Abstract
The goal of this study was to gain insight into the diversity of culturable actinobacteria in desert soil crusts and to determine the physiological characteristics of the predominant actinobacterial group in these crusts. Culture-dependent method was employed to obtain actinobacterial strains from desert soil samples collected from Shapotou National Desert Ecological Reserve (NDER) located in Tengger Desert, China. A total of 376 actinobacterial strains were isolated and 16S rRNA gene sequences analysis indicated that these isolates belonged to 29 genera within 18 families, among which the members of the family Geodermatophilaceae were predominant. The combination of 16S rRNA gene information and the phenotypic data allowed these newly-isolated Geodermatophilaceae members to be classified into 33 “species clusters,” 11 of which represented hitherto unrecognized species. Fermentation broths from 19.7% of the isolated strains showed activity in at least one of the six screens for antibiotic activity. These isolates exhibited bio-diversity in enzymatic characteristics and carbon utilization profiles. The physiological characteristics of the isolates from different types of crusts or bare sand samples were specific to their respective micro-ecological environments. Our study revealed that members of the family Geodermatophilaceae were ubiquitous, abundant, and diverse in Shapotou NDER, and these strains may represent a new major group of potential functional actinobacteria in desert soil.
Keywords: Geodermatophilaceae, 16S rRNA, diversity, physiological characteristics, desert
Introduction
It has become increasingly clear that the overuse of antibiotics and the subsequent rise in antibiotic-resistant pathogens will force us to search for new antibiotics to meet urgent clinical needs (Talbot et al., 2006). Previous studies have indicated that environments considered to be extreme habitats are rich sources of novel actinobacteria (Subramani and Aalbersberg, 2013). It has been hypothesized that unusual climate conditions and ecological factors may endow the organisms in such habitats with the unique capacity to produce novel bioactive compounds (Bull et al., 2005; Okoro et al., 2008).
The Shapotou desert region (latitude 36°39′-37°41′N, elevation 104°25′-105°40′E) is recognized as the first “National Desert Ecological Reserve” (NDER) in China. This NDER is renowned worldwide as a teaching and scientific research base for studying controlled desertification. It is located on the southeast edge of the Tengger Desert, south of the Yellow River, in the northwest part of China. This region is at an altitude of 1300–1700 m, has an annual average precipitation of 186.2 mm, an annual mean temperature of 9.7°C, and an annual average wind speed of 2.8 m/s with a typical temperate desert climate.
In desert regions, microbiotic crusts play a significant role in controlling desertification by providing surface stability. Microbiotic crusts are important in stabilization of the sandy surface, soil formation, and in carbon and nitrogen assimilation (Evans and Johansen, 1999). Microbiotic crusts in Shapotou NDER are generally categorized into the following three typical types: Cyanobacteria-dominated crusts (CC), Moss-dominated crusts (MC), and Lichen-dominated crusts (LC). Samples were therefore, collected from these three types of crusts and bare sands. Culture-dependent method was employed to evaluate the diversity of culturable actinobacteria in Shapotou NDER, and to explore the potential functional actinobacterial resources from this extreme environment.
Actinobacterial strains were discovered and identified from the three types of soil crusts and bare sands samples from the Shapotou NDER. We found that the members of the family Geodermatophilaceae were ubiquitous in the different types of crusts, as well as the bare sands samples. Based on the physiological characteristics of these diverse Geodermatophilaceae members, we characterized the influence of micro-ecological niche environments on the phenotypic characteristics of these isolates.
Materials and methods
Sample collection
A total of 50 samples for isolation of actinobacteria were collected from four different micro-ecological environments in Shapotou NDER (latitude 36°39′-37°41′N, elevation 104°25′-105°40′E). The detailed information regarding the sample number, type of sample, and specific collection location of the 50 samples is displayed in Table 1. All the samples were placed in sterilized envelopes following collection and taken to the laboratory within 1 week of collection. All samples were immediately processed for isolation after arriving at the laboratory.
Table 1.
Samples collected in the Shapotou region.
| Sample number | Sample type | Site information | Sample number | Sample type | Site information | Sample number | Sample type | Site information |
|---|---|---|---|---|---|---|---|---|
| BCL12001 | BS | 37°25′38.72″N | MSY12018 | MS | 37°27′38.50″N | BSY12035 | BS | 37°27′38.66″N |
| 104°35′8.13″E | 104°59′58.79″E | 104°59′58.52″E | ||||||
| 1701 mH | 1329 mH | 1329 mH | ||||||
| MCL12002 | MS | 37°25′37.85″N | MSY12019 | MS | 37°27′38.36″N | CSY12036 | CC | 37°27′38.12″N |
| 104°35′8.26″E | 104°59′59.10″E | 104°59′58.79″E | ||||||
| 1701 mH | 1329 mH | 1329 mH | ||||||
| BCL12003 | BS | 37°25′38.89″N | MSY12020 | MS | 37°27′38.18″N | CSY12037 | CC | 37°27′37.81″N |
| 104°35′9.01″E | 104°59′59.54″E | 104°59′59.47″E | ||||||
| 1701 mH | 1329 mH | 1329 mH | ||||||
| CCL12004 | CC | 37°25′38.65″N | MSY12021 | MS | 37°27′38.04″N | CSY12038 | CC | 37°27′37.76″N |
| 104°35′7.67″E | 104°59′59.84″E | 104°59′59.72″E | ||||||
| 1701 mH | 1329 mH | 1329 mH | ||||||
| MCL12005 | MS | 37°25′39.09″N | MSY12022 | MS | 37°27′37.86″N | CSY12039 | CC | 37°27′37.60″N |
| 104°35′7.97″E | 105°00′0.17″E | 104°59′59.72″E | ||||||
| 1701 mH | 1329 mH | 1329 mH | ||||||
| CYW12006 | CC | 37°25′30.86″N | MSY12023 | MS | 37°27′37.89″N | CSY12040 | CC | 37°27′37.40″N |
| 104°43′52.00″E | 105°00′0.51″E | 104°59′59.86″E | ||||||
| 1698 mH | 1329 mH | 1329 mH | ||||||
| LYW12007 | LC | 37°25′30.76″N | MSY12024 | MS | 37°27′38.09″N | CSY12041 | CC | 37°27′37.45″N |
| 104°43′53.52″E | 105°00′0.28″E | 105°00′0.13″E | ||||||
| 1698 mH | 1329 mH | 1329 mH | ||||||
| LYW12008 | LC | 37°25′29.83″N | MSY12025 | MS | 37°27′38.29″N | BSY12042 | BS | 37°27′37.29″N |
| 104°43′53.65″E | 105°00′0.04″E | 104°59′59.86″E | ||||||
| 1698 mH | 1329 mH | 1329 mH | ||||||
| BYW12009 | BS | 37°25′30.14″N | BSY12026 | BS | 37°27′38.63″N | BSY12043 | BS | 37°27′37.32″N |
| 104°43′51.19″E | 104°59′59.68″E | 104°59′59.59″E | ||||||
| 1698 mH | 1329 mH | 1329 mH | ||||||
| MYW12010 | MS | 37°25′31.17″N | CSY12027 | CC | 37°27′38.87″N | CSY12044 | CC | 37°27′37.34″N |
| 104°43′51.13″E | 104°59′59.44″E | 104°59′59.18″E | ||||||
| 1698 mH | 1329 mH | 1329 mH | ||||||
| BYW12011 | BS | 37°25′31.35″N | MSY12028 | MS | 37°27′39.09″N | BSY12045 | BS | 37°27′37.36″N |
| 104°43′52.09″E | 104°59′59.24″E | 104°59′58.79″E | ||||||
| 1698 mH | 1329 mH | 1329 mH | ||||||
| BHW12012 | BS | 37°27′3.06″N | MSY12029 | MS | 37°27′39.16″N | CSY12046 | CC | 37°27′37.55″N |
| 104°47′41.19″E | 104°59′59.63″E | 104°59′58.97″E | ||||||
| 1619 mH | 1329 mH | 1329 mH | ||||||
| BHW12013 | BS | 37°27′3.45″N | CSY12030 | CC | 37°27′39.03″N | CSY12047 | CC | 37°27′37.76″N |
| 104°47′42.21″E | 104°59′59.89″E | 104°59′58.65″E | ||||||
| 1619 mH | 1329 mH | 1329 mH | ||||||
| MHW12014 | MS | 37°27′3.67″N | MSY12031 | MS | 37°27′38.83″N | BSY12048 | BS | 37°27′37.96″N |
| 104°47′40.92″E | 105°00′0.15″E | 104°59′58.32″E | ||||||
| 1619 mH | 1329 mH | 1329 mH | ||||||
| LHW12015 | LC | 37°27′4.05″N | BSY12032 | BS | 37°27′38.54″N | BSY12049 | BS | 37°27′37.90″N |
| 104°47′41.63″E | 105°00′0.61″E | 104°59′58.03″E | ||||||
| 1619 mH | 1329 mH | 1329 mH | ||||||
| LHW12016 | LC | 37°27′3.24″N | MSY12033 | MS | 37°27′38.38″N | CSY12050 | CC | 37°27′37.99″N |
| 104°47′41.36″E | 105°00′0.81″E | 104°59′59.11″E | ||||||
| 1619 mH | 1329 mH | 1329 mH | ||||||
| MSY12017 | MS | 37°27′38.52″N | BSY12034 | BS | 37°27′38.17″N | |||
| 104°59′59.89″E | 105°00′0.06″E | |||||||
| 1329 mH | 1329 mH |
CC, Cyanobacteria-dominated soil crusts; MC, Moss-dominated soil crusts; LC, Lichen-dominated soil crusts; BS, Bare sand.
Actinobacteria isolation and maintenance
The following four types of media were prepared to isolate the actionbacterial strains. The main components of the media were as follows: M1 (g l−1): glucose 10, yeast extract 1, beef extract 1, casein (enzymatic hydrolysate) 2, agar 15; M2 (g l−1): 1/5 strength R2A (Difco); M3 (g l−1): cellobiose 2, yeast extract 5, CaCO32, K2HPO4 1, MgSO4·7H2O 0.5, agar 15; M4 (g l−1): sodium propionate 2, NH4NO3 0.1, KCl 0.1, MgSO4·7H2O 0.05, agar 15. The isolation media were adjusted to pH 7.2–7.5 using 1 M NaOH and/or 1 M HCl. In addition, betaine (0.125% w/v), sodium pyruvate (0.125% w/v), compound trace salts solution (0.1% v/v), and compound vitamins (0.1% w/v) were added to the media to facilitate the isolation of strains that are difficult to culture (Yue et al., 2006). Aztreonam (25 mg l−1) and potassium dichromate (50 mg l−1) were also added to the media to prevent or stymie the growth of Gram-stain negative bacteria and fungi that may be present.
The procedure for actinobacteria isolation was carried out as described in Zhang et al. (2010). Briefly, 0.3 ml of 10−3 soil suspension was spread on each isolation plate and the plates were incubated at 28°C for 3 weeks. Single colonies were transferred to freshly prepared PYG plates [(g l−1) (peptone 3, yeast extract 5, glycerol 10, glycine betaine 1.25, sodium pyruvate 1.25, agar 15, pH 7.5), supplemented with compound trace salts solution (FeSO4·7 H2O 0.2 g, MnCl·2 H2O 0.1 g, ZnSO4·7 H2O 0.1 g, 0.1% v/v) and compound vitamins (vitamin B1 1 mg, vitamin B2 1 mg, vitamin B3 1 mg, vitamin B6 1 mg, phenylalanine 1 mg, biotin 1 mg, alanine 0.3 mg, 0.1% w/v)] and subsequently purified. The pure cultures were maintained on PYG slants at 4°C and also as glycerol suspensions (20%, v/v) at −80°C.
Identification of Geodermatophilaceae
Purified isolates were transferred to PYG medium and International Streptomyces Project (ISP) medium 2 for observation of the morphological characteristics. Extraction of genomic DNA and PCR amplification of the 16S rRNA gene were performed as described in the methods section of Xu et al. (2003). The purified PCR products were sequenced with an ABI PRISM automatic sequencer. The sequences obtained were compared with available 16S rRNA gene sequences from GenBank using the EzTaxon-e server (http://eztaxon-e.ezbiocloud.net; Kim et al., 2012). The server was used to determine an approximate phylogenetic affiliation of each strain. Multiple alignments with sequences of the related strains and calculations of levels of sequence similarities were carried out using MEGA version 5 (Tamura et al., 2011). A phylogenetic tree was constructed using the neighbor-joining method described in Saitou and Nei (1987). The topology of the phylogenetic tree was evaluated by the bootstrap resampling method of Felsenstein (1985) with 1000 replicates.
Bioactivity screening
Antimicrobial activities of the isolated strains were investigated by using media containing Enterococcus faecalis HH22, Klebsiella pneumonia ATCC 700603, Mycobacterium smegmatis CPCC240556, Sporobolomyces salmonicolor SS04, and Xanthomonas campestris pv. oryzae PXO99A, respectively, all at a concentration of 108 colony forming units (CFU) per ml. The anti-viral activity against the human immunodeficiency virus (HIV) was investigated using the procedure described in Yang et al. (2013). Results were considered positive if the HIV inhibition ratio was above 90% and at least 80% of the cells survived. This assay was performed under conditions where the sample concentration was 1% (v/v).
Physiological characteristics determination
From the 70 newly-isolated Geodermatophilaceae members, the physiological characteristics were determined for 34 representative strains. Carbohydrate utilization tests were carried out using API 50 CH test kits (bioMérieux) and Biolog GEN III MicroPlates (Biolog Inc.) according to the manufacturer's instructions. Enzymatic activities were determined using API ZYM test kits (bioMérieux) according to the manufacturer's instructions. Bacterial growth was tested at 4, 10, 20, 28, 30, 32, 37, 40, and 45°C on PYG agar medium incubated for 15–30 days. The ability of the strains to grow at different concentrations of NaCl was tested at the following concentrations: 0, 1, 3, and 5–20%, w/v, with 5–20% being tested at intervals of 1.0%. Growth ability in this experiment was determined according to the protocol described by Wang et al. (2001). The pH tolerance was assayed in PYG medium at pH values from 5.0 to 11.0 at intervals of 0.5 pH units. Other physiological and biochemical tests were performed according to the methods established by Williams et al. (1983) and Kämpfer et al. (1991).
The sensitivity of the bacteria to 33 different antibiotics was tested on PYG agar using the following concentrations: amikacin (1500 μg/ml), ampicillin (510 μg/ml), aztreonam (1500 μg/ml), cephalothin (1500 μg/ml), cefazolin (1500 μg/ml), cefepime (1500 μg/ml), cefoperazone (3700 μg/ml), cefotaxime (1500 μg/ml), ceftazidime (1500 μg/ml), ceftriaxone (1500 μg/ml), cefuroxime (1500 μg/ml), chloromycetin (1500 μg/ml), ciprofloxacin (250 μg/ml), clarithromycin (750 μg/ml), clindamycin (100 μg/ml), erythromycin (765 μg/ml), gentamycin (515 μg/ml), gentamycin (6000 μg/ml), levofloxacin (250 μg/ml), macrodantin (15,000 μg/ml), minocycline (1500 μg/ml), norfloxacin (500 μg/ml), ofloxacin (250 μg/ml), oxacillin (50 μg/ml), penicillin G (500 μg/ml), piperacillin (5000 μg/ml), streptomycin (540 μg/ml), streptomycin (15,000 μg/ml), sulfamethoxazole/trimethoprim (1187.5 μg/ml and 62.5 μg/ml), sulfanilamide (15,000 μg/ml), tetracycline (1500 μg/ml), tobramycin (500 μg/ml), and vancomycin (1500 μg/ml).
Numerical comparative analysis of the physiological and biochemical characteristics tested was performed using the NTSYSpc package (version 2.2 for Windows; Exeter Software) (Rohlf, 2000). A binary 0/1 matrix was created based on the positive or negative respective values of 173 physiological characteristics, some of which are described above.
Results
Isolation of actinobacteria
A total of 470 purified isolates were obtained in the present study. The 16S rRNA gene sequences revealed that 376 actinobacterial strains were isolated from the 50 samples. These isolates belonged to 18 families and 29 genera, among which the members of Geodermatophilaceae were predominant, including 70 strains of three genera. (Supplementary Figure S1). Among the four types of isolation media, M2 resulted in the most successful isolation of actinobacterial strains. Specifically, 35% of the actinobacterial strains were obtained from M2. While 29, 26, and 10% of the actinobacterial isolates were purified from M1, M4, and M3, respectively (Supplementary Figure S2).
The actinobacterial strains, measured in number of isolates per sample, accounted for 35, 30, 24, and 11%, from cyanobacteria-dominated soil crusts, lichen-dominated soil crusts, moss-dominated soil crusts, and bare sands respectively. At the genus level, the diversity of the isolates from the lichen-dominated soil crusts was higher (33%) than cyanobacteria-dominated soil crusts (30.8%) moss-dominated soil crusts (23.6%), and bare sands (12.6%).
Diversity of Geodermatophilaceae
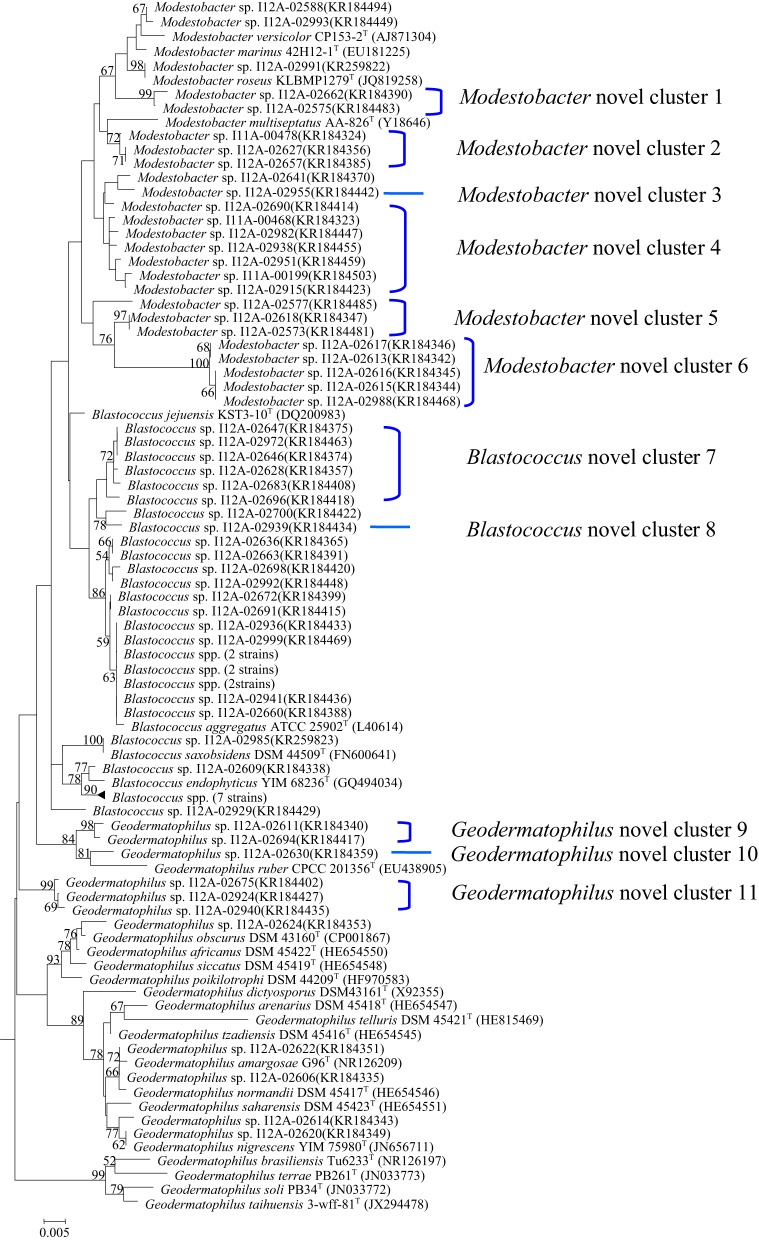
In total, 70 Geodermatophilaceae strains, including 34 Blastococcus spp., 11 Geodermatophilus spp., and 25 Modestobacter spp. were collected from the 50 samples (Table 2). In the phylogenetic dendrogram based on 16S rRNA gene sequences analysis of the isolates and the type stains of 25 validly described species in the family Geodermatophilaceae, these 70 newly-isolated members of the family Geodermatophilaceae fell into 23 “species clusters,” with the 16S rRNA gene sequence similarity below 98.65% to the closest homolog as the threshold for differentiating two species (Kim et al., 2014) (Figure 1). As indicated in the phylogenetic dendrogram, six Modestobacter “species clusters,” two Blastococcus “species clusters” and three Geodermatophilus “species clusters” may represent hitherto unrecognized species.
Table 2.
Newly-isolated Geodermatophilaceae members.
| Strain number | Accession number | The closest homolog | 16S rRNA gene similarity with the closest homolog (%) | Sample number | Sample type | Geographical location | Isolationmedium |
|---|---|---|---|---|---|---|---|
| I12A-02628 | KR184357 | Blastococcus aggregatus ATCC 25902(T) | 98.1 | MSY12029 | MC | G4 | M1 |
| I12A-02647 | KR184375 | Blastococcus aggregatus ATCC 25902(T) | 98.3 | MYW12010 | MC | G2 | M1 |
| I12A-02683 | KR184408 | Blastococcus aggregatus ATCC 25902(T) | 98.4 | CSY12047 | CC | G4 | M3 |
| I12A-02696 | KR184418 | Blastococcus aggregatus ATCC 25902(T) | 98.5 | CSY12044 | CC | G4 | M2 |
| I12A-02698 | KR184420 | Blastococcus aggregatus ATCC 25902(T) | 99.4 | CSY12040 | CC | G4 | M3 |
| I12A-02663 | KR184391 | Blastococcus aggregatus ATCC 25902(T) | 99.4 | MSY12019 | MC | G4 | M3 |
| I12A-02636 | KR184365 | Blastococcus aggregatus ATCC 25902(T) | 99.4 | MSY12028 | MC | G4 | M1 |
| I12A-02691 | KR184415 | Blastococcus aggregatus ATCC 25902(T) | 99.5 | CSY12030 | CC | G4 | M3 |
| I12A-02992 | KR184448 | Blastococcus aggregatus ATCC 25902(T) | 99.5 | CSY12040 | CC | G4 | M1 |
| I12A-02653 | KR184381 | Blastococcus aggregatus ATCC 25902(T) | 99.7 | BYW12011 | BS | G2 | M2 |
| I12A-02672 | KR184399 | Blastococcus aggregatus ATCC 25902(T) | 99.7 | CSY12038 | CC | G4 | M3 |
| I12A-02689 | KR184413 | Blastococcus aggregatus ATCC 25902(T) | 99.7 | CSY12040 | CC | G4 | M3 |
| I12A-02692 | KR184416 | Blastococcus aggregatus ATCC 25902(T) | 99.7 | CCL12004 | CC | G1 | M2 |
| I12A-02936 | KR184433 | Blastococcus aggregatus ATCC 25902(T) | 99.7 | CSY12040 | CC | G4 | M2 |
| I12A-02941 | KR184436 | Blastococcus aggregatus ATCC 25902(T) | 99.7 | CSY12040 | CC | G4 | M1 |
| I12A-02999 | KR184469 | Blastococcus aggregatus ATCC 25902(T) | 99.7 | CSY12040 | CC | G4 | M2 |
| I12A-02654 | KR184382 | Blastococcus aggregatus ATCC 25902(T) | 99.7 | LHW12015 | LC | G3 | M2 |
| I12A-02660 | KR184388 | Blastococcus aggregatus ATCC 25902(T) | 99.7 | MHW12014 | MC | G3 | M2 |
| I12A-02639 | KR184368 | Blastococcus aggregatus ATCC 25902(T) | 99.7 | MSY12025 | MC | G4 | M3 |
| I12A-02626 | KR184355 | Blastococcus aggregatus ATCC 25902(T) | 99.7 | MSY12029 | MC | G4 | M3 |
| I12A-02666 | KR184394 | Blastococcus endophyticus YIM 68236(T) | 98.9 | BHW12013 | BS | G3 | M1 |
| I12A-02971 | KR184446 | Blastococcus endophyticus YIM 68236(T) | 98.9 | MCL12005 | MC | G1 | M3 |
| I12A-02953 | KR184441 | Blastococcus endophyticus YIM 68236(T) | 99 | CSY12027 | CC | G4 | M2 |
| I12A-02649 | KR184377 | Blastococcus endophyticus YIM 68236(T) | 99.1 | BHW12012 | BS | G3 | M2 |
| I12A-02599 | KR184501 | Blastococcus endophyticus YIM 68236(T) | 99.1 | MSY12019 | MC | G4 | M1 |
| I11A-00338 | KR184318 | Blastococcus endophyticus YIM 68236(T) | 99.1 | MSY12025 | MC | G4 | M2 |
| I12A-02986 | KR184467 | Blastococcus endophyticus YIM 68236(T) | 99.2 | BSY12034 | BS | G4 | M1 |
| I12A-02609 | KR184338 | Blastococcus endophyticus YIM 68236(T) | 99.6 | LHW12015 | LC | G3 | M2 |
| I12A-02939 | KR184434 | Blastococcus jejuensis KST3-10(T) | 98.6 | CSY12040 | CC | G4 | M2 |
| I12A-02700 | KR184422 | Blastococcus jejuensis KST3-10(T) | 98.8 | CSY12040 | CC | G4 | M1 |
| I12A-02972 | KR184463 | Blastococcus jejuensis KST3-10(T) | 98.8 | CSY12040 | CC | G4 | M2 |
| I12A-02929 | KR184429 | Blastococcus jejuensis KST3-10(T) | 98.8 | MSY12029 | MC | G4 | M2 |
| I12A-02646 | KR184374 | Blastococcus jejuensis KST3-10(T) | 98.8 | MYW12010 | MC | G2 | M1 |
| I12A-02985 | KR259823 | Blastococcus saxobsidens BC448(T) | 99.7 | CSY12040 | CC | G4 | M3 |
| I12A-02622 | KR184351 | Geodermatophilus amargosae G12(T) | 99.8 | CSY12050 | CC | G4 | M1 |
| I12A-02606 | KR184335 | Geodermatophilus normandii CF5/3(T) | 99.8 | MYW12010 | MC | G2 | M3 |
| I12A-02614 | KR184343 | Geodermatophilus nigrescens YIM 75980(T) | 99.5 | CSY12030 | CC | G4 | M4 |
| I12A-02620 | KR184349 | Geodermatophilus nigrescens YIM 75980(T) | 100 | CSY12044 | CC | G4 | M2 |
| I12A-02675 | KR184402 | Geodermatophilus obscurus DSM 43160(T) | 97.8 | CSY12039 | CC | G4 | M2 |
| I12A-02940 | KR184435 | Geodermatophilus obscurus DSM 43160(T) | 97.8 | CSY12040 | CC | G4 | M1 |
| I12A-02924 | KR184427 | Geodermatophilus obscurus DSM 43160(T) | 98 | CCL12004 | CC | G1 | M2 |
| I12A-02624 | KR184353 | Geodermatophilus obscurus DSM 43160(T) | 99.1 | MSY12029 | MC | G4 | M3 |
| I12A-02694 | KR184417 | Geodermatophilus ruber CPCC 201356(T) | 97.8 | CSY12030 | CC | G4 | M3 |
| I12A-02611 | KR184340 | Geodermatophilus ruber CPCC 201356(T) | 97.8 | LHW12016 | LC | G3 | M3 |
| I12A-02630 | KR184359 | Geodermatophilus ruber CPCC 201356(T) | 98.4 | CSY12050 | CC | G4 | M1 |
| I12A-02982 | KR184447 | Modestobacter marinus 42H12-1(T) | 97.8 | CSY12040 | CC | G4 | M1 |
| I11A-00468 | KR184323 | Modestobacter marinus 42H12-1(T) | 97.8 | CSY12040 | CC | G4 | M1 |
| I12A-02690 | KR184414 | Modestobacter marinus 42H12-1(T) | 98.1 | CSY12030 | CC | G4 | M1 |
| I12A-02938 | KR184455 | Modestobacter marinus 42H12-1(T) | 98.1 | CSY12040 | CC | G4 | M1 |
| I12A-02915 | KR184423 | Modestobacter marinus 42H12-1(T) | 98.1 | MSY12017 | MC | G4 | M2 |
| I12A-02627 | KR184356 | Modestobacter marinus 42H12-1(T) | 98.2 | CSY12027 | CC | G4 | M1 |
| I12A-02662 | KR184390 | Modestobacter marinus 42H12-1(T) | 98.2 | MCL12002 | MC | G1 | M3 |
| I12A-02951 | KR184459 | Modestobacter marinus 42H12-1(T) | 98.3 | CSY12040 | CC | G4 | M1 |
| I11A-00199 | KR184503 | Modestobacter marinus 42H12-1(T) | 98.3 | CSY12050 | CC | G4 | M2 |
| I12A-02657 | KR184385 | Modestobacter marinus 42H12-1(T) | 98.6 | MHW12014 | MC | G3 | M1 |
| I12A-02575 | KR184483 | Modestobacter marinus 42H12-1(T) | 98.6 | MSY12029 | MC | G4 | M4 |
| I12A-02993 | KR184449 | Modestobacter marinus 42H12-1(T) | 99.4 | LYW12008 | LC | G2 | M2 |
| I12A-02588 | KR184494 | Modestobacter marinus 42H12-1(T) | 99.5 | BSY12032 | BS | G4 | M4 |
| I12A-02613 | KR184342 | Modestobacter multiseptatus AA-826(T) | 96.1 | CSY12040 | CC | G4 | M2 |
| I12A-02616 | KR184345 | Modestobacter multiseptatus AA-826(T) | 96.3 | CSY12040 | CC | G4 | M2 |
| I12A-02615 | KR184344 | Modestobacter multiseptatus AA-826(T) | 97 | CSY12040 | CC | G4 | M1 |
| I12A-02617 | KR184346 | Modestobacter multiseptatus AA-826(T) | 97.1 | CSY12040 | CC | G4 | M2 |
| I12A-02988 | KR184468 | Modestobacter multiseptatus AA-826(T) | 97.2 | CCL12004 | CC | G1 | M2 |
| I12A-02577 | KR184485 | Modestobacter multiseptatus AA-826(T) | 97.5 | CSY12027 | CC | G4 | M1 |
| I12A-02573 | KR184481 | Modestobacter multiseptatus AA-826(T) | 97.8 | BSY12026 | BS | G4 | M2 |
| I12A-02618 | KR184347 | Modestobacter multiseptatus AA-826(T) | 97.8 | MSY12029 | MC | G4 | M2 |
| I11A-00478 | KR184324 | Modestobacter multiseptatus AA-826(T) | 98.8 | MSY12029 | MC | G4 | M1 |
| I12A-02991 | KR259822 | Modestobacter roseus KLBMP 1279(T) | 100 | MSY12025 | MC | G4 | M4 |
| I12A-02955 | KR184442 | Modestobacter versicolor CP153-2(T) | 98.6 | MHW12014 | MC | G3 | M1 |
| I12A-02641 | KR184370 | Modestobacter versicolor CP153-2(T) | 98.8 | BYW12009 | BS | G2 | M4 |
CC, Cyanobacteria-dominated soil crusts; MC, Moss-dominated soil crusts; LC, Lichen-dominated soil crusts; BS, Bare sand. G1, 37°25′37″−37°25′40″N, 104°35′7″−104°35′10″ E, ~1700 mH; G2, 37°25′29″−37°25′32″N, 104°43′51″−104°43′54″ E, ~1700 mH; G3, 37°27′3″−37°27′5″N, 104°47′40″−104°47′43″ E, ~1620 mH; G4, 37°27′37″−37°27′40″N, 104°59′58″−105°0′1″ E, ~1330 mH.
Figure 1.
Phylogenetic dendrogram based on the 16S rRNA gene sequences of the newly-isolated strains and 25 type strains of Geodermatophilaceae. The sequence of Kineosporia aurantiaca 14067T was used as the outgroup. Numbers on branch nodes are bootstrap values. Bar, 1.0% sequence divergence.
Bioactivities of newly-isolated strains
Among the 70 Geodermatophilaceae strains, 3 exhibited activity against Enterococcus faecalis (4.3%), 3 against Klebsiella pneumonia (4.3%), 4 against Mycobacterium smegmatis (5.7%), 6 against Sporobolomyces salmonicolor (8.6%), 2 against Xanthomonas campestris pv. oryzae PXO99A (2.9%), and 6 against HIV (8.6%), respectively. Additionally, 9 of the isolates exhibited activities in more than one of these screening models. In total, 19.7% of the newly-isolated Geodermatophilaceae strains showed activity in at least one of the six antibiotic screens.
Physiological characteristics of newly-isolated strains
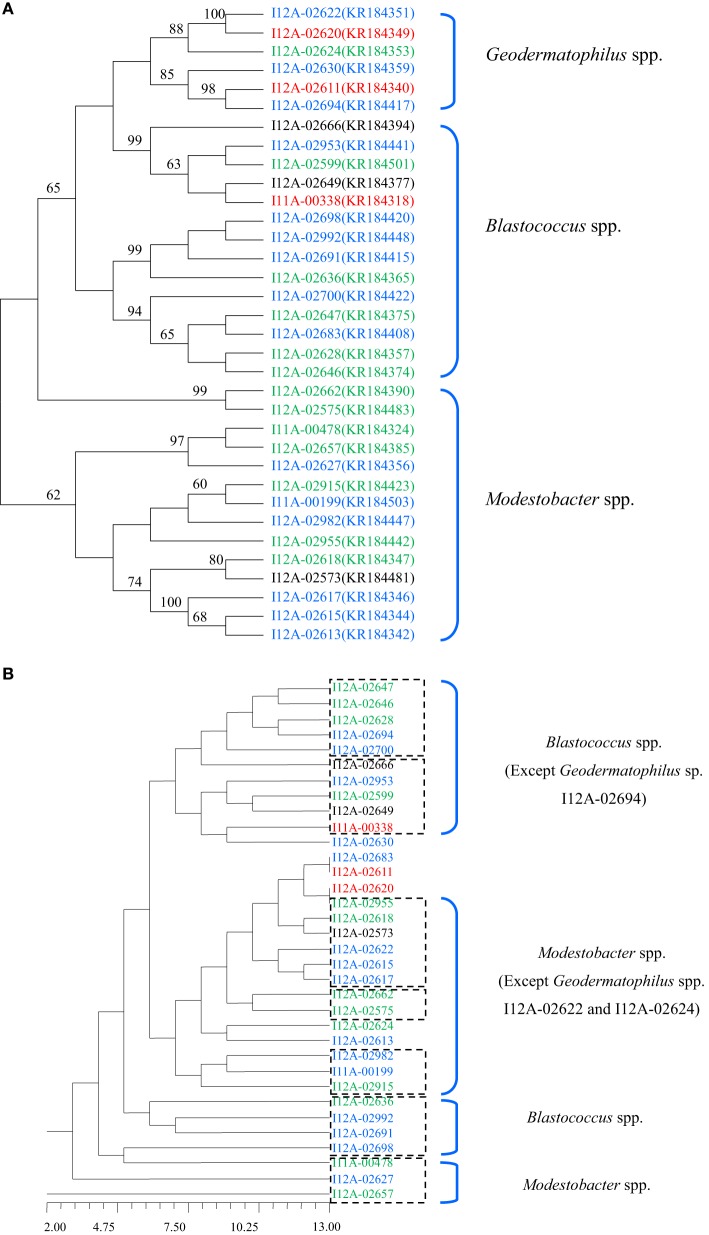
The strains assayed for physiological characteristics were similar in their physiological characteristic profiles in the following capacity: more than 60% of the strains tested could utilize dextrin, D-fructose, D-fructose-6-PO4, D-galactose, α-D-glucose, glucuronamide, α-keto-glutaric acid, D-malic acid, D-maltose, D-mannose, D-trehalose, D-turanose and sucrose as their sole carbon source, and 91% of the strains tested assimilated esculin ferric citrate and potassium 5-ketogluconate and produced acid. In the API ZYM assay, none of the strains tested was positive for β-fucosidase, N-acetyl-β-glucosaminidase, or α-mannosidase. Twenty-nine strains showed the enzymatic activities of acid phosphatase, alkaline phosphatase, esterase (C4), esterase lipase (C8), leucine arylamidase, lipase (C14), and valine arylamidase. Most of the tested strains were resistant to aztreonam (1500 μg/ml), sulfanilamide (15,000 μg/ml), and sulfamethoxazole/trimethoprim (1187.5 μg/ml and 62.5 μg/ml). The phylogenetic dendrogram based on 173 physiological characteristics of the tested strains showed that the micro-ecological environment from which the strains were isolated was an important factor correlating with the physiological characteristic profiles of the isolates. The strains exhibited characteristics specific to the micro-ecological environment where they were found (Figure 2).
Figure 2.
(A) Dendrogram based 16S rRNA gene sequences analysis of the tested strains. (B) Dendrogram based on the physiological characteristics profiles of the tested strains. Different colors denote the strains isolated from different types of samples. Blue, Cyanobacteria-dominated soil crusts; Green, Moss-dominated soil crusts; Red, Lichen-dominated soil crusts; Black, Bare sand.
Discussion
The family Geodermatophilaceae is a newly-established actinobacterial taxon. Normand et al. (1996) proposed the family Geodermatophilaceae in 1996, which was regarded as an invalid taxon at that time. In 2006, based on the common characteristics of the genera Geodermatophilus, Blastococcus, and Modestobacter, Normand (2006) summarized the typical characteristics of Geodermatophilaceae. Subsequently, the family Geodermatophilaceae was finally accommodated as a validly described taxon in the phylum Actinobacteria. To date, the family Geodermatophilaceae consists of three genera: Geodermatophilus, Blastococcus, and Modestobacter, that includes 25 validly described species.
The members of Geodermatophilaceae were found from various environments, including soil samples (Zhang et al., 2011; Jin et al., 2013), soil crusts (Reddy et al., 2007), deep sub-seafloor sediment (Ahrens and Moll, 1970), even stone habitats (Salazar et al., 2006; Chouaia et al., 2012; Gtari et al., 2012; Normand et al., 2012), dry-hot valley (Nie et al., 2012), and arid sand from desert (Montero-Calasanz et al., 2012, 2013a,b,c). In this study, we found Geodermatophilaceae members ubiquitously in desert soil samples, and we obtained Geodermatophilaceae cultures from three different types of desert soil crusts, as well as from the bare sands. These four environments represent typical micro-ecological environments in the Shapotou region. As we have observed, most Geodermatophilaceae members could form tiny motile spores or dormant spores, allowing them to spread around and survive long periods of desiccation. Moreover, most of the Geodermatophilaceae members we tested formed pink to black colonies on different types of agar plates. The pigmentation, cell wall composition and a high G+C content may increase protection of these strains from UV damage in the desert environments, where the UV transparency is often high.
The abundance and ubiquitous distribution of the Geodermatophilaceae in desert environments exhibited in relation to their resident microbiota, and in turn, the micro-ecological environments endowed the microorganisms with some specific metabolic characteristics. We found that the abundance and diversity of the Geodermatophilaceae in lichen- and cyanobacteria-dominated soil crusts were much higher than those of the bacteria found in moss-dominated soil crusts or bare sands. In the desert, the moisture, organic, and nitrogen content of the soil were the vital factors in determining physiological characteristics of the organisms. The lichen- and cyanobacteria-dominated soil crusts may contain a much higher proportion of clay and humic colloidal material, which can markedly affect the physiological activities of the strains from different micro-ecological environments.
The assayed physiological characteristics of the Geodermatophilaceae also showed a probable relationship with the resident microbes of the respective micro-ecological environments. In the dendrogram based on 173 physiological characteristics of the 34 tested Geodermatophilaceae strains, strains from the same micro-ecological environment were more likely to gather closely. The clusters shown in the phylogenetic dendrogram based on 16S rRNA gene sequences were interrupted in the dendrogram based on the physiological characteristics profile, which indicated that the micro-ecological environments where the strains were isolated significantly influenced the physiological characteristic profiles of the isolates (Figure 2).
Compared to our previous study and other related studies in the literature, we discovered many interesting diverse bioactivities for rare actinobactieria, which may be caused by characteristics of the extreme environments where these strains were found. Isolation and analysis of the bioactive compounds underlying these bioactivities will provide more detailed information on the mechanism of these activities. In this context, the members of the family Geodermatophilaceae are found to be the biological pioneers in extreme environments, especially in extreme arid environments. Further study on the cultures in this family will be advantageous to those seeking to understand mechanisms of environmental stress resistance, desertification control, and environmental remediation. In addition, studying these organisms will aid in the discovery of novel metabolic compounds.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by the National Infrastructure of Microbial Resources (NIMR-2014-3), the National Natural Science Foundation of China (NSFC) (31170041; 81173026; 81441093), the National S&T Major Special Project on Major New Drug Innovation (2012ZX09301002-003) and 863 Program (2014AA021504).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.01059
Phylogenetic dendrogram based on 16S rRNA gene sequences analysis of the isolates. Bootstrap values >50% (based on 1000 resampled datasets) are shown at branch nodes. Bar, 0.02 substitutions per site.
The number of actinobacteria in the different samples. Sample numbers were ordered from left to right (x-axis) according to the sample number in Table 1, and numbers of actinobacterial colonies from bottom to top (y-axis) were estimated and ordered according to the colony numbers that appeared on isolation medium of M4, M3, M2, and M1.
References
- Ahrens R., Moll G. (1970). Ein neues knospendes Bakterium aus der Ostsee (A new budding bacterium from the Baltic Sea). Arch. Microbiol. 70, 243–265. 10.1007/bf00407714 [DOI] [PubMed] [Google Scholar]
- Bull A. T., Stach J. E. M., Ward A. C., Goodfellow M. (2005). Marine actinobacteria: perspectives, challenges, future directions. Antonie Van Leeuwenhoek 87, 259–276. 10.1007/s10482-005-3678-4 [DOI] [PubMed] [Google Scholar]
- Chouaia B., Crotti E., Brusetti L., Daffonchio D., Essoussi I., Nouioui I., et al. (2012). Genome sequence of Blastococcus saxobsidens DD2, a stone-inhabiting bacterium. J. Bacteriol. 194, 2752–2753. 10.1128/JB.00320-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans R. D., Johansen J. R. (1999). Microbiotic crusts and ecosystem processes. Crit. Rev. Plant Sci. 18, 183–225. 10.1016/S0735-2689(99)00384-6 [DOI] [Google Scholar]
- Felsenstein J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. 10.2307/2408678 [DOI] [PubMed] [Google Scholar]
- Gtari M., Essoussi I., Maaoui R., Sghaier H., Boujmil R., Gury J., et al. (2012). Contrasted resistance of stone-dwelling Geodermatophilaceae species to stresses known to give rise to reactive oxygen species. FEMS. Microbiol. Ecol. 80, 566–577. 10.1111/j.1574-6941.2012.01320.x [DOI] [PubMed] [Google Scholar]
- Jin L., Lee H. G., Kim H. S., Ahn C. Y., Oh H. M. (2013). Geodermatophilus soli sp. nov. and Geodermatophilus terrae sp. nov., two novel actinobacteria isolated from grass soil. Int. J. Syst. Evol. Microbiol. 63, 2625–2629. 10.1099/ijs.0.048892-0 [DOI] [PubMed] [Google Scholar]
- Kämpfer P., Kroppenstedt R. M., Dott W. (1991). A numerical classification of the genera Streptomyces and Streptoverticillium using miniaturized physiological tests. J. Gen. Microbiol. 137, 1831–1891. 10.1099/00221287-137-8-1831 [DOI] [Google Scholar]
- Kim M., Oh H. S., Park S. C., Chun J. (2014). Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 64, 346–351. 10.1099/ijs.0.059774-0 [DOI] [PubMed] [Google Scholar]
- Kim O. S., Cho Y. J., Lee K., Yoon S. H., Kim M., Na H., et al. (2012). Introducing EzTaxone: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721. 10.1099/ijs.0.038075-0 [DOI] [PubMed] [Google Scholar]
- Montero-Calasanz M. C., Göker M., Pötter G., Rohde M., Spröer C., Schumann P., et al. (2012). Geodermatophilus arenarius sp. nov., a xerophilic actinomycete isolated from Saharan desert sand in Chad. Extremophiles 16, 903–909. 10.1007/s00792-012-0486-4 [DOI] [PubMed] [Google Scholar]
- Montero-Calasanz M. C., Göker M., Pötter G., Rohde M., Spröer C., Schumann P., et al. (2013a). Geodermatophilus africanus sp. nov., a halotolerant actinomycete isolated from Saharan desert sand. Antonie Van Leeuwenhoek 104, 207–216. 10.1007/s10482-013-9939-8 [DOI] [PubMed] [Google Scholar]
- Montero-Calasanz M. C., Göker M., Pötter G., Rohde M., Spröer C., Schumann P., et al. (2013b). Geodermatophilus saharensis sp. nov., isolated from sand of the Saharan desert in Chad. Arch. Microbiol. 195, 153–159. 10.1007/s00203-012-0860-8 [DOI] [PubMed] [Google Scholar]
- Montero-Calasanz M. C., Göker M., Pötter G., Rohde M., Spröer C., Schumann P., et al. (2013c). Geodermatophilus normandii sp. nov., isolated from Saharan desert sand. Int. J. Syst. Evol. Microbiol. 63, 3437–3443. 10.1099/ijs.0.051201-0 [DOI] [PubMed] [Google Scholar]
- Nie G. X., Ming H., Li S., Zhou E. M., Cheng J., Yu T. T., et al. (2012). Geodermatophilus nigrescens sp. nov., isolated from a dry-hot valley. Antonie Van Leeuwenhoek 101, 811–817. 10.1007/s10482-012-9696-0 [DOI] [PubMed] [Google Scholar]
- Normand P. (2006). Geodermatophilaceae fam. nov., a formal description. Int. J. Syst. Evol. Microbiol. 56, 2277–2278. 10.1099/ijs.0.64298-0 [DOI] [PubMed] [Google Scholar]
- Normand P., Gury J., Pujic P., Chouaia B., Crotti E., Brusetti L., et al. (2012). Genome sequence of radio-resistant Modestobacter marinus strain BC501, a representative actinobacterium thriving on calcareous stone surfaces. J. Bacteriol. 194, 4773. 10.1128/JB.01029-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normand P., Orso S., Cournoyer B., Jeannin P., Chapelon C., Dawson J., et al. (1996). Molecular phylogeny of the genus Frankia and related genera and emendation of the family Frankiaceae. Int. J. Syst. Bacteriol. 46, 1–9. [DOI] [PubMed] [Google Scholar]
- Okoro C. K., Brown R., Jones A. L., Andrews B. A., Asenjo J. A., Goodfellow M., et al. (2008). Diversity of culturable actinomycetes in hyper-arid soils of the Atacama Desert, Chile. Antonie Van Leeuwenhoek 95, 121–133. 10.1007/s10482-008-9295-2 [DOI] [PubMed] [Google Scholar]
- Reddy G. S. N., Potrafka R. M., Garcia-Pichel F. (2007). Modestobacter versicolor sp. nov., an actinobacterium from biological soil crusts that produces melanins under oligotrophy, with emended descriptions of the genus Modestobacter and Modestobacter multiseptatus Mevs et al. 2000. Int. J. Syst. Evol. Microbiol. 57, 2014–2020. 10.1099/ijs.0.64932-0 [DOI] [PubMed] [Google Scholar]
- Rohlf F. J. (2000). NTSYSpc, Numerical Taxonomy and Multivariate Analysis System Version 2.1. Stony Brook, NY: Department of Ecology and Evolution, State University of New York. [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor–joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Salazar O., Valverde A., Genilloud O. (2006). Real-time PCR for the detection and quantification of Geodermatophilaceae from stone samples and identification of new members of the genus Blastococcus. Appl. Environ. Microbiol. 72, 346–352. 10.1128/AEM.72.1.346-352.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani R., Aalbersberg W. (2013). Culturable rare actinomycetes: diversity, isolation and marine natural product discovery. Appl. Microbiol. Biotechnol. 97, 9291–9321. 10.1007/s00253-013-5229-7 [DOI] [PubMed] [Google Scholar]
- Talbot G. H., Bradley J., Edwards J. E., Jr., Gilbert D., Scheld M., Bartlett J. G. (2006). Bad bugs need drugs: an update of the development pipeline from the antimicrobial task force of the Infectious Diseases Society of America. Clin. Infect. Dis. 42, 657–668. 10.1086/499819 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. M., Zhang Z. S., Xu X. L., Ruan J. S., Wang Y. (2001). Actinopolymorpha singaporensis gen. nov., sp. nov., a novel actinomycete from the tropical rainforest of Singapore. Int. J. Syst. Evol. Microbiol. 51, 467–473. 10.1099/00207713-51-2-467 [DOI] [PubMed] [Google Scholar]
- Williams S. T., Goodfellow M., Alderson G., Wellington E. M. H., Sneath P. H. A., Sackin M. J. (1983). Numerical classification of Streptomyces and related genera. J. Gen. Microbiol. 129, 1743–1813. 10.1099/00221287-129-6-1743 [DOI] [PubMed] [Google Scholar]
- Xu P., Li W. J., Xu L. H., Jiang C. L. (2003). A microwave–based method for genomic DNA extraction from actinomycetes. Microbiology 30, 82–84. 10.3969/j.issn.0253-2654.2003.04.020 [DOI] [Google Scholar]
- Yang Z. J., Ding J. W., Ding K. S., Chen D. J., Cen S., Ge M. (2013). Phomonaphthalenone A: a novel dihydronaphthalenone with anti-HIV activity from Phomopsis sp. HCCB04730. Phytochem. Lett. 6, 257–260. 10.1016/j.phytol.2013.02.003 [DOI] [Google Scholar]
- Yue X. J., Yu L. Y., Li Q. P., Wei Y. Z., Guan Y., Zhang Y. Q. (2006). Study of methods to isolate viable but non-culturable microorganisms from natural environment. Microbiology 33, 77–81. 10.3969/j.issn.0253-2654.2006.03.015 [DOI] [Google Scholar]
- Zhang Y. Q., Chen J., Liu H. Y., Zhang Y. Q., Li W. J., Yu L. Y. (2011). Geodermatophilus ruber sp. nov., isolated from rhizosphere soil of a medicinal plant. Int. J. Syst. Evol. Microbiol. 61, 190–193. 10.1099/ijs.0.020610-0 [DOI] [PubMed] [Google Scholar]
- Zhang Y. Q., Liu H. Y., Chen J., Yuan L. J., Sun W., Zhang L. X., et al. (2010). Diversity of culturable actinobacteria from Qinghai–Tibet plateau, China. Antonie Van Leeuwenhoek 98, 213–223. 10.1007/s10482-010-9434-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic dendrogram based on 16S rRNA gene sequences analysis of the isolates. Bootstrap values >50% (based on 1000 resampled datasets) are shown at branch nodes. Bar, 0.02 substitutions per site.
The number of actinobacteria in the different samples. Sample numbers were ordered from left to right (x-axis) according to the sample number in Table 1, and numbers of actinobacterial colonies from bottom to top (y-axis) were estimated and ordered according to the colony numbers that appeared on isolation medium of M4, M3, M2, and M1.