Abstract
The fields of pharmacogenetics and pharmacogenomics have become increasingly promising regarding the clinical application of genetic data to aid in prevention of adverse reactions. Specific screening tests can predict which animals express modified proteins or genetic sequences responsible for adverse effects associated with a drug. Among the genetic variations that have been investigated in dogs, the multidrug resistance gene (MDR) is the best studied. However, other genes such as CYP1A2 and CYP2B11 control the protein syntheses involved in the metabolism of many drugs. In the present study, the MDR-1, CYP1A2 and CYP2B11 genes were examined to identify SNP polymorphisms associated with these genes in the following four canine breeds: Uruguayan Cimarron, Border Collie, Labrador Retriever and German Shepherd. The results revealed that several SNPs of the CYP1A2 and CYP2B11 genes are potential targets for drug sensitivity investigations.
Keywords: dog, drug resistance, pharmacogenomics, SNP polymorphisms
Introduction
There is increasing evidence of a clear genetic link between the phenotypic characteristics of dogs and adverse drug reactions. The fields of pharmacogenetics and pharmacogenomics have become increasingly promising regarding the clinical application of genetic data to aid in prevention of adverse reactions, prediction of the behaviour of drugs and discovery of new drug targets for study.
The introduction of a new parasiticide in the 1980s revealed a pre-existing mutation in dogs that predisposes the animals to potentially fatal neurotoxicosis. The drug, ivermectin, exerts its antiparasitic action by potentiating ligand-gated chloride ion channels in the peripheral nervous system of several invertebrate phyla [24]. The mdr1-1Δ mutation in the multidrug resistance gene (MDR-1) causes ivermectin intoxication in carriers. This mutation results in altered expression of p-glycoprotein and altered behaviour of the drug in Collies and other related dog breeds [2,16,24,28]. The MDR-1 and CYP2D15 genes have been studied in the Uruguayan Cimarron dog breed [8,9,10,11]; however, no mutations were identified in the 36 Uruguayan Cimarron animals analysed in these studies.
Several recent pharmacogenetic findings have been shown to be clinically relevant for patients of veterinary clinics, and the dog population is one of the most suitable models for examination of population genetics [15,22]. Clinically, Collies and other related dog breeds have been observed to be more susceptible to the effects of ivermectin on the central nervous system (CNS). The clinical signs of these effects include tremors, salivation, coma, depression and ataxia. Moreover, very small doses (1/100-1/200 standard) cause acute and severe reactions in some, but not all Collie breeds. Collies also exhibit adverse reactions with higher concentrations of ivermectin in the brain [2,16,22,23,24].
Approximately 75% of Collies in the United States, France and Australia have a mutant allele for the expression of modified p-glycoprotein [2,23]. Furthermore, the affected breeds have a similar lineage that includes other sheepdog breeds, such as Old English Sheepdogs, Australian Shepherds, Shelties, English Shepherds, Border Collies, German Shepherds, Longhaired Whippets and Silken Windhounds [22]. Breeds that suffer from a deletion of the MDR-1 gene are more likely to experience adverse reactions in response to low doses of the drug [22,23].
The CYP2B6 enzyme system (CYP2B6 is a member of the cytochrome P450 group of enzymes) is encoded by the CYP2B6 gene in humans and CYP2B11 in dogs [4]. This class of enzymes is responsible for the metabolism of a wide variety of drugs. While the risk of drug-drug interactions involving human CYP2B enzymes appears to be low due to minimal involvement in drug oxidation and low hepatic expression [4,26], canine CYP2B11 has exhibited surprisingly high levels of activity in vitro toward drugs that are used in dogs, such as benzodiazepines [4]. These enzymes have also been shown to be induced or inhibited by specific drugs. CYP1A11 was shown to be deficient in 10% of a small population of Beagles [4,12,26].
The CYP1A2 enzyme is a member of cytochrome P450 that is encoded by the CYP1A2 gene. Cytochrome P450 (CYP) is a superfamily of enzymes that plays an important role in the oxidative metabolism of a wide variety of xenobiotics and endogenous compounds [27]. The expression of CYP1A2 is induced by a variety of dietary constituents [4,14]. Additionally, this gene is constitutively expressed in human and dog livers and involved in the metabolism of many drugs, including caffeine, phenacetin, teophylline and tracing drugs [7,29].
Single nucleotide polymorphisms (SNPs) have been identified as deficiencies in the canine CYP1A2 gene [3,14,21,26,27,30,31]. These deficiencies have been shown to significantly alter the pharmacokinetic behaviour of two drugs and associated with large inter-individual differences in the kinetic behaviour of a third drug [3,31]. The resulting deficiency of CYP1A2 has been found to cause significant kinetic variations in AC3933, YM-64277, and a few other drugs in Beagles. However, the significance of the effects of genetic polymorphisms of other canine CYPs have not yet been fully explored [21].
In contrast, SNP genetic markers are considered to be the most recent generation of molecular markers. These markers are identified based on the substitution of one nucleotide for another. Any of the four nucleotides may be present at any position in the genome; therefore, it must be assumed that each SNP has four alleles. This is theoretically possible, but in practice, the majority of SNP variants have only two alleles, the original sequence and a single mutated version. This is because of the way in which they appear and are distributed in a population.
This study was conducted to gain further insight into the characteristics of a breed or population and identify the SNP genetic polymorphisms associated with the three genes (MDR-1, CYP1A2 and CYP2B11) involved in the metabolism of drugs used for medical treatments in several animals belonging to four different canine breeds.
As several authors have noted [19,21], further studies are required to understand the regulatory effects of the polymorphisms and their potential clinical relevance. The rationale was to identify genetic polymorphisms in the genes that encode proteins and enzymes involved in drug transport, metabolism and action that can predict the usefulness of a particular drug to increase the numbers of responders and decrease the numbers of subjects affected by adverse drug reactions.
The future of pharmacogenetics and its veterinary applications will have lasting effects on clinical decisions made in the future. The goal of drug therapy is to maximise the therapeutic effects while minimising the adverse effects associated with drugs and drug interactions.
Materials and Methods
A total of 106 different animals belonging to four different canine breeds were studied; 25 Uruguayan Cimarrons, 23 Border Collies, 29 Labrador Retrievers and 29 German Shepherds. For analysis, blood was extracted under aseptic conditions from dogs while they were with their owners in their houses. Blood extraction was performed in a fashion to cause minimal stress to the animal.
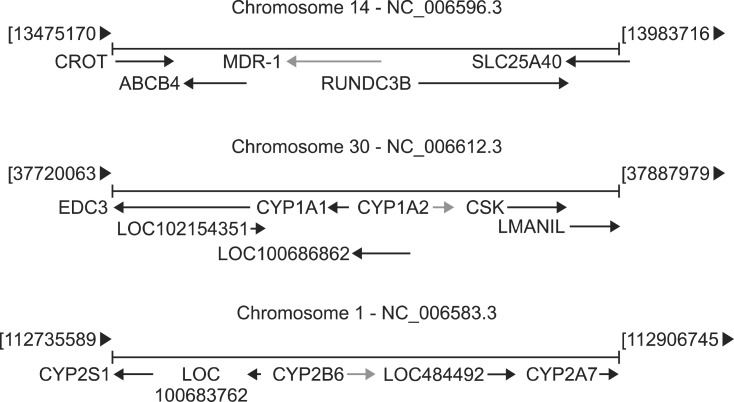
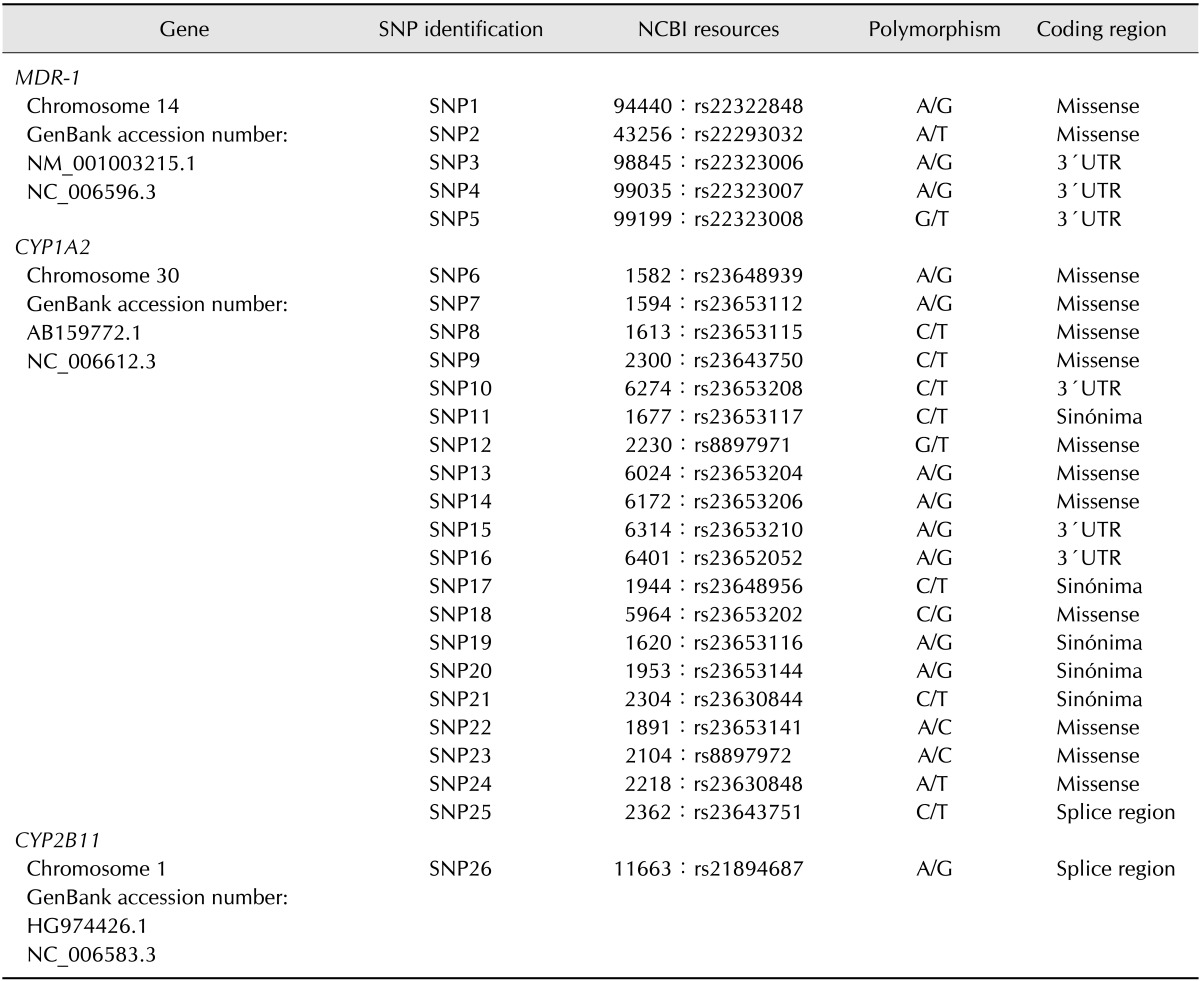
DNA was extracted from the blood samples using a DNeasy Tissue Kit (Qiagen, the Netherlands) according to the manufacturer's instructions, after which the DNA quality and purity were evaluated using a Nanodrop ND1000 spectrophotometer. Twenty-six SNPs belonging to three different genes, MDR-1 (5 SNP), CYP1A2 (20 SNP) and CYP2B11 (one SNP), were investigated according to the results obtained by several authors [18], with reference to the SNP Cluster Report of the dbSNP (NCBI) GenBank (Fig. 1 and Table 1). Samples were sent out to GeneSeek (Neogen, USA) for SNP testing. A total of 5,512 sequences were analysed in the 26 loci (two sequences for each locus in the 106 animals).
Fig. 1. Locations of the three studied genes.
Table 1. Locations and polymorphisms of the 26 SNPs studied in the three genes.
The obtained sequences were aligned using the BioEdit programme [13] and manually corrected. The resulting sequences were then analysed using the BLAST programme [1] to search for the most similar sequences in GenBank (National Center for Biotechnology Information, USA). Sequences were then aligned with closely matching sequences using the CLUSTALW programme [20]. SNP polymorphisms were analysed using GENETIX [5], ARLEQUIN [6], multivariate descriptive statistical analyses (correspondence analyses) [17] and the SNPStat programmes [25].
The analysis of correspondence [17] is an interesting method of measuring genetic diversity. This analysis allows investigation of a qualitative variable (breed) in terms of other qualitative variables (alleles) and the classical term of inertia can be assimilated into diversity. This method is conceptually similar to principal component analysis, but applies to categorical rather than continuous data. In a manner similar to principal component analysis, this method provides a means of displaying or summarising a set of data in three-dimensional graphical form. The analysis is conducted from a contingency table according to breed in which the rows are composed of the dependent variable (breed) and the columns are the set of explanatory variables (alleles). The analysis mode is multiple correspondence. The results allow acquisition of the relative importance (inertias) of both breeds and alleles. A graphical representation is created through a system of points in Euclidean space, and the degree of their robustness is verified.
The SNPStat programme [25] has been designed to conduct genetic association studies using SNPs and to analyse moderate numbers of SNPs (26 DNPs in this paper). This programme provides allele and genotype frequencies, a test for Hardy-Weinberg equilibrium, analysis of the association with a response variable based on linear or logistic regression, analysis of interactions, linkage disequilibrium statistics, haplotype frequency estimation, analysis of the associations of haplotypes with the response and analysis of interactions (haplotypes-covariate). Descriptive statistics revealed the relative frequency of each estimated haplotype. The cumulative frequency is also known to facilitate selection of the threshold cut-off point with which rare haplotypes are grouped for further analysis. The association analyses of haplotypes are similar to those of genotypes in that logistic regression results are shown as either ORs and 95% CIs or as linear regression results with differences in means and 95% CIs. The most frequent haplotype is automatically selected as the reference category and rare haplotypes are pooled together in a group. The analysis of haplotypes assumes an additive model.
Results
The 26 SNPs examined in the 106 animals exhibited different frequencies in each breed (Table 2). Each SNP was described in terms of its allelic and genotype frequencies. An exact test for Hardy-Weinberg equilibrium was performed, and it was confirmed in the Uruguayan Cimarron population. Indeed, this population was in equilibrium for all loci (SNP12, SNP14, SNP15, SNP16 and SNP18 0.05 < p < 0.10) and was therefore used as the control population. As shown in Table 2, SNP1, SNP2, SNP3, SNP4 and SNP 5 exhibited allelic fixation in the Uruguayan Cimarron and Labrador Retriever breeds. Additionally, the Cimarron population exhibited 10 loci without heterozygotes.
Table 2. Genotype frequencies in the four canine breeds studied.
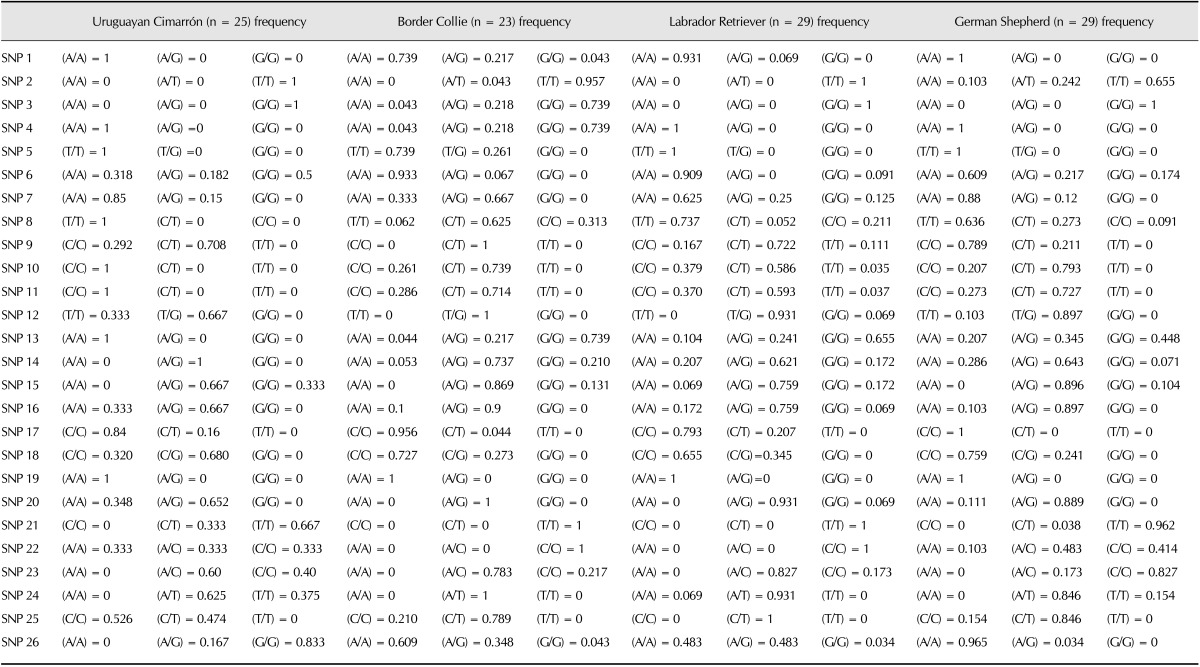
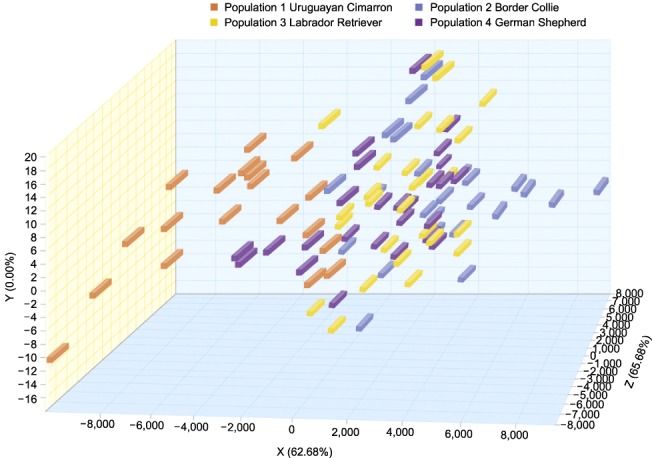
The Fis values are described in Table 3. All of the obtained Fis values were negative, indicating a lack of inbreeding in the examined animals. The genetic distances of the four studied breeds are shown in Fig. 2. The Cimarron population (Population 1) exhibited a greater genetic distance with respect to the Border Collie (Population 2), Labrador Retriever (Population 3) and German Shepherd (Population 4) breeds. Correspondence analysis indicated that the Border Collie breed exhibited 60% inertia, which was the greatest inertia observed in all four breeds, while the Uruguayan Cimarron breed exhibited the least (8%). The first axis, which explained 65.68% of the total inertia, clearly differentiated the Cimarron breed from the other breeds. Axis 2 explained only 10.42% of the total inertia, and the third axis, which explained 23.90%, did not clearly discriminate any of the breeds. All genetic distances for the animals and breeds are represented in Fig. 3. The Uruguayan Cimarron breed exhibited the greatest genetic difference with respect to the other studied breeds.
Table 3. Fis values for the different canine breeds studied.
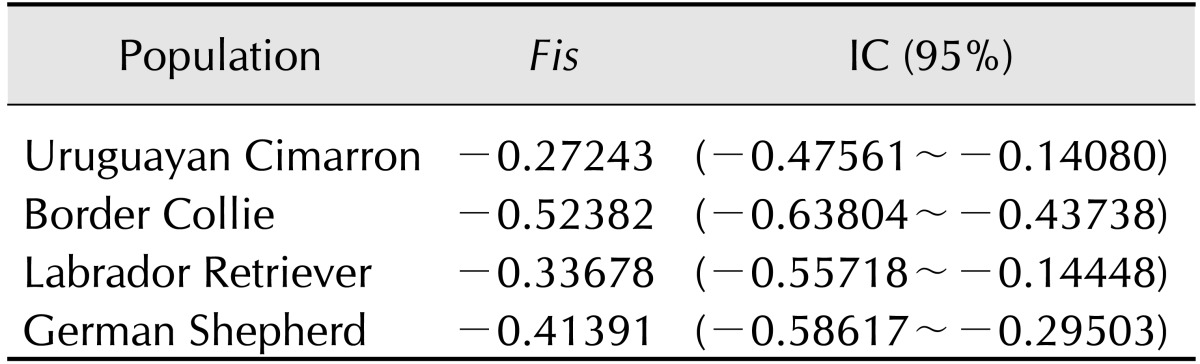
Fig. 2. Genetic distances between the four studied breeds.
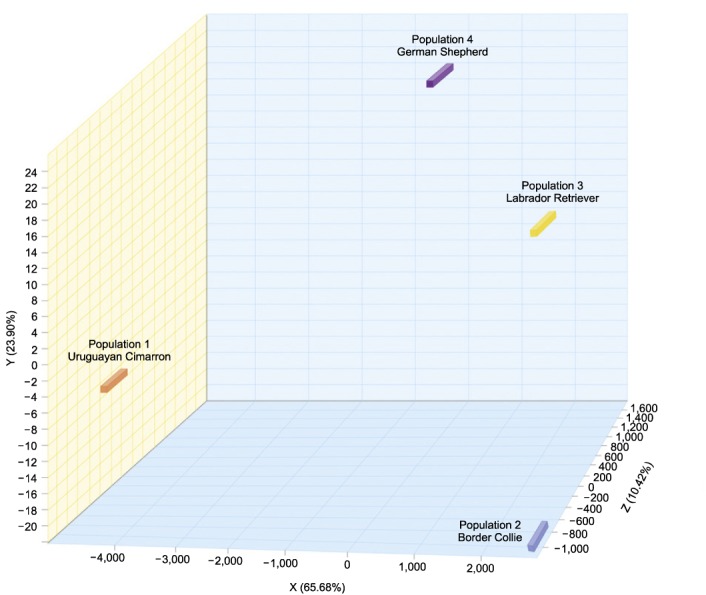
Fig. 3. Genetic distances for animals and breeds when all animals are represented.
The haplotypes for all individuals were analysed using the SNPStat programme, which enabled estimation of the haplotype frequencies. Although the number of potential combinations of haplotypes can be very high (226), the most frequent haplotypes (those with frequencies greater than 1%) included 12 in the Cimarron population, 19 in the Border Collie population, 22 in the Labrador Retriever population and 18 in the German Shepherd population. These haplotypes were compared to those of the control population, while those that exhibited frequencies below 1% are included in the rare haplotypes section. Descriptive statistics revealed the estimated relative frequency of each haplotype (Table 4). The programme selected the most frequent haplotype as the reference, while rare haplotypes were pooled together into a group. In each breed, the most frequent haplotype was termed haplotype number 1. In the Cimarron breed, 34% of the individuals exhibited haplotype number 1, while 21% exhibited haplotype number 2 and the remaining haplotypes had frequencies below 10%. Border Collies exhibited haplotype number 1 at 32%, while the remaining haplotypes occurred at frequencies below 10%. The Labrador Retriever breed exhibited haplotype number 1 at 36%, while the German Shepherd breed exhibited haplotype 1 at 16% and haplotype number 2 at 13%. The remaining haplotypes exhibited frequencies below 10%.
Table 4. Comparisons and haplotype frequencies in the breeds.
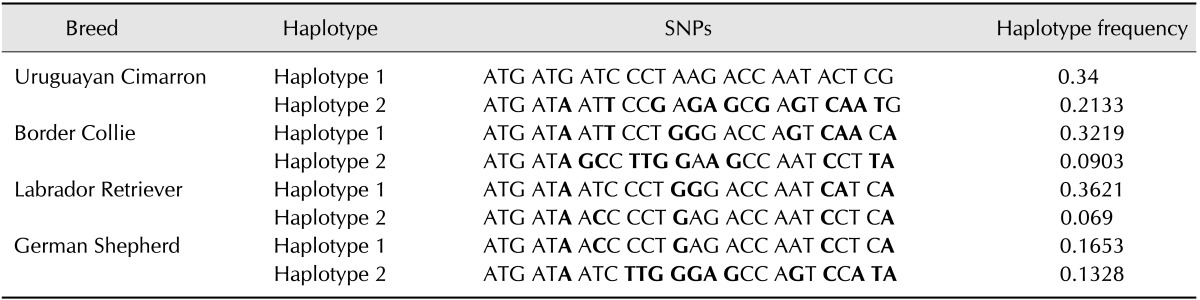
*Bold characters differ from haplotype number 1.
Analysis of the associations of each SNP were performed for the binary response variable, and logistic regression analysis provided a summary of the genotype frequencies, proportions, odds ratios and 95% confidence intervals. All of these analytical parameters were obtained for all animals and all studied loci. We used the null hypothesis of similar effects in all studied dog breeds to determine if significant differences existed among breeds. An example of locus SNP22 is shown in Table 5, in which the SNP marker exhibited a significant difference from the control population (p < 0.0001). In this case, an association with the effects on the CYP1A2 gene was shown.
Table 5. The SNP22 association with response status (n = 81, crude analysis).
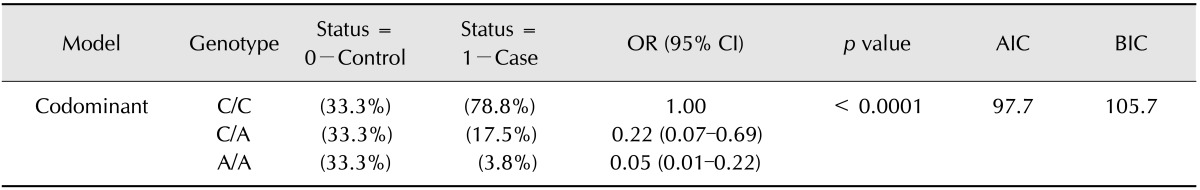
OR: odds ratios, CI: confidence interval, AIC: Akaike information criterion, BIC: Bayesian information criterion.
According to the obtained results, the following 10 of the 26 SNPs exhibited significant differences and were associated with drug sensitivity: SNP6, SNP12, SNP15, SNP16, SNP20, SNP21, SNP22, SNP24 and SNP25 of the CYP1A2 gene, and SNP26 of the CYP2B11 gene. All of these SNPs are potential candidates for studies of drug sensitivity in these breeds.
Discussion
The results obtained in this study revealed that three breeds (Border Collie, Labrador Retrievers and German Shepherd) exhibited several candidate alleles useful for investigation of genes that control drug sensitivity.
The correspondence analysis indicated that the Border Collie breed exhibited greater genetic variability and reduced inbreeding characteristics (-0.52382 Fis; Fis values range from -1 to +1). In contrast, the Uruguayan Cimarron breed exhibited less genetic variability and fewer inbreeding characteristics (-0.27243 Fis value) and was also free of inbreeding problems.
SNPs have been used as markers for diagnoses of specific features that are abundant in the genome, genetically stable and easily analysed. The underlying principle is based on identifying associations between a gene or genes that affect a variable (e.g., drug resistance) and the SNP markers. The individuals are classified by genotype for a marker. If the application of a statistical programme (i.e., SNPStat in the present study) reveals significant differences between genotypic classes, it can be concluded that there is an association between that marker and the studied characteristic. Our results indicated that the Border Collie, Labrador Retriever and German Shepherd breeds exhibited significant differences from the Uruguayan Cimarron breed in 10 SNPs of the CYP1A2 and CYP2B11 genes.
Different studies have examined the frequencies of several single nucleotide substitutions of canine genes that are associated with product resistance and drug sensitivity to survey other purebred populations that might be genetically at risk [3,24,29]. Our results are concordant with those obtained by other authors who have examined other canine breeds. The CYP1A2 gene of the Beagle breed has been examined using the cSNP method [14,21] and a non-functional allele that indicated that interindividual differences in the pharmacokinetics of CYP1A2 substrates should be examined in this breed was discovered. Additionally, one SNP identified in CYP1A2 causes a protein deletion and deficiencies in toxicological evaluations [27].
Most common diseases involve complex genetic traits, and multiple genetic and environmental components contribute to susceptibility. It has been proposed that common genetic variants, including single nucleotide polymorphisms (SNPs), influence the susceptibilities to common disease. This proposal has begun to be tested in numerous studies of the associations between the genetic variations in these common DNA polymorphisms and the variations in disease susceptibilities. Our preliminary results indicate that several polymorphisms were present in the studied genes and that these polymorphic loci are potential candidates for investigations of drug resistance. The analysis of these genetic marker SNP candidates might be very useful before subjecting animals to drugs and other medical treatments because they are easily identified, and the methodology is simple and inexpensive. The results presented herein will contribute to efficient and reliable studies of drug candidates in dogs.
Pharmacogenetics provides the opportunity to clinically practice the best possible drug therapy. This technology can detect animals that might be at risk and verify their genotypic characteristics to personalise drug therapy at the individual level rather than the population level. With advances that provide clear genetic responses, we can identify new drug targets and more precisely apply drug therapy to minimise adverse effects and maximise therapeutic benefits. Academic and clinical research efforts in pharmacogenetics and pharmacogenomics might be beneficial to the entirety of veterinary medicine in the coming years.
Acknowledgments
This work was partially supported by the Aragón Government, DGA-Fondo Social Europeo (FSE).
Footnotes
Conflict of Interest: There is no conflict of interest.
References
- 1.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alves L, Hülsmeyer V, Jaggy A, Fischer A, Leeb T, Drögemüller M. Polymorphisms in the ABCB1 gene in phenobarbital responsive and resistant idiopathic epileptic Border Collies. J Vet Intern Med. 2011;25:484–489. doi: 10.1111/j.1939-1676.2011.0718.x. [DOI] [PubMed] [Google Scholar]
- 3.Aretz JS, Geyer J. Detection of the CYP1A2 1117C > T polymorphism in 14 dog breeds. J Vet Pharmacol Ther. 2011;34:98–100. doi: 10.1111/j.1365-2885.2010.01222.x. [DOI] [PubMed] [Google Scholar]
- 4.Baratta MT, Zaya MJ, White JA, Locuson CW. Canine CYP2B11 metabolizes and is inhibited by anesthetic agents often co-administered in dogs. J Vet Pharmacol Ther. 2009;33:50–55. doi: 10.1111/j.1365-2885.2009.01101.x. [DOI] [PubMed] [Google Scholar]
- 5.Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. GENETIX 4.05, Windows TM soft for population genetics. Laboratoire Génome, Populations, Interactions, Université de Montpellier II, Montpellier: 1996-2004. [Google Scholar]
- 6.Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 7.Fontana RJ, Lown KS, Paine MF, Fortlage L, Santella RM, Felton JS, Knize MG, Greenberg A, Watkins PB. Effects of a chargrilled meat diet on expression of CYP3A, CYP1A, and P-glycoprotein levels in healthy volunteers. Gastroenterology. 1999;117:89–98. doi: 10.1016/s0016-5085(99)70554-8. [DOI] [PubMed] [Google Scholar]
- 8.Gagliardi R, García C, Llambí S, Arruga M. Analysis of mdr1-1Δ mutation of MDR-1 gene in the "Cimarron Uruguayo" dog. Rev MVZ Córdoba. 2013;18:3480–3483. [Google Scholar]
- 9.Gagliardi R, Llambí S, García C, Arruga M. Preliminary studies of the genetic structure of "Cimarron uruguayo" dog using microsatellite markers. Rev MVZ Córdoba. 2010;15:2234–2239. [Google Scholar]
- 10.Gagliardi R, Llambí S, García CB, Arruga MV. Microsatellite characterization of Cimarron Uruguayo dogs. Genet Mol Biol. 2011;34:165–168. doi: 10.1590/S1415-47572010005000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gagliardi R, Llambí S, Arruga MV. Molecular study of gene CYP2D15 (Cytochrome P450 2D15) in Cimarron Uruguayo dog. Acta Iberoam Conserv Anim. 2011;1:313–315. [Google Scholar]
- 12.Graham RA, Downey A, Mudra D, Krueger L, Carroll K, Chengelis C, Madan A, Parkinson A. In vivo and in vitro induction of cytochrome P450 enzymes in beagle dogs. Drug Metab Dispos. 2002;30:1206–1213. doi: 10.1124/dmd.30.11.1206. [DOI] [PubMed] [Google Scholar]
- 13.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 14.Kamimura H. Genetic polymorphism of cytochrome P450s in beagles: possible influence of CYP1A2 deficiency on toxicological evaluations. Arch Toxicol. 2006;80:732–738. doi: 10.1007/s00204-006-0100-6. [DOI] [PubMed] [Google Scholar]
- 15.Karriker M. Genetic predisposition to adverse reactions drug in dogs. Vet Focus (Boulogne) 2007;17:11–17. [Google Scholar]
- 16.Lankas GR, Cartwright ME, Umbenhaur D. P-glycoprotein deficiency in subpopulation of CF-1 mice enhances avermectin-induced neurotoxicity. Toxicol Appl Pharmacol. 1997;143:357–365. doi: 10.1006/taap.1996.8086. [DOI] [PubMed] [Google Scholar]
- 17.Lebart L, Morineau A, Warwick KM. Multivariate Descriptive Statistical Analysis: Correspondence Analysis and Related Techniques for Large Matrices (Probability & Mathematical Statistics) John Wiley & Sons, Chichester; 1984. [Google Scholar]
- 18.Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ, 3rd, Zody MC, Mauceli E, Xie X, Breen M, Wayne RK, Ostrander EA, Ponting CP, Galibert F, Smith DR, DeJong PJ, Kirkness E, Alvarez P, Biagi T, Brockman W, Butler J, Chin CW, Cook A, Cuff J, Daly MJ, DeCaprio D, Gnerre S, Grabherr M, Kellis M, Kleber M, Bardeleben C, Goodstadt L, Heger A, Hitte C, Kim L, Koepfli KP, Parker HG, Pollinger JP, Searle SM, Sutter NB, Thomas R, Webber C, Baldwin J, Abebe A, Abouelleil A, Aftuck L, Ait-Zahra M, Aldredge T, Allen N, An P, Anderson S, Antoine C, Arachchi H, Aslam A, Ayotte L, Bachantsang P, Barry A, Bayul T, Benamara M, Berlin A, Bessette D, Blitshteyn B, Bloom T, Blye J, Boguslavskiy L, Bonnet C, Boukhgalter B, Brown A, Cahill P, Calixte N, Camarata J, Cheshatsang Y, Chu J, Citroen M, Collymore A, Cooke P, Dawoe T, Daza R, Decktor K, DeGray S, Dhargay N, Dooley K, Dooley K, Dorje P, Dorjee K, Dorris L, Duffey N, Dupes A, Egbiremolen O, Elong R, Falk J, Farina A, Faro S, Ferguson D, Ferreira P, Fisher S, FitzGerald M, Foley K, Foley C, Franke A, Friedrich D, Gage D, Garber M, Gearin G, Giannoukos G, Goode T, Goyette A, Graham J, Grandbois E, Gyaltsen K, Hafez N, Hagopian D, Hagos B, Hall J, Healy C, Hegarty R, Honan T, Horn A, Houde N, Hughes L, Hunnicutt L, Husby M, Jester B, Jones C, Kamat A, Kanga B, Kells C, Khazanovich D, Kieu AC, Kisner P, Kumar M, Lance K, Landers T, Lara M, Lee W, Leger JP, Lennon N, Leuper L, LeVine S, Liu J, Liu X, Lokyitsang Y, Lokyitsang T, Lui A, Macdonald J, Major J, Marabella R, Maru K, Matthews C, McDonough S, Mehta T, Meldrim J, Melnikov A, Meneus L, Mihalev A, Mihova T, Miller K, Mittelman R, Mlenga V, Mulrain L, Munson G, Navidi A, Naylor J, Nguyen T, Nguyen N, Nguyen C, Nguyen T, Nicol R, Norbu N, Norbu C, Novod N, Nyima T, Olandt P, O'Neill B, O'Neill K, Osman S, Oyono L, Patti C, Perrin D, Phunkhang P, Pierre F, Priest M, Rachupka A, Raghuraman S, Rameau R, Ray V, Raymond C, Rege F, Rise C, Rogers J, Rogov P, Sahalie J, Settipalli S, Sharpe T, Shea T, Sheehan M, Sherpa N, Shi J, Shih D, Sloan J, Smith C, Sparrow T, Stalker J, Stange-Thomann N, Stavropoulos S, Stone C, Stone S, Sykes S, Tchuinga P, Tenzing P, Tesfaye S, Thoulutsang D, Thoulutsang Y, Topham K, Topping I, Tsamla T, Vassiliev H, Venkataraman V, Vo A, Wangchuk T, Wangdi T, Weiand M, Wilkinson J, Wilson A, Yadav S, Yang S, Yang X, Young G, Yu Q, Zainoun J, Zembek L, Zimmer A, Lander ES. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 19.Mancinelli L, Cronin M, Sadée W. Pharmacogenomics: the promise of personalized medicine. AAPS PharmSci. 2000;2:324–329. doi: 10.1208/ps020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, Cowley AP, Lopez R. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 2013;41(W1):W597–W600. doi: 10.1093/nar/gkt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mise M, Hashizume T, Matsumoto S, Terauchi Y, Fujii T. Identification of non-functional allelic variant of CYP1A2 in dogs. Pharmacogenetics. 2004;14:769–773. doi: 10.1097/00008571-200411000-00008. [DOI] [PubMed] [Google Scholar]
- 22.Mizukami K, Yabuki A, Chang HS, Uddin MM, Rahman MM, Kushida K, Kohyama M, Yamato O. High frequency of a single nucleotide substitution (c.-6-180T>G) of the canine MDR1/ABCB1 gene associated with phenobarbital-resistant idiopathic epilepsy in Border Collie dogs. Dis Markers. 2013;35:669–672. doi: 10.1155/2013/695918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muñana KR, Nettifee-Osborne JA, Bergman RL, Jr, Mealey KL. Association between ABCB1 genotype and seizure outcome in Collies with epilepsy. J Vet Intern Med. 2012;26:1358–1364. doi: 10.1111/j.1939-1676.2012.01006.x. [DOI] [PubMed] [Google Scholar]
- 24.Neff MW, Robertson KR, Wong AK, Safra N, Broman KW, Slatkin M, Mealey KL, Pedersen NC. Breed distribution and history of canine mdr1-1Δ, a pharmacogenetic mutation that marks the emergence of breeds from the collie lineage. Proc Natl Acad Sci U S A. 2004;101:11725–11730. doi: 10.1073/pnas.0402374101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solé X, Guinó E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22:1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 26.Talakad JC, Wilderman PR, Davydov DR, Kumar S, Halpert JR. Rational engineering of cytochromes P450 2B6 and 2B11 for enhanced stability: insights into structural importance of residue 334. Arch Biochem Biophys. 2010;494:151–158. doi: 10.1016/j.abb.2009.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tenmizu D, Endo Y, Noguchi K, Kamimura H. Identification of the novel canine CYP1A2 1117 C > T SNP causing protein deletion. Xenobiotica. 2004;34:835–846. doi: 10.1080/00498250412331285436. [DOI] [PubMed] [Google Scholar]
- 28.Tomiyasu H, Goto-Koshino Y, Fujino Y, Ohno K, Tsujimoto H. Epigenetic regulation of the ABCB1 gene in drugsensitive and drug-resistant lymphoid tumour cell lines obtained from canine patients. Vet J. 2014;199:103–109. doi: 10.1016/j.tvjl.2013.10.022. [DOI] [PubMed] [Google Scholar]
- 29.Walsky RL, Astuccio AV, Obach RS. Evaluation of 227 drugs for in vitro inhibition of cytochrome P450 2B6. J Clin Pharmacol. 2006;46:1426–1438. doi: 10.1177/0091270006293753. [DOI] [PubMed] [Google Scholar]
- 30.Whiterock VJ, Delmonte TA, Hui LE, Orcutt TL, Sinz MW. Frequency of CYP1A2 polymorphism in beagle dogs. Drug Metab Lett. 2007;1:163–165. doi: 10.2174/187231207780363688. [DOI] [PubMed] [Google Scholar]
- 31.Whiterock VJ, Morgan DG, Lentz KA, Orcutt TL, Sinz MW. Phenacetin pharmacokinetics in CYP1A2-deficient beagle dogs. Drug Metab Dispos. 2012;40:228–231. doi: 10.1124/dmd.111.041848. [DOI] [PubMed] [Google Scholar]