Abstract
A new sorting scheme based on ferrofluid hydrodynamics (ferrohydrodynamics) was used to separate mixtures of particles and live cells simultaneously. Two species of cells, including Escherichia coli and Saccharomyces cerevisiae, as well as fluorescent polystyrene microparticles were studied for their sorting throughput and efficiency. Ferrofluids are stable magnetic nanoparticles suspensions. Under external magnetic fields, magnetic buoyancy forces exerted on particles and cells lead to size-dependent deflections from their laminar flow paths and result in spatial separation. We report the design, modeling, fabrication and characterization of the sorting device. This scheme is simple, low-cost and label-free compared to other existing techniques.
Keywords: Cell sorting, Ferrohydrodynamics, Continuous-flow, Ferrofluid, Microfluidics
1. Introduction
Microfluidic particle and cell sorting plays an important role in environmental monitoring (Liu et al. 2004; Beyor et al. 2008; Dharmasiri et al. 2010), disease diagnostics (Nagrath et al. 2007; Adams et al. 2008; Hoshino et al. 2011), and therapeutics (Toner and Irimia 2005; Yung et al. 2009). Compared to high-specificity and label-based cell sorting techniques such as 0fluorescence-activated cell sorter (FACS) (Bonner et al. 1972) and magnetic-activated cell sorter (MACS) (Miltenyi et al. 1990), microfluidic sortings are mostly label-free, relying on cells’ intrinsic properties such as size, shape, density, deformability, electric and magnetic properties for manipulation specificity (Pamme 2007; Tsutsui and Ho 2009; Gossett et al. 2010; Lenshof and Laurell 2010). When applicable, microfluidic sortings are favored over label-based ones, because they are inexpensive and require minimal user training for operation (Gossett et al. 2010). Among them, those based on channel design including pinched flow fractionation (Yamada et al. 2004) and deterministic lateral displacement (Huang et al. 2004; Davis et al. 2006) combine laminar flows with mechanical structures to direct particles of different sizes into separate streamlines. Continuous inertial separation uses balance between inertial lift force and Dean drag force in curved channels for size-dependent sorting of particles and cells (Di Carlo 2009). On the other hand, external energy inputs such as acoustic, electric and magnetic forces have also been used to manipulate cells in microfluidic systems. Depending on the application, their simpler channel geometry and faster manipulation speed may outweigh the complications of integrating electrodes in their designs. For example, acoustophoresis can separate particles and cells according to their size, density, as well as compressibility (Laurell et al. 2007; Shi et al. 2009; Wang and Zhe 2011). Dielectrophoresis (DEP), arising from interactions of cells’ dipoles and their surrounding electric fields, can realize low-cost and integrated devices for cell manipulation (Voldman 2006). Magnetophoresis (MAP) takes advantages of paramagnetic nature of red blood cells and magnetotactic bacteria and applies non-uniform magnetic fields to separate them from non-magnetic objects (Zborowski et al. 2003; Lee et al. 2004). However, most applications of magnetophoresis use functionalized magnetic beads for labeling (Pamme 2006; Liu et al. 2009; Gijs et al. 2010). The label-based methods are manually intensive and time-consuming. The magnetic moments of these beads, even from the same batch, can vary dramatically due to their manufacturing procedure, making scaling of the method scaling difficult (Hafeli et al. 1997; Miller et al. 2001; Rife et al. 2003; Mihajlovic et al. 2007; Shevkoplyas et al. 2007).
To address problems with label-based magnetophoresis, a label-free technique that uses reverse magnetophoresis to manipulate and sort cells has been developed recently based on ferrofluid hydrodynamics (ferrohydrodynamics) (Yellen et al. 2005; Kose et al. 2009; Zhu et al. 2010; Zhu et al. 2011a; Kose and Koser 2012). Ferrofluids are colloidal suspensions of magnetic nanoparticles, typically magnetite (Fe3O4) with approximately 10 nm diameters (Rosensweig 1985). They are covered by either electrostatic or steric surfactants to keep them from agglomeration due to van der Waals force and in suspension within a water or oil medium. Ferrohydrodynamics studies mechanics of ferrofluid motion under external magnetic fields (Rosensweig 1985; Odenbach and Editor 2002). Its applications in microfluidics, recently reviewed by Nguyen (Nguyen 2012), include miniaturized polymerase chain reaction (PCR) (Sun et al. 2007; Sun et al. 2008), traveling-wave magnetic field pumping (Mao and Koser 2006; Mao et al. 2011), micro-scale mixing (Mao and Koser 2007), micropump (Hatch et al. 2001; Love et al. 2004), and droplet manipulation (Nguyen et al. 2006; Zhang et al. 2011b, a).
In applications of cell manipulation, the purpose of using ferrofluids is to induce effective magnetic dipole moments within cells. Under non-uniform magnetic fields, cells will experience in the weaker field direction a magnetic buoyancy force, analogous to buoyancy force, as magnitude of the force is proportional to the volume of cell (Rosensweig 1985). Many groups have been working on adapting this principle to particles and cells sorting. For example, Whitesides’ group separated synthetic particles according to their densities’ difference using paramagnetic salt solutions (Winkleman et al. 2007; Mirica et al. 2009). Pamme’s group demonstrated continuous particle and cell manipulation using paramagnetic salt solution in microfluidic devices (Peyman et al. 2009; Rodriguez-Villarreal et al. 2011). Xuan’s group studied the transport of particles in both paramagnetic solutions and ferrofluids through a rectangular microchannel embedded with permanent magnets (Liang et al. 2011; Zhu et al. 2012). Park’s group recently sorted human histolytic lymphoma monocytes cells from red blood cells using gadolinium diethylenetriamine pentaacetic acid (Gd-DTPA) solution (Shen et al. 2012). However, magnetic susceptibility of paramagnetic salt solutions is inherently small, about 5 orders of magnitude weaker than that of a ferrofluid (Krebs Melissa et al. 2009), rendering slower manipulation speed and low throughput. As a result of the higher susceptibility of ferrofluids, Koser’s group was able to use an integrated microfluidic platform for sorting of microparticles and live cells within a citrate stabilized cobalt-ferrite ferrofluid in static flow conditions (Kose et al. 2009). The same device was also applied to continuous-flow frequency-adjustable particles separation (Kose and Koser 2012). Our group developed high-efficiency and high-throughput continuous-flow particle separation and focusing devices using commercial ferrofluids and hand-held permanent magnets (Zhu et al. 2010; Zhu et al. 2011b; Zhu et al. 2011a). Permanent magnet based devices are low-cost and easy to operate; their operations do not generate heat. Magnetic fields produced by permanent magnets are substantially larger than the ones by current-carrying electrodes.
High throughput, label-free and selective cell sorting realized in a single automated device can have profound impacts on environmental monitoring, diagnostics and therapeutics. Although continuous-flow ferrohydrodynamic sorting has been demonstrated with microparticles, it has not previously been reported with live cells (Zhu et al. 2010). The potential for live cell applications of continuous-flow ferrohydrodynamic sorting motivates the study presented here. We developed a microfluidic device that could continuously sort cells of different sizes based on ferrohydrodynamics, which involved manipulation of cells within ferrofluids via external non-uniform magnetic fields. When cell mixtures and ferrofluids were injected into the channel by a pressure-driven flow, deflections of cells from their laminar flow paths would occur because of the magnetic field gradient and resulting magnetic buoyance force. This deflection will lead to spatial separation of cells of different sizes at the end of channel.
In the following sections, we first summarize materials and methods used in this study, followed by results from a three-dimensional theoretical study of cells’ transport in the microfluidic device. Cell viabilities of Escherichia coli and Saccharomyces cerevisiae in a commercial ferrofluid are then discussed. Afterwards, calibration of the sorting device with fluorescent polystyrene microparticles is performed. Escherichia coli and Saccharomyces cerevisiae are sorted in the device, and cells distribution is analyzed on samples collected from channel outlets. In the end we will discuss outlook of ferrohydrodynamic sorting.
2. Materials and Methods
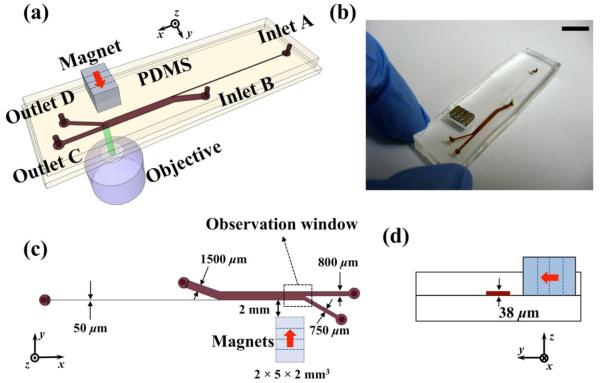
The prototype polydimethylsiloxane (PDMS) microfluidic device was fabricated through a standard soft-lithography approach and attached to a flat surface of another piece of PDMS, as shown in Figures 1(a) and (b). A mask of the device pattern was created using AutoCAD 2008 (Autodesk Inc., San Rafael, CA) and printed by a commercial photo-plotting company (CAD/Art Services Inc, Bandon, OR). Dimensions of the microfluidic channel are listed in Figures 1(c) and 1(d). Thickness of the device was measured to be 38 μm by a profilometer (Dektak 150, Veeco Instruments Inc., Chadds Ford, PA). Before attachment, PDMS surfaces were treated with plasma (PDC-32G plasma cleaner, Harrick Plasma, Ithaca, NY) at 11.2 Pa O2 partial pressure with 18 W power for 1 minute. A stack of four NdFeB permanent magnets was embedded into PDMS channel with their magnetization direction vertical to the channel during curing stage. Each magnet is 2 mm in width, 5 mm in length and 2 mm in thickness. The magnet stack was placed 2 mm away from the channel. Flux density at the center of magnets stack’s surface was measured to be 470 mT by a Gauss meter (Model 5080, Sypris, Orlando, FL) and an axial probe with 0.381 mm diameter of circular active area. Before liquid injection, the device was treated with plasma for 10 minutes to render PDMS surfaces hydrophilic. This step ensured both cells and microparticles would not attach onto PDMS surfaces during sorting.
Figure 1.
(a) Schematic representation of the sorting device with permanent magnets and a microfluidic channel. (b) An image of protype device. Scale bar is 10 mm. (c) Topview of the device and relevant dimensions. Red arrows indicate direction of magnets’ magnetization. (d) Cross-section of the device.
We used a commercial water-based, pH ~7 magnetite ferrofluid coated with anionic surfactants (EMG 408, Ferrotec Co., NH). Volume fraction of magnetite particles in this ferrofluid is 1.1%. Mean diameter of nanoparticles has been determined from Transmission Electron Microscopy (TEM) images to be ~10 nm. Initial magnetic susceptibility was measured to be 0.26; saturation magnetization was 60 Gauss; viscosity was 1.2×10−3 kg/m·s. Escherichia coli (strain MG1655) and Saccharomyces cerevisiae (Baker’s yeast), and two fluorescent microparticles (green 1.0 μm diameter, Thermo Fisher Scientific Inc., Waltham, MA, and red 7.3 μm diameter, Bangs Laboratories Inc., Fishers, IN) were used in sorting. Ferrofluid and particles/cells mixture injected into microchannel were maintained at tunable flow rates using a syringe pump (Nexus 3000, Chemyx Inc., Stafford, TX). Sorting was conducted on the stage of an inverted microscope (Zeiss Axio Observer, Carl Zeiss Inc., Germany). Micrographs of cells and particles were recorded through either a green fluorescent filter set (41001 FITC, Chroma Technology Corp., Rockingham, VT), or a red filter set (43HE, Carl Zeiss Inc., Germany), and a CCD camera (SPOT RT3, Diagnostic Instruments, Inc., Sterling Heights, MI). Cell samples collected from channel outlets were pipetted onto microscope slides and analyzed using a high-resolution CCD camera (AxioCam HR, Carl Zeiss Inc., Germany) for size distributions to quantitatively evaluate efficiency of this approach. ImageJ® software was used to count the number of cells.
Saccharomyces cerevisiae (Baker’s yeast) cells were first grown in a 10 ml test tube containing 2 ml of YPG medium (10 g/l yeast extract, 20 g/l glucose, 20 g/l glucose) overnight. They were then transferred into a 100 ml shake flask containing 20 ml of YPG medium. After 4 h growth at 30°C and 250 rpm, cells in the flask were stained with fluorophores. Escherichia coli (strain MG1655) cells were first grown in a 10 ml test tube containing 2 ml of Luria-Bertani (LB) medium overnight. They were then transferred into a 100 ml shake flask containing 20 ml of LB medium (25 g/l LB). After 4 h growth at 37°C and 250 rpm, cells were stained with fluorophores. Nucleic acid stains SYTO9 (green) and SYTO17 (red) (Molecular Probes Inc., Eugene, OR) were used in cell staining.
To study of viability of Escherichia coli and Saccharomyces cerevisiae cells exposed to EMG 408 ferrofluids, nominally 2×109 cells Escherichia coli and 2×107 cells Saccharomyces cerevisiae grown as described above were centrifuged twice at 4°C and washed in defined M9 medium (6.78 g/l Na2HPO4, 3.0 g/l KH2PO4, 0.5 g/l NaCl, 1.0 g/l NH4Cl) without carbon source. For either cell type in duplicate, the washed cell pellet from centrifugation was combined with either 2 ml of EMG 408 ferrofluid or 2 ml M9 medium as a control. After 2 hours of incubation at room temperature in these fluids, cell density was determined in triplicate using standard microbial serial dilutions (106 dilution for Escherichia coli, and 104 dilution for Saccharomyces cerevisiae), with the transferring of known volumes to Petri plates and counting of Colony Forming Units (CFU) after 24 hours.
3. Theory and Simulation
Previously, we reported both two-dimensional (2D) and three-dimensional (3D) analytical models for microfluidic transports of microparticles in ferrofluids (Zhu et al. 2011a; Zhu et al. 2011b). In this work, we applied the 3D analytical model to predict cells’ sorting in permanent magnet based device. Briefly, we obtained cells’ trajectories by first calculating magnetic buoyancy force on cells using a 3D analytical model of magnetic fields (Furlani and Sahoo 2006) and a nonlinear magnetization model of ferrofluids (Rosensweig 1985), and then solving governing equations of motion for cells in laminar flow condition (Brody et al. 1996). All relevant parameters used in our simulation are listed in Figure 1 and Materials and Methods section. In addition, we calculated volume of a single rod-shape Escherichia coli cell with short axis of 0.5 – 1 μm and long axis of 2 – 4 μm to be 2.1 – 16.7 μm3 (Kaya and Koser 2009), and volume of a single sphere-shape Saccharomyces cerevisiae cell with diameter of 7 – 9 μm to be 180 – 382 μm3 (Jorgensen et al. 2002).
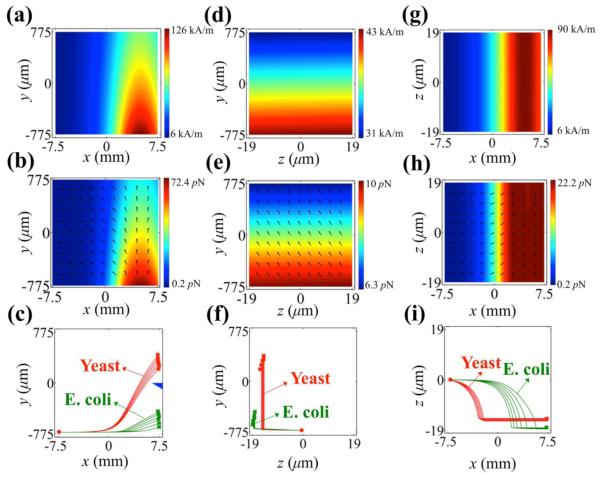
Figure 2 summarizes simulated distribution of magnetic fields and magnetic buoyance forces in the sorting channel, as well as 3D trajectories of Escherichia coli and Saccharomyces cerevisiae cells. The surface plot in Figure 2(a) shows magnitude of magnetic fields of x-y plane at z = 0. Magnetic fields decayed rather quickly from the surface of the magnet and formed a gradient that resulted in magnetic buoyance force on cells in both x and y directions, as indicated in Figure 2(b). Consequently, cells experiencing such force when entering the sorting channel would decelerate in x direction and accelerate in y direction. Force computed on a spherical microparticle of 7.3 μm diameter, with its total volume (~200 μm3) close to that of a single Saccharomyces cerevisiae cell, is on the order of 10 pN. Cell mixtures were quickly sorted by magnetic buoyancy force towards the end of channel, as shown in Figure 2(c) with simulated cells’ trajectories considering their natural size variations. All Escherichia coli cells, having much smaller size and volume compared to Saccharomyces cerevisiae cells, exited the channel through Outlet D, while all Saccharomyces cerevisiae cells migrated towards Outlet C. Figures 2(d)-(f) illustrate distribution of magnetic fields and forces, as well as trajectories of cells of y-z plane at x = 0; Figures 2(g)-(i) depict the cases of x-z plane at y = 0. We are interested in 3D trajectories of cells, in part due to the opaqueness of ferrofluids and difficulty in recording cells’ weak fluorescence in the channel, especially the red fluorescent from Saccharomyces cerevisiae cells, as shown later in the results. In a concentrated ferrofluid (~10% v/v), particles and cells are visible only when they are very close (~1 μm) to the surface of channel (Zhu et al. 2011b). Visibility was a less of a problem when diluted ferrofluids (~1% v/v) and thin microchannel were used in our device. Simulation results from Figures 2(f) and (i) indicated in our current setup all cells were pushed towards the channel bottom surface, which would enhance visibility of stained cells.
Figure 2.
Analytical three-dimensional simulation of magnetic field and force distributions in microfluidic channel, and trajectories of cells. Simulation parameters match exact experimental conditions. (a)-(c) x-y plane (z = 0), (d)-(f) y-z plane (x = 0), (g)-(i) x-z plane (y = 0) of magnetic field strength (surface plot) (a, d, g), magnetic force (surface plot: force magnitude; arrow plot: force direction) (b, e, h), and particles’ trajectories (c, f, i). Dots indicate starting points, while crosses indicate ending points of cells’ trajectories. E.coli cell has volume range of 2.1 – 16.7 μm3 and Yeast cell has volume range of 180 – 382 μm3, resulting in a distribution of trajectories for each type of cell. Blue triangle in (c) indicates boundary between Outlets C and D.
4. Results and Discussions
4.1. Cell Viability
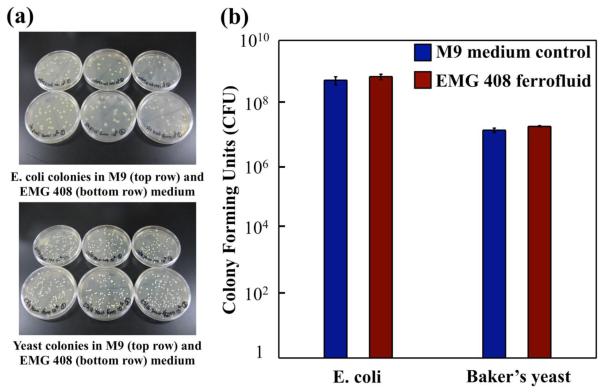
Figure 3(a) shows the CFU in both M9 medium and EMG 408 ferrofluids after incubation. Counts of CFU for each case were averaged over 3 plates and plotted in Figure 3(b). We observed a slight increase in cell density after 2 hours of incubation in the ferrofluid compared to the M9 medium control for both cell types, suggesting a possibility that either the EMG 408 ferrofluid acted as a cell protectant or the cells continued to grow in this ferrofluid during incubation. Nonetheless, this ferrofluid was not detrimental to the viability of both cell types after 2 hours of exposure, which allowed enough time to carry out the sorting procedure.
Figure 3.
Cell viability test of Escherichia coli and Saccharomyces cerevisiae. (a) Top and bottom photos show Escherichia coli and Yeast colonies formed in M9 medium and EMG 408 ferrofluids after 106 dilution from initial growth, respectively. (b) Colony Forming Unites (CFU) count of Escherichia coli and Saccharomyces cerevisiae using initial growth cell concentration.
4.2 Cells Sorting
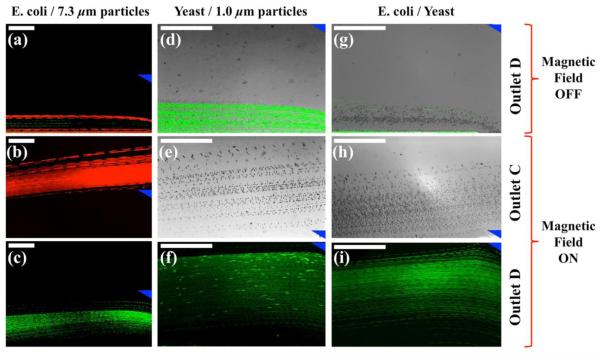
We first calibrated the sorting device using a mixture of Escherichia coli cells and red fluorescent 7.3 μm particles, which have similar total volume of Saccharomyces cerevisiae cells. Washed Escherichia coli cell pellet from centrifugation as described above was stained with 1 μl of green nucleic acid stain SYTO9. Both particles and cells have concentrations of ~107 counts/ml. We introduced microparticles/cells mixture into microfluidic channel Inlet A at a constant flow rate of 1.5 μl/min. The mixture was hydrodynamically focused into a narrow stream by sheath flow from Inlet B at a flow rate of 6 μl/min. The observation window was located right before the channel outlets, as indicated in Figure 1(c). When magnetic fields were off, particles and cells were observed in fluorescent mode flowing together near sidewall of the channel and exiting through Outlet D, as shown in composite micrograph of Figure 4(a). When magnetic fields were on, magnetic buoyancy forces deflected particles from their laminar flow paths towards Outlet C, as shown in Figure 2(b). On the other hand, forces on smaller Escherichia coli cells were inadequate to deflect them to Outlet C; therefore they exited the channel through Outlet D still, as shown in Figure 2(c). This resulted in spatial separation of particles/cells mixture at the end of channel. We were able to separate ~106 particles from ~106 cells per hour with 1.5 μl/min flow rate. Simply increasing the flow rate can further increase sorting throughput. Current microfluidic sorting schemes use flow rates ranging between ~10 μl/min and ~1 ml/min (Gossett et al. 2010). With such flow rates and 107 – 108 cells/ml concentration, maximum sorting throughput of our device in theory can go up to 109 cells per hour.
Figure 4.
Experimental composite micrographs of sorting process. (a), (d), (g) were particles/cells mixture ((a): Escherichia coli (green) and 7.3 μm particles (red); (d): Saccharomyces cerevisiae (red and bright-field) and 1.0 μm particles (green); (g): Escherichia coli (green) and Saccharomyces cerevisiae (red and bright-field) before magnetic fields were applied. (b), (e), (h) were micrographs of Outlet C after magnetic fields were applied, and (c), (f), (i) were micrographs of Outlet D. Blue triangles indicate boundary between Outlets C and D. Scale bars represent 200 μm.
Secondly, we calibrated the device using a mixture of Saccharomyces cerevisiae cells and green fluorescent 1.0 μm particles, which have similar volume as Escherichia coli cells. Saccharomyces cerevisiae were stained with red nucleic acid stain SYTO17. Both particles and cells again have concentrations of ~107 counts/ml. Due to weak red fluorescence from SYTO17 in our setup, we chose to use a combination of bright-field and fluorescent modes microscopy to record the sorting process. Figure 4(d) shows merged composite micrograph of green fluorescent 1.0 μm particles and bright-field particles/Saccharomyces cerevisiae mixture, both of which exited channel through Outlet D when magnetic fields were off. Sorting of this mixture was achieved as soon as magnetic fields were on, as depicted in Figures 4(e) and 4(f). Cells distribution analysis presented in the following section confirmed a close to 100% sorting efficiency. Sorting throughput was ~106 cells per hour. Here we demonstrated that combination of bright-field and fluorescent microscopy can successfully circumvent recording issues originating from opaqueness of ferrofluids and weak fluorescence from stained live cells.
Finally, sorting of Escherichia coli and Saccharomyces cerevisiae cells were carried out in the same device at the same time. Escherichia coli cells were stained with green fluorescence while Saccharomyces cerevisiae were stained with red fluorescence. Both types of cells were adjusted to ~107 cells/ml concentration in initial mixture. It is clearly shown in Figure 4(g) that all cells exited from the channel through Outlet D when there was no magnetic field. Both bright-field and fluorescent mode micrographs of cells were recorded and merged to form Figure 4(g). Saccharomyces cerevisiae cells were successfully sorted from the initial cell mixture with the application of magnetic fields, as demonstrated in Figures 4(h) and 4(i).
4.3 Cell Sorting Efficiency
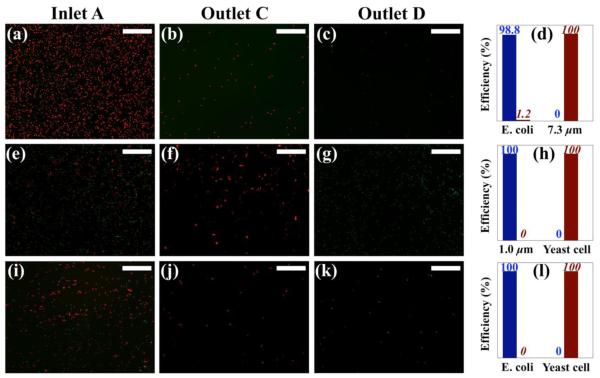
In order to precisely evaluate sorting efficiency, we collected samples from both Outlets C and D and analyzed them for size distributions off chip. We stained cells in distinctive fluorescence and counted them using ImageJ® software. Specifically, in first calibration, Escherichia coli cells were green and 7.3 μm particles were red; in second calibration, Saccharomyces cerevisiae cells were red and 1.0 μm particles were green; in cells sorting, Saccharomyces cerevisiae cells were red and Escherichia coli cells were green. Fluorescent mode was chosen for distribution analysis to avoid miscounting of cell types in bright-field micrographs. A magnetic field was applied to push all particles and cells onto a surface of glass slide to increase visibility. We define remaining efficiency as ratio of number of particles or cells exiting from Outlet D after magnetic field application to their initial number before magnetic field application. Similarly, sorting efficiency is defined as the ratio of number of particles or cells exiting from Outlet C after magnetic field application to their initial number before magnetic field application. Figure 5(a) shows a representative composite micrograph of Escherichia coli cells and 7.3 μm particles collected from Inlet A before sorting. 100% of 7.3 μm particles migrated to Outlet C and 98.8% Escherichia coli cells remained in Outlet D, as depicted in Figures 5(b) and 5(c). Remaining and separation efficiencies for both particles are plotted in Figure 5(d). Figures 3(e) – 3(h) and Figures 3(i) – 3(l) show micrographs and efficiencies for Saccharomyces cerevisiae cells/1.0 μm particles mixture sorting and Saccharomyces cerevisiae cells/Escherichia coli cells mixture sorting, respectively. Both cases have 100% efficiencies. It should be noted that samples collected from Outlets C and D were greatly diluted by ferrofluid sheath flow from Inlet B, rendering much lower particles and cells concentration for distribution analysis. A possible solution to this problem is integration of cell focusing (Zhu et al. 2011a) and sorting steps on one chip.
Figure 5.
Experimental composite micrographs of size distribution analysis, including micrographs of particles/cells mixture collected before sorting at Inlet A and after separation at Outlets C and D, and remaining and separation efficiencies. (a) – (d) were for Escherichia coli and 7.3 μm particles mixture; (e) – (h) were for Saccharomyces cerevisiae and 1.0 μm particles mixture; (i) – (l) were for Escherichia coli and Saccharomyces cerevisiae mixture. Blue bar with normal number on top shows remaining efficiency, while red bar with italic number on top shows separation efficiency. Scale bars represent 200 μm.
4.4 Outlooks of Ferrohydrodynamic Sorting
Ferrohydrodynamic cell sorting offers the potential for high throughput (~107 cells/hour in this study and ~109 cells/hour in theory) and high separation efficiency (~100%) that are comparable to existing microfluidic sorting techniques but without the use of labels. The associated device is inexpensive and simple, only requiring a channel and hand-held permanent magnets. Sorting specificity of this approach is not limited to size difference only; it is also sensitive to cells’ shape and deformability (Kose et al. 2009). In adapting it to miniaturized flow cytometry, ferrohydrodynamic manipulation can first focus cells into single cell streams before sorting, eliminating needs for excessive sheath flow and preventing sample dilution (Zhu et al. 2011a). Compared to paramagnetic solution based sorting, ferrofluid offers much higher magnetic susceptibility, eliminating needs for either microfabricated ferromagnetic structures to enhance field gradient or hypertonic concentrations of paramagnetic salts that are not biocompatible for live cell manipulation.
On the other hand, using water-based ferrofluids for cell manipulation is a work in progress. Diagnostic and research applications directed towards simply purifying or isolating cells of interest from complex mixtures such as blood and exfoliated cytology specimens are exciting. For instance, blood cells obscure the detection of the larger but rare abnormal cervical cells from Pap test specimens and metastatic epithelial tumor cells circulating in blood (Moriarty et al. 2009; Yu et al. 2011). Misinterpreted cervical cytology ranks third among causes of medical negligence claims against pathologist (Frable 2007). A simple, low-cost tumor cell enrichment platform would benefit cancer screening. However, two issues, cell visibility and biocompatibility of mammalian cells in ferrofluids, limit applications of ferrohydrodynamic manipulation. Ferrofluids are opaque due to light diffraction from their high concentration of magnetic nanoparticles. Fluorescent cells need to be close to channel surface for microscopic recording. In order to address this issue, ferrofluids with low solid content, as well as shallow microfluidic channel, are favored for cell manipulation. In addition, magnetic fields can be used to push cells onto channel surface, increasing visibility of cells in fluorescent mode. In this study, we used a combination of both bright-field and fluorescent modes microscopy to circumvent the opaqueness issue. Cells were readily visible in a shallow channel in bright-field micrographs. Another potential issue is biocompatibility of ferrofluids. Our next step is to extend this methodology to mammalian cells, particularly human specimens such as blood and other bodily fluids, exfoliated musical cells, and tumor aspirates. The requirements of mammalian cells may differ from Escherichia coli and Saccharomyces cerevisiae. For cell manipulation, materials, pH value, and surfactants of ferrofluids need to be rendered biocompatible, at the same time the overall colloidal system of ferrofluids must be maintained. Typically, nanoparticles within ferrofluids for cell applications are made of magnetite (Pankhurst et al. 2003). pH value of ferrofluids needs to be compatible with cell culture and maintained at 7.4. Salt concentration, tonicity, and surfactant must be carefully chosen close to physiological conditions to reduce cell death. Although these are stringent requirements, progress has been made towards synthesizing biocompatible ferrofluids. For example, Koser’s group used citrate to stabilize cobalt-ferrite nanoparticles for live red blood cell and Escherichia coli cell sorting (Kose et al. 2009). Yellen’s group used Bovine Serum Albumin (BSA) to stablize magnetite nanoparticles for human umbilical vein endothelial cells manipulation (Krebs Melissa et al. 2009). Viability tests from both studies have shown cells were able to retain their viability for up to several hours in ferrofluids. In our study, a commercially available pH ~7 magnetite ferrofluid was able to sustain viability of both Escherichia coli and Saccharomyces cerevisiae cells for at least 2 hours.
5. Conclusion
In conclusion, we have developed a label-free and continuous-flow ferrohydrodynamic cell sorting device and applied it in separating Escherichia coli and Saccharomyces cerevisiae cells. A commercial magnetite ferrofluid was used to separate particle and cell mixtures. Construction of our device is simple and low-cost; we choose to use permanent magnets instead of integrated electrodes to eliminate complex microfabrication process and auxiliary power supply. Current sorting throughput is 107 cells/hour, and sorting efficiency is close to 100%. We envision this device can achieve up to two orders higher throughput while still maintaining current sorting efficiency. This device can also be used for mammalian cells sorting and enrichment with a biocompatible ferrofluid.
Acknowledgments
This work is in part supported by the National Science Foundation CAREER Award (ECCS-1150042), American Society for Microbiology/Centers for Disease Control and Prevention Postdoctoral Research Fellowship, Centers for Disease Control and Prevention and University of Georgia Seed Award Program.
Footnotes
Disclaimer: The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the supporting agencies.
References
- Adams AA, Okagbare PI, Feng J, Hupert ML, Patterson D, Gottert J, McCarley RL, Nikitopoulos D, Murphy MC, Soper SA. Highly efficient circulating tumor cell isolation from whole blood and label-free enumeration using polymer-based microfluidics with an integrated conductivity sensor. J Am Chem Soc. 2008;130(27):8633–8641. doi: 10.1021/ja8015022. doi:Doi 10.1021/Ja8015022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyor N, Seo TS, Liu P, Mathies RA. Immunomagnetic bead-based cell concentration microdevice for dilute pathogen detection. Biomed Microdevices. 2008;10(6):909–917. doi: 10.1007/s10544-008-9206-3. doi:Doi 10.1007/S10544-008-9206-3. [DOI] [PubMed] [Google Scholar]
- Bonner WA, Sweet RG, Hulett HR, Herzenbe La. Fluorescence Activated Cell Sorting. Review of Scientific Instruments. 1972;43(3):404. doi: 10.1063/1.1685647. [DOI] [PubMed] [Google Scholar]
- Brody JP, Yager P, Goldstein RE, Austin RH. Biotechnology at low Reynolds numbers. Biophys J. 1996;71(6):3430–3441. doi: 10.1016/S0006-3495(96)79538-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JA, Inglis DW, Morton KJ, Lawrence DA, Huang LR, Chou SY, Sturm JC, Austin RH. Deterministic hydrodynamics: Taking blood apart. P Natl Acad Sci USA. 2006;103(40):14779–14784. doi: 10.1073/pnas.0605967103. doi:Doi 10.1073/Pnas.0605967103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri U, Witek MA, Adams AA, Osiri JK, Hupert ML, Bianchi TS, Roelke DL, Soper SA. Enrichment and Detection of Escherichia coli O157:H7 from Water Samples Using an Antibody Modified Microfluidic Chip. Analytical Chemistry. 2010;82(7):2844–2849. doi: 10.1021/ac100323k. doi:Doi 10.1021/Ac100323k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Carlo D. Inertial microfluidics. Lab on a Chip. 2009;9(21):3038–3046. doi: 10.1039/b912547g. doi:Doi 10.1039/B912547g. [DOI] [PubMed] [Google Scholar]
- Frable WJ. Error reduction and risk management in cytopathology. Semin Diagn Pathol. 2007;24(2):77–88. doi: 10.1053/j.semdp.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Furlani EP, Sahoo Y. Analytical model for the magnetic field and force in a magnetophoretic microsystem. J Phys D Appl Phys. 2006;39(9):1724–1732. doi:Doi 10.1088/0022-3727/39/9/003. [Google Scholar]
- Gijs MAM, Lacharme F, Lehmann U. Microfluidic Applications of Magnetic Particles for Biological Analysis and Catalysis. Chem Rev. 2010;110(3):1518–1563. doi: 10.1021/cr9001929. [DOI] [PubMed] [Google Scholar]
- Gossett DR, Weaver WM, Mach AJ, Hur SC, Tse HTK, Lee W, Amini H, Di Carlo D. Label-free cell separation and sorting in microfluidic systems. Analytical and Bioanalytical Chemistry. 2010;397(8):3249–3267. doi: 10.1007/s00216-010-3721-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafeli U, Schutt W, Teller J, Zborowski M. Scientific and clinical applications of magnetic carriers. Springer; New York: 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch A, Kamholz AE, Holman G, Yager P, Bohringer KF. A ferrofluidic magnetic micropump. Microelectromechanical Systems, Journal of. 2001;10(2):215–221. [Google Scholar]
- Hoshino K, Huang YY, Lane N, Huebschman M, Uhr JW, Frenkel EP, Zhang XJ. Microchip-based immunomagnetic detection of circulating tumor cells. Lab on a Chip. 2011;11(20):3449–3457. doi: 10.1039/c1lc20270g. doi:Doi 10.1039/C1lc20270g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LR, Cox EC, Austin RH, Sturm JC. Continuous particle separation through deterministic lateral displacement. Science. 2004;304(5673):987–990. doi: 10.1126/science.1094567. doi:10.1126/science.1094567304/5673/987 [pii] [DOI] [PubMed] [Google Scholar]
- Jorgensen P, Nishikawa JL, Breitkreutz BJ, Tyers M. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002;297(5580):395–400. doi: 10.1126/science.1070850. doi:Doi 10.1126/Science.1070850. [DOI] [PubMed] [Google Scholar]
- Kaya T, Koser H. Characterization of Hydrodynamic Surface Interactions of Escherichia coli Cell Bodies in Shear Flow. Physical Review Letters. 2009;103(13) doi: 10.1103/PhysRevLett.103.138103. doi:Artn 138103 Doi 10.1103/Physrevlett.103.138103. [DOI] [PubMed] [Google Scholar]
- Kose AR, Fischer B, Mao L, Koser H. Label-free cellular manipulation and sorting via biocompatible ferrofluids. P Natl Acad Sci USA. 2009;106(51):21478–21483. doi: 10.1073/pnas.0912138106. doi:Doi 10.1073/Pnas.0912138106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kose AR, Koser H. Ferrofluid mediated nanocytometry. Lab on a Chip. 2012;12(1):190–196. doi: 10.1039/c1lc20864k. doi:Doi 10.1039/C1lc20864k. [DOI] [PubMed] [Google Scholar]
- Krebs Melissa D, Erb Randall M, Yellen Benjamin B, Samanta B, Bajaj A, Rotello Vincent M, Alsberg E. Formation of ordered cellular structures in suspension via label-free negative magnetophoresis. Nano Lett. 2009;9(5):1812–1817. doi: 10.1021/nl803757u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurell T, Petersson F, Nilsson A. Chip integrated strategies for acoustic separation and manipulation of cells and particles. Chem Soc Rev. 2007;36(3):492–506. doi: 10.1039/b601326k. doi:Doi 10.1039/B601326k. [DOI] [PubMed] [Google Scholar]
- Lee H, Purdon AM, Chu V, Westervelt RM. Controlled assembly of magnetic nanoparticles from magnetotactic bacteria using microelectromagnets arrays. Nano letters. 2004;4(5):995–998. doi:Doi 10.1021/Nl049562x. [Google Scholar]
- Lenshof A, Laurell T. Continuous separation of cells and particles in microfluidic systems. Chem Soc Rev. 2010;39(3):1203–1217. doi: 10.1039/b915999c. doi:Doi 10.1039/B915999c. [DOI] [PubMed] [Google Scholar]
- Liang LT, Zhu JJ, Xuan XC. Three-dimensional diamagnetic particle deflection in ferrofluid microchannel flows. Biomicrofluidics. 2011;5(3) doi: 10.1063/1.3618737. doi:Artn 034110 Doi 10.1063/1.3618737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CX, Stakenborg T, Peeters S, Lagae L. Cell manipulation with magnetic particles toward microfluidic cytometry. J Appl Phys. 2009;105(10) doi:Artn 102014 Doi 10.1063/1.3116091. [Google Scholar]
- Liu RH, Yang JN, Lenigk R, Bonanno J, Grodzinski P. Self-contained, fully integrated biochip for sample preparation, polymerase chain reaction amplification, and DNA microarray detection. Analytical Chemistry. 2004;76(7):1824–1831. doi: 10.1021/ac0353029. doi:Doi 10.1021/Ac0353029. [DOI] [PubMed] [Google Scholar]
- Love LJ, Jansen JF, McKnight TE, Roh Y, Phelps TJ. A magnetocaloric pump for microfluidic applications. Ieee T Nanobiosci. 2004;3(2):101–110. doi: 10.1109/tnb.2004.828265. doi:Doi 10.1109/Tnb.2004.828265. [DOI] [PubMed] [Google Scholar]
- Mao L, Koser H. Overcoming the Diffusion Barrier: Ultra-Fast Micro-Scale Mixing Via Ferrofluids; 14th International Conference on Solid-State Sensors, Actuators and Microsystems; Lyon, France. 2007. pp. 1829–1832. [Google Scholar]
- Mao LD, Elborai S, He XW, Zahn M, Koser H. Direct observation of closed-loop ferrohydrodynamic pumping under traveling magnetic fields. Phys Rev B. 2011;84(10) doi:Artn 104431 Doi 10.1103/Physrevb.84.104431. [Google Scholar]
- Mao LD, Koser H. Towards ferrofluidics for mu-TAS and lab on-a-chip applications. Nanotechnology. 2006;17(4):S34–S47. doi: 10.1088/0957-4484/17/4/007. doi:Doi 10.1088/0957-4484/17/4/007. [DOI] [PubMed] [Google Scholar]
- Mihajlovic G, Aledealat K, Xiong P, Von Molnar S, Field M, Sullivan GJ. Magnetic characterization of a single superparamagnetic bead by phase-sensitive micro-Hall magnetometry. Applied Physics Letters. 2007;91(17) doi:Artn 172518 Doi 10.1063/1.2802732. [Google Scholar]
- Miller MM, Sheehan PE, Edelstein RL, Tamanaha CR, Zhong L, Bounnak S, Whitman LJ, Colton RJ. A DNA array sensor utilizing magnetic microbeads and magnetoelectronic detection. J Magn Magn Mater. 2001;225(1-2):138–144. [Google Scholar]
- Miltenyi S, Muller W, Weichel W, Radbruch A. High-Gradient Magnetic Cell-Separation with Macs. Cytometry. 1990;11(2):231–238. doi: 10.1002/cyto.990110203. [DOI] [PubMed] [Google Scholar]
- Mirica KA, Shevkoplyas SS, Phillips ST, Gupta M, Whitesides GM. Measuring Densities of Solids and Liquids Using Magnetic Levitation: Fundamentals. Journal of the American Chemical Society. 2009;131(29):10049–10058. doi: 10.1021/ja900920s. doi:Doi 10.1021/Ja900920s. [DOI] [PubMed] [Google Scholar]
- Moriarty AT, Clayton AC, Zaleski S, Henry MR, Schwartz MR, Eversole GM, Tench WD, Fatheree LA, Souers RJ, Wilbur DC. Unsatisfactory reporting rates: 2006 practices of participants in the college of american pathologists interlaboratory comparison program in gynecologic cytology. Arch Pathol Lab Med. 2009;133(12):1912–1916. doi: 10.5858/133.12.1912. doi:2008-0793-CPR1 [pii] 10.1043/1543-2165-133.12.1912. [DOI] [PubMed] [Google Scholar]
- Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450(7173):1235–1239. doi: 10.1038/nature06385. doi:nature06385 [pii] 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen NT. Micro-magnetofluidics: interactions between magnetism and fluid flow on the microscale. Microfluidics and Nanofluidics. 2012;12(1-4):1–16. doi:Doi 10.1007/S10404-011-0903-5. [Google Scholar]
- Nguyen NT, Ng KM, Huang XY. Manipulation of ferrofluid droplets using planar coils. Applied Physics Letters. 2006;89(5) doi:Artn 052509 Doi 10.1063/1.2335403. [Google Scholar]
- Odenbach S, editor. Ferrofluids: Magnetically Controllable Fluids and Their Applications. 2002 [Google Scholar]
- Pamme N. Magnetism and microfluidics. Lab Chip. 2006;6(1):24–38. doi: 10.1039/b513005k. doi:Doi 10.1039/B513005k. [DOI] [PubMed] [Google Scholar]
- Pamme N. Continuous flow separations in microfluidic devices. Lab Chip. 2007;7(12):1644–1659. doi: 10.1039/b712784g. doi:Doi 10.1039/B712784g. [DOI] [PubMed] [Google Scholar]
- Pankhurst QA, Connolly J, Jones SK, Dobson J. Applications of magnetic nanoparticles in biomedicine. Journal of Physics D: Applied Physics. 2003;(13):R167. [Google Scholar]
- Peyman SA, Iwan EY, Margarson O, Iles A, Pamme N. Diamagnetic repulsion-A versatile tool for label-free particle handling in microfluidic devices. J Chromatogr A. 2009;1216(52):9055–9062. doi: 10.1016/j.chroma.2009.06.039. doi:Doi 10.1016/J.Chroma.2009.06.039. [DOI] [PubMed] [Google Scholar]
- Rife JC, Miller MM, Sheehan PE, Tamanaha CR, Tondra M, Whitman LJ. Design and performance of GMR sensors for the detection of magnetic microbeads in biosensors. Sensors and Actuators a-Physical. 2003;107(3):209–218. doi:Doi 10.1016/S0924-4247(03)00380-7. [Google Scholar]
- Rodriguez-Villarreal AI, Tarn MD, Madden LA, Lutz JB, Greenman J, Samitier J, Pamme N. Flow focussing of particles and cells based on their intrinsic properties using a simple diamagnetic repulsion setup. Lab on a Chip. 2011;11(7):1240–1248. doi: 10.1039/c0lc00464b. doi:Doi 10.1039/C0lc00464b. [DOI] [PubMed] [Google Scholar]
- Rosensweig RE. Ferrohydrodynamics. Cambridge University Press; Cambridge: 1985. [Google Scholar]
- Shen F, Hwang H, Hahn YK, Park J-K. Label-Free Cell Separation Using a Tunable Magnetophoretic Repulsion Force. Analytical Chemistry. 2012 doi: 10.1021/ac201505j. doi:10.1021/ac201505j. [DOI] [PubMed] [Google Scholar]
- Shevkoplyas SS, Siegel AC, Westervelt RM, Prentiss MG, Whitesides GM. The force acting on a superparamagnetic bead due to an applied magnetic field. Lab Chip. 2007;7(10):1294–1302. doi: 10.1039/b705045c. doi:Doi 10.1039/B705045c. [DOI] [PubMed] [Google Scholar]
- Shi JJ, Huang H, Stratton Z, Huang YP, Huang TJ. Continuous particle separation in a microfluidic channel via standing surface acoustic waves (SSAW) Lab on a Chip. 2009;9(23):3354–3359. doi: 10.1039/b915113c. doi:Doi 10.1039/B915113c. [DOI] [PubMed] [Google Scholar]
- Sun Y, Kwok YC, Nguyen NT. A circular ferrofluid driven microchip for rapid polymerase chain reaction. Lab on a Chip. 2007;7(8):1012–1017. doi: 10.1039/b700575j. doi:Doi 10.1039/B700575j. [DOI] [PubMed] [Google Scholar]
- Sun Y, Nguyen NT, Kwok YC. High-throughput polymerase chain reaction in parallel circular loops using magnetic actuation. Analytical Chemistry. 2008;80(15):6127–6130. doi: 10.1021/ac800787g. doi:Doi 10.1021/Ac800787g. [DOI] [PubMed] [Google Scholar]
- Toner M, Irimia D. Blood-on-a-chip. Annu Rev Biomed Eng. 2005;7:77–103. doi: 10.1146/annurev.bioeng.7.011205.135108. doi:Doi 10.1146/Annurev.Bioeng.7.011205.135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui H, Ho CM. Cell separation by non-inertial force fields in microfluidic systems. Mechanics Research Communications. 2009;36(1):92–103. doi: 10.1016/j.mechrescom.2008.08.006. doi:Doi 10.1016/J.Mechrescom.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voldman J. Electrical forces for microscale cell manipulation. Annu Rev Biomed Eng. 2006;8:425–454. doi: 10.1146/annurev.bioeng.8.061505.095739. doi:Doi 10.1146/Annurev.Bioeng.8.061505.095739. [DOI] [PubMed] [Google Scholar]
- Wang ZC, Zhe JA. Recent advances in particle and droplet manipulation for lab-on-a-chip devices based on surface acoustic waves. Lab on a Chip. 2011;11(7):1280–1285. doi: 10.1039/c0lc00527d. doi:Doi 10.1039/C0lc00527d. [DOI] [PubMed] [Google Scholar]
- Winkleman A, Perez-Castillejos R, Gudiksen KL, Phillips ST, Prentiss M, Whitesides GM. Density-based diamagnetic separation: Devices for detecting binding events and for collecting unlabeled diamagnetic particles in paramagnetic solutions. Analytical Chemistry. 2007;79(17):6542–6550. doi: 10.1021/ac070500b. doi:Doi 10.1021/Ac070500b. [DOI] [PubMed] [Google Scholar]
- Yamada M, Nakashima M, Seki M. Pinched flow fractionation: Continuous size separation of particles utilizing a laminar flow profile in a pinched microchannel. Analytical Chemistry. 2004;76(18):5465–5471. doi: 10.1021/ac049863r. doi:Doi 10.1021/Ac049863r. [DOI] [PubMed] [Google Scholar]
- Yellen BB, Hovorka O, Friedman G. Arranging matter by magnetic nanoparticle assemblers. P Natl Acad Sci USA. 2005;102(25):8860–8864. doi: 10.1073/pnas.0500409102. doi:Doi 10.1073/Pnas.0500409102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating tumor cells: approaches to isolation and characterization. J Cell Biol. 2011;192(3):373–382. doi: 10.1083/jcb.201010021. doi:jcb.201010021 [pii] 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung CW, Fiering J, Mueller AJ, Ingber DE. Micromagnetic-microfluidic blood cleansing device. Lab Chip. 2009;9(9):1171–1177. doi: 10.1039/b816986a. doi:Doi 10.1039/B816986a. [DOI] [PubMed] [Google Scholar]
- Zborowski M, Ostera GR, Moore LR, Milliron S, Chalmers JJ, Schechter AN. Red blood cell magnetophoresis. Biophys J. 2003;84(4):2638–2645. doi: 10.1016/S0006-3495(03)75069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Liang QL, Ai XN, Hu P, Wang YM, Luo GA. Comprehensive Two-Dimensional Manipulations of Picoliter Microfluidic Droplets Sampled from Nanoliter Samples. Analytical Chemistry. 2011a;83(20):8029–8034. doi: 10.1021/ac2017458. doi:Doi 10.1021/Ac2017458. [DOI] [PubMed] [Google Scholar]
- Zhang K, Liang QL, Ai XN, Hu P, Wang YM, Luo GA. On-demand microfluidic droplet manipulation using hydrophobic ferrofluid as a continuous-phase. Lab on a Chip. 2011b;11(7):1271–1275. doi: 10.1039/c0lc00484g. doi:Doi 10.1039/C0lc00484g. [DOI] [PubMed] [Google Scholar]
- Zhu JJ, Liang LT, Xuan XC. On-chip manipulation of nonmagnetic particles in paramagnetic solutions using embedded permanent magnets. Microfluidics and Nanofluidics. 2012;12(1-4):65–73. doi:Doi 10.1007/S10404-011-0849-7. [Google Scholar]
- Zhu TT, Cheng R, Mao LD. Focusing microparticles in a microfluidic channel with ferrofluids. Microfluidics and Nanofluidics. 2011a;11(6):695–701. doi:Doi 10.1007/S10404-011-0835-0. [Google Scholar]
- Zhu TT, Lichlyter DJ, Haidekker MA, Mao LD. Analytical model of microfluidic transport of non-magnetic particles in ferrofluids under the influence of a permanent magnet. Microfluidics and Nanofluidics. 2011b;10(6):1233–1245. doi:Doi 10.1007/S10404-010-0754-5. [Google Scholar]
- Zhu TT, Marrero F, Mao LD. Continuous separation of non-magnetic particles inside ferrofluids. Microfluidics and Nanofluidics. 2010;9(4-5):1003–1009. [Google Scholar]