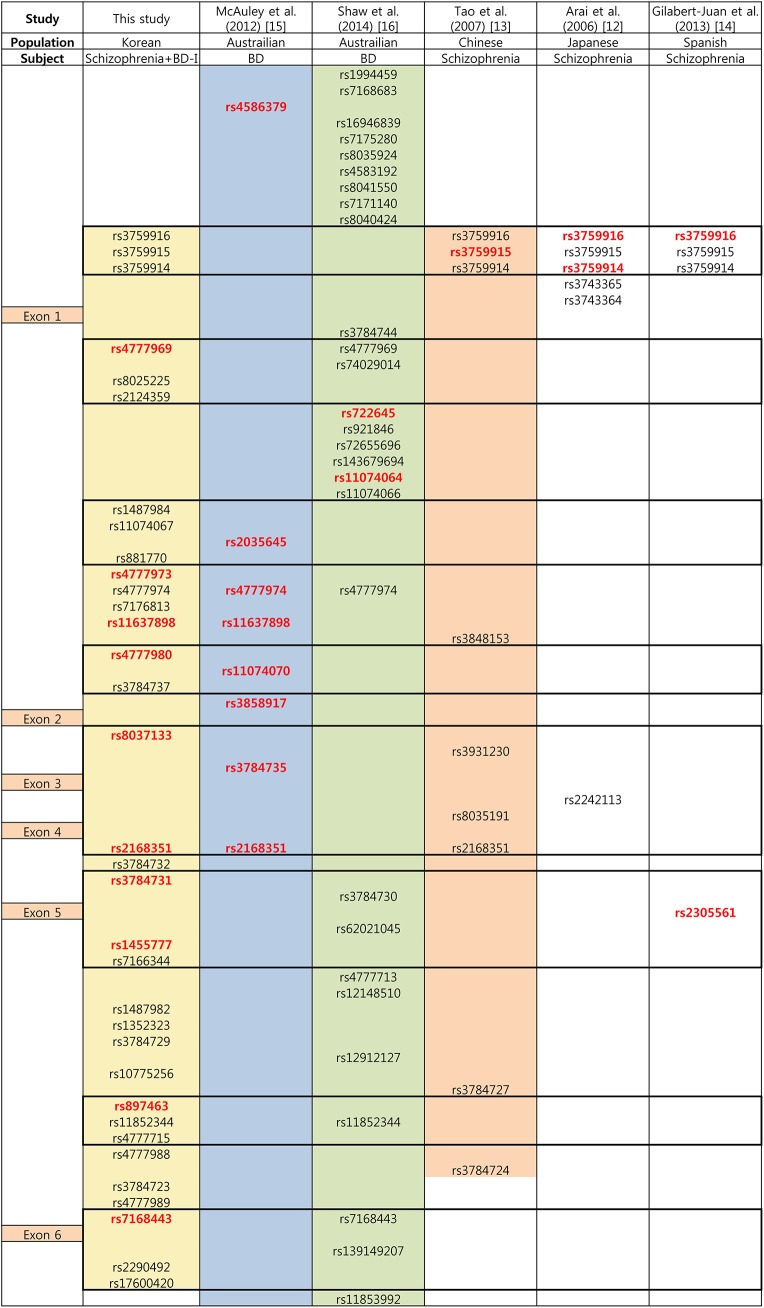
In the present study, we found suggestive associations between ST8SIA2 and schizophrenia and BD-I in the Korean population. Through fine mapping of the gene with tag SNPs and additional candidate SNPs, we identified 14 associated variants with at least a nominal level of significance in a diagnostic group, however none of them survived after multiple testing correction; these association trends were strongest in the combined schizophrenia and BD-I group. The strongest association was observed between rs11637898 and schizophrenia and BD-I under the dominant model, and the association with combined schizophrenia and the BD-I group remained significant after correcting for multiple testing. These results are consistent with a previous study of the Australian population revealing an association trend between several SNPs in ST8SIA2 and BD [15]. Two of these SNPs (rs11637898 and rs2168351) overlapped with SNPs showing association signals in the present study. Consistency between the two studies was also observed for two more pairs of SNPs with high linkage disequilibrium, i.e., rs4777980–rs11074070 and rs8037133–rs3784735. Fig 2 presents the relative positions of the ST8SIA2 SNPs that have been studied in schizophrenia and/or BD.
Fig 2. Relative positions of the ST8SIA2 single nucleotide polymorphisms (SNPs) analyzed in the current study and previous studies reporting positive associations with schizophrenia or bipolar disorder.
BD-I, bipolar I disorder; BD, bipolar disorder. Relative positions of the exons are displayed in the left column. Colored area is the covered area of the gene in the corresponding study. Box with bold outline indicates high linkage disequilibrium block (D’> 0.9) generated by Haploview v4.0 (http://www.broad.mit.edu/mpg/haploview) using the control group data of the current study (n = 502). SNPs with red letter indicate a significant association with nominal p-values < 0.05.
Three SNPs selected from a putative promoter region (rs3759916, rs3759915, and rs3759914) showed an association with schizophrenia in the Japanese, Chinese, and Spanish populations [12–14]. In contrast, rs3759915 showed a nominally significant association with only the BD-I group in the current study.
A remarkable finding of the present study was that schizophrenia and only BD-I shared the same association pattern with ST8SIA2 SNPs. This pattern was not observed in BD-II. As previous ST8SIA2 genetic studies did not analyze BD-II separately, this result needs to be replicated in a future study. According to the DSM-IV or DSM-5, BD-I and BD-II are distinguished by the presence of manic or hypomanic episode. The criteria for mania and hypomania have the same symptom profile and differ only in the duration and severity of the episode. Thus, BD-II may be regarded as a milder form of BD-I in a spectrum of illnesses having different thresholds on a continuum of the same underlying multi-factorial vulnerability [35]. However, recent investigations suggest that BD-II could be a discrete diagnostic category from BD-I in genetic, biological, clinical, and pharmacological aspects [36–39]. Results of the present study seem to support differences in genetic make-up between patients with BD-I and BD-II.
In contrast, BD-I and schizophrenia showed the same ST8SIA association pattern. When we combined these two groups for the analysis, the significance of association became stronger. This finding is consistent with previous GWAS results suggesting common vulnerability genes in patients with schizophrenia and BD crossing the diagnostic boundary[40, 41]. Considering gene function, ST8SIA2 could be related to the putative pathological mechanism of both schizophrenia and BD. Animal studies have shown that the amount of polySia synthesized by polysialyltransferase encoded by ST8SIA2 is crucial during brain development [42, 43]. ST8SIA2 knockout mice showed misguided infrapyramidal mossy fibers and formed ectopic synapses in the hippocampus [44]. These mice exhibited increased aggressive behavior and hyperactivity with impaired social behavior that could be seen in human patients with schizophrenia and BD [45]. In the context of gene-gene and gene-environment interactions, schizophrenia and BD-I may have some common vulnerability genes, and the susceptibility of each disorder could be established after the complex interplay of these genes with diagnosis-specific genes and environmental factors.
Another possible explanation for the common susceptibility between schizophrenia and BD-I is the existence of symptoms-specific genes rather than diagnosis-specific genes. Many attempts have been made to disassemble the diagnoses of schizophrenia and BD and reassess their symptoms in multiple phenomenological dimensions [46]. In the future, genetic studies targeting vulnerability genes for common clinical symptoms crossing the Kraepelinian dichotomy, such as psychotic features, mood dysregulation, and cognitive deficits, are needed.
The limitations of the current study are as follows. First, our sample size was relatively small, particularly for BD-II, which did not show the same significant association as BD-I. However, this negative result does not seem to be a false negative. The significant associations found in BD-I disappeared or became weaker when we analyzed the combined BD-I and BD-II group (S1 Table). Second, because we evaluated only diagnostic categories as phenotypes, we could not reveal possible associations of the gene with common clinical symptoms of schizophrenia and BD-I.
Considering both functional aspects and recent genetic study results including those of the present study, ST8SIA2 may be a susceptibility gene for both schizophrenia and BD beyond the boundary of diagnosis. Schizophrenia and BD could be a clinical syndrome composed of various biological subgroups with heterogeneous genetic backgrounds and could share some genetic risks that affect early neurodevelopment. Reconstructing the phenotypes toward broader clinical categories crossing the current diagnoses, and toward symptom-based dimensions could guide us to novel findings and an understanding of the background behind major mental disorders.
Supporting Information
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by a grant of the Korean Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (A120887). The work of T Park and IS H was supported by National Research Foundation (NRF) of Korea grant funded by the Korea government (MSIP) (No. 2012R1A3A2026438). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Tamminga CA, Ivleva EI, Keshavan MS, Pearlson GD, Clementz BA, Witte B, et al. Clinical phenotypes of psychosis in the Bipolar-Schizophrenia Network on Intermediate Phenotypes (B-SNIP). Am J Psychiatry. 2013;170: 1263–1274. 10.1176/appi.ajp.2013.12101339 [DOI] [PubMed] [Google Scholar]
- 2.Sadock BJ, Sadock VA, Ruiz P. Synopsis of psychiatry; behavioral sciences/clinical psychiatry, 11th edition. 2014: 23.
- 3. Andreasen NC, Arndt S, Alliger R, Miller D, Flaum M. Symptoms of schizophrenia: methods, meanings, and mechanisms. Archives of general psychiatry. 1995;52: 341–351. [DOI] [PubMed] [Google Scholar]
- 4. Gonzalez-Pinto A, Gutierrez M, Mosquera F, Ballesteros J, Lopez P, Ezcurra J, et al. First episode in bipolar disorder: misdiagnosis and psychotic symptoms. Journal of affective disorders. 1998;50: 41–44. [DOI] [PubMed] [Google Scholar]
- 5. Robinson LJ, Thompson JM, Gallagher P, Goswami U, Young AH, Ferrier IN, et al. A meta-analysis of cognitive deficits in euthymic patients with bipolar disorder. Journal of affective disorders. 2006;93: 105–115. [DOI] [PubMed] [Google Scholar]
- 6. Krabbendam L, Arts B, van Os J, Aleman A. Cognitive functioning in patients with schizophrenia and bipolar disorder: a quantitative review. Schizophrenia research. 2005;80: 137–149. [DOI] [PubMed] [Google Scholar]
- 7. Lichtenstein P, Yip BH, Björk C, Pawitan Y, Cannon TD, Sullivan PF, et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. The Lancet. 2009;373: 234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Valles V, Van Os J, Guillamat R, Gutierrez B, Campillo M, Gento P, et al. Increased morbid risk for schizophrenia in families of in-patients with bipolar illness. Schizophrenia research. 2000;42: 83–90. [DOI] [PubMed] [Google Scholar]
- 9. Maziade M, Roy M, Chagnon Y, Cliche D, Fournier J, Montgrain N, et al. Shared and specific susceptibility loci for schizophrenia and bipolar disorder: a dense genome scan in Eastern Quebec families. Molecular psychiatry. 2005;10: 486–499. [DOI] [PubMed] [Google Scholar]
- 10. McAuley E, Blair I, Liu Z, Fullerton J, Scimone A, Van Herten M, et al. A genome screen of 35 bipolar affective disorder pedigrees provides significant evidence for a susceptibility locus on chromosome 15q25-26. Molecular psychiatry. 2009;14: 492–500. 10.1038/sj.mp.4002146 [DOI] [PubMed] [Google Scholar]
- 11. Lee M, Chen C, Lee C, Chen C, Chong M, Ouyang W, et al. Genome-wide association study of bipolar I disorder in the Han Chinese population. Molecular psychiatry. 2011;16: 548–556. 10.1038/mp.2010.43 [DOI] [PubMed] [Google Scholar]
- 12. Arai M, Yamada K, Toyota T, Obata N, Haga S, Yoshida Y, et al. Association Between Polymorphisms in the Promoter Region of the Sialyltransferase 8B (SIAT8B) Gene and Schizophrenia. Biological psychiatry. 2006;59: 652–659. [DOI] [PubMed] [Google Scholar]
- 13. Tao R, Li C, Zheng Y, Qin W, Zhang J, Li X, et al. Positive association between SIAT8B and schizophrenia in the Chinese Han population. Schizophrenia research. 2007;90: 108–114. [DOI] [PubMed] [Google Scholar]
- 14. Gilabert-Juan J, Nacher J, Sanjuán J, Moltó MD. Sex-specific association of the ST8SIAII gene with schizophrenia in a Spanish population. Psychiatry research. 2013;210: 1293–1295. 10.1016/j.psychres.2013.09.001 [DOI] [PubMed] [Google Scholar]
- 15. McAuley EZ, Scimone A, Tiwari Y, Agahi G, Mowry BJ, Holliday EG, et al. Identification of sialyltransferase 8B as a generalized susceptibility gene for psychotic and mood disorders on chromosome 15q25-26. PloS one. 2012;7: e38172 10.1371/journal.pone.0038172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shaw AD, Tiwari Y, Kaplan W, Heath A, Mitchell PB, Schofield PR, et al. Characterisation of Genetic Variation in ST8SIA2 and Its Interaction Region in NCAM1 in Patients with Bipolar Disorder. PloS one. 2014;9: e92556 10.1371/journal.pone.0092556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bonfanti L. PSA-NCAM in mammalian structural plasticity and neurogenesis. Progress in neurobiology. 2006;80: 129–164. [DOI] [PubMed] [Google Scholar]
- 18. Gómez-Climent MÁ, Guirado R, Castillo-Gómez E, Varea E, Gutierrez-Mecinas M, Gilabert-Juan J, et al. The polysialylated form of the neural cell adhesion molecule (PSA-NCAM) is expressed in a subpopulation of mature cortical interneurons characterized by reduced structural features and connectivity. Cerebral Cortex. 2011;21: 1028–1041. 10.1093/cercor/bhq177 [DOI] [PubMed] [Google Scholar]
- 19. Hu H, Tomasiewicz H, Magnuson T, Rutishauser U. The role of polysialic acid in migration of olfactory bulb interneuron precursors in the subventricular zone. Neuron. 1996;16: 735–743. [DOI] [PubMed] [Google Scholar]
- 20. Seki T, Rutishauser U. Removal of polysialic acid–neural cell adhesion molecule induces aberrant mossy fiber innervation and ectopic synaptogenesis in the hippocampus. The Journal of neuroscience. 1998;18: 3757–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cremer H, Lange R, Christoph A, Plomann M, Vopper G, Roes J, et al. Inactivation of the N-CAM gene in mice results in size reduction of the olfactory bulb and deficits in spatial learning. Nature. 1994;367: 455–459. [DOI] [PubMed] [Google Scholar]
- 22. Eckhardt M, Bukalo O, Chazal G, Wang L, Goridis C, Schachner M, et al. Mice deficient in the polysialyltransferase ST8SiaIV/PST-1 allow discrimination of the roles of neural cell adhesion molecule protein and polysialic acid in neural development and synaptic plasticity. The Journal of Neuroscience. 2000;20: 5234–5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Senkov O, Sun M, Weinhold B, Gerardy-Schahn R, Schachner M, Dityatev A. Polysialylated neural cell adhesion molecule is involved in induction of long-term potentiation and memory acquisition and consolidation in a fear-conditioning paradigm. The Journal of neuroscience. 2006;26: 10888–109898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lewis DA, Levitt P. Schizophrenia as a disorder of neurodevelopment. Annual review of Neuroscience. 2002;25: 409–432. [DOI] [PubMed] [Google Scholar]
- 25. Sanches M, Keshavan MS, Brambilla P, Soares JC. Neurodevelopmental basis of bipolar disorder: a critical appraisal. Progress in Neuro-Psychopharmacology and Biological Psychiatry. 2008;32: 1617–1627. 10.1016/j.pnpbp.2008.04.017 [DOI] [PubMed] [Google Scholar]
- 26. Ryu S, Won H-H, Oh S, Kim J-W, Park T, Cho E-Y, et al. Genome-wide linkage scan of quantitative traits representing symptom dimensions in multiplex schizophrenia families. Psychiatry research. 2013;210: 756–760. 10.1016/j.psychres.2013.08.015 [DOI] [PubMed] [Google Scholar]
- 27. Association AAP. Diagnostic and statistical manual of mental disorders (DSM-IV) American Psychiatric Association; 1994. [Google Scholar]
- 28. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21: 263–265. [DOI] [PubMed] [Google Scholar]
- 29. de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nature genetics. 2005;37: 1217–1223. [DOI] [PubMed] [Google Scholar]
- 30. Akaike H. A new look at the statistical model identification. IEEE Transactions on Automatic Control. 1974;19: 716–723. [Google Scholar]
- 31.Clayton D. snpStats: SnpMatrix and XSnpMatrix classes and methods. R package version 1.18.0. 2014.
- 32. Holmans P, Zubenko GS, Crowe RR, DePaulo JR, Scheftner WA, Weissman MM, et al. Genomewide significant linkage to recurrent, early-onset major depressive disorder on chromosome 15q. The American Journal of Human Genetics. 2004;74: 1154–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Verma R, Holmans P, Knowles JA, Grover D, Evgrafov OV, Crowe RR, et al. Linkage disequilibrium mapping of a chromosome 15q25-26 major depression linkage region and sequencing of NTRK3. Biological psychiatry. 2008;63: 1185–1189. 10.1016/j.biopsych.2008.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Anney R, Klei L, Pinto D, Regan R, Conroy J, Magalhaes TR, et al. A genome-wide scan for common alleles affecting risk for autism. Hum Mol Genet. 2010;19: 4072–4082. 10.1093/hmg/ddq307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gershon ES, Hamovit J, Guroff JJ, Dibble E, Leckman JF, Sceery W, et al. A family study of schizoaffective, bipolar I, bipolar II, unipolar, and normal control probands. Archives of General psychiatry. 1982;39: 1157–1167. [DOI] [PubMed] [Google Scholar]
- 36. Vieta E, Gasto C, Otero A, Nieto E, Vallejo J. Differential features between bipolar I and bipolar II disorder. Comprehensive psychiatry. 1997;38: 98–101. [DOI] [PubMed] [Google Scholar]
- 37. Andreasen NC, Rice J, Endicott J, Coryell W, Grove WM, Reich T. Familial rates of affective disorder: a report from the National Institute of Mental Health Collaborative Study. Archives of General Psychiatry. 1987;44: 461–469. [DOI] [PubMed] [Google Scholar]
- 38. Baek JH, Park DY, Choi J, Kim JS, Choi JS, Ha K, et al. Differences between bipolar I and bipolar II disorders in clinical features, comorbidity, and family history. Journal of affective disorders. 2011;131: 59–67. 10.1016/j.jad.2010.11.020 [DOI] [PubMed] [Google Scholar]
- 39. Merikangas K, Cui L, Heaton L, Nakamura E, Roca C, Ding J, et al. Independence of familial transmission of mania and depression: results of the NIMH family study of affective spectrum disorders. Molecular psychiatry. 2014;19: 214–219. 10.1038/mp.2013.116 [DOI] [PubMed] [Google Scholar]
- 40. Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460: 748–752. 10.1038/nature08185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Consortium C-DGotPG. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. The Lancet. 2013;381: 1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hildebrandt H, Mühlenhoff M, Oltmann-Norden I, Röckle I, Burkhardt H, Weinhold B, et al. Imbalance of neural cell adhesion molecule and polysialyltransferase alleles causes defective brain connectivity. Brain. 2009;132: 2831–2838. 10.1093/brain/awp117 [DOI] [PubMed] [Google Scholar]
- 43. Hildebrandt H, Mühlenhoff M, Weinhold B, Gerardy‐Schahn R. Dissecting polysialic acid and NCAM functions in brain development. Journal of neurochemistry. 2007;103: 56–64. [DOI] [PubMed] [Google Scholar]
- 44. Angata K, Long JM, Bukalo O, Lee W, Dityatev A, Wynshaw-Boris A, et al. Sialyltransferase ST8Sia-II assembles a subset of polysialic acid that directs hippocampal axonal targeting and promotes fear behavior. Journal of Biological Chemistry. 2004;279: 32603–32613. [DOI] [PubMed] [Google Scholar]
- 45. Calandreau L, Márquez C, Bisaz R, Fantin M, Sandi C. Differential impact of polysialyltransferase ST8SiaII and ST8SiaIV knockout on social interaction and aggression. Genes, Brain and Behavior. 2010;9: 958–967. [DOI] [PubMed] [Google Scholar]
- 46. Keshavan MS, Morris DW, Sweeney JA, Pearlson G, Thaker G, Seidman LJ, et al. A dimensional approach to the psychosis spectrum between bipolar disorder and schizophrenia: the Schizo-Bipolar Scale. Schizophrenia research. 2011;133: 250–254. 10.1016/j.schres.2011.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.