Abstract
Hepatitis C virus (HCV) cell entry is a complex, multistep process requiring numerous host cell factors, including the tight junction protein claudin-1 (CLDN1). It is not known whether CLDN1 and the HCV glycoproteins physically interact. Therefore, the focus of this work was to study genetic interactions between CLDN1 and HCV. We used CRISPR technology to generate CLDN1 knockout (KO) Huh-7.5 cells, which could not be infected by genotype 2a Jc1 HCV unless CLDN1 expression was restored. Passage of Jc1-transfected CLDN1 KO cells resulted in the selection of a virus that could infect these cells. This virus encoded a single mutation, H316N (numbered relative to the HCV polyprotein), in the E1 glycoprotein. Whereas Jc1 H316N efficiently infected cells lacking CLDN1, such infection was blocked by an antibody targeting CLDN6, another member of the claudin family that is expressed in these cells. Furthermore, HuH6 cells, which express CLDN6, but not CLDN1, were infectable only with the mutant virus. Thus, this mutant virus adapted to the loss of CLDN1 by developing the capacity to utilize other CLDNs. Indeed, CLDN1/CLDN6 double-KO Huh-7.5 cells supported infection by the mutant virus only when CLDN1, CLDN6, or CLDN9 was expressed. Finally, this phenotype was not genotype dependent, given that the H316N mutation rendered a Japanese fulminant hepatitis 1 chimeric HCV genome encoding the genotype 5a glycoproteins able to utilize CLDN6 for host cell entry.
Conclusion
These data demonstrate plasticity of HCV virus-host interactions, where a previously CLDN1-dependent virus was capable of evolving to use CLDN6. They also reveal a role for E1 in determining entry factor usage and imply a direct, physical interaction between E1 and CLDNs.
Hepatitis C virus (HCV) is a major global health problem, with more than 180 million people currently infected worldwide.1 Chronic HCV infection can result in severe liver disease, including cirrhosis and hepatocellular carcinoma, making HCV the leading cause of liver transplants in the Western hemisphere.2 HCV cell entry is a complex, multistep process requiring the two viral envelope glycoproteins, E1 and E2, and many host factors (reviewed in a previous work3). Many of these host factors cannot be classified as classical receptors because a physical association with HCV has not been demonstrated. The aim of this study was to provide genetic evidence for an interaction between the tight junction protein claudin-1 (CLDN1) and the HCV glycoproteins.
CLDN1 is an integral membrane protein with four transmembrane domains, intracellular termini, and two extracellular loops (EL1 and EL2). Residues located in EL1 modulate HCV cell-entry functionality.4 CLDN usage is also influenced by viral determinants; whereas all genotypes of the virus can use CLDN1, some HCV genotypes can also use CLDN6 and CLDN9 as HCV cell-entry factors.5-8 Physical binding between the HCV glycoproteins and CLDN1 have been difficult to explore because of the lack of purified, soluble forms of CLDN1 and the HCV E1 glycoprotein. Whereas the capacity for CLDN1 to associate with E1 or E2 has been demonstrated by coimmunoprecipitation,9 CLDN1 mutations that impair HCV cell-entry functionality have not been shown to affect such interactions, and this assay does not reveal whether HCV interactions with CLDN1 are direct or mediated through additional proteins. Thus, it remains to be determined whether CLDN1 and the HCV glycoproteins functionally interact.
To better understand how HCV uses CLDN1 to enter cells, and to provide evidence for potential physical interactions between this host protein and the virus, we sought to identify a genetic interaction between HCV and CLDN1. By selecting viruses capable of entering CLDN1 knockout (KO) cells, we identified a single-amino-acid change in HCV E1 that confers the ability of a previously solely CLDN1-dependent virus to utilize CLDN6. This genetic interaction implies a physical interaction between HCV E1 and CLDN1.
Materials and Methods
Plasmid Construction
To perform CRISPR-mediated gene KO, we generated expression plasmids encoding U6 promoter-driven CLDN1- or CLDN6-specific guide RNAs.10 Two rounds of overlapping polymerase chain reaction (PCR) were performed by amplifying a guide RNA-encoding plasmid (provided by George Church, Harvard University, Boston, MA; Addgene plasmid no. 41819): In the first round, PCR products were generated encompassing the U6 promoter into the 5′ end of the guide RNA (consisting of the specific target sequence) with the ME-O-1122 oligo (5′ CGGGCCCCCCCTCGAGTGTACAAAAAAGCAGGCT) and a CLDN1 target sequence-specific reverse oligo (ME-O-1139; 5′ GAAGGCGAGAATGAAGCCCGGTGTTTCGTCCTTTCC) or a CLDN6 target sequence-specific reverse oligo (ME-O-1342; 5′ ATGTGGAAGGTGACCGCTTTCGGTGTTTCGTCCTTTCC). PCR products were also generated encompassing a region from the CLDN1 or CLDN6 target sequence through the end of the guide RNA-coding sequence with a forward-direction CLDN1 target sequence-specific oligo (ME-O-1138; 5′ GCTTCATTCTCGCCTTCCGTTTTAGAGCTAGAAATA) or a forward-direction CLDN6 target sequence-specific oligo (ME-O-1341; 5′ AAAGCGGTCACCTTCCACATGTTTTAGAGCTAGAAATA) and a guide RNA-specific reverse oligo ME-O-1123 (5′ CGGGCTGCAGGAATTCTAATGCCAACTTTGTACA). These products were then reamplified with only the outside oligos, ME-O-1122 and –1123, to produce single PCR products flanked by XhoI and EcoRI sites at the 5′ and 3′ ends, respectively, that were cloned into pBlueScript at these sites.
As previously described,11 all CLDN proteins were expressed by lentiviral transduction from the context of pTRIP,12,13 which is a self-inactivating lentiviral provirus that expresses no HIV proteins, but instead employs an internal cytomegalovirus promoter to express cloned genes. TRIP-green fluorescent protein (GFP)-CLDN3 has been previously described.4 The parental TRIP-GFP-linker lentiviral plasmid, in which wild-type (WT) CLDN1, CLDN1 I32M/E48K, CLDN6, and CLDN9 were cloned, has been previously described.14 The open reading frames (ORFs) of these CLDNs were PCR amplified and cloned into the TRIP-GFP-linker plasmid at the BstBI and SalI sites. To generate TRIP-GFP-CLDN1 linker, the CLDN1 ORF was amplified with forward oligo ME-O-540 (5′ TTCGAAATGGCCAACGCGGGGCTGCAG) and reverse oligo ME-O-541 (5′ GTCGACTCACACGTAGTCTTTCCCG CT). TRIP-GFP-CLDN1 I32M/E48K-linker was generated through two rounds of overlapping PCR with the outside oligos, ME-O-540 and ME-O-541, and internal oligos that encode the I32M and E48K mutations (ME-O-603; 5′ GGCGGTCACGATGTTGTCGCCGGCATAGGAGTACATCCTCCACTGGGGCAGGGCAGT and ME-O-604; 5′ GGCGACAACATCGTGACCGCCCAGGCCATGTACAAGGGGCTGTGGATGTCCTGCGTG). To generate TRIP-GFP-CLDN6-linker, the CLDN6 ORF was amplified with forward oligo ME-O-1173 (5′ CTAGCACTAGTTTCGAAATGGCCTCTGCCGGAATG) and reverse oligo ME-O-1174 (5′ TCTCGAGCTAGTCGACTCAGACGTAATTCTTGGT). To generate TRIP-GFP-CLDN9-linker, the CLDN9 ORF was amplified with forward oligo ME-O-1565 (5′ CTAGCACTAGTTTCGAAATGGCTTCGACCGGCTTAG) and reverse oligo ME-O-1566 (5′ TCTCGAGCTAGTCGACTCACACGTAGTCCCTCTTGTC).
To generate the H316N mutant nonreporter Jc1 HCV, HCV E1-E2 genes were amplified as an AfeI-BsaBI fragment from a reverse-transcriptase (RT)-PCR product containing only the H316N mutation using ME-O-956 (5′ GGCCCAACAAGATGAGCATCCATAAGC) and ME-O-1068 (5′ TATCTTCTTGCTAGCGCTGCTGTCCTGCATC). The resulting PCR product was cloned into the AfeI and BsaBI sites of a version of Jc1, termed Jc1 ABS, in which silent mutations were made to introduce a unique AfeI site at the junction of the core and E1 genes. To generate the H316N mutant Gaussia luciferase (GLuc) expressing Jc1 HCV, the mutant glycoprotein was cloned as a BspDI fragment into the reporter virus. The 5a H316N GLuc-expressing virus was generated through two rounds of overlapping PCR with the outside oligos, ME-O-1576 (5′ TGGGTAAGGTCATCGATACCCTGACGTGCGGATTCG) and ME-O-1570 (5′ ACGGGGATGAAATCGATGGATTTGGCCACGCCCCGA), and internal oligos that encode the H316N mutation (ME-O-1577; 5′ ATCACCGGCAACCGGATGGCATGGGACATGATG and ME-O-1578; 5′ TGCCATCCGGTTGCCGGTGATGTGGCCACTGTA).
All PCR-amplified sequences and cloning junctions were verified by sequence analysis.
Cell Culture and Cell Lines
293T and Huh-7.5 (provided by Charles Rice, Rockefeller University, New York, NY)15 and HuH6 cells (provided by Thomas Pietschmann, TWINCORE Center for Experimental and Clinical Infection Research Hannover, Germany) were grown in Dulbecco’s modified Eagle’s medium (Sigma-Aldrich, St. Louis, MO) with 100 U/mL of penicillin, 100 mg/mL of streptomycin (Corning Life Sciences, Tewksbury, MA), and 5% fetal bovine serum (Gibco BRL Life Technologies, Gaithersburg, MD).
CRISPR-Mediated Gene KO
Huh-7.5 cells were transiently transfected with expression plasmids encoding a human codon-optimized Cas9 protein from Streptococcus pyogenes (provided by George Church, Harvard University; Addgene plasmid no. 41815)10 and either a CLDN1- or CLDN6-specific guide RNA. Cas9-mediated cleavage was directed by the CLDN1 targeting sequence (5′ GGGCTTCATTCTCGCCTTCC) to base 44 relative to the start of the CLDN1 ORF, or by the CLDN6 targeting sequence (5′ AAAGCGGTCACCTTCCACAT) to base 124 in the CLDN6 ORF, which is located immediately before the EL1 coding sequence.
Transfected cells were passaged for 1-2 weeks to allow the turnover of previously translated target protein. Cells were then fluorescence-activated cell sorting (FACS) sorted for loss of the respective target protein after staining with either an anti-CLDN1 monoclonal antibody (mAb; clone 5.16v5) provided by Isidro Hotzel (Genentech, South San Francisco, CA)16 or an anti-CLDN6 antibody (Ab; clone 342927; R&D Systems, Minneapolis, MN) and a goat anti-human or -mouse Alexa-647 Ab (Invitrogen, Carlsbad, CA). KO efficiency was between 4% and 6%. Single-cell clones were derived by dilution cloning in 96-well plates.
Virus Generation and Infection
Lentiviral production, infection, and assays were performed as previously described.11 Plasmids encoding Jc1 genotype 2a chimeric HCV and Jc1 GLuc viruses were provided by Charles Rice (Rockefeller University; reviewed in a previous work17). The plasmid encoding genotype 5a chimeric HCV GLuc was provided by Jens Bukh (Copenhagen University Hospital, Copenhagen, Denmark).18 HCV stocks were produced as previously described.19 HCV titers were determined by nonstructural (NS)5A staining limiting dilution assay on Huh-7.5 cells, as previously described,20 and HuH6 cells by infecting 3.2 × 104 cells per 48-well tissue culture plate at a maximum multiplicity of infection (MOI) of 0.2. Staining was performed using the clone 9E10 anti-NS5A Ab (provided by Charles Rice, Rockefeller University) and a goat anti-mouse peroxidase-conjugated Ab (Invitrogen).
2′-C-methyladenosine (2′CMA)21 was provided by Timothy Tellinghuisen (Scripps Research Institute, Jupiter, FL). The anti-CD81 Ab (clone JS-81) was purchased from BD Pharmingen (San Diego, CA). The anti-CLDN6 Ab (clone 342927) was purchased from R&D Systems. HCV-GLuc and HCV nonreporter infections were performed at an MOI of 0.005 or 0.005 to 0.4, respectively. For multicycle growth curves, HCV infections were initiated by infection of 5 × 104 cells per 24-well tissue culture plate at an MOI of 0.01, and luciferase was measured in culture supernatants as previously reported.11
Immunoblotting Analysis
Immunoblotting analysis was performed as previously described,11 with primary Abs against CLDN1 (2H10D10; Invitrogen), GFP (ab290; Abcam, Cambridge, MA), CLDN6 (A-4; Santa Cruz Biotechnology, Santa Cruz, CA), and actin (AC-15; Sigma-Aldrich); horseradish peroxidase (HRP) conjugated anti-mouse or -rabbit secondary Abs (GE Healthcare, Little Chalfont, UK); and Immobilon Chemiluminescent HRP detection reagent (Millipore, Billerica, MA).
RT-PCR
Total RNA was extracted from infected cells using the PureLink RNA Mini kit (Invitrogen), according to the manufacturer’s protocol. To sequence the HCV glycoproteins, RNA was reverse transcribed with random hexamers (SuperScript III; Invitrogen), and the resulting complementary DNA was amplified with HCV specific oligos (ME-O-1270; 5′ GGTTCCCGTCCCTCTTGG and ME-O-229; 5′ GCCAGTGAGGGAATAGGTAG) that anneal in the core and p7 genes, respectively.
Statistical Analysis
Data were analyzed for statistical significance using Prism software (GraphPad Software Inc., La Jolla, CA) using an unpaired Student t test. A P value ≤0.05 was considered significant. Values in graphs represent the mean and standard error (SE) of greater than two independent experiments, each performed in triplicate or quadruplicate.
Results
Generation of CLDN1-Deficient Huh-7.5 Cells
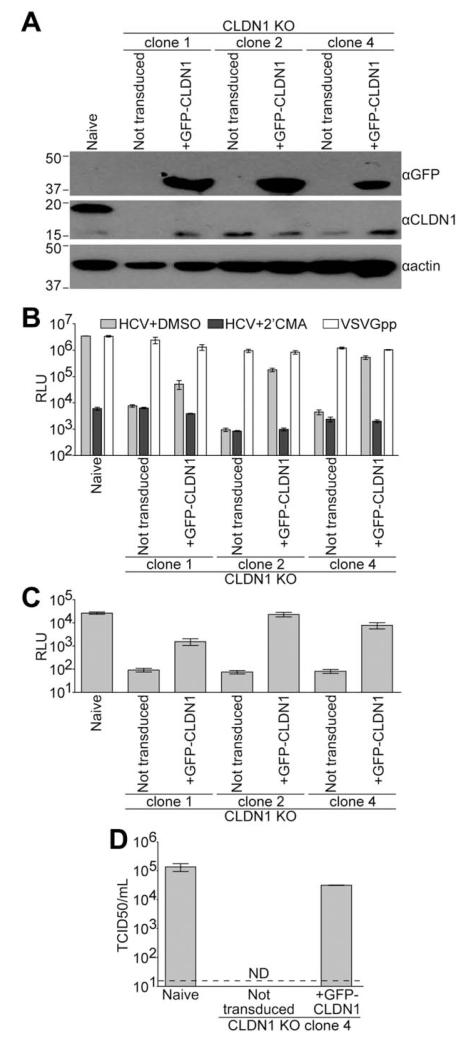
We generated Huh-7.5 CLDN1 KO single-cell clones using CRISPR technology.10,22,23 CLDN1 expression was undetectable by immunoblotting in these clones (Fig. 1A). CLDN1 expression was complemented in each clone by transduction with lentiviral vectors expressing CLDN1 fused to GFP (Fig. 1A). To determine their capacity to support HCV cell entry, non-transduced and CLDN1-complemented CLDN1 KO clones were challenged with GLuc-expressing Jc1 virus, which is derived from a genotype 2a chimeric HCV genome bearing the structural proteins of the HC-J6 isolate and the nonstructural proteins from Japanese fulminant hepatitis 1 (JFH-1).24 HCV infections were performed in the presence or absence of the HCV polymerase inhibitor, 2′CMA, to demonstrate background of the assay. Jc1 HCV infection was severely impaired in each CLDN1 KO clone, and this phenotype was restored by CLDN1 expression (Fig. 1B). As a positive control, these cells were also challenged with vesicular stomatitis virus glycoprotein-bearing lentiviral particles (VSVGpp) that also express GLuc. VSVGpp infection was not dependent on CLDN1 expression (Fig. 1B), demonstrating a specific block to HCV infection in CLDN1 KO cells that was overcome by CLDN1 complementation. To determine whether these clones supported the full HCV life cycle, supernatants from HCV infections were collected and used to infect naïve Huh-7.5 cells. All three CLDN1 KO clones were capable of producing infectious virus (Fig. 1C).
Fig. 1.
Generation of CLDN1-deficient Huh-7.5 cells. (A) Immunoblotting analysis for GFP, CLDN1, or actin was performed on lysates from naïve Huh-7.5 cells or CLDN1 KO Huh-7.5 cell clones either not expressing a transgene (not transduced) or transduced with lentiviral particles to express GFP-CLDN1. Approximate molecular mass (kDa) marker positions are indicated to the left of each blot. (B) Above cells were infected with GLuc-expressing Jc1 in the presence of DMSO (vehicle, light gray columns) or the HCV polymerase inhibitor, 2′CMA (dark gray columns). Parallel VSVGpp infections were performed as a positive control (white columns). Luciferase values are expressed as RLU. Mean and SE of two independent experiments, each performed in triplicate, are shown. (C) Naïve Huh-7.5 cells were infected with supernatants from the HCV+DMSO infections shown in (B). (D) Relative titers of non-reporter Jc1 virus were determined by limiting dilution assay on naïve and CLDN1 KO clone 4 Huh-7.5 cells either not transduced or expressing GFP-CLDN1. Values represent means and SE of three independent virus preparations and are expressed as TCID50/mL. Limit of detection is indicated by a dashed line. Abbreviations: DMSO, dimethyl sulfoxide; ND, not detected; RLU, relative light units; TCID50/mL, 50% tissue culture infectious dose per milliliter.
We chose CLDN1 KO clone 4 for our downstream experiments. To determine the absolute HCV susceptibility of CLDN1 KO clone 4 cells in a more sensitive assay, virus was titered by limiting dilution assay on these cells. Infection of CLDN1 KO clone 4 cells complemented with CLDN1 was within the range of naïve Huh-7.5 cells (Fig. 1D). However, no infectious events were detected in CLDN1 KO clone 4 cells, even at the highest concentration of inoculum tested. Thus, knocking out CLDN1 in these cells resulted in at least a 2,000-fold decrease in HCV susceptibility.
Adaptation of HCV to Infect CLDN1 KO Huh-7.5 Cells
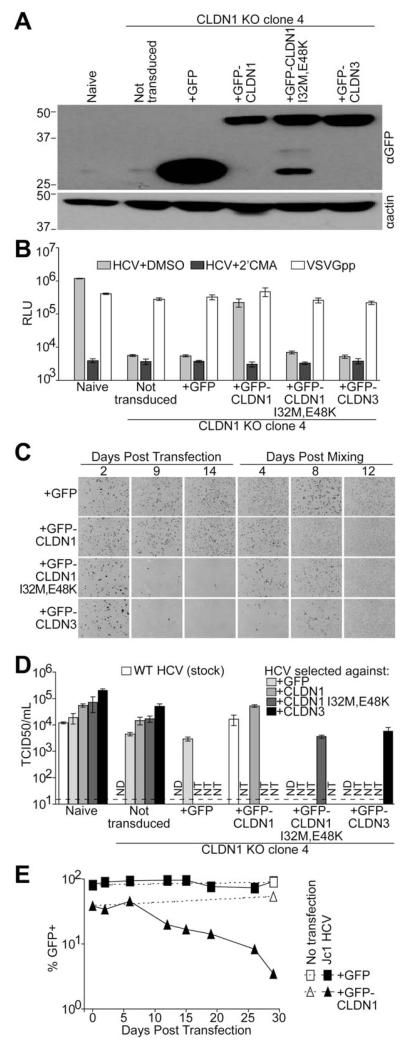
We next sought to derive HCV mutants that can infect cells expressing CLDN variants that do not efficiently mediate HCV cell entry. To provide target proteins for virus selection, CLDN1 KO clone 4 cells were transduced to express either GFP alone, GFP-CLDN1, or GFP-tagged CLDNs that have been previously demonstrated as nonfunctional HCV cell-entry factors, CLDN1 I32M/E48K and CLDN3.4 Each CLDN protein was similarly expressed, as determined by immunoblotting (Fig. 2A). As shown above, GFP-CLDN1-expressing cells supported HCV infection nearly as efficiently as naïve Huh-7.5 cells (Fig. 2B). However, CLDN1 KO clone 4 cells expressing GFP, GFP-CLDN1 I32M/E48K, or GFP-CLDN3 were poorly infectable with HCV (Fig. 2B). Again, each cell line was equally infectable with VSVGpp regardless of the protein expressed (Fig. 2B), demonstrating a specific block to HCV infection of cells expressing HCV cell-entry impaired CLDNs or GFP alone. This indicated that CLDN1 I32M/E48K and CLDN3 are suitable targets for gain-of-function experiments.
Fig. 2.
Adaptation of HCV to infect CLDN1 KO Huh-7.5 cells. (A) Immunoblotting analysis for either GFP or actin was performed on lysates from naïve or CLDN1 KO clone 4 Huh-7.5 cells either not expressing a transgene (not transduced) or transduced with lentiviral particles to express GFP alone or the indicated GFP-CLDN fusion proteins. Approximate molecular mass (kDa) marker positions are indicated to the left of each blot. (B) Above cells were infected with GLuc expressing Jc1 in the presence of DMSO (vehicle, light gray columns) or 2′CMA (dark gray columns). Parallel VSVGpp infections were performed as a positive control (white columns). Luciferase values are expressed as RLU. Mean and SE of two independent experiments, each performed in triplicate, are shown. (C) HCV persistence in serially passaged Jc1 HCV-RNA-transfected CLDN1 KO clone 4 cells expressing either GFP alone or GFP-tagged CLDNs was monitored at the indicated days post-transfection and postmixing with fresh target cells. Shown are representative fields of 80%-100% confluent monolayers of cells stained for the HCV NS5A protein (dark cells). At Day 12 postmixing essentially all cells stained NSSA positive. (D) A stock of WT Jc1, as a reference point, and supernatants containing virus passed in the cell populations indicated in the figure key were titered by limiting dilution assay on naïve Huh-7.5 cells and CLDN1 KO clone 4 cells either not transduced or expressing the indicated transgene. Means and SEs of three independent TCID50 assays are shown. Limit of detection is indicated by a dashed line. (E) CLDN1 KO clone 4 cells expressing either GFP alone or GFP-CLDN1 were either not transfected (open symbols and dotted lines) or transfected with Jc1 HCV RNA (filled symbols and solid lines). Cells were passaged and analyzed for GFP expression by FACS at the indicated days post-transfection to monitor transgene expression. Abbreviations: DMSO, dimethyl sulfoxide; ND, not detected; NT, not tested; RLU, relative light units; TCID50/mL, 50% tissue culture infectious dose per milliliter.
To adapt HCV to infect CLDN1 KO clone 4 cells expressing the transgenes described above, we used a strategy analogous to one used to select an HCV mutant that can use a wide range of species’ CD81 proteins.25 The above cell populations were transfected with Jc1 HCV RNA to bypass their respective entry blocks and establish viral RNA replication in approximately 30% of each culture (Fig. 2C, 2 days post-transfection). HCV-RNA replication is extremely error prone, with approximately one mutation per genome per round of synthesis.26 With thousands of copies of viral RNA per cell, a large pool of viable viral mutants is present in any HCV-positive cell population. The transfected cell populations were passed every 4-5 days, and HCV persistence, determined by staining for the HCV NS5A protein in parallel cultures, was used to gauge the capacity of the circulating virus to spread between these cells.
In CLDN1 KO cells complemented with WT CLDN1, viral spread was relatively efficient and thus the majority of cells became HCV positive by 9 days post-transfection (Fig. 2C). In CLDN1 I32M/E48K– and CLDN3-expressing cells, the percentage of HCV-infected cells in the population decreased over time, reflecting the inability of those proteins to support WT HCV infection. Conversely, the proportion of HCV-positive cells expressing GFP alone was similar to that of WT CLDN1 cells, at all time points. Because these cells are refractory to HCV infection, this result suggested that a viral mutation occurred early after transfection that enhanced viral persistence. At 14 days post-transfection, infected cells were mixed with uninfected CLDN1 KO clone 4 cells expressing the respective transgene to provide new target cells to amplify circulating virus. The proportion of HCV-infected cells continued to be high in CLDN1- and GFP-expressing cells and rebounded in CLDN1 I32M/E48K– and CLDN3-expressing cells (Fig. 2C), which suggested that we had selected mutant viruses capable of spreading within these cell populations.
Passaged Viruses Can Infect CLDN1-Deficient Cells
To gauge whether selection for viruses with enhanced fitness in each cell population had occurred, supernatants from infected cultures were titered by limiting dilution assay on naïve Huh-7.5, CLDN1 KO clone 4 cells, and CLDN1 KO clone 4 cells expressing the respective transgene from which each viral population was derived (Fig. 2D). As observed above, a primary stock of WT Jc1 could not infect CLDN1 KO cells expressing GFP, CLDN1 I32M/E48K, or CLDN3. Viruses passed in each cell line were able to efficiently infect both naïve Huh-7.5 cells and those expressing the respective transgene. However, passed viruses from all cultures were also able to infect untransduced CLDN1 KO cells, indicating that these viruses may not have acquired the capacity to specifically use entry-activity–impaired CLDNs, but instead adapted to infect CLDN1-deficient cells. We then sequenced the viral glycoproteins from the serially passaged viruses and identified a single conserved amino acid change, H316N, in the E1 coding sequence of all passaged viruses.
It was not immediately clear why the H316N mutation was also found in virus passed in WT GFP-CLDN1-expressing CLDN1 KO clone 4 cells. These cells are efficiently infected with WT HCV; thus, there should not be pressure to evolve viruses with altered CLDN1 usage in this cell population. Closer examination of the parent CLDN1-expressing cells showed that a substantial fraction of these cells were not transduced to express GFP-CLDN1 (Fig. 2E; GFP-CLDN1, day 0). We hypothesized that during HCV spread within these cultures, the cytotoxic effects of viral replication might have killed infected GFP-CLDN1-expressing cells, leaving untransduced CLDN1-negative cells as target cells for HCV selection. To test this hypothesis, we transfected either GFP- or GFP-CLDN1-expressing CLDN1 KO clone 4 cells with HCV RNA, passed these infected cells over the course of 4 weeks, and periodically fixed cells for subsequent FACS analysis of GFP expression. The percentage of GFP-CDLN1-expressing cells decreased after HCV transfection (Fig. 2E, solid triangles). On the other hand, the percentage of cells transduced with GFP alone was maintained throughout the course of long-term HCV infection (Fig. 2E, solid squares). At 29 days post-transfection, the H316N mutation was found in viruses passed in both GFP- and GFP-CLDN1-expressing cells. This result confirmed our hypothesis that CLDN1-expressing cells are negatively selected by HCV infection, leaving CLDN1-negative cells in the culture, thus putting pressure on the virus to evolve a CLDN1-independent cell-entry mechanism.
The H316N Mutation Alters Jc1 CLDN Usage
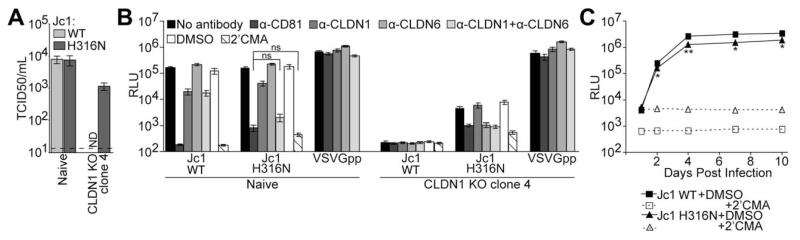
To confirm that the H316N mutation rendered HCV able to infect cells in a CLDN1-independent manner, this mutation was cloned into the parental Jc1 genome, and mutant virus stocks were produced by transfection of Huh-7.5 cells. Whereas both WT and H316N virus readily infected naïve Huh-7.5 cells, only the H316N virus infected CLDN1 KO clone 4 cells, albeit at a level that was 7-fold less than naïve Huh-7.5 cells (Fig. 3A). This result verified that the H316N mutation confers the ability of Jc1 to infect CLDN1 KO cells.
Fig. 3.
The H316N mutation renders Jc1 HCV able to infect CLDN1 KO cells. (A) Relative viral titers of nonreporter WT or H316N Jc1 were determined by limiting dilution assay on naïve and CLDN1 KO clone 4 Huh-7.5 cells. Shown are means and SEs of infections with three independent preparations of each virus. Limit of detection is indicated by a dashed line. (B) Naïve and CLDN1 KO clone 4 Huh-7.5 cells were challenged with the same number of infectious units of GLuc-expressing WT or H316N Jc1, or with VSVGpp as a negative control, in the presence or absence of the indicated inhibitors. Luciferase values are expressed as RLU. Mean and SE of two independent experiments, each performed in quadruplicate, are shown. (C) Multicycle growth analysis of GLuc-expressing WT or H316N Jc1 in naïve Huh-7.5 cells was monitored by quantifying luciferase production at the indicated days postinfection. Mean and SE of quadruplicate infections performed in the presence of 2′CMA (open symbols and dotted lines) or DMSO control (filled symbols and solid lines) are shown. Asterisks represent statistically significant differences between Jc1 WT+DMSO and Jc1 H316N+DMSO at the indicated time points. *P < 0.05; **P < 0.01 (Student t test). Abbreviations: DMSO, dimethyl sulfoxide; ND, not detected; RLU, relative light units.
We then wanted to probe the mechanism by which Jc1 H316N infected CLDN1 KO cells. We hypothesized that this mutant virus could enter cells using the CLDN6 protein, which is also expressed in our Huh-7.5 cells. As described above, whereas some HCV genotypes can use CLDN6 as a cell-entry factor, the genotype 2a glycoproteins of the Jc1 isolate can only use CLDN1.5-8 Naïve Huh-7.5 and CLDN1 KO cells were infected with either WT or H316N virus in the presence of no inhibitor, an Ab targeting the required HCV cell-entry factor CD81 as a positive entry-inhibitor control, a CLDN1 Ab, a CLDN6 Ab, both the CLDN1 and CLDN6 Abs, or the HCV polymerase inhibitor, 2′CMA. Infection of naïve Huh-7.5 cells by the WT virus was reduced 8.4-fold by the CLDN1 Ab, similar to inhibition observed with this Ab previously,27 but infection was not inhibited by the CLDN6 Ab (Fig. 3B). Combining the CLDN1 and CLDN6 Abs did not enhance inhibition, confirming that the WT virus does not utilize CLDN6 for infection. Infection of these cells by the H316N virus was reduced only 3.8-fold by the CLDN1 Ab and was not inhibited by the CLDN6 Ab. However, when the CLDN1 and CLDN6 Abs were combined, inhibition of infection was enhanced to 79-fold, a level that is not a statistically significant difference from inhibition achieved with the CD81 Ab or 2′CMA, indicating that the mutant virus requires either CLDN1 or CLDN6 for infection. As shown above, the H316N virus, but not WT Jc1, could infect CLDN1 KO clone 4 cells. Furthermore, infection of these cells with the H316N virus was not inhibited by the CLDN1 Ab, but was equally inhibited by the CD81 and CLDN6 Ab. This result provides additional evidence that the H316N mutant virus can utilize CLDN6 to enter host cells.
To assess the relative fitness of the H316N mutant virus, naïve Huh-7.5 cells were infected with the same titer of WT or H316N Jc1 reporter viruses (MOI of 0.005), and luciferase production was quantified over the course of 10 days. Both WT and H316N Jc1 infected these cells efficiently, as revealed by an increase in luciferase production over time, compared to infections performed in the presence of 2′CMA (Fig. 3C). Between 4 and 10 days postinfection, the luciferase signal from cells infected with WT virus was only 2-fold higher than in cells infected with the H316N mutant virus, indicating that the H316N virus is only slightly less fit than the WT virus.
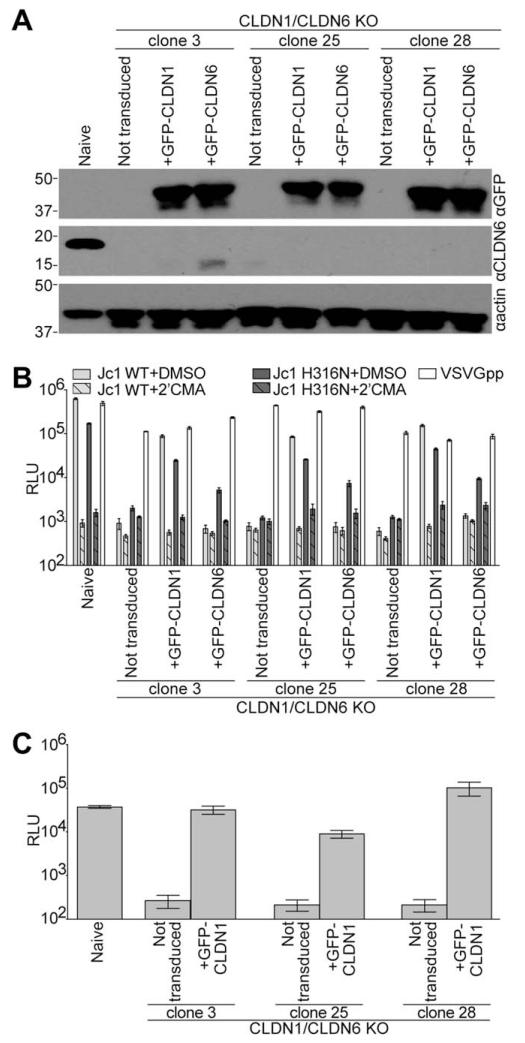
To generate a CLDN1 and CLDN6 null background to test CLDN usage by WT and H316N Jc1, we used CRISPR technology to knock out CLDN6 in the CLDN1 KO clone 4 cells. We chose three single-cell clones in which CLDN6 expression was undetectable by immunoblotting (Fig. 4A). These CLDN1/CLDN6 KO cells were highly impaired for infection with either WT or H316N Jc1 reporter viruses (Fig. 4B, not transduced). These cells were transduced with lentiviruses to express either GFP-CLDN1 or GFP-CLDN6 (Fig. 4A). GFP-CLDN1 expression rendered these cells susceptible to infection with both viruses (Fig. 4B). Conversely, GFP-CLDN6 transduction only enhanced infection with the H316N virus. Similar to the results shown above, H316N Jc1 infection of GFP-CLDN6-expressing cells was 3.5- to 4.8-fold less efficient than when GFP-CLDN1 was present. VSVGpp infection was not influenced by CLDN KO or by expression of either GFP-CLDN transgene (Fig 4B, white filled bars). To determine whether these clones supported the full HCV life cycle, supernatants from WT HCV infections of naïve Huh-7.5 cells and CLDN1/CLDN6 KO clones complemented with CLDN1 were collected and used to infect naïve Huh-7.5 cells. All three CLDN1/CLDN6 KO clones were capable of producing infectious virus (Fig. 4C).
Fig. 4.
The H316N mutation in Jc1 alters CLDN usage. (A) Immunoblotting analysis for either GFP, CLDN6, or actin was performed on lysates from naïve Huh-7.5 cells or CLDN1/CLDN6 KO clones either not expressing a transgene (not transduced) or transduced with lentiviral particles to express GFP-CLDN1 or GFP-CLDN6. Approximate molecular mass (kDa) marker positions are indicated to the left of each blot. (B) Above cells were infected with GLuc-expressing WT (light gray columns) or H316N (dark gray columns) Jc1 in the presence of DMSO (vehicle, solid columns) or 2′CMA (hashed columns). Parallel VSVGpp infections were performed as a positive control (white columns). Luciferase values are expressed as RLU. Mean and SE of two independent experiments, each performed in triplicate, are shown. (C) Naïve Huh-7.5 cells were infected with supernatants from the HCV+DMSO infections of naïve Huh-7.5 cells and not transduced or GFP-CLDN1 expressing CLDN1/CLDN6 KO cells shown in (B). Abbreviations: DMSO, dimethyl sulfoxide; RLU, relative light units.
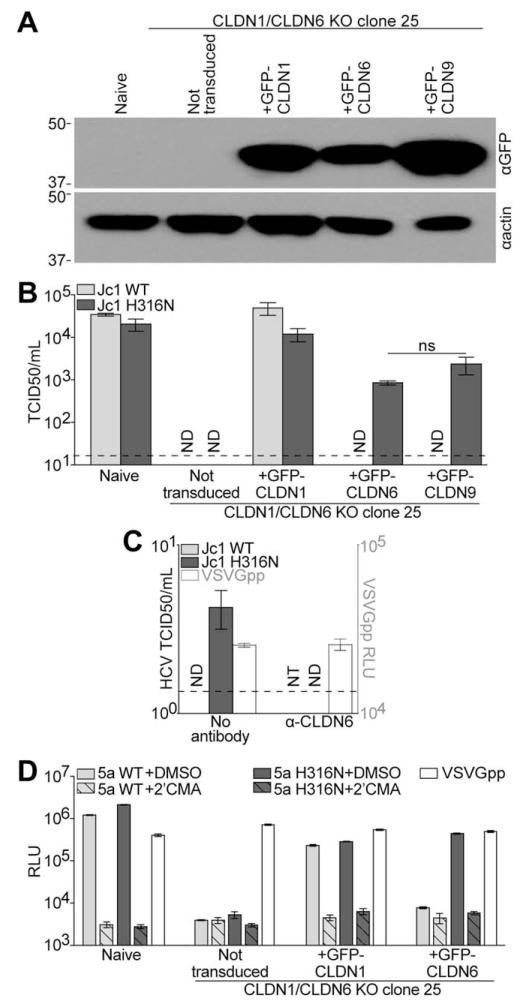
Next, we sought to probe the ability of CLDN9, the only other CLDN protein demonstrated to be a functional HCV cell-entry factor,5-7 to support infection by the mutant virus. We hypothesized that CLDN9 would also support infection by Jc1 H316N, given that CLDN6 and CLDN9 differ by only one amino acid in EL1. To test that hypothesis, CLDN1/CLDN6 KO clone 25 Huh-7.5 cells were transduced to express GFP-CLDN1, GFP-CLDN6, or GFP-CLDN9, and each CLDN protein was similarly expressed, as determined by immunoblotting (Fig. 5A). The relative titers of WT and H316N Jc1 were then determined by limiting dilution assay. No infection of either WT or H316N virus was detected in the double-KO cells (Fig. 4B). Again, expression of GFP-CLDN1 enhanced infection with both viruses. GFP-CLDN6- and GFP-CLDN9-expressing double-KO cells were only infectable with the mutant virus, although 15- and 5-fold less efficiently than in GFP-CLDN1 cells, respectively (Fig. 5B). These results show that the H316N mutant virus requires either CLDN1, CLDN6, or CLDN9 for cell entry, whereas entry of WT virus is dependent on CLDN1 expression.
Fig. 5.
The H316N mutation alters CLDN usage in other cell types and by other HCV genotypes. (A) Immunoblotting analysis for either GFP or actin was performed on lysates from naïve Huh-7.5 cells or CLDN1/CLDN6 KO clone 25 cells either not expressing a transgene (not transduced) or transduced with lentiviral particles to express GFP-CLDN1, GFP-CLDN6, or GFP-CLDN9. Approximate molecular mass (kDa) marker positions are indicated to the left of each blot. (B) Relative viral titers of nonreporter WT or H316N Jc1 were determined by limiting dilution assay on naïve Huh-7.5 cells and CLDN1/CLDN6 KO clone 25 cells either not transduced or transduced to express GFP-CLDN1, GFP-CLDN6, or GFP-CLDN9. Shown are means and SEs of infections with three independent preparations of each virus. Limit of detection is indicated by a dashed line. (C) Relative viral titers (left axis) of nonreporter WT or H316N Jc1 were determined by limiting dilution assay on naïve HuH6 cells in the presence or absence of a CLDN6 Ab. Means and SEs of three independent assays are shown. Limit of detection is indicated by a dashed line. Naïve HuH6 cells were challenged with VSVGpp in the presence or absence of the CLDN6 Ab. Luciferase values are expressed as RLU (right axis). Mean and SE of quadruplicate infections are shown. (D) CLDN1/CLDN6 KO Huh-7.5 cells were infected with GLuc-expressing WT (light gray columns) or H316N (dark gray columns) 5a HCV in the presence of DMSO (vehicle, solid columns) or 2′CMA (hashed columns). Parallel VSVGpp infections were performed as a positive control (white columns). Luciferase values are expressed as RLU. Mean and SE of two independent experiments, each performed in triplicate, are shown. Abbreviations: DMSO, dimethyl sulfoxide; ND, not detected; NT, not tested; RLU, relative light units.
To provide additional evidence for the ability of Jc1 H316N to utilize CLDN6 for host cell entry, we titered WT and H316N Jc1 on HuH6 cells, a human hepatoblastoma cell line that lacks CLDN1, but does express CLDN6.8,28 We found that HuH6 cells were not infectable with WT Jc1, as previously reported, but these cells were infectable with Jc1 H316N (Fig. 5C). This infection was 200-fold lower than Jc1 H316N infection of GFP-CLDN6-expressing CLDN1/CLDN6 KO Huh-7.5 cells, reflecting the reduced capacity for HuH6 cells to support HCV replication, compared to Huh-7.5 cells.8,28,29 When HuH6 cells were challenged with Jc1 H316N in the presence of a CLDN6 Ab, infection could no longer be observed, whereas infection of HuH6 cells with VSVGpp was not impacted by the CLDN6 Ab. This result further confirmed the ability of the mutant virus to utilize CLDN6 for host cell entry.
To assess the impact of the H316N mutation on the ability of other HCV genotypes to utilize CLDN6, this mutation was introduced into a GLuc-expressing, JFH-1-based chimeric HCV genome bearing the structural proteins of the genotype 5a S13 HCV isolate.18 As was previously reported for genotype 5a HCV,8 this virus was dependent on CLDN1 for infection and only infected CLDN1/CLDN6 KO Huh-7.5 cells when GFP-CLDN1 was expressed (Fig. 5D). Conversely, the H316N 5a reporter virus efficiently infected these cells when either GFP-CLDN1 or GFP-CLDN6 were expressed. This result demonstrated the ability of the H316N mutation to render other CLDN1-dependent HCV isolates capable of also utilizing CLDN6 for host cell entry. In contrast to Jc1 H316N, 5a H316N utilized CLDN6 and CLDN1 for infection with equal efficiency, which suggested that additional viral determinants affect the efficiency of CLDN1 usage.
Discussion
The aim of this research was to probe genetic interactions between HCV and CLDN1, which would imply and direct investigation of a physical interaction between these proteins. To achieve this goal, we first established Huh-7.5 single cell clones where CLDN1 expression was knocked out using CRISPR technology. We were unable to demonstrate any infection of these cells with a genotype 2a Jc1 HCV virus. However, when CLDN1 was reconstituted, the capacity to support HCV cell entry was restored. Furthermore, CLDN1-complemented KO clones could support HCV-RNA replication and infectious virus release, indicating that any potential CRISPR off-target effects are irrelevant to subsequent HCV experiments. This CRISPR approach was pivotal to the success of our forward genetic studies. Our earlier attempts to stably silence CLDN1 in Huh-7.5 cells by short hairpin RNA expression only partially and transiently reduced CLDN1 expression and thus resulted in just moderate reductions in HCV susceptibility (data not shown). We found that CLDN1 KO Huh-7.5 cells are at least 2,000-fold less susceptible to HCV than naïve Huh-7.5 cells, which provided a strong selection pressure for mutant viruses with altered receptor usage.
The original design of this project was to select viruses that could use either CLDN1 I32M/E48K or CLDN3 to enter CLDN1 KO cells. Instead, long-term culturing and passage of Jc1 HCV through CLDN1 KO cells resulted in the selection of a virus with a single-amino-acid change in E1 (H316N) that allowed this virus to use CLDN6 for host cell entry. These data point to a role for E1 in the HCV cell-entry process as a modulator of entry factor usage. In further experiments, we were unable to select HCV mutants that could infect CLDN1/CLDN6 KO cells expressing entry-activity–impaired CLDN1 mutants or CLDN3 (data not shown), suggesting that there are limits to the ability of HCV to evolve to overcome cell-entry barriers.
Because many HCV isolates can naturally use both CDLN1 and CLDN6 for host cell entry,5-8 it is not entirely surprising that a CLDN6 using virus was identified. Of note, the H316N mutation does not create a glycosylation consensus sequence. Whereas the number of E1 glycosylation sites varies between HCV genotypes (reviewed in a previous work30), the number of E1 glycosylation sites does not predict the ability to use CLDN6 as a cell-entry factor, suggesting that glycosylation does not play a role in determining CLDN usage. Additionally, a search of the Los Alamos National Laboratory HCV database revealed that, whereas some HCV isolates encode a glutamine residue at position 316, the amino acid present at this position does not predict the ability to use CLDN6. This indicates that we have identified a novel mechanism by which changes in the viral glycoproteins influence CLDN usage. However, residues other than that at position 316 must play a role in CLDN usage given that, unlike Jc1 H316N, 5a H316N could utilize CLDN6 as efficiently as CLDN1 for host cell entry (Fig. 5D).
Although our results imply a functional physical interaction between the HCV glycoproteins and CLDNs, we were unable to convincingly experimentally demonstrate such an interaction. Whereas we were able to show that the viral glycoproteins can coimmunoprecipitate with either CLDN1 or CLDN6, we could not demonstrate an impact of the H316N mutation in this assay (data not shown). However, our assertion that E1 determinants affect CLDN interactions is supported by research from other groups. Douam et al. recently published their finding that mutations in E1 that affect the infectivity of pseudoparticles bearing the HCV glycoproteins also modulated the binding of these particles to CLDN1-expressing cells.31 A direct, physical interaction between E1 and CLDN is the simplest, and therefore most compelling, interpretation of ours and these previous results. However, the possibility remains that E1 mutations may be functioning indirectly by influencing how E2 interacts with cell-entry factors. Furthermore, it remains possible that another host factor indirectly bridges CLDNs and the HCV glycoproteins, although it is difficult to imagine a manner in which E1 mutations, such as H316N, would impact such an indirect utilization of specific CLDNs. Purified and properly folded forms of these proteins will be required to conclusively demonstrate direct interactions between E1 and CLDNs.
Ultimately, this work demonstrates plasticity in HCV-host interactions, given that this virus was able to evolve to use a new host protein for the cell-entry process. However, the capacity of HCV to evolve to use CLDN6 or CLDN9 is likely not relevant in vivo because these proteins appear not to be expressed in hepatocytes. CLDN6 messenger RNA (mRNA) levels are very low in liver biopsies from patients chronically infected with HCV,8 as are the mRNA levels of CLDN6 and CLDN9 in primary human hepatocytes (PHHs) from healthy donors7 and in liver tissue.5 Additionally, whereas CLDN1 expression can be detected in PHHs by western blotting, CLDN6 and CLDN9 expression is not detectable.6 Furthermore, CLDN9 protein could not be detected by flow cytometry analysis of PHHs, and CLDN6 protein could not be detected by immunohistochemistry analysis of liver sections.7 Therefore, using CLDN1-blocking Abs16,32-34 to prevent HCV infection may remain a viable strategy. Ultimately, in vivo experiments will be required to conclusively demonstrate that virus cannot escape CLDN1 blockage by evolving to use CLDN6 or CLDN9.
Acknowledgments
The authors thank Matthew Chambers and Norma Hopcraft for their assistance in editing the manuscript, Maria Michta and Benjamin Israelow for their technical assistance, Tien-Huei Hsu for her technical assistance and plasmid construction, and Marion Sourisseau for her technical assistance and editing of the manuscript. The authors are grateful to George Church for providing CRISPR plasmids, to Timothy Tellinghuisen for providing 2′CMA, to Isidro Höotzel for providing the CLDN1 mAb, and to Charles Rice for providing cell lines, HCV and pseudoparticle plasmids, and the NS5A mAb. The authors also thank Thomas Pietschmann for providing HuH6 cells and Jens Bukh for providing the genotype 5a HCV plasmid.
This work was supported, in part, by award number R01DK095125 (to M.J.E.) from the National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases and R00AI077800 and R56AI091792 from the National Institutes of Health/National Institute of Allergy and Infectious Diseases (to M.J.E.), a USPHS Institutional Research Training Award AI07647 (to S.E.H.), a Research Scholar Grant (RSG-12-176-01-MPC) from the American Cancer Society, and the Pew Charitable Funds (to M.J.E.).
Abbreviations
- Ab
antibody
- CLDN1
claudin-1
- CMA
2′-C-methyladenosine
- FACS
fluorescence-activated cell sorting
- GLuc
Gaussia luciferase
- GFP
green fluorescent protein
- HCV
hepatitis C virus
- HRP
horseradish peroxidase
- JFH-1
Japanese fulminant hepatitis 1
- KO
knockout
- mAb
monoclonal antibody
- MOI
multiplicity of infection
- mRNA
messenger RNA
- NS
nonstructural
- ORF
open reading frame
- PHHs
primary human hepatocytes
- RT-PCR
reverse-transcriptase polymerase chain reaction
- SE
standard error
- VSVGpp
vesicular stomatitis virus glycoprotein-bearing lentiviral particles
- WT
wild type
Footnotes
Potential conflict of interest: Nothing to report.
References
- 1.Messina JP, Humphreys I, Flaxman A, Brown A, Cooke GS, Pybus OG, Barnes E. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology. 2015;61:77–87. doi: 10.1002/hep.27259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown RS., Jr. Hepatitis C and liver transplantation. Nature. 2005;436:973–978. doi: 10.1038/nature04083. [DOI] [PubMed] [Google Scholar]
- 3.Ploss A, Evans MJ. Hepatitis C virus host cell entry. Curr Opin Virol. 2012;2:14–19. doi: 10.1016/j.coviro.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wolk B, et al. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- 5.Zheng A, Yuan F, Li Y, Zhu F, Hou P, Li J, et al. Claudin-6 and claudin-9 function as additional coreceptors for hepatitis C virus. J Virol. 2007;81:12465–12471. doi: 10.1128/JVI.01457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meertens L, Bertaux C, Cukierman L, Cormier E, Lavillette D, Cosset FL, Dragic T. The tight junction proteins claudin-1, -6, and -9 are entry cofactors for hepatitis C virus. J Virol. 2008;82:3555–3560. doi: 10.1128/JVI.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fofana I, Zona L, Thumann C, Heydmann L, Durand SC, Lupberger J, et al. Functional analysis of claudin-6 and claudin-9 as entry factors for hepatitis C virus infection of human hepatocytes by using monoclonal antibodies. J Virol. 2013;87:10405–10410. doi: 10.1128/JVI.01691-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haid S, Grethe C, Dill MT, Heim M, Kaderali L, Pietschmann T. Isolate-dependent use of claudins for cell entry by hepatitis C virus. Hepatology. 2014;59:24–34. doi: 10.1002/hep.26567. [DOI] [PubMed] [Google Scholar]
- 9.Yang W, Qiu C, Biswas N, Jin J, Watkins SC, Montelaro RC, et al. Correlation of the tight junction-like distribution of Claudin-1 to the cellular tropism of hepatitis C virus. J Biol Chem. 2008;283:8643–8653. doi: 10.1074/jbc.M709824200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michta ML, Hopcraft SE, Narbus CM, Kratovac Z, Israelow B, Sourisseau M, Evans MJ. Species-specific regions of occludin required by hepatitis C virus for cell entry. J Virol. 2010;84:11696–11708. doi: 10.1128/JVI.01555-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sirven A, Ravet E, Charneau P, Zennou V, Coulombel L, Guetard D, et al. Enhanced transgene expression in cord blood CD34(1)-derived hematopoietic cells, including developing T cells and NOD/SCID mouse repopulating cells, following transduction with modified trip lentiviral vectors. Mol Ther. 2001;3:438–448. doi: 10.1006/mthe.2001.0282. [DOI] [PubMed] [Google Scholar]
- 13.Zennou V, Petit C, Guetard D, Nerhbass U, Montagnier L, Charneau P. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell. 2000;101:173–185. doi: 10.1016/S0092-8674(00)80828-4. [DOI] [PubMed] [Google Scholar]
- 14.Sabo MC, Luca VC, Prentoe J, Hopcraft SE, Blight KJ, Yi M, et al. Neutralizing monoclonal antibodies against hepatitis C virus E2 protein bind discontinuous epitopes and inhibit infection at a postattachment step. J Virol. 2011;85:7005–7019. doi: 10.1128/JVI.00586-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for hepatitis C virus genomic and subgenomic RNA replication. J Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hotzel I, Chiang V, Diao J, Pantua H, Maun HR, Kapadia SB. Efficient production of antibodies against a mammalian integral membrane protein by phage display. Protein Eng Des Sel. 2011;24:679–689. doi: 10.1093/protein/gzr039. [DOI] [PubMed] [Google Scholar]
- 17.Vieyres G, Pietschmann T. Entry and replication of recombinant hepatitis C viruses in cell culture. Methods. 2013;59:233–248. doi: 10.1016/j.ymeth.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 18.Gottwein JM, Scheel TK, Jensen TB, Lademann JB, Prentoe JC, Knudsen ML, et al. Development and characterization of hepatitis C virus genotype 1-7 cell culture systems: role of CD81 and scavenger receptor class B type I and effect of antiviral drugs. Hepatology. 2009;49:364–377. doi: 10.1002/hep.22673. [DOI] [PubMed] [Google Scholar]
- 19.Ploss A, Evans MJ, Gaysinskaya VA, Panis M, You H, de Jong YP, Rice CM. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457:882–886. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 21.Carroll SS, Tomassini JE, Bosserman M, Getty K, Stahlhut MW, Eldrup AB, et al. Inhibition of hepatitis C virus RNA replication by 2′-modified nucleoside analogs. J Biol Chem. 2003;278:11979–11984. doi: 10.1074/jbc.M210914200. [DOI] [PubMed] [Google Scholar]
- 22.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pietschmann T, Kaul A, Koutsoudakis G, Shavinskaya A, Kallis S, Steinmann E, et al. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc Natl Acad Sci U S A. 2006;103:7408–7413. doi: 10.1073/pnas.0504877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bitzegeio J, Bankwitz D, Hueging K, Haid S, Brohm C, Zeisel MB, et al. Adaptation of hepatitis C virus to mouse CD81 permits infection of mouse cells in the absence of human entry factors. PLoS Pathog. 2010;6:e1000978. doi: 10.1371/journal.ppat.1000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bukh J, Miller RH, Purcell RH. Genetic heterogeneity of hepatitis C virus: Quasispecies and genotypes. Semin Liver Dis. 1995;15:41–63. doi: 10.1055/s-2007-1007262. [DOI] [PubMed] [Google Scholar]
- 27.Sourisseau M, Michta ML, Zony C, Israelow B, Hopcraft SE, Narbus CM, et al. Temporal analysis of hepatitis C virus cell entry with occludin directed blocking antibodies. PLoS Pathog. 2013;9:e1003244. doi: 10.1371/journal.ppat.1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haid S, Windisch MP, Bartenschlager R, Pietschmann T. Mouse-specific residues of claudin-1 limit hepatitis C virus genotype 2a infection in a human hepatocyte cell line. J Virol. 2010;84:964–975. doi: 10.1128/JVI.01504-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chhatwal P, Bankwitz D, Gentzsch J, Frentzen A, Schult P, Lohmann V, Pietschmann T. Bile acids specifically increase hepatitis C virus RNA-replication. PLoS One. 2012;7:e36029. doi: 10.1371/journal.pone.0036029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goffard A, Dubuisson J. Glycosylation of hepatitis C virus envelope proteins. Biochimie. 2003;85:295–301. doi: 10.1016/s0300-9084(03)00004-x. [DOI] [PubMed] [Google Scholar]
- 31.Douam F, Dao Thi VL, Maurin G, Fresquet J, Mompelat D, Zeisel MB, Baumert TF, et al. Critical interaction between E1 and E2 glycoproteins determines binding and fusion properties of hepatitis C virus during cell entry. Hepatology. 2014;59:776–788. doi: 10.1002/hep.26733. [DOI] [PubMed] [Google Scholar]
- 32.Krieger SE, Zeisel MB, Davis C, Thumann C, Harris HJ, Schnober EK, et al. Inhibition of hepatitis C virus infection by anti-claudin-1 antibodies is mediated by neutralization of E2-CD81-claudin-1 associations. Hepatology. 2010;51:1144–1157. doi: 10.1002/hep.23445. [DOI] [PubMed] [Google Scholar]
- 33.Fofana I, Krieger SE, Grunert F, Glauben S, Xiao F, Fafi-Kremer S, et al. Monoclonal anti-claudin 1 antibodies prevent hepatitis C virus infection of primary human hepatocytes. Gastroenterology. 2010;139:953–964. 964.e1–4. doi: 10.1053/j.gastro.2010.05.073. [DOI] [PubMed] [Google Scholar]
- 34.Mailly L, Xiao F, Lupberger J, Wilson GK, Aubert P, Duong FH, et al. Clearance of persistent hepatitis C virus infection in humanized mice using a claudin-1-targeting monoclonal antibody. Nat Biotechnol. 2015 doi: 10.1038/nbt.3179. doi: 10.1038/nbt.3179 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]