SUMMARY
Recent discoveries of regulated cell death in bacteria have led to speculation about possible benefits that apoptosis-like pathways may confer to single-celled organisms. However, establishing how these pathways provide increased ecological fitness has remained difficult to determine. Here, we report a pathway in Bacillus subtilis in which regulated cell death maintains the fidelity of sporulation through selective removal of cells that misassemble the spore envelope. The spore envelope, which protects the dormant spore’s genome from environmental insults, uses the protein SpoIVA as a scaffold for assembly. We found that disrupting envelope assembly activates a cell death pathway wherein the small protein CmpA acts as an adaptor to the AAA+ ClpXP protease to degrade SpoIVA, thereby halting sporulation and resulting in lysis of defective sporulating cells. We propose that removal of unfit cells from a population of terminally differentiating cells protects against evolutionary deterioration, and ultimately loss, of the sporulation program.
Graphical Abstract
INTRODUCTION
Maintaining the fidelity of developmental programs is essential for ensuring the correct morphology of an organism. As such, key factors that determine morphogenesis are often subject to multiple layers of regulation. The alternative developmental pathway of bacterial endospore formation (“sporulation”) provides a genetically tractable system to study cellular morphogenesis and mechanisms that maintain the robustness of differentiation programs (Higgins and Dworkin, 2012; Tan and Ramamurthi, 2014). In nutrient-rich conditions, the bacterium Bacillus subtilis divides symmetrically to yield two genetically and morphologically identical daughter cells. However, upon nutrient deprivation, B. subtilis initiates sporulation by dividing asymmetrically, resulting in two genetically identical, but morphologically distinct daughter cells (the larger “mother cell” and the smaller “forespore”) that undergo different cell fates (Fig. 1A). Next, the mother cell engulfs the forespore; as a result, the forespore resides as a double membrane-bound cell within the mother cell. The forespore is then encased by the spore envelope which protects it from environmental insults and consists of two concentric shells whose assemblies are largely mediated by the mother cell (Henriques and Moran, 2007). The inner shell (the cortex) is made of a specialized peptidoglycan that assembles between the two membranes surrounding the forespore and eventually protects the spore from heat and helps maintain the dehydrated state of the spore core (Leggett et al., 2012; Popham et al., 1996). The outer shell (the coat) is composed of ~70 different proteins produced in the mother cell and is responsible for protecting the spore from chemical and enzymatic assaults (McKenney et al., 2013; Setlow, 2006). Ultimately, the mother cell lyses, thereby releasing the mature spore into the environment.
Figure 1.
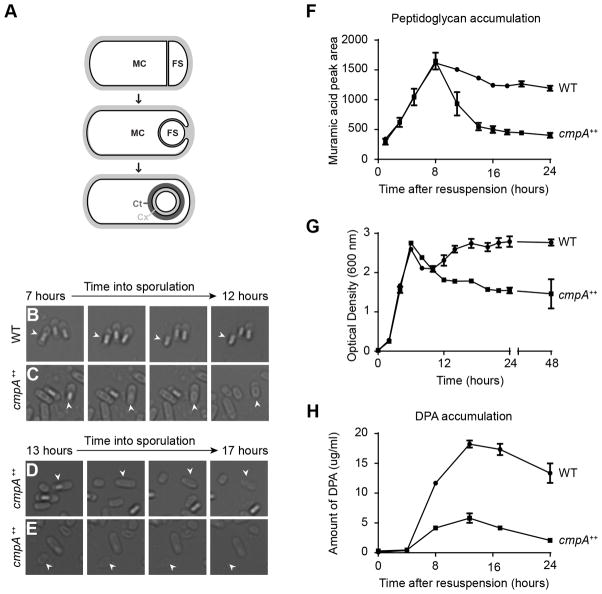
Overexpression of cmpA causes cell lysis and defects in cortex maintenance. (A) Schematic of sporulation in Bacillus subtilis. Asymmetric division (top) results in the formation of the larger mother cell (MC) and smaller forespore (FS) compartments, which are genetically identical. Next, the mother cell engulfs the forespore (middle). Eventually, the forespore resides in the mother cell cytosol (bottom). The forespore is encased in two concentric shells: the proteinaceous coat (Ct, dark gray), and the cortex (Cx, light gray), made of a specialized peptidoglycan. Membranes are depicted in black; cell wall material is depicted in light gray. (BE) Time-lapse microscopy of sporulating WT (C; strain PY79) and cmpA overexpressing (D–F; IT478) cells. Time after induction of sporulation is indicated above. Fate of phase bright forespores in WT (B) and cmpA overexpressing (C) cells. Fate of (D) phase gray forespores in cmpA overexpressing cells while still in the mother cell, or phase gray spores (E) after release into the medium. Arrowheads indicate phase gray forespore or released spore. (F) Accumulation of peptidoglycan during sporulation in WT (●; PY79) and cmpA overexpressing (■; IT478) cells. (G) Growth curves, as measured by optical density of the cultures grown in sporulation media (DSM), of WT (●; PY79) and cmpA overexpressing (■; IT478) strains. (H) Accumulation of dipicolinic acid (DPA) during sporulation in WT (●; PY79) and cmpA overexpressing (■ IT478) cells. Symbols represent mean values obtained from three independent measurements; error bars represent standard error of the mean. Strain genotypes are listed in Table S8. See also Figure S1 and Table S1–S2.
Assembly of the coat begins with the localization of a small 26-amino acid protein, SpoVM (Levin et al., 1993), which recognizes the outer forespore membrane by preferentially adsorbing onto the forespore’s positively curved membrane surface (Gill et al., 2015; Ramamurthi et al., 2009). SpoVM recruits the structural protein SpoIVA (Price and Losick, 1999; Ramamurthi et al., 2006; Roels et al., 1992; Wu et al., 2015) which polymerizes irreversibly in an ATP-dependent manner around the forespore surface (Castaing et al., 2013; Ramamurthi and Losick, 2008). Deletion of either spoVM or spoIVA results in a misassembled coat that is not anchored to the forespore surface (Levin et al., 1993; Roels et al., 1992). Thus, SpoVM and SpoIVA are required for proper assembly of the basement layer, atop which other coat proteins are deposited (McKenney et al., 2010). Interestingly, deletion of spoVM or spoIVA, but not other major coat proteins, also abolishes assembly of the cortex (Levin et al., 1993; Roels et al., 1992) suggesting that SpoVM and SpoIVA play unique roles in orchestrating coat and cortex assembly.
Previously, we identified a small (37 codon) unannotated ORF in B. subtilis (which we termed “cmpA”), whose deletion suppressed the sporulation defect imposed by a mutant allele of spoVM that permitted initiation of coat assembly, but abrogated cortex assembly (Ebmeier et al., 2012). CmpA is produced in the mother cell during sporulation and overexpression of cmpA in wild type cells reduced sporulation efficiency. Whereas CmpA was degraded in wild type cells at a late stage of sporulation (when phase-bright forespores were elaborated), CmpA persisted in cells harboring the mutant allele of spoVM. We proposed that CmpA participates with SpoVM and SpoIVA in coordinating spore envelope assembly by inhibiting cortex assembly until coat assembly occurred, but the mechanism remained unclear. Here, we report that CmpA ensures proper envelope assembly by participating in a quality control pathway that selectively removes defective sporulating cells through regulated cell death to ultimately preserve the integrity of the sporulation program in the population. Using classical genetics we identified the AAA+ protease ClpXP (Baker and Sauer, 2012; Gottesman, 1996) as an additional participant in this pathway and propose that CmpA is an adaptor (Zhou et al., 2001) that delivers SpoIVA for degradation by ClpXP specifically in those cells in which the spore envelope has misassembled. Accordingly, we found that amino acid substitutions in CmpA, SpoIVA, or ClpX that disrupted complex formation led to restoration of SpoIVA stability in cells that had misassembled the spore envelope. We also found that deletion of cmpA permitted the completion of the sporulation program by cells harboring mutant alleles of spoVM and spoIVA, resulting in the formation of viable spores despite containing envelope assembly defects. We propose that CmpA persistence in mutant cells harboring an envelope defect promotes degradation of SpoIVA, which destabilizes the forespore and leads to cell lysis, thereby specifically removing defective cells and ensuring the fidelity of the sporulation program.
RESULTS
cmpA overexpression causes spore maturation defects and cell lysis
Overexpression of cmpA inhibits cortex assembly (Ebmeier et al., 2012), but the mechanism of inhibition remained unclear. To understand the impact of CmpA overproduction, we performed single-cell time lapse microscopy of wild type (WT) and cmpA-overexpressing (cmpA++) cells (in which cmpA expression was driven by a constitutive promoter at an ectopic chromosomal locus) and monitored the fate of forespores that had achieved the phase bright state, an indicator of the spore core’s dehydration (Imae and Strominger, 1976). Forespores in both WT and cmpA++ cells progressed to the phase bright state. However, while 74% (n=498) of WT phase bright forespores were ultimately released upon mother cell lysis during the monitoring period (Fig. 1B, arrowhead), in cmpA++ cells 15% (n=222) of phase bright forespores were released and the remaining 85% relapsed to a phase-gray state (Fig. 1C, arrowhead), suggesting that they were largely unable to maintain core dehydration. In sporangia harboring relapsed phase-gray forespores, both mother cell and phase gray forespore lysed (Fig. 1D); additionally, released phase gray spores also ultimately lysed (Fig. 1E), suggesting that cmpA overexpression inhibited the cell’s ability to maintain core dehydration, leading to cell lysis.
Since cell shape and osmotic stability are governed by cell wall integrity (Holtje, 1998), we next examined the effect of cmpA-overexpression on cortex peptidoglycan (PG) levels and composition. We harvested soluble PG precursors and assembled PG from WT and cmpA++ cells at various time points during sporulation. Both strains initially (~t0–t8) accumulated soluble PG precursors, and depleted this pool coincident with cortex assembly (Fig. S1). During this initial period, both strains also accumulated assembled PG, indicating that cortex assembly was occurring normally (Fig. 1F). WT cells subsequently maintained steady levels of assembled PG after t8 (the slight reduction after t8 is due to mother cell lysis upon completion of sporulation), indicating the presence of a stable cortex. However, recovered PG from cmpA++ cells decreased after t8 (Fig. 1F), presumably due to an inability to harvest cells that lysed (Fig. 1D–E). To confirm that cmpA++ cells were lysing we monitored the optical density (OD600) of sporulating cmpA++ cultures. In sporulation medium, OD600 of WT cultures increased; upon stationary phase (~t6), OD600 remained roughly constant, after a transient decrease coincident with the entry into sporulation (Fig. 1G). In contrast, OD600 of cmpA++ cultures initially increased in density similar to WT, but decreased in stationary phase, indicative of cell lysis (Fig. 1G).
Consistent with a defect in cortex maintenance, we observed that cmpA++ sporangia did not accumulate dipicolinic acid (DPA; Fig. 1H), a small molecule that displaces water and is required for core dehydration (Paidhungat et al., 2000). Interestingly, the inability to maintain a stable cortex after normal initial assembly and the inability to accumulate DPA of cmpA++ cells was reminiscent of the phenotypes associated with a spoIVA cortex-deficient point mutant (Catalano et al., 2001), suggesting that cmpA++ is abrogating spoIVA’s cortex assembly function.
Two mechanisms could possibly contribute to the instability of the assembled cortex PG: (1) mechanical instability of the cortex due to defective envelope assembly, or (2) premature activation of cortex lytic hydrolases that degrade the cortex during germination. To rule out the second scenario, we deleted sleB and cwlJ, which encode the two known germination cortex lytic hydrolases (Heffron et al., 2009), and measured sporulation efficiency. Deletion of sleB and cwlJ did not suppress the cmpA++ sporulation defect (Table S1, Strain D) indicating that cmpA’s inhibition of cortex maturation is not due to premature germination, and instead suggesting that cmpA++ prevents the maintenance of a stable cortex.
To determine if cell lysis caused by cmpA++ is mediated by factors involved in the programmed mother cell lysis that releases the mature spore, we examined sporulation efficiencies of cmpA++ cells in the absence of the three known sporulation-specific cell wall hydrolases. cmpA++ cells exhibited an almost 200-fold decrease in sporulation efficiency (Table S2, Strain B), while ΔcwlC, ΔcwlH, ΔlytC, and ΔlytC ΔcwlC cells (Smith and Foster, 1995) sporulated similarly to WT (Table S2, Strains C, E, G, I). However, deletion of cwlC, cwlH, lytC, or lytC and cwlC, did not rescue the sporulation defect imposed by CmpA overproduction (Table S2, Strains D, F, H, J), suggesting that CmpA-mediated lysis and cortex inhibition is largely independent of known enzymes involved in programmed mother cell lysis.
Identification of other factors interacting with CmpA
To understand how CmpA inhibits cortex stability we sought to identify other factors that participate in this pathway. Deletion of cmpA was identified as an extragenic suppressor of the cortex-deficient spoVMI15A allele, and CmpA abnormally persisted late into sporulation in the spoVMI15A mutant (Ebmeier et al., 2012). We therefore selected for additional suppressors that corrected the spoVMI15A sporulation defect. To prevent isolating cmpA null mutations, we performed the selection in a strain harboring two chromosomal copies of cmpA. Two of the suppressors mapped to spoIVA (Table S3, Strain C and D). The first changed Glu423 to Gly, and the second changed Leu424 to Phe. The proximity of these two substitutions indicated that this region of SpoIVA may be important for interacting with CmpA or SpoVM and suggested that SpoVM, SpoIVA and CmpA participate together to orchestrate coat and cortex assembly.
We previously reported that proper localization of the largely functional CmpA-GFP is directly or indirectly dependent on spoVM [(Ebmeier et al., 2012); Fig. 2B)]; however, the mislocalization phenotype in the absence of SpoVM (increased CmpA-GFP accumulation as a focus on the forespore, reminiscent of the mislocalization phenotype of GFP-SpoIVA in the absence of SpoVM; Fig. 2B, arrowhead), together with the isolation of suppressor mutations in spoIVA, suggested that SpoIVA may be interacting with CmpA. Accordingly, in the absence of SpoIVA or SpoIVA and SpoVM, we found that CmpA-GFP was principally mislocalized in the mother cell cytosol (Fig. 2C–D), suggesting that SpoIVA is the major localization determinant for CmpA.
Figure 2.
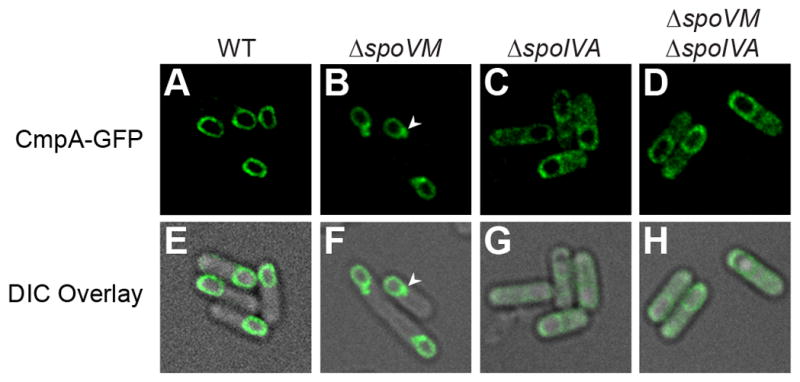
CmpA-GFP localization is dependent on spoIVA. (A–D) Localization of CmpA-GFP in WT (A; SE364), ΔspoVM (B; IT86), ΔspoIVA (C; IT114) and ΔspoVM ΔspoIVA (D; IT113) cells. Arrowheads (B, F) indicate mislocalized focus of CmpA-GFP. (E–H) Overlay of DIC and GFP fluorescence from A–D, respectively. See also Table S3.
Next, we exploited the heat sensitivity caused by cmpA overexpression (Table S4) to select for suppressors that would correct this phenotype. This selection yielded four suppressors. Three suppressors mapped to clpX, which encodes the ATPase subunit of the ClpXP protease [(Gottesman et al., 1993; Wojtkowiak et al., 1993); Table S3, Strains C–E]. All three suppressors (D21Y, I34M, and E44G) resulted in N-terminal substitutions in ClpX, the domain implicated in substrate binding (Wojtyra et al., 2003). A fourth suppressor mapped to clpP, which encodes the proteolytic subunit of ClpXP (Table S4, Strain F). This mutation changed Asp187 to Asn near the hydrophobic pocket of ClpP that interacts with the “IGF loop” of ClpX (Kim et al., 2001), thereby possibly affecting interactions between ClpX and ClpP.
ClpX is required for entry into sporulation [(Liu et al., 1999); Table S5, strain B], and overproduction of ClpX did not significantly affect sporulation (Table S5, strain E). However, co-overproduction of ClpX and CmpA resulted in a sporulation efficiency of 7.6 × 10−5 relative to WT (Table S5, strain G), which was worse than the ~200-fold defect caused by overproduction of CmpA alone (Table S5, strain C). To test if suppression by the clpX mutations were due to a loss of function, we deleted the N-terminus of ClpX and determined whether it suppressed the cmpA++ defect. Cells overproducing ClpXΔ1-53 (the endogenous copy of clpX was present in this strain to ensure entry into sporulation) and CmpA sporulated at near WT levels (Table S5, strain I), suggesting that the suppressor mutations decreased a function of ClpX’s N-terminus. We conclude that the loss of ClpX’s ability to bind a substrate overcomes the sporulation defect imposed by CmpA overproduction.
In addition to being the ATPase subunit of ClpXP protease, ClpX can function alone as a folding chaperone for some substrates (Schirmer et al., 1996). To test if the clpPD187N suppressor was a loss-of-function mutation, we constructed a ClpXF270W variant, which disrupts the IGF loop and diminishes ClpX interaction with ClpP (Kim et al., 2001). Similar to ClpXΔ1-53, co-overproduction of ClpXF270W with CmpA restored sporulation efficiency to near WT levels (Table S5, strain I), suggesting that abrogating the interaction between ClpX and ClpP corrects the cmpA++ defect. Taken together, the genetic data are consistent with a model in which binding and degradation of one or more substrates by ClpXP is responsible for the sporulation defect imposed by CmpA overproduction.
CmpA is a ClpXP adaptor for binding SpoIVA
Since CmpA overproduction inhibited cortex maturation and ClpX acted synergistically with CmpA to inhibit sporulation (Table S5, Strain G), we hypothesized that CmpA is an adaptor that delivers a protein required for cortex maturation to ClpXP for degradation. To isolate a nonfunctional CmpA variant for use as a negative control, we performed alanine scanning mutagenesis of the 37 codon cmpA ORF, monitored the subcellular localization of the resulting variants fused to GFP (Fig. S2) and assessed each variant’s function by measuring their ability to suppress the spoVMI15A sporulation defect (Table S6). In the presence of cmpAP2A, cells harboring spoVMI15A sporulated similarly to cells harboring ΔcmpA (Table S6, row 4, strain IT139), suggesting that substitution of Pro2 with Ala abolished CmpA function. Nonetheless, CmpAP2A-GFP localized properly to the forespore surface (Fig. S2).
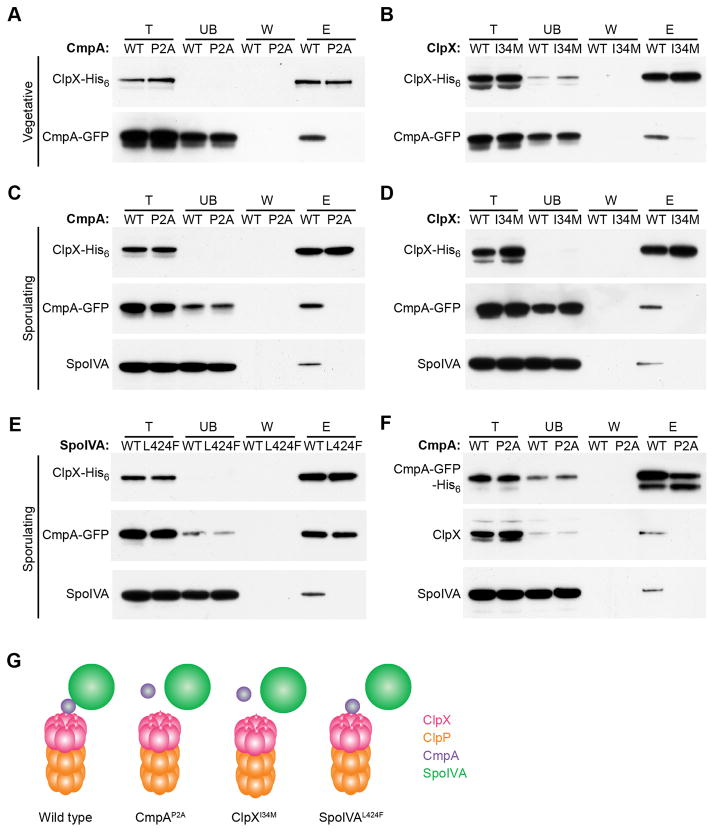
To test if CmpA and ClpX interact we produced ClpX harboring a C-terminal His6 tag. To promote stabilization of the hexameric form of ClpX that would bind adaptors and substrates, but would be unable to degrade bound substrates, we introduced a substitution (E182A) in ClpX’s Walker B motif that disrupts ATP hydrolysis, but not ATP binding (Baker and Sauer, 2012). We then overproduced ClpXE182A-His6 (hereafter, simply ClpX-His6) in cells that also produced low levels of WT ClpX to enable entry into sporulation. Next, we purified ClpX-His6 from vegetative or sporulating cells that also produced CmpA-GFP or CmpAP2A-GFP (Fig. 3). In vegetative cells, CmpA-GFP, but not CmpAP2A-GFP, co-purified with ClpX-His6 (Fig. 3A). We next purified ClpXI134M-His6 (also harboring a Walker B disruption) from vegetative cells that produced CmpA-GFP. Whereas ClpX-His6 co-purified with CmpA-GFP, the amount of CmpA-GFP that co-purified with ClpXI34M-His6 was diminished (Fig. 3B). Together, the data indicated that ClpX and CmpA interact in the absence of sporulation factors in a manner requiring Pro2 of CmpA and Ile34 of ClpX.
Figure 3.
ClpX, CmpA and SpoIVA form a complex. (A–B) Extracts of vegetative B. subtilis producing CmpA-GFP and ClpX-His6 (A; IT735) or ClpXI34M-His6 (B; T751) were applied to a Ni2+-agarose affinity column and eluted. The presence of ClpX and CmpA-GFP in the total (T), unbound (UB), wash (W), or eluate (E) fractions was monitored by immunoblotting. (C–F) Extracts of sporulating B. subtilis producing CmpAP2A-GFP and ClpX-His6 (C; IT760); ClpXI34M-His6 and CmpA-GFP (D; IT751); ClpX-His6, CmpA-GFP, and SpoIVAL424F (E; IT818); or CmpA-GFP-His6 (F; IT784) were applied to a Ni2+-agarose affinity chromatography and eluted. The presence of ClpX, CmpA-GFP, and SpoIVA were monitored in each fraction by immunoblotting. (G) Schematic summarizing the interactions between variants of ClpX, CmpA, and SpoIVA (ClpX, pink; ClpP, orange; CmpA, purple; SpoIVA, green). See also Figure S2 and Table S4–S6.
We next wondered if SpoIVA would bind to ClpX and CmpA. In sporulating cells, SpoIVA co-purified with ClpX-His6 and CmpA-GFP, but not in the presence of CmpAP2A-GFP (Fig. 3C). Additionally, while SpoIVA co-purified with ClpX-His6, SpoIVA did not co-purify with ClpXI134M-His6 (Fig. 3D). Further, whereas SpoIVA co-purified with ClpX-His6 and CmpA-GFP, SpoIVAL424F did not (Fig. 3E). Consistent with the results from the co-purification performed in vegetative cells (Fig. 3A), CmpA-GFP co-purified with ClpX-His6 even in the presence of SpoIVAL424F (Fig. 3E). Finally, to test if all three proteins exist in a single complex, we repeated the co-purification experiment with the affinity tag placed on CmpA-GFP. In sporulating cells producing low levels of ClpX and overproducing ClpXE182A, purification of CmpA-GFP-His6, but not CmpAP2A-GFP-His6, led to the co-purification of ClpX and SpoIVA (Fig. 3F). Taken together, we conclude that ClpX, CmpA, and SpoIVA form a complex, SpoIVA is not required for ClpX-CmpA interaction, and disruption of the N-terminal substrate binding domain of ClpX, Pro2 in CmpA, or Leu424 in SpoIVA abolishes formation of the ternary complex (Fig. 3G). The data thus far are consistent with a model in which CmpA delivers SpoIVA to ClpXP for degradation.
Spore envelope defects cause CmpA-driven SpoIVA degradation
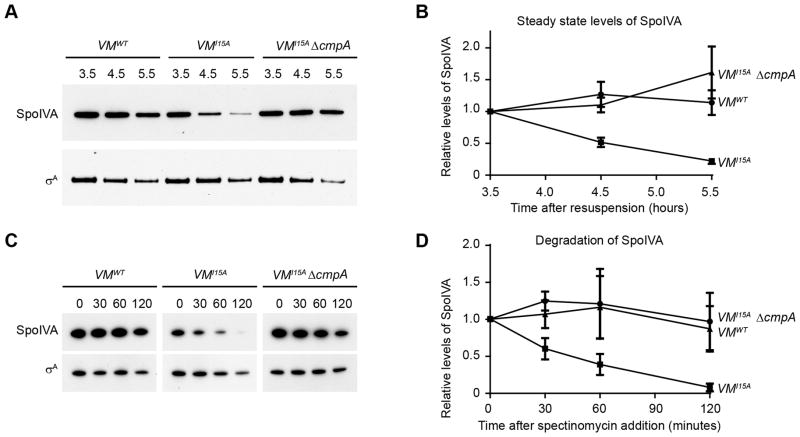
To test if SpoIVA is a substrate for degradation by ClpXP, we examined the steady state levels of SpoIVA by immunoblotting cell extracts harvested from cells containing spoVMI15A, which activates the CmpA pathway. SpoIVA was detected in WT cells at t3.5 of sporulation, and similar levels of SpoIVA were maintained through t5.5 (Fig. 4A–B). In the presence of spoVMI15A, SpoIVA levels rapidly diminished by t5.5, but SpoIVA levels were restored upon cmpA deletion, even in the presence of spoVMI15A (Fig. 4A–B). To test if the decrease in SpoIVA levels was due to degradation, we inhibited translation by addition of spectinomycin and measured SpoIVA levels. Whereas WT strains maintained steady levels of synthesized SpoIVA even 2 h after inhibition of translation, in strains expressing spoVMI15A SpoIVA was rapidly depleted and almost undetectable after 2 h (Fig. 4C–D). Deletion of cmpA, though, prevented depletion of SpoIVA in cells expressing spoVMI15A, suggesting that in cells harboring mutations causing spore envelope defects (e.g. spoVMI15A) SpoIVA is degraded in a CmpA-dependent manner.
Figure 4.
SpoIVA is degraded in a cmpA dependent manner in the spoVMI15A mutant. (A) Cell extracts were prepared from sporulating cells of ΔspoIVA (KP73), or strains harboring WT spoVM (KR103), spoVMII15A (KR322), or spoVMII15 ΔcmpA (SE181) at times (h) indicated after induction of sporulation. SpoIVA and, as a control, σA were detected by immunoblotting. (B) Quantification of SpoIVA band intensities of each strain in (A) relative to σA levels at each time point and represented as a fraction compared to the level at t3.5 (●, spoVMWT; ■, spoVMII15A; ▲, spoVMII15 △cmpA). (C) Strains KR103, KR322, and SE181 were induced to sporulate. At t4.5, spectinomycin (200 μg ml−1) was added to inhibit translation and cell extracts were prepared from aliquots taken at the times (min) indicated. SpoIVA and σA were detected by immunoblotting. (D) Quantification of SpoIVA band intensities in (C) relative to σA levels at each time point and represented as a fraction compared to the level at t0 after addition of spectinomycin (●, spoVMWT;■, spoVMII15A; ▲, spoVMII15 △cmpA). Symbols represent mean values obtained from four (B) or three (D) independent measurements; error bars represent standard error of the mean.
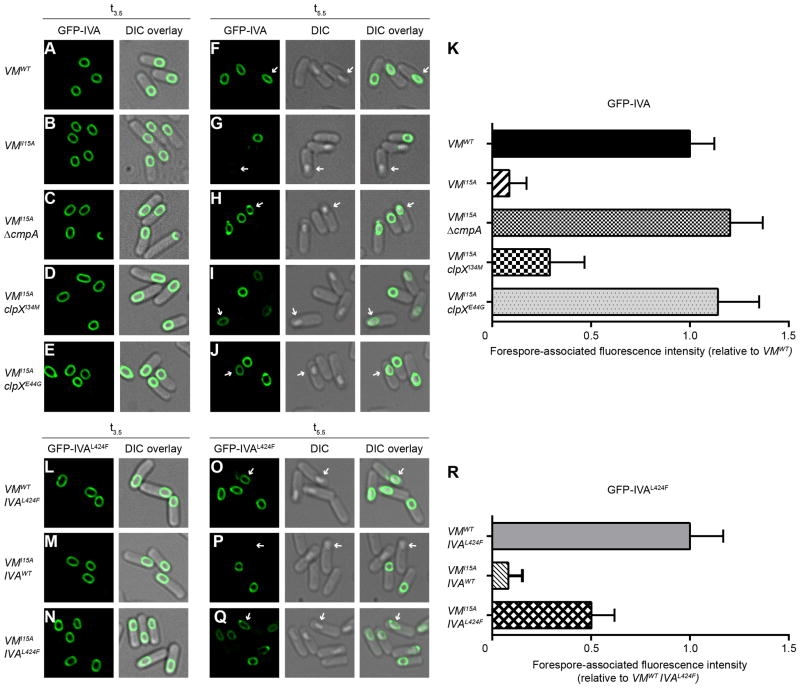
To determine if SpoIVA is degraded in a particular subpopulation of sporulating cells, we examined the stability of GFP-SpoIVA in cells expressing spoVMI15A using fluorescence microscopy (Fig. 5A–K). At t3.5 of sporulation, localization and levels of GFP-SpoIVA were similar in WT and spoVMI15A cells (Fig. 5A–B). At t5.5, when most sporangia had elaborated a phase-bright forespore, WT cells maintained similar amounts of GFP-SpoIVA (Fig. 5F, 5K), but in the presence of spoVMI15A, sporangia containing a phase-bright forespore were largely devoid of GFP-SpoIVA (Fig. 5G, 5K). When cmpA was deleted, GFP-SpoIVA levels returned to near WT levels despite the presence of spoVMI15A (Fig. 5H, 5K). We then reasoned that disruption of the CmpA-ClpX-SpoIVA complex through the use of the clpX point mutants should also restore SpoIVA stability at t5.5 and found that this was the case (Fig. 5D–E, I–K).
Figure 5.
CmpA-dependent degradation of GFP-SpoIVA in sporangia harboring spoVMI15A that elaborate phase bright forespores. (A–J) GFP-SpoIVA in cells harboring spoVMWT (A and F, KR165) spoVMI15A (B and G, KRC77), spoVMI15A ΔcmpA (C and H, IT839), spoVMI15A clpXI34M (D and I, IT884) or spoVMI15A clpXE44G (E and J, IT883) at t3.5 (A–E) and t5.5 (F–J) after induction of sporulation. (K) Quantification of GFP-SpoIVA fluorescence in strains shown in (A–J). (L–Q) GFP-SpoIVAL424F in cells harboring spoVMWT spoIVAL424F(L and O, IT869), spoVMI15A spoIVAWT (M and P, KRC77) or spoVMI15A spoIVAL424F (N and Q, IT852) at t3.5 (L–N) and t5.5 (O–Q) after induction of sporulation. (R) Quantification of GFP-SpoIVAL424F forespore-associated fluorescence in strains shown in (L–Q). All bars represent mean values of >80 phase bright forespores; error bars represent standard deviation. Arrows indicate a phase bright forespore. See also Figure S3.
We also examined the persistence of GFP-SpoIVAL424F (Fig. 5L–R), with the expectation that it would be resistant to degradation since it did not bind to ClpX (Fig. 3E). In WT cells GFP-SpoIVAL424F behaved similarly to GFP-SpoIVA in its localization to the forespore and persistence in phase bright forespores (Fig. 5L, 5R). In the presence of spoVMI15A and spoIVA at the native locus, GFP-SpoIVAL424F was degraded (Fig. 5P, 5R), but when the native locus harbored spoIVAL424F instead, GFP-SpoIVAL424F was largely resistant to degradation (Fig. 5Q, 5R). The recessive nature of the spoIVAL424F allele may be explained by the ability of SpoIVA to polymerize (Ramamurthi and Losick, 2008), whereby SpoIVA that are degraded in a CmpA-dependent manner could recruit interacting SpoIVAL424F molecules to ClpX that would not otherwise bind to CmpA and be subject to degradation. We conclude that SpoIVA is degraded in a CmpA-dependent manner in cells harboring a mutation (e.g. spoVMI15A) that causes improper spore envelope assembly.
Additional factors under σK control are required for degradation of SpoIVA and CmpA
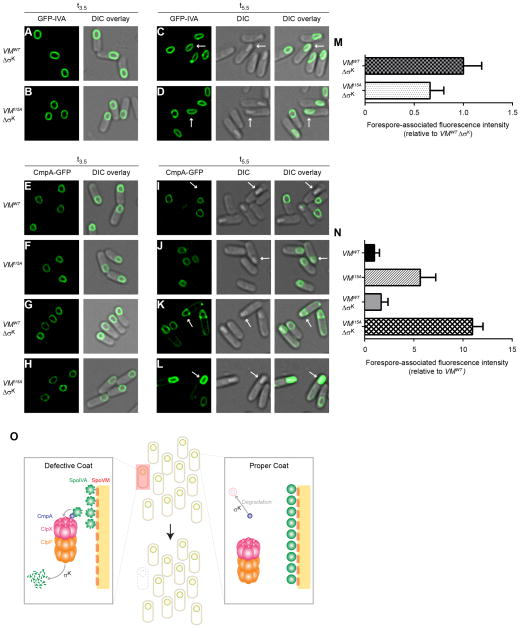
Our observation that degradation of GFP-SpoIVA only occurred in cells that had reached the phase bright forespore state led us to wonder whether some factor was preventing degradation at an earlier time point or if an additional factor was required for SpoIVA degradation at a later time point. We therefore overproduced CmpA and ClpX at an early time point (t3 of sporulation), added spectinomycin after 15 min to prevent further translation, and monitored SpoIVA levels and observed no significant degradation of SpoIVA (Fig. S3), indicating that ClpX and CmpA were not sufficient for SpoIVA degradation. The appearance of phase bright spores is coincident with activation of the mother cell-specific sigma factor, σK, so we wondered whether the additional factor(s) were produced by σK. If so, we would expect GFP-SpoIVA degradation to be inhibited by preventing σK activation. Thus, we monitored GFP-SpoIVA levels in sporulating cells harboring a deletion in spoIVCA, which is required for producing functional σK (Kunkel et al., 1990). At an early time point, GFP-SpoIVA stability and localization were similar in WT cells and in cells harboring spoVMI15A in the absence of σK (Fig. 6A, B). However, at a later time point, the absence of σK prevented degradation of GFP-SpoIVA in phase-bright forespores of the spoVMI15A mutant (Fig. 6D) suggesting that at least one factor produced by σK is required for CmpA/ClpXP-mediated degradation of SpoIVA.
Figure 6.
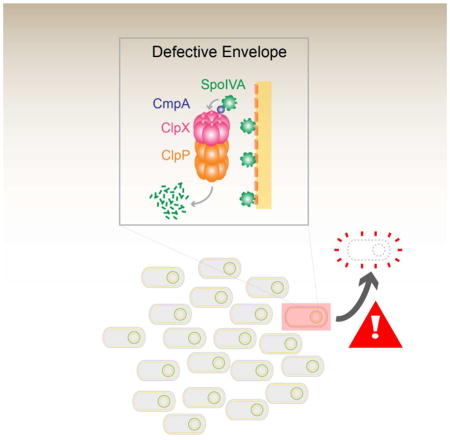
Additional factor(s) under σK control are required for SpoIVA and CmpA-GFP degradation. (A–D) GFP-SpoIVA in cells harboring spoVMWT ΔσK (A and C, IT891) or spoVMI15A 3σK (B and D, IT892) at t3.5 (A–B) or t5.5 (C–D) after induction of sporulation. (E–L) CmpA-GFP in cells harboring spoVMWT(E and I, IT897), spoVMI15A (F and J, IT896), spoVMWT 3σK (G and K, IT904) or spoVMI15A 3σK (H and L, IT903) at t3.5 (E–H) or t5.5 (I–L) after induction of sporulation. Arrows indicate a phase bright forespore. (M–N) Quantification of forespore-associated GFP fluorescence intensities shown in (C–D) and (I–L), respectively. All bars represent mean values of 14–101 phase bright forespores; error bars represent standard deviation. See also Figure S4 and Table S7. (O) Model of the CmpA-mediated regulated cell death pathway. Depicted is a population of sporulating B. subtilis cells (center, top) in which a single cell has elaborated a defective spore envelope (highlighted in red) and, as a consequence, is removed from the population by cell lysis (center, bottom). Left: the CmpA pathway in a cell elaborating a defective spore envelope. CmpA persists in this cell and acts as an adaptor of ClpXP to mediate degradation of SpoIVA in a σK-dependent manner. Right: in cells that successfully initiate spore envelope assembly, CmpA is degraded upon activation of σK and, in the absence of SpoIVA degradation, the sporulation program proceeds. SpoIVA, green; SpoVM, red; CmpA, purple; ClpX, pink; ClpP, orange.
Since CmpA-GFP is also degraded specifically in WT cells elaborating phase bright forespores (Ebmeier et al., 2012) we next tested if CmpA-GFP stability was affected by the absence of σK. We induced cmpA-gfp expression at t3 of sporulation; at t3.5 CmpA-GFP localized to the forespore in all tested strains (Fig. 6E–H). At t5.5, CmpA-GFP was no longer detectable in WT cells containing a phase-bright forespore (Fig. 6I), but persisted in the spoVMI15A mutant [(Ebmeier et al., 2012); Fig. 6J]. In the absence of σK, CmpA-GFP persisted not only in cells harboring spoVMI15A (Fig. 6L), but also in WT cells that elaborated phase bright forespores (Fig. 6K), indicating that degradation of CmpA requires a factor under the control of σK. The data suggest that σK controls two parts of the pathway. In WT cells that have successfully initiated spore envelope assembly, σK directs CmpA degradation so that sporulation may proceed. In cells that are unable to initiate spore envelope assembly, at least one additional factor produced under control of σK is required for CmpA/ClpXP-mediated degradation of SpoIVA.
CmpA is part of a quality control mechanism that maintains the integrity of a sporulating population
To determine if CmpA participates in a quality control mechanism that removes cells harboring incorrectly assembled spore envelopes from the population, we examined if deletion of cmpA would permit cells harboring other spore envelope defects to proceed past this checkpoint. To this end, we investigated the effect of cmpA deletion on a spoVMK2A mutant that behaves similarly to the spoVMI15A mutant (Fig. S4). Cells harboring spoVMK2A produced ~100-fold fewer heat resistant spores and ~150-fold fewer lysozyme resistant spores relative to WT cells (Table 1, Strain B), but deletion of cmpA suppressed these defects (Table 1, Strain C). Sporangia harboring SpoIVA variants that fail to polymerize, either due to an inability to bind (SpoIVAK30A) or hydrolyze (SpoIVAT70A-T71A) ATP, do not assemble a coat and produce 104–106-fold fewer heat resistant spores than WT (Table 1, Strains F and H). Although deletion of cmpA did not suppress the sporulation defect of ΔspoIVA [and so is unable to bypass the requirement for SpoIVA altogether (Table 1, Strains J–K)] ΔcmpA suppressed the polymerization-defective spoIVA alleles, raising the sporulation efficiency (heat resistance) ~200–3000 -fold (Table 1, Strains G and I). ΔcmpA also partially suppressed the lysozyme sensitivity of these mutants, indicating that some coat assembly was permitted to occur (Table 1, Strains G and I).
Table 1.
Sporulation efficiencies (as measured by heat or lysozyme resistance) of spoVM and spoIVA mutants with and without deletion of cmpA. Standard deviation from mean is reported in parentheses (n=3).
| Straina | Mutation | cmpA | Sporulation Efficiency (relative to WT) | |
|---|---|---|---|---|
| Heat Resistance | Lysozyme Resistance | |||
| A | WT | - | 1 | 1 |
| B | spoVMK2A | 0.01 (0.007) | 0.007 (0.003) | |
| C | spoVMK2A | Δ | 0.25 (0.05) | 0.47 (0.03) |
| D | spoVMI15A | 5 x 10−6 (1.7 x 10−6) | 1.8 x 10−6 (1.1 x 10−6) | |
| E | spoVMI15A | Δ | 0.16 (0.09) | 0.05 (0.01) |
| F | spoIVAK30A | 3.8 x 10−4 (1.7 x 10−4) | 3 x 10−4 (3.2 x 10−4) | |
| G | spoIVAK30A | Δ | 0.08 (0.01) | 0.03 (0.02) |
| H | spoIVAT70A-T71A | 5.8 x 10−6 (3 x 10−6) | 2 x 10−6 (1 x 10−6) | |
| I | spoIVAT70A-T71A | Δ | 0.02 (0.01) | 0.002 (7 x 10−4) |
| J | ΔspoIVA | <10−8 | <10−8 | |
| K | ΔspoIVA | Δ | <10−8 | ND |
Strain A: PY79; B: KRC102; C:IT102; D: KR322; E:SE181; F:KR367; G:IT880; H: JPC221; I: IT882; J: KP73; K: IT895. Genotypes are listed in Table S9. ND, not determined.
To test if the presence of CmpA in cells harboring envelope defects resulted in the production of fewer viable spores, or a high level of viable, but heat sensitive, spores, we determined the total viable spore count for cells containing spoVMI15A or spoIVAT70A-T71A in the presence and absence of CmpA by isolating viable spores by gradient centrifugation, then subjecting them to high heat. Cells harboring spoVMI15A produced just 347 ± 141 viable spores ml−1, compared to 8.6 × 107 ± 4.1 × 107 spores ml−1 produced by WT, of which 79% and 44%, respectively, were heat resistant (Table S7, strains A–B). Deletion of cmpA in cells harboring spoVMI15A resulted in the production of 6.1×106 ± 4.0 × 106 spores ml−1, of which nearly half were heat resistant (Table S7, strain C). Similar results were obtained in the suppression of the spoIVAT70A-T71A allele by deletion of cmpA (Table S7, strains D–E). Taken together, we conclude that deletion of cmpA allows mutants harboring spore envelope defects to bypass a quality control checkpoint, escape cell lysis, and continue with sporulation to produce large quantities of viable spores leading to the persistence and propagation of such mutations in the population. In contrast, the presence of CmpA in envelope-defective cells prevents completion of the sporulation program and removes those cells from the population.
DISCUSSION
The use of checkpoints ensures that steps in a developmental program properly finish prior to progression into the next stage of the program (Hartwell and Weinert, 1989). Here, we discovered that, during sporulation in B. subtilis, the small protein CmpA participates with the ClpXP proteolytic machinery in a quality control checkpoint during a terminal differentiation program to monitor proper spore envelope assembly. We propose a model (Fig. 6O) in which CmpA is an adaptor to ClpXP to mediate the degradation of the coat morphogenetic protein SpoIVA specifically in cells that improperly assemble the spore envelope. Degradation of SpoIVA is linked to destabilization of defective sporangia and ultimately results in their lysis, thereby removing defective spores from the population. In cells that properly construct the spore envelope, degradation of CmpA signals successful spore envelope assembly and prevents SpoIVA proteolysis, permitting these cells to continue through the sporulation program. The model suggests that deleting cmpA, or disrupting the interactions between CmpA, ClpXP, and SpoIVA bypasses the checkpoint and permits mutant cells elaborating defective spore envelopes to continue through the sporulation program and persist in the population.
Our model is consistent with several lines of genetic, biochemical, and cytological evidence. Disruption of sporulation by overexpression of cmpA was suppressed by three different loss-of-function mutations in clpX that changed residues in the substrate and adaptor-binding N-terminus of ClpX. A fourth spontaneous mutation in clpP, that altered a residue near the hydrophobic binding pocket that mediates ClpP’s interaction with ClpX, also suppressed the sporulation defect caused by cmpA overexpression. Involvement of SpoIVA as a target for degradation by ClpXP arose from two mutations in adjacent codons of spoIVA that altered residues between the Middle and C-terminal domain of SpoIVA (Castaing et al., 2014), suppressed the sporulation defect of the spoVMI15A allele (which arrests sporulation in a CmpA-dependent manner) and led to stabilization of SpoIVA.
Biochemical support for the model came from co-purification experiments which showed that CmpA, ClpX, and SpoIVA can exist in complex. Consistent with our genetic data, amino acid substitutions in CmpA, ClpX, or SpoIVA that bypassed the CmpA checkpoint in vivo also failed to form the ternary complex in vitro. We also found that CmpA interacts with the N-terminus of ClpX, and that this interaction can occur in the absence of SpoIVA. However, the interaction between SpoIVA and ClpX absolutely required CmpA. Together, this behavior is consistent with an adaptor-like function for CmpA, similar to that which has been described for ClpXP adaptors in other systems (Battesti and Gottesman, 2013).
Finally, epifluorescence microscopy of individual cells revealed that degradation of GFP-SpoIVA occurred specifically in mutant cells that had progressed to the phase bright forespore state, which is the stage in sporulation at which CmpA-GFP abnormally persists in the spoVMI15A mutant. However, disrupting CmpA/ClpX/SpoIVA complex formation in envelope-assembly mutants resulted in persistence of GFP-SpoIVA, completion of the sporulation program, and production of high levels of viable spores. The eventual disappearance of CmpA-GFP in in WT cells suggests that regulation of the checkpoint may be controlled via degradation of CmpA. Unlike other early-acting sporulation checkpoints that monitor ploidy, or fidelity and translocation of the genome prior to commitment to the onset of sporulation (Bejerano-Sagie et al., 2006; Burkholder et al., 2001; Fiche et al., 2013; Veening et al., 2009), CmpA is part of a late stage quality control mechanism that monitors the integrity of the developing spore. In this pathway, we propose that spore envelope defects arising at a late stage (so-called “Stage IV–V”) result in lysis of the sporangium linked to SpoIVA degradation by CmpA and ClpXP. Consistent with this timing, we also report that the switch to initiate this pathway resides under control of σK, the final compartment-specific sporulation sigma factor which is also required for activating cortex synthesis (Vasudevan et al., 2007). We found that at least one unidentified factor produced by σK is required to activate CmpA/ClpXP-mediated degradation of SpoIVA in mutant cells, and that σK is required for degradation of CmpA in properly sporulating cells. It is possible that the additional factor required for SpoIVA degradation is responsible for a post-translational modification of SpoIVA that marks it as a substrate for degradation by CmpA/ClpXP, or that recruitment of SpoIVA to ClpXP requires a second factor, in addition to CmpA (Lau et al., 2015).
We previously reported that ΔcmpA cells attain heat resistance slightly faster than WT (Ebmeier et al., 2012), suggesting that CmpA may perform a still poorly-defined role in slowing down the sporulation program, perhaps to ensure that proper spore envelope assembly occurs before the cell moves on to subsequent steps. In this report, we demonstrate that CmpA also acts upon mutant cells that fail to elaborate a proper envelope, and removes them from the population entirely. In this capacity, we propose that sporulating cells provide a window between σE and σK activation for the spore envelope to initiate assembly properly. Upon σK activation, if the spore envelope has properly assembled, the cell degrades CmpA and sporulation proceeds. However, if the envelope improperly assembles, CmpA instead persists, SpoIVA is degraded, and the cell lyses. Since SpoIVA and SpoVM are the only coat proteins that are also required for cortex assembly (heat resistance), it is appropriate that targeting a single protein, SpoIVA, for regulated degradation may result in destabilization of the entire spore envelope.
Recent reports of “apoptotic-like” cell death in bacterial cells have added to the changing view of bacterial communities as more than simply a collection of independently operating cells, leading to speculation that unicellular organisms may initiate cell death to benefit the population as a whole (Bos et al., 2012; Dwyer et al., 2012; McFarland et al., 2015). Although specific mechanisms for cell death in these cases have been well characterized, the exact benefit of such an act to a single-celled organism has sometimes been difficult to explicitly demonstrate. The use of the term “apoptosis” in bacteria has therefore remained controversial (Galluzzi et al., 2012; Hacker, 2013). Unlike regulated cell death pathways described in actively dividing cells, the CmpA pathway functions during a terminal differentiation program that produces a spore, a specialized cell type that preserves the cell’s genetic material in a manner functionally similar to that of germ cells in multicellular organisms. The construction of an envelope that is resistant to environmental insults is critical for the spore’s survival in harsh environmental conditions. As such, removal of cells harboring mutations that produce defective spore envelopes confers an obvious advantage to a population that needs to protect the integrity of its germ line’s genome and ensures the survival of future generations (Gartner et al., 2008; Setlow, 2006). In the absence of CmpA, cells run the risk of producing large numbers of spores that harbor mutations that may not confer the resistance properties required to protect the cell’s genome.
Why is it more beneficial to completely remove defective spores from a population, rather than allowing spores that harbor mutations, to persist? Sporulating cells employ a bet-hedging strategy whereby a fraction of the population does not initiate sporulation in order to retain the ability to rapidly respond to a return to favorable growth conditions (Veening et al., 2008). During continued favorable growth conditions, the selective pressure to maintain sporulation becomes relaxed, resulting in the enrichment of sporulation-defective cells (Maughan et al., 2007; Sastalla et al., 2010). In the absence of the CmpA pathway, cells harboring mutations that diminish spore envelope assembly can complete sporulation and persist in the population. When these mutant cells resume vegetative growth, and eventually re-sporulate, a fraction of them may employ the bet-hedging strategy and avoid initiating sporulation. In this scenario, this group of non-sporulating cells has the capacity to quickly flourish, upon a sudden increase in nutrient availability, and out-compete the population of WT cells that initiate sporulation. Therefore, it is important for the population to actively remove cells that accumulate detrimental mutations. We propose that the CmpA pathway ensures that mutations diminishing the capacity to produce a robust spore do not become incorporated into the bacterium’s “germ line” and ensures that faulty spores, which may be prone to accumulate additional mutations leading to the ultimate deterioration of the sporulation program, are removed by lysis.
Interestingly, the degradation of SpoIVA, which forms static polymers to encase the forespore, is reminiscent of the degradation of the nuclear lamins (static intermediate filaments that maintain the integrity of the nuclear envelope) during apoptosis in metazoans. While degradation of lamins is a feature of apoptosis, incorrect assembly of lamins to form the nuclear envelope can also induce apoptosis (Burke, 2001). Similarly, SpoIVA may not only be a target for degradation, but its incorrect assembly appears to induce CmpA-dependent cell lysis. Thus, the ability of morphogenetic proteins that serve to maintain the integrity of cellular envelopes to play these dual roles in regulated cell death appears to have arisen in evolutionarily and mechanistically distinct systems.
EXPERIMENTAL PROCEDURES
For a full explanation of experimental protocols, please see the Supplemental Experimental Procedures.
Strain construction
Strains are otherwise isogenic derivatives of B. subtilis PY79 (Youngman et al., 1984). Point mutant variants of cmpA, cmpA-gfp, clpX and clpX-His6 were created using integration vectors containing the appropriate inserts as templates for Quikchange site-directed mutagenesis (Agilent). All plasmids were integrated into the B. subtilis chromosome by double recombination at the specified ectopic locus.
General methods
Sporulation efficiency was measured by inducing sporulation in Difco sporulation medium (DSM) for at least 24 h at 37°C and subjecting spores to heat treatment (80°C for 20 min) or lysozyme treatment (250 μg ml−1 for 1 h at 37°C). Cultures were then serially diluted and colony-forming units (cfu) that survived were determined and reported relative to the cfu obtained in a parallel culture of the WT PY79 strain. Viable spore counts were determined by first purifying spores using 30%–70% metrizoic acid density gradient centrifugation, then determining cfu ml−1 by serial dilution. PG was harvested from 1 ml cultures by acid hydrolysis and analyzed by HPLC; DPA was harvested from 5 ml cultures and analyzed by a colorimetric assay. Spontaneous suppressor mutations were identified by enriching for mutants that grew after multiple rounds of sporulation and heat treatment followed by whole genome sequencing.
Microscopy
Cells were induced to sporulate by resuspension in SM medium (Sterlini and Mandelstam, 1969). When required, IPTG was added (1 mM final concentration) to induce expression of wild type or variants of clpX or cmpA. At various time points 1 ml of culture was harvested and resuspended in 100 μl PBS. 5 μl was spotted on a glass bottom culture dish (Mattek Corp.) and covered with a 1% agarose pad made with distilled water. Cells were viewed at 25°C with a DeltaVision Core microscope system (Applied Precision/GE Healthcare) equipped with a Photometrics CoolSnap HQ2 camera and an environmental chamber. Seventeen planes were acquired every 200 nm and the data were deconvolved using SoftWorx software as described previously (Eswaramoorthy et al., 2014).
Co-purification
Overnight cultures were grown in CH media and induced to sporulate in 20 ml resuspension medium. IPTG (1 mM final concentration) was added to induce expression of tagged or untagged versions of WT and mutant CmpA and ClpX 2 h after resuspension. Cells were harvested and protoplasted in protoplast buffer (0.5 M sucrose, 20 mM MgCl2, 10 mM potassium phosphate at pH 6.8, 0.1 mg ml−1 lysozyme) for 25 min at 37°C. Protoplasts were collected by centrifugation and resuspended in 1 ml of ice cold 0.5X PD buffer (Kim et al., 2001) without ATP regenerating system (hereafter referred to as 0.5X PD buffer). Phenylmethylsulfonyl fluoride (1mM final concentration) was added to cell lysate and cell debris was removed by centrifugation. The supernatant was collected and incubated with Ni-NTA agarose beads (Qiagen) equilibrated in 0.5X PD buffer for 10 min at 4°C with inversion. Beads were washed with 0.5X PD buffer containing 20mM imidazole. Bound proteins were eluted with 250 μl of 0.5X PD buffer containing 250mM imidazole. Fractions were analyzed by immunoblotting using rabbit antisera raised against purified E. coli ClpX (gift from Sue Wickner), GFP, or SpoIVA.
Cell lysate harvesting
Overnight cultures were grown in CH medium and induced to sporulate by the resuspension method (described above). 1 ml of culture was harvested at various time points. Cells were harvested and protoplasted as described above. Protoplasts were resuspended in 500 μl 5% tricholoroacetic acid (TCA) to lyse cells and precipitate proteins. After incubation on ice for 15 min, precipitate was collected by centrifugation and washed three times with 1 ml acetone. Precipitated pellet was air-dried overnight at room temperature, then resuspended in 300 μl 1× LDS NuPAGE Buffer (Invitrogen) and heated at 55°C f or 10 min with shaking. Fractions were analyzed by immunoblotting using rabbit antisera raised against SpoIVA or σA.
Supplementary Material
HIGHLIGHTS.
During sporulation misassembly of the spore envelope causes CmpA-mediated lysis
CmpA is a ClpXP adaptor for degradation of the morphogenetic protein SpoIVA
The CmpA pathway helps ensure evolutionary maintenance of the sporulation program
Acknowledgments
We thank V. Bliskovsky (Genomics Core facility, NCI) for whole genome sequencing of spontaneous suppressors; P. Setlow, R. Losick, D. Kearns, S. Ebmeier, and D. Ziegler (BGSC) for strains; S. Wickner for E. coli ClpX antiserum; S. Gottesman, S. Wickner, M. Maurizi, G. Storz, M. Lichten, and Y.-S. Lee for discussion; and A. Hitchcock-Camp and C. Ellermeier for comments on the manuscript. This work was funded by NIH grant AI088298 (DLP) and the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research (KSR).
Footnotes
AUTHOR CONTRIBUTIONS
Conceptualization, IST. and KSR.; Methodology, IST and KSR; Formal analysis, IST, CAW, DLP, and KSR.; Investigation, IST, CAW, DLP, and KSR; Writing-Original Draft, IST and KSR; Writing-Review and Editing, IST, CAW, DLP, and KSR; Funding Acquisition, DLP and KSR.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baker TA, Sauer RT. ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim Biophys Acta. 2012;1823:15–28. doi: 10.1016/j.bbamcr.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battesti A, Gottesman S. Roles of adaptor proteins in regulation of bacterial proteolysis. Curr Opin Microbiol. 2013;16:140–147. doi: 10.1016/j.mib.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano-Sagie M, Oppenheimer-Shaanan Y, Berlatzky I, Rouvinski A, Meyerovich M, Ben-Yehuda S. A checkpoint protein that scans the chromosome for damage at the start of sporulation in Bacillus subtilis. Cell. 2006;125:679–690. doi: 10.1016/j.cell.2006.03.039. [DOI] [PubMed] [Google Scholar]
- Bos J, Yakhnina AA, Gitai Z. BapE DNA endonuclease induces an apoptotic-like response to DNA damage in Caulobacter. Proc Natl Acad Sci U S A. 2012;109:18096–18101. doi: 10.1073/pnas.1213332109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke B. Lamins and apoptosis: a two-way street? J Cell Biol. 2001;153:F5–7. doi: 10.1083/jcb.153.3.f5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkholder WF, Kurtser I, Grossman AD. Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cell. 2001;104:269–279. doi: 10.1016/s0092-8674(01)00211-2. [DOI] [PubMed] [Google Scholar]
- Castaing JP, Lee S, Anantharaman V, Ravilious GE, Aravind L, Ramamurthi KS. An autoinhibitory conformation of the Bacillus subtilis spore coat protein SpoIVA prevents its premature ATP-independent aggregation. FEMS Microbiol Lett. 2014;358:145–153. doi: 10.1111/1574-6968.12452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaing JP, Nagy A, Anantharaman V, Aravind L, Ramamurthi KS. ATP hydrolysis by a domain related to translation factor GTPases drives polymerization of a static bacterial morphogenetic protein. Proc Natl Acad Sci U S A. 2013;110:E151–160. doi: 10.1073/pnas.1210554110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalano FA, Meador-Parton J, Popham DL, Driks A. Amino acids in the Bacillus subtilis morphogenetic protein SpoIVA with roles in spore coat and cortex formation. J Bacteriol. 2001;183:1645–1654. doi: 10.1128/JB.183.5.1645-1654.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer DJ, Camacho DM, Kohanski MA, Callura JM, Collins JJ. Antibiotic-induced bacterial cell death exhibits physiological and biochemical hallmarks of apoptosis. Mol Cell. 2012;46:561–572. doi: 10.1016/j.molcel.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebmeier SE, Tan IS, Clapham KR, Ramamurthi KS. Small proteins link coat and cortex assembly during sporulation in Bacillus subtilis. Mol Microbiol. 2012;84:682–696. doi: 10.1111/j.1365-2958.2012.08052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswaramoorthy P, Winter PW, Wawrzusin P, York AG, Shroff H, Ramamurthi KS. Asymmetric division and differential gene expression during a bacterial developmental program requires DivIVA. PLoS Genet. 2014;10:e1004526. doi: 10.1371/journal.pgen.1004526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiche JB, Cattoni DI, Diekmann N, Langerak JM, Clerte C, Royer CA, Margeat E, Doan T, Nollmann M. Recruitment, assembly, and molecular architecture of the SpoIIIE DNA pump revealed by superresolution microscopy. PLoS Biol. 2013;11:e1001557. doi: 10.1371/journal.pbio.1001557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Vitale I, Abrams JM, Alnemri ES, Baehrecke EH, Blagosklonny MV, Dawson TM, Dawson VL, El-Deiry WS, Fulda S, et al. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2012;19:107–120. doi: 10.1038/cdd.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner A, Boag PR, Blackwell TK. Germline survival and apoptosis. WormBook. 2008:1–20. doi: 10.1895/wormbook.1.145.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill RL, Jr, Castaing JP, Hsin J, Tan IS, Wang X, Huang KC, Tian F, Ramamurthi KS. Structural basis for the geometry-driven localization of a small protein. Proc Natl Acad Sci U S A. 2015;112:E1908–1915. doi: 10.1073/pnas.1423868112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S. Proteases and their targets in Escherichia coli. Annu Rev Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- Gottesman S, Clark WP, de Crecy-Lagard V, Maurizi MR. ClpX, an alternative subunit for the ATP-dependent Clp protease of Escherichia coli. Sequence and in vivo activities. J Biol Chem. 1993;268:22618–22626. [PubMed] [Google Scholar]
- Hacker G. Is there, and should there be, apoptosis in bacteria? Microbes Infect. 2013;15:640–644. doi: 10.1016/j.micinf.2013.05.005. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Heffron JD, Orsburn B, Popham DL. Roles of germination-specific lytic enzymes CwlJ and SleB in Bacillus anthracis. J Bacteriol. 2009;191:2237–2247. doi: 10.1128/JB.01598-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques AO, Moran CP., Jr Structure, assembly, and function of the spore surface layers. Annu Rev Microbiol. 2007;61:555–588. doi: 10.1146/annurev.micro.61.080706.093224. [DOI] [PubMed] [Google Scholar]
- Higgins D, Dworkin J. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol Rev. 2012;36:131–148. doi: 10.1111/j.1574-6976.2011.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imae Y, Strominger JL. Relationship between cortex content and properties of Bacillus sphaericus spores. J Bacteriol. 1976;126:907–913. doi: 10.1128/jb.126.2.907-913.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YI, Levchenko I, Fraczkowska K, Woodruff RV, Sauer RT, Baker TA. Molecular determinants of complex formation between Clp/Hsp100 ATPases and the ClpP peptidase. Nat Struct Biol. 2001;8:230–233. doi: 10.1038/84967. [DOI] [PubMed] [Google Scholar]
- Kunkel B, Losick R, Stragier P. The Bacillus subtilis gene for the development transcription factor sigma K is generated by excision of a dispensable DNA element containing a sporulation recombinase gene. Genes Dev. 1990;4:525–535. doi: 10.1101/gad.4.4.525. [DOI] [PubMed] [Google Scholar]
- Lau J, Hernandez-Alicea L, Vass RH, Chien P. A Phosphosignaling Adaptor Primes the AAA+ Protease ClpXP to Drive Cell Cycle-Regulated Proteolysis. Mol Cell. 2015;59:104–116. doi: 10.1016/j.molcel.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggett MJ, McDonnell G, Denyer SP, Setlow P, Maillard JY. Bacterial spore structures and their protective role in biocide resistance. J Appl Microbiol. 2012;113:485–498. doi: 10.1111/j.1365-2672.2012.05336.x. [DOI] [PubMed] [Google Scholar]
- Levin PA, Fan N, Ricca E, Driks A, Losick R, Cutting S. An unusually small gene required for sporulation by Bacillus subtilis. Mol Microbiol. 1993;9:761–771. doi: 10.1111/j.1365-2958.1993.tb01736.x. [DOI] [PubMed] [Google Scholar]
- Liu J, Cosby WM, Zuber P. Role of lon and ClpX in the post-translational regulation of a sigma subunit of RNA polymerase required for cellular differentiation in Bacillus subtilis. Mol Microbiol. 1999;33:415–428. doi: 10.1046/j.1365-2958.1999.01489.x. [DOI] [PubMed] [Google Scholar]
- Maughan H, Masel J, Birky CW, Jr, Nicholson WL. The roles of mutation accumulation and selection in loss of sporulation in experimental populations of Bacillus subtilis. Genetics. 2007;177:937–948. doi: 10.1534/genetics.107.075663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland KA, Dolben EL, LeRoux M, Kambara TK, Ramsey KM, Kirkpatrick RL, Mougous JD, Hogan DA, Dove SL. A self-lysis pathway that enhances the virulence of a pathogenic bacterium. Proc Natl Acad Sci U S A. 2015;112:8433–8438. doi: 10.1073/pnas.1506299112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney PT, Driks A, Eichenberger P. The Bacillus subtilis endospore: assembly and functions of the multilayered coat. Nat Rev Microbiol. 2013;11:33–44. doi: 10.1038/nrmicro2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney PT, Driks A, Eskandarian HA, Grabowski P, Guberman J, Wang KH, Gitai Z, Eichenberger P. A distance-weighted interaction map reveals a previously uncharacterized layer of the Bacillus subtilis spore coat. Curr Biol. 2010;20:934–938. doi: 10.1016/j.cub.2010.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paidhungat M, Setlow B, Driks A, Setlow P. Characterization of spores of Bacillus subtilis which lack dipicolinic acid. J Bacteriol. 2000;182:5505–5512. doi: 10.1128/jb.182.19.5505-5512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popham DL, Helin J, Costello CE, Setlow P. Analysis of the peptidoglycan structure of Bacillus subtilis endospores. J Bacteriol. 1996;178:6451–6458. doi: 10.1128/jb.178.22.6451-6458.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price KD, Losick R. A four-dimensional view of assembly of a morphogenetic protein during sporulation in Bacillus subtilis. J Bacteriol. 1999;181:781–790. doi: 10.1128/jb.181.3.781-790.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthi KS, Clapham KR, Losick R. Peptide anchoring spore coat assembly to the outer forespore membrane in Bacillus subtilis. Mol Microbiol. 2006;62:1547–1557. doi: 10.1111/j.1365-2958.2006.05468.x. [DOI] [PubMed] [Google Scholar]
- Ramamurthi KS, Lecuyer S, Stone HA, Losick R. Geometric cue for protein localization in a bacterium. Science. 2009;323:1354–1357. doi: 10.1126/science.1169218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthi KS, Losick R. ATP-driven self-assembly of a morphogenetic protein in Bacillus subtilis. Mol Cell. 2008;31:406–414. doi: 10.1016/j.molcel.2008.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roels S, Driks A, Losick R. Characterization of spoIVA, a sporulation gene involved in coat morphogenesis in Bacillus subtilis. J Bacteriol. 1992;174:575–585. doi: 10.1128/jb.174.2.575-585.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastalla I, Rosovitz MJ, Leppla SH. Accidental selection and intentional restoration of sporulation-deficient Bacillus anthracis mutants. Appl Environ Microbiol. 2010;76:6318–6321. doi: 10.1128/AEM.00950-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer EC, Glover JR, Singer MA, Lindquist S. HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem Sci. 1996;21:289–296. [PubMed] [Google Scholar]
- Setlow P. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J Appl Microbiol. 2006;101:514–525. doi: 10.1111/j.1365-2672.2005.02736.x. [DOI] [PubMed] [Google Scholar]
- Smith TJ, Foster SJ. Characterization of the involvement of two compensatory autolysins in mother cell lysis during sporulation of Bacillus subtilis 168. J Bacteriol. 1995;177:3855–3862. doi: 10.1128/jb.177.13.3855-3862.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterlini JM, Mandelstam J. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem J. 1969;113:29–37. doi: 10.1042/bj1130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan IS, Ramamurthi KS. Spore formation in Bacillus subtilis. Environ Microbiol Rep. 2014;6:212–225. doi: 10.1111/1758-2229.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan P, Weaver A, Reichert ED, Linnstaedt SD, Popham DL. Spore cortex formation in Bacillus subtilis is regulated by accumulation of peptidoglycan precursors under the control of sigma K. Mol Microbiol. 2007;65:1582–1594. doi: 10.1111/j.1365-2958.2007.05896.x. [DOI] [PubMed] [Google Scholar]
- Veening JW, Murray H, Errington J. A mechanism for cell cycle regulation of sporulation initiation in Bacillus subtilis. Genes Dev. 2009;23:1959–1970. doi: 10.1101/gad.528209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening JW, Stewart EJ, Berngruber TW, Taddei F, Kuipers OP, Hamoen LW. Bet-hedging and epigenetic inheritance in bacterial cell development. Proc Natl Acad Sci U S A. 2008;105:4393–4398. doi: 10.1073/pnas.0700463105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtkowiak D, Georgopoulos C, Zylicz M. Isolation and characterization of ClpX, a new ATP-dependent specificity component of the Clp protease of Escherichia coli. J Biol Chem. 1993;268:22609–22617. [PubMed] [Google Scholar]
- Wojtyra UA, Thibault G, Tuite A, Houry WA. The N-terminal zinc binding domain of ClpX is a dimerization domain that modulates the chaperone function. J Biol Chem. 2003;278:48981–48990. doi: 10.1074/jbc.M307825200. [DOI] [PubMed] [Google Scholar]
- Wu IL, Narayan K, Castaing JP, Tian F, Subramaniam S, Ramamurthi KS. A versatile nano display platform from bacterial spore coat proteins. Nat Commun. 2015;6:6777. doi: 10.1038/ncomms7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman P, Perkins JB, Losick R. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid. 1984;12:1–9. doi: 10.1016/0147-619x(84)90061-1. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Gottesman S, Hoskins JR, Maurizi MR, Wickner S. The RssB response regulator directly targets sigma(S) for degradation by ClpXP. Genes Dev. 2001;15:627–637. doi: 10.1101/gad.864401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.