Abstract
The first 12 cleavage divisions in Xenopus embryos provide a natural experiment in size scaling, as cell radius decreases ∼16-fold with little change in biochemistry. Analyzing both natural cleavage and egg extract partitioned into droplets revealed that mitotic spindle size scales with cell size, with an upper limit in very large cells. We discuss spindle-size scaling in the small- and large-cell regimes with a focus on the “limiting-component” hypotheses. Zygotes and early blastomeres show a scaling mismatch between spindle and cell size. This problem is solved, we argue, by interphase asters that act to position the spindle and transport chromosomes to the center of daughter cells. These tasks are executed by the spindle in smaller cells. We end by discussing possible mechanisms that limit mitotic aster size and promote interphase aster growth to cell-spanning dimensions.
The size of the mitotic spindle is generally proportional to the size of the cell. In large cells (e.g., early blastomeres), spindle size is limited, and interphase asters may perform tasks normally executed by the spindle.
How components and processes within cells scale in size and rate with the size of the cell has become a topic of considerable interest in recent years (reviewed in Chan and Marshall 2012; Goehring and Hyman 2012; Levy and Heald 2012). For molecular machines with precise architectures (e.g., ribosomes), size is invariant, but rates of assembly and function, which depend on regulation and energy, might scale. For assemblies whose dimensions are not hard wired (e.g., cytoskeleton assemblies and organelles), both size and rate might scale. For pathways involving distributed biochemical change (e.g., the cell-cycle oscillator), size is not well defined, but rate might scale in interesting ways. Here, we will address only size scaling, and refer the reader to interesting recent progress on cell-cycle timing in early Xenopus embryos (Chang and Ferrell 2013; Tsai et al. 2014).
Size-scaling relationships, which are part of the science of allometry, have long informed on whole organism physiology. Explicitly seeking them at the subcellular level is a newer endeavor, which in our mind holds two kinds of promise. It can inform on mechanism at the level of integrated cell physiology (e.g., on establishment of cleavage plane geometry). It can also inform on molecular processes involved in assembly growth and dynamics, and perhaps help us discern logic in often frustratingly complex molecular architectures. It is not obvious, for example, why ∼100 protein complexes are required to build a mitotic spindle in higher eukaryotes (Hutchins et al. 2010), when bacteria can segregate plasmids with far fewer (Salje et al. 2010). Part of the answer is the need for higher fidelity in the eukaryotic process. Gerhart and Kirschner (1997) also emphasized the need for highly adaptable processes in the evolution of higher eukaryotes. At least part of the complexity of subcellular assemblies might reflect the need for adaptable scaling of size, shape, and timing.
Vertebrate embryos derived from large eggs provide a natural experiment in size scaling (Fig. 1). A Xenopus laevis egg, for example, is ∼1.2 mm in diameter. Following fertilization, it cleaves completely ∼12 times at an approximately constant rate of ∼2 divisions/h (most rates in early development are temperature dependent, and can vary up to about eightfold over the tolerated range). These divisions generate a quasispherical array of quasispherical cells that are, on average, smaller by 212-fold in volume, or 24-fold in radius. The first 12 divisions occur with little gene expression and little change in cell physiology, and it may be reasonable to assume approximately constant biochemistry (discussed below), other than periodic cell-cycle regulation. After the 12th division, cell physiology changes dramatically as part of the midblastula transition (MBT) (discussed below), which provides a natural cut-off for size-scaling investigations. An interesting and potentially informative complication is that cleaving amphibian embryos develop a gradient in blastomere sizes, with larger cells at the vegetal pole where yolk is more abundant (evident in Fig. 1C,D). Larger blastomeres tend to divide more slowly, which gradually eliminates division synchrony (Gerhart 1980).
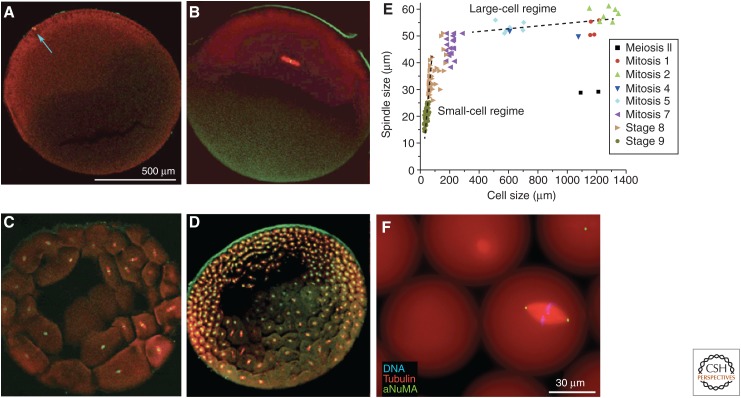
Figure 1.
Spindle-size scaling in Xenopus laevis. A–D show confocal images of eggs and early embryos fixed at different stages, stained for tubulin (red) and DNA (green), cleared and imaged by confocal microscopy. Embryos containing metaphase spindles were selected for analysis. (A) Unfertilized egg with meiosis-II spindle (blue arrow). (B) First mitosis. Note scaling mismatch between the spindle and egg. (C,D) Cleavage stages. (E) Spindle lengths and cell lengths derived from confocal images like A–D. Note spindle length is approximately constant in the large-cell regime and scales with cell size in the small-cell regime. (F) Spindle assembled in a droplet of unfertilized egg extract containing fluorescent probes suspended in oil and imaged live. aNuMA, anti-nuclear mitotic apparatus. (A–E from Wühr et al. 2008; adapted, with permission, from the author; F is an unpublished image provided by Jesse Gatlin, University of Wyoming, which is similar to images in Hazel et al. 2013.)
Embryos from different species have pros and cons for experimental analysis of size scaling during early divisions. Amphibian eggs provide a large dynamic range in cell size, complete division, and quasispherical geometry of both cells and embryos. In the minus column, they are opaque unless fixed and cleared and difficult to manipulate using genetics. Undiluted, cell-free extracts from Xenopus eggs and early embryos provide access to live imaging and molecular analysis and recapitulate the biology of intact eggs, including scaling relationships (Wilbur and Heald 2013), but it is important to go back to the intact embryo to check validity of key findings where possible. Zebrafish eggs provide a transparent, genetically tractable vertebrate system with very large cells but incomplete cleavage at early stages. Caenorhabditis elegans and Drosophila embryos have excellent imaging and genetics, which are advantages for scaling analysis, especially rate scaling (e.g., Carvalho et al. 2009; Hara and Kimura 2013), but these embryos start smaller, so they provide a lower dynamic range for analyzing size-scaling behavior.
SIZE AND RATIO SCALING
As cell size decreases during cleavage, two important ratios, DNA/cytoplasm and surface-area/volume, increase. These three changes are naturally linked and likely contribute in important but different ways to size scaling of cell physiology. Initial DNA/cytoplasm ratios can be changed fourfold or more in amphibians using haploid or polyploid embryos. This perturbation generated important discoveries in cell-size scaling effects at the tissue level (Fankhauser 1945). In a discussion of early embryo scaling effects, the most important impact of DNA/cytoplasm ratio is to trigger MBT around the 12th division (Newport and Kirschner 1982a). As this ratio approaches somatic values, division rate slows, G1 and G2 stages are added to the cell cycle, and rapid transcription commences (Newport and Kirschner 1982b). Certain proteins involved in regulating DNA replication were recently identified whose titration by DNA appears to cause MBT in Xenopus (Collart et al. 2013). This is an important breakthrough, although its generality remains to be determined. Important microtubule binding proteins, notably the spindle assembly factors TPX2 and HURP, are nuclear import substrates in interphase (Karsenti and Vernos 2001; Silljé et al. 2006). DNA/cytoplasm ratio might affect the degree to which such proteins are sequestered inside the nucleus during the short embryonic cell cycle. This potential source of size-scaling effects on interphase microtubule dynamics has not been investigated.
Surface-area/volume ratio plays a critical role in size scaling of whole organism physiology, and is one reason smaller animals have faster metabolic rates. It is presumably important in embryos; for example, it might influence metabolic physiology, or the influence of cortical actomyosin on interior cytomechanics, but this topic has been little investigated. When investigating nuclear size scaling in Xenopus, Levy and Heald (2010) found that the cytoplasmic concentration of the nuclear import factor importin-α decreased during cleavage. They proposed that this was caused by proportionally increased sequestration of importin by the plasma membrane. Importin-α is a positive factor in nuclear growth, but also a negative factor in spindle assembly, where it sequesters spindle assembly factors (Karsenti and Vernos 2001; Silljé et al. 2006). More work is needed to elucidate the biochemistry of importin binding to membranes, but this line of research suggests a general mechanism by which the sizes of subcellular assemblies are controlled by the cell’s surface/volume ratio.
STEADY-STATE VERSUS KINETIC DETERMINATION OF ASSEMBLY SIZE
A crucial question in any discussion of assembly size is whether size is measured at steady state or at some point in a time-varying trajectory. Implications for size-determining mechanisms are quite different, but it is sometimes difficult to know whether a steady state has been reached. Early frog and fish embryos divide frequently (every ∼20–40 min), so the spindle persists for only minutes in mitosis, and the nucleus for only tens of minutes in interphase. One could compare these durations to the time needed for typical proteins to diffuse through the cytoplasm (∼10 sec for somatic cells, potentially hours for large eggs). This means that nuclei in large blastomeres do not have time to import all of the nuclear proteins in the cell during the brief interphase, at least if diffusion limits import. Rather, we suspect that they grow continually, by nuclear import, until mitosis causes them to break down. Kinetic determination might explain the relatively constant size of nuclei in early embryos (Gerhart 1980), and their continual growth, albeit at nonlinear rates, in interphase egg extracts (Levy and Heald 2010).
Mitotic spindles and interphase asters likely differ in whether their size is determined kinetically or reaches a steady state. Interphase asters grow continuously, starting from either fertilization (for the sperm aster) or anaphase onset (for sister aster pairs derived from mitotic spindles). This growth continues until they reach a boundary, either the cell cortex or the interaction zone between sister asters at the cell center (Mitchison et al. 2012; Ishihara et al. 2014). Metaphase spindles are usually thought to reach a steady state in length. This is surely true of the egg meiosis-II spindle, which can persist for hours in unfertilized eggs, and at least 2 h in extracts from unfertilized eggs. It is less clear for zygote and blastomere mitotic spindles, which enter anaphase shortly after they assemble. Spindle length increased continuously from prophase to anaphase onset during of first mitosis in Xenopus (Wühr et al. 2008). Astral spindles assembled in embryo extracts arrested in mitosis do appear to reach a steady state in length (Wühr et al. 2008; Wilbur and Heald 2013), and there is good reason to believe that aster radius and microtubule length are bounded during mitosis (discussed below). Thus, we believe the upper limit to spindle size in early embryos is at or close to a steady state.
BIOCHEMICAL CONSTANCY IN EMBRYOS AND DROPLETS
A simplifying assumption when interpreting scaling measurements is that the biochemistry of the amphibian embryo is approximately constant through the first 12 divisions, other than changes caused by cell-cycle regulation and scaling of the DNA/cytoplasm and surface area/volume ratios. Validity of this assumption is currently being assessed using various approaches. The Kirschner and Gygi groups are using proteomics to quantify the concentration changes of ∼7000 proteins during early developmental stages. Preliminary results suggest that rather few protein levels change significantly in pre-MBT stages (M Wühr, pers. comm.), although this analysis might miss key regulatory proteins and, in its current form, does not address posttranslational regulation. Wilbur and Heald (2013) made concentrated extracts from egg and cleaving embryos at different stages, and found that extracts from later stages assembled smaller spindles that depended more on nucleation from centrosomes. These data suggest developmentally regulated changes in size-determining biochemistry during early embryogenesis, and revealed specific biochemical changes during early development that influenced spindle size invalidating the biochemical constancy assumption. However, the spindle length regulation they recapitulated in extract (about twofold change between extracts from 4 and 4000 cell stage) was less than that in intact embryos. Furthermore, caution is required interpreting data from extracts made from different stages, because they could differ for technical as well as developmental reasons. For example, later-stage embryos contain many nuclei, whose removal during extract preparation could deplete nuclear proteins. Certain nuclear proteins, including RCC1 and TPX2, are implicated in spindle-size control. In addition, the amount of water trapped between blastomeres, which is included in the extract following embryo rupture, and the presence or absence of proteins that are secreted after fertilization, might affect spindle assembly. It seems likely that spindle length is determined in part by size-scaling effects alone, and in part by developmental changes in biochemistry. Comparative proteomics applied to different-stage embryos, and extracts derived from them, should help disentangle these contributions.
An elegant recent approach explored spindle-size scaling using microfluidics, and, by using extracts from a single developmental stage, circumvented potential complications from biochemical change during early development (Good et al. 2013; Hazel et al. 2013). Extract from unfertilized eggs was partitioned into variable-sized droplets suspended in fluorocarbon oil, thus mimicking different blastomere sizes. Spindles were assembled (see Fig. 1F), and size scaling between spindle and droplet investigated. This approach revealed scaling that was quantitatively comparable to that between spindle and cell in cleaving embryos, with an almost linear relationship between spindle length and droplet radius in small droplets, and an upper limit in large droplets. These findings open a versatile new approach to size-scaling questions, and support the interpretation that spindle scaling in cleaving embryos depends in large part on cell size, rather than biochemical change. Both articles mainly used unfertilized egg extract, which generated anastral spindles resembling egg meiosis-II spindles, rather than astral blastomere spindles, although Good et al. (2013) also used extracts from stage 8 embryos, and obtained similar scaling results. It will be exciting to apply the nanodroplet technology to investigate size scaling of other subcellular assemblies.
Biochemical constancy cannot be assumed for between-species comparisons. The Heald laboratory took advantage of this to analyze molecular mechanisms underlying egg meiosis-II spindle-size scaling between X. laevis and the smaller frog Xenopus tropicalis, which has smaller eggs and smaller spindles. Using comparative genomics, they identified an amino acid change in the microtubule-severing protein katanin that altered its regulation by a mitotic kinase, and provided evidence that this genetic change partly accounts for between-species size scaling (Loughlin et al. 2011). This was the first time, to our knowledge, that species variation in the size of a cellular assembly had been attributed to a specific amino acid change. Following the logic of Loughlin et al. (2011), one might imagine that many amino acid changes would be required to allow evolution of different-sized cells, at least one for every organelle or assembly whose size changes. Gerhart and Kirschner (1997) argued the opposite, that the intrinsic adaptability of eukaryotic assemblies allows them to scale to different cell sizes and shapes without immediate need for mutations to accommodate the change, and this adaptability facilitates rapid morphological evolution. How evolution uses and acts on scaling relationships for subcellular assemblies is a fascinating topic for future research.
MITOTIC SPINDLE SCALING: POSSIBLE ROLES OF COMPONENT LIMITATION
Metaphase spindle length in early Xenopus embryos is near constant at 50–60 µm for the first five divisions, then scales somewhat linearly with cell size in divisions 8–12, such that spindle length approximately equals the cell radius (Fig. 1). We will refer to the two extremes as the large- and small-cell regimes, and discuss them separately. Size-scaling experiments in droplets did not explore the full range seen in embryos, but showed the same trends, with linear scaling in small droplets, and evidence for an upper limit in large droplets. Following previous investigators, we will discuss spindle-size scaling in light of a “limiting-component” hypothesis, which holds that that one or more components are limiting for the growth of an assembly, and the amount of those components per cell sets assembly size (Goehring and Hyman 2012). We note that a limiting component more naturally sets spindle “mass” than “length,” and that mass is arguably the more fundamental unit for scaling discussions. Length is, however, easier to measure by microscopy, more naturally related to spindle biology, and has, so far, dominated the discussion.
Small-Cell Regime
It is natural to propose that spindle size in the small-cell regime is determined by limiting amounts of one or more components, so that when cell volume halves, the amount per cell of these components also halves, giving rise to linear size scaling (Fig. 2A). This limiting component(s) could be tubulin, one or more spindle proteins, or something else. Complete knockdown or depletion experiments, which dominate the molecular literature, do not address potential size-limiting roles of spindle components. Rather, the precise relationship between amount of protein and spindle mass or length must be measured. A semisystematic reduction of five spindle proteins in C. elegans embryos—γ-tubulin, Tac-1 (similar to TACC3), Zyg9 (similar to XMAP215), KLP-7 (similar to MCAK), and TPXL (similar to TPX2)—concluded that only TPXL fulfilled the criteria to be length limiting, although depletion of others affected microtubule mass (Greenan et al. 2010). TPX2 is an important effector between Ran-GTP and microtubule assembly, suggesting that length scaling might ultimately depend on activities emanating from chromatin. More work is required to test the hypothesis that limiting amounts of one or more spindle proteins limit spindle size in small cells and to identify limiting proteins. Proteomics might provide an unbiased screen, and the droplet system will be useful to test candidates.
Figure 2.
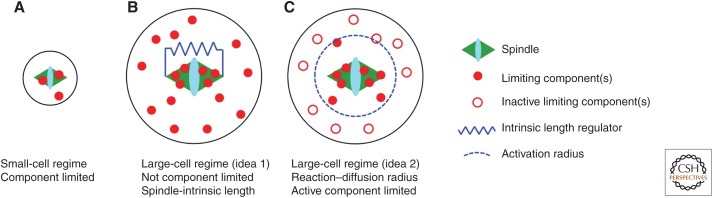
Hypothetical roles of component limitation in spindle-size scaling. (A) Small-cell regime. Spindle size is limited by depletion of one or more limiting components, drawn as a red dot, from the whole cell. (B) Large-cell regime hypothesizing spindle size is not component limited. Some spindle-intrinsic mechanisms, drawn as a blue spring, limits length. Candidate mechanisms include bounded single-microtubule lengths, opposed motor activities, and a mechanochemical switch at the pole (see text for references). (C) Large-cell regime hypothesizing spindle size is component limited. The limiting component is locally activated by a reaction–diffusion system centered on chromosomes. The spindle depletes the activated component within the activation radius, leading to component limitation. A candidate reaction–diffusion system is RCC1-Ran-RanGAP1, which activates two potentially limiting spindle components, TPX2 and HURP.
Large-Cell Regime
Uncoupling of spindle and cell size in this regime would appear to rule out the limiting component model. Rather, spindle size might by be determined by processes that are intrinsic to the spindle (Fig. 2B). Candidate processes include microtubule length limitation by bounded dynamic instability (Reber et al. 2013), opposed motor activities acting on poles (Goshima and Scholey 2010), and a mechanochemical switch regulating kinetochore fiber depolymerization at poles (Dumont and Mitchison 2009a). These models were reviewed in Dumont and Mitchison (2009b), and we will not discuss them further here. Rather, we suggest that a modified version of the limiting component model should still be considered in the large-cell regime. The necessary modification is that spindle growth is limited by the amount of the “activated” form of one or more spindle proteins, as opposed to the total amount, and activation only occurs in a limited radius around spindles (Fig. 2C). The spindle assembly factors TPX2 and HURP must be released from importin α/β by Ran-GTP to exert their assembly-promoting activities (Gruss et al. 2001; Silljé et al. 2006), and are, thus, interesting candidates for limiting spindle growth by the mechanism proposed in Figure 3C. Ran-GTP is generated locally at chromatin by RCC1 and inactivated globally by RANGAP1, leading to a reaction-diffusion gradient at steady state with a length scale in the tens of microns in Xenopus meiosis-II spindles (Kalab and Heald 2008). The radius of the resulting sphere of Ran-GTP-positive cytoplasm—denoted by the dotted blue line in Figure 3C—is, thus, much smaller than the cell in the large-cell regime. We note that the activation process shown in Figure 3C might cause a flux of unactivated factor across the boundary, with the spindle acting as a sink, in which case spindles would grow over time. In this sense, it might not generate a true steady-state spindle length.
Figure 3.
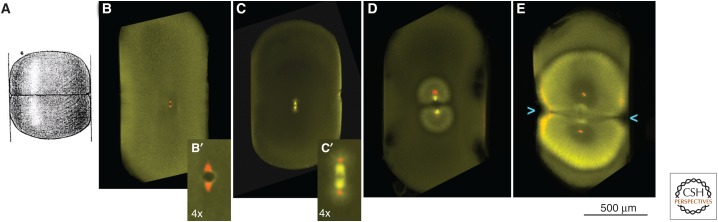
Aster scaling and centrosome orientation in compressed eggs. (A) In a famous experiment, Hertwig compressed amphibian eggs after fertilization, which oriented the first cleavage plane to cut across the short axis of the cell. (Image from Hertwig 1893.) (B–E) Repeat of Hertwig’s egg compression experiment in Xenopus laevis followed by fixation at different stages and staining for tubulin (yellow–green) and the microtubule nucleation factor γ-tubulin (red). (From Wühr et al. 2010; adapted, with permission, from the authors.) (B,B′) Prophase of first mitosis, just before nuclear envelope breakdown. The remains of the cell-spanning sperm aster can be faintly seen near the egg periphery. Note that centrosomes (red) are already oriented correctly (N–S) to define the future division plane (E–W). (C, C′) Metaphase of first mitosis. The scaling mismatch between spindle and cell is evident. Astral microtubules are poorly visualized, but appear short compared with egg radius, even in the compressed egg. (D) Early interphase. Sister asters from the mitotic spindle are growing out toward the cortex. Their periphery grows outward at 20–30 µm/min, the centrosomes and nuclei at their centers move apart at about half this rate. (E) Early cytokinesis. The expanding sister asters have now grown to touch the cortex. The cleavage furrow has just started to ingress at the overlap zone between the two asters (blue arrowheads). In this example, the furrow is imperfectly aligned relative to the long axis of the compressed cell (by ∼7°) (see Wühr et al. 2010 for alignment statistics).
SCALING MISMATCH PROBLEMS AND SOLUTIONS
In small somatic cells, the plus ends of astral microtubules reach the cortex during mitosis, allowing metaphase spindles to position and orient themselves, which in turn controls cleavage geometry (Kiyomitsu and Cheeseman 2013). Spindle length is approximately half of cell length, so spindles can separate sister chromosomes by a large fraction of cell length. In large embryo cells, metaphase spindle length is small compared with cell radius, and astral microtubules are too short, at metaphase, to position the spindle or to move chromosomes to the center of incipient daughter cells. In other words, a scaling mismatch exists between the metaphase spindle and the cell. How, then, are spindles positioned and oriented so as to control cleavage geometry, and how are chromosomes segregated? We argued that both problems are solved by interphase asters, which grow to cell-spanning dimensions, and execute tasks that are the job of metaphase microtubules in smaller cells. Specifically, prophase centrosomes are positioned and oriented by dynein-pulling forces acting on cell-spanning astral microtubules in the previous cell cycle, and sister nuclei move far apart, to a position midway between the cell center and the cortex, using the same pulling force, now acting on growing telophase asters (Wühr et al. 2010). These points are illustrated in an experiment, in which the egg was compressed after fertilization to orient the first cleavage furrow (see Fig. 3). Note that centrosomes are already positioned at prophase, presumably by dynein forces acting on the sperm aster, and that asters only grow out to the cortex, where they position the cleavage furrow, in interphase. The nature of the dynein-mediated forces that pull on microtubules from bulk cytoplasm to position centrosomes has been discussed elsewhere (Wühr et al. 2010; Kimura and Kimura 2011; Minc et al. 2011).
The potential for a scaling mismatch between metaphase asters and cell radius generalizes beyond embryos. Astral microtubules in mitotic tissue culture cells are normally just long enough to touch the cortex and capture chromosomes, although how this length is determined is not known. Compressing mitotic cells, or preventing them from rounding up by increasing cell-substrate adhesion, increased cell radius sufficiently to cause a scaling mismatch (Lancaster et al. 2013). Spindles were mispositioned, and astral microtubules were no longer long enough to capture chromosomes in compressed cells. Major defects in chromosome segregation ensued. According to this analysis, even somatic cells are perilously close to a scaling mismatch between mitotic aster and cell radius. Lancaster et al. argued that mitotic rounding in animal tissues evolved in part to prevent this mismatch. By rendering the cell more spherical, rounding brings chromosomes and the cortex closer to plus ends of mitotic aster microtubules, allowing mitosis to proceed without error. Speculating on the basis of these observations, mild scaling mismatches between mitotic aster and cell radius might cause chromosome-segregation defects in cancer cells, especially those that overexpress the polymerization-inhibiting protein Stathmin/Op18, which shortens astral microtubules and can cause aneuploidy (Holmfeldt et al. 2006).
MITOTIC VERSUS INTERPHASE ASTER SCALING
How is aster growth bounded in mitosis, and how do interphase asters grow to fill the cell? We lack definitive answers to these questions, but will discuss current knowledge in light of three models shown in Figure 4. The mitotic aster length scale problem is related to the metaphase spindle length problem, because both assemblies are built from highly dynamic mitotic microtubules and microtubule length can determine spindle length (Reber et al. 2013). The aster problem is, however, simpler, because we can ignore the complexities of sliding motors and kinetochore fibers. The Karsenti and Leibler groups first pointed out that dynamic instability naturally generates a regime in which microtubule growth is bounded, and provided evidence that mitotic aster microtubules in egg extract are in this regime (Verde et al. 1992). If we believe that the plus ends of microtubules as undergoing biased random walks where the minus end is the origin, the bounded regime corresponds to bias back toward the origin. Microtubules only exist in the bounded regime because they are renucleated continuously after their length fluctuates to zero (blue arrows in Fig. 4A). More complex mechanisms for bounding microtubule growth, in which a plus end–directed motor transports a catastrophe factor (which could be intrinsic to the motor) to the microtubule tip, have also been proposed (Varga et al. 2009). Whether by dynamic instability alone or by some more complex mechanism, mitotic aster radius is likely set by bounded growth of individual microtubules (Fig. 4A). What determines the relevant dynamics parameters and, thus, scales the aster is an important future question.
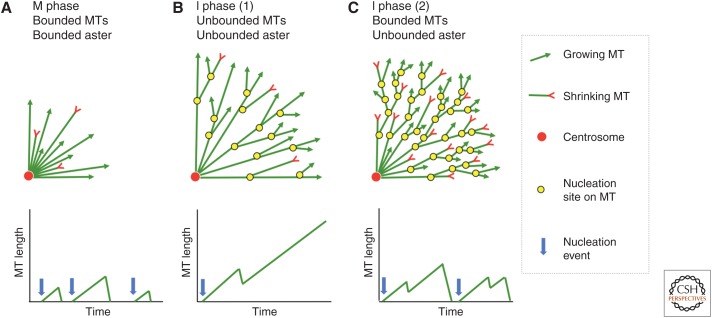
Figure 4.
Models for aster and microtubule (MT) length scaling. The top row illustrates a model for aster growth in mitosis (A) and two related models for growth in interphase (B,C). A quadrant of the aster is shown. The bottom row illustrates individual microtubule dynamics for each model, in which length fluctuations are caused by dynamic instability of plus ends. (A) Bounded mitotic aster made from bounded microtubules. Nucleation is restricted to the centrosome. Aster radius is determined by the length scale of individual microtubules. A similar model with unbounded microtubules would suffice for interphase asters in small cells. (B) Unbounded interphase aster made from unbounded microtubules. Both the aster and individual microtubules grow to cell-spanning dimensions. Nucleation away from centrosomes maintains a high density of microtubules at the periphery of the growing aster. (C) Unbounded interphase aster made from bounded microtubules. Individual microtubules are short compared with aster radius. Nucleation away from centrosomes is required at a faster rate than in B to promote aster growth.
When cells exit mitosis, the activity of the Cdk1 kinase abruptly decreases, and its mitotic substrates are dephosphorylated at different rates (Hunt 2013). Many microtubule-binding proteins are regulated by Cdk1, although the full impact of change in Cdk1 activity on microtubule dynamics has yet to be elucidated (Niethammer et al. 2007). What are the overall consequences of this regulation and, specifically, how do interphase asters grow to span the cell? These questions can be asked of the aster as a network, and also of the microtubules within it. In small cells, it is probably sufficient to modify the mitotic model (Fig. 4A) by changing dynamic instability parameters to make individual microtubule growth unbounded at least as far as the cortex, where catastrophes are triggered by an unknown mechanism. Aster radius is directly coupled to maximum microtubule length in this model. We believe, however, that this standard model does not scale to large interphase asters in eggs and early embryos, either in principle or in reality. If all minus ends anchor at the centrosome, microtubule density per surface area at the periphery of a spherical aster should scale as ∼1/r2, where r is aster radius. This would lead to a very low density at the cortex in large cells. Imaging in frog and fish embryos showed that density at the aster periphery is approximately constant as radius increases (Mitchison et al. 2012). This presumably requires microtubule nucleation away from centrosomes, illustrated as yellow circles in Figure 4B and 4C.
We recently asked how large interphase asters grow in frog eggs using the extract system (Ishihara et al. 2014). Asters grew to hundreds of microns in radius, and by tracking growing plus ends using an EB1 probe, we found strong evidence for nucleation remote from the centrosome. Microtubules also moved outward by dynein-powered sliding, but sliding was not essential for aster growth. How microtubules are nucleated away from the centrosome in interphase asters is not clear. A logical possibility is that microtubules are nucleated from the sides of preexisting microtubules, as they are in plants (Murata et al. 2005), via recruitment of γ-tubulin complexes by Augmin/Haus complexes (Petry et al. 2013). However, in preliminary experiments, immunodepletion of augmin did not block aster growth (K Ishihara and TJ Mitchison, unpubl.). We concluded that interphase asters in large egg and blastomere cells likely grow outward as an unbounded network of cross-linked microtubules, using an unknown mechanism to nucleate remote from centrosomes (Fig. 4B,C).
Growth of interphase asters as a network of microtubules is unbounded, but is the growth of individual microtubules within them also unbounded? Do some microtubules extend all the way from the centrosome to the cortex by unbounded growth (Fig. 4B), or are large asters built from a cross-linked array of short, bounded microtubules (Fig. 4C)? Old measurements of polymerization dynamics in extract suggested unbounded growth in interphase (Verde et al. 1992), but with frequent catastrophes and only a fairly small net bias toward polymerization (Belmont et al. 1990). When large interphase asters grew in egg extract, we observed a relatively constant density of growing plus ends inside the aster (Ishihara et al. 2014). This observation is more consistent with short, bounded microtubules (Fig. 4C) than long, unbounded microtubules (Fig. 4B). If individual microtubule growth was unbounded, then either the aster interior would become depleted of growing plus ends over time, or the microtubule density in the interior would steadily increase. Because neither was observed, we tentatively conclude that millimeter-scale egg-spanning interphase asters are built from an expanding network of short microtubules by constant nucleation at the aster periphery (Fig. 4C). Nucleation at the periphery, we hypothesize, “rectifies” dynamic instability, and allows the aster to grow even though individual plus ends eventually shrink (Ishihara et al. 2014). We are currently pursuing mathematical models and additional microscopy to test this model.
Minus-end dynamics are also of considerable interest in interphase asters, especially in light of the recently discovered CAMSAP/Patronin family of minus-end stabilizing proteins (Meng et al. 2008; Goodwin and Vale 2010; Tanaka et al. 2012). In frog eggs, we invariably observe a lower microtubule density in the center compared with the periphery by immunofluorescence (e.g., Fig. 3D,E). This could be a staining artifact, but we also observe interphase asters “hollowing out” at later time points in extract experiments. The internal dynamics of large interphase asters, and how asters transition from interphase back to mitosis, are interesting topics for future study, and might reflect regulated minus-end dynamics.
AFTERWORD—SIZES, RATES, AND MOLECULES
How subcellular assemblies scale with cell size is a rich question, and work on this aspect of microtubule organization has so far only brushed the surface of the problem. There has been more progress on length than mass, and on size than rates, although all are interesting. Compression experiments on cancer cells revealed, unexpectedly, that even small, somatic cells face microtubule-scaling challenges that might be significant for pathophysiology (Lancaster et al. 2013). This exciting work suggests that it will be profitable to seek size-scaling relationships and their breakdown even in small tissue cells. We clearly need more progress on linking mesoscopic scaling behavior to microscopic molecular mechanism. Elucidating such links holds great hope for understanding the logic and evolution of the complex molecular networks that support growth and dynamics of cellular assemblies.
ACKNOWLEDGMENTS
Work from our group is supported by National Institutes of Health (NIH) Grant GM39565 and research fellowships from the Marine Biological Laboratory (MBL), Woods Hole. Microscopy was performed in part at the Nikon Imaging Center at Harvard Medical School (HMS), and we thank Nikon for microscopy support at MBL. M.W. is supported by the Charles A. King Trust Postdoctoral Fellowship Program and R01GM103785.
Footnotes
Editors: Rebecca Heald, Iswar K. Hariharan, and David B. Wake
Additional Perspectives on Size Control in Biology: From Organelles to Organisms available at www.cshperspectives.org
REFERENCES
- Belmont LD, Hyman AA, Sawin KE, Mitchison TJ. 1990. Real-time visualization of cell cycle–dependent changes in microtubule dynamics in cytoplasmic extracts. Cell 62: 579–589. [DOI] [PubMed] [Google Scholar]
- Carvalho A, Desai A, Oegema K. 2009. Structural memory in the contractile ring makes the duration of cytokinesis independent of cell size. Cell 137: 926–937. [DOI] [PubMed] [Google Scholar]
- Chan YHM, Marshall WF. 2012. How cells know the size of their organelles. Science 337: 1186–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JB, Ferrell JE. 2013. Mitotic trigger waves and the spatial coordination of the Xenopus cell cycle. Nature 500: 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart C, Allen GE, Bradshaw CR, Smith JC, Zegerman P. 2013. Titration of four replication factors is essential for the Xenopus laevis midblastula transition. Science 341: 893–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont S, Mitchison TJ. 2009a. Compression regulates mitotic spindle length by a mechanochemical switch at the poles. Curr Biol 19: 1086–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont S, Mitchison TJ. 2009b. Force and length in the mitotic spindle. Curr Biol 19: R749–R761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser G. 1945. Maintenance of normal structure in heteroploid salamander larvae, through compensation of changes in cell size by adjustment in cell number and shape. J Exp Zool 100: 445–455. [DOI] [PubMed] [Google Scholar]
- Gerhart JC. 1980. Mechanisms regulating pattern formation in the amphibian egg and embryo. In Biological regulation and development (ed. Goldberger R) pp. 133–316. Plenum, New York. [Google Scholar]
- Gerhart J, Kirschner M. 1997. Cells, embryos, and evolution: Towards a cellular and developmental understanding of phenotypic variation and evolutionary adaptability. Blackwell, London. [Google Scholar]
- Goehring NW, Hyman AA. 2012. Organelle growth control through limiting pools of cytoplasmic components. Curr Biol 22: R330–R339. [DOI] [PubMed] [Google Scholar]
- Good MC, Vahey MD, Skandarajah A, Fletcher DA, Heald R. 2013. Cytoplasmic volume modulates spindle size during embryogenesis. Science 342: 856–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin SS, Vale RD. 2010. Patronin regulates the microtubule network by protecting microtubule minus ends. Cell 143: 263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Scholey JM. 2010. Control of mitotic spindle length. Annu Rev Cell Dev Biol 26: 21–57. [DOI] [PubMed] [Google Scholar]
- Greenan G, Brangwynne CP, Jaensch S, Gharakhani J, Jülicher F, Hyman AA. 2010. Centrosome size sets mitotic spindle length in Caenorhabditis elegans embryos. Curr Biol 20: 353–358. [DOI] [PubMed] [Google Scholar]
- Gruss OJ, Carazo-Salas RE, Schatz CA, Guarguaglini G, Kast J, Wilm M, Le Bot N, Vernos I, Karsenti E, Mattaj IW. 2001. Ran induces spindle assembly by reversing the inhibitory effect of importin α on TPX2 activity. Cell 104: 83–93. [DOI] [PubMed] [Google Scholar]
- Hara Y, Kimura A. 2013. An allometric relationship between mitotic spindle width, spindle length, and ploidy in Caenorhabditis elegans embryos. Mol Biol Cell 24: 1411–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazel J, Krutkramelis K, Mooney P, Tomschik M, Gerow K, Oakey J, Gatlin JC. 2013. Changes in cytoplasmic volume are sufficient to drive spindle scaling. Science 342: 853–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertwig O. 1893. Ueber den Werth der ersten Furchungszellen für die Organbildung des Embryo. Experimentelle Studien am Frosch- und Tritonei [About the role of the first cleavage cells on the formation of organs in the embryo: Experimental studies on the frog and newt egg]. Arch Mikr Anat 42: 662–807. [Google Scholar]
- Holmfeldt P, Brännström K, Stenmark S, Gullberg M. 2006. Aneugenic activity of Op18/stathmin is potentiated by the somatic Q18→E mutation in leukemic cells. Mol Biol Cell 17: 2921–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt T. 2013. On the regulation of protein phosphatase 2A and its role in controlling entry into and exit from mitosis. Adv Biol Regul 53: 173–178. [DOI] [PubMed] [Google Scholar]
- Hutchins JRA, Toyoda Y, Hegemann B, Poser I, Hériché JK, Sykora MM, Augsburg M, Hudecz O, Buschhorn BA, Bulkescher J, et al. 2010. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 328: 593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara K, Nguyen PA, Groen AC, Field CM, Mitchison TJ. 2014. Microtubule nucleation remote from centrosomes may explain how asters span large cells. Proc Natl Acad Sci 111: 17715–17722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P, Heald R. 2008. The RanGTP gradient—A GPS for the mitotic spindle. J Cell Sci 121: 1577–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenti E, Vernos I. 2001. The mitotic spindle: A self-made machine. Science 294: 543–547. [DOI] [PubMed] [Google Scholar]
- Kimura K, Kimura A. 2011. Intracellular organelles mediate cytoplasmic pulling force for centrosome centration in the Caenorhabditis elegans early embryo. Proc Natl Acad Sci 108: 137–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T, Cheeseman IM. 2013. Cortical dynein and asymmetric membrane elongation coordinately position the spindle in anaphase. Cell 154: 391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster OM, Le Berre M, Dimitracopoulos A, Bonazzi D, Zlotek-Zlotkiewicz E, Picone R, Duke T, Piel M, Baum B. 2013. Mitotic rounding alters cell geometry to ensure efficient bipolar spindle formation. Dev Cell 25: 270–283. [DOI] [PubMed] [Google Scholar]
- Levy DL, Heald R. 2010. Nuclear size is regulated by importin α and Ntf2 in Xenopus. Cell 143: 288–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DL, Heald R. 2012. Mechanisms of intracellular scaling. Annu Rev Cell Dev Biol 28: 113–135. [DOI] [PubMed] [Google Scholar]
- Loughlin R, Wilbur JD, McNally FJ, Nédélec FJ, Heald R. 2011. Katanin contributes to interspecies spindle length scaling in Xenopus. Cell 147: 1397–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng W, Mushika Y, Ichii T, Takeichi M. 2008. Anchorage of microtubule minus ends to adherens junctions regulates epithelial cell–cell contacts. Cell 135: 948–959. [DOI] [PubMed] [Google Scholar]
- Minc N, Burgess D, Chang F. 2011. Influence of cell geometry on division-plane positioning. Cell 144: 414–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T, Wühr M, Nguyen P, Ishihara K, Groen A, Field CM. 2012. Growth, interaction, and positioning of microtubule asters in extremely large vertebrate embryo cells. Cytoskeleton (Hoboken) 69: 738–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T, Sonobe S, Baskin TI, Hyodo S, Hasezawa S, Nagata T, Horio T, Hasebe M. 2005. Microtubule-dependent microtubule nucleation based on recruitment of γ-tubulin in higher plants. Nat Cell Biol 7: 961–968. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M. 1982a. A major developmental transition in early Xenopus embryos: I. Characterization and timing of cellular changes at the midblastula stage. Cell 30: 675–686. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M. 1982b. A major developmental transition in early Xenopus embryos. II: Control of the onset of transcription. Cell 30: 687–696. [DOI] [PubMed] [Google Scholar]
- Niethammer P, Kronja I, Kandels-Lewis S, Rybina S, Bastiaens P, Karsenti E. 2007. Discrete states of a protein interaction network govern interphase and mitotic microtubule dynamics. PLoS Biol 5: e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry S, Groen AC, Ishihara K, Mitchison TJ, Vale RD. 2013. Branching microtubule nucleation in Xenopus egg extracts mediated by augmin and TPX2. Cell 152: 768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reber SB, Baumgart J, Widlund PO, Pozniakovsky A, Howard J, Hyman AA, Jülicher F. 2013. XMAP215 activity sets spindle length by controlling the total mass of spindle microtubules. Nat Cell Biol 15: 1116–1122. [DOI] [PubMed] [Google Scholar]
- Salje J, Gayathri P, Löwe J. 2010. The ParMRC system: Molecular mechanisms of plasmid segregation by actin-like filaments. Nat Rev Microbiol 8: 683–692. [DOI] [PubMed] [Google Scholar]
- Silljé HHW, Nagel S, Körner R, Nigg EA. 2006. HURP is a Ran-importin β-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr Biol 16: 731–742. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Meng W, Nagae S, Takeichi M. 2012. Nezha/CAMSAP3 and CAMSAP2 cooperate in epithelial-specific organization of noncentrosomal microtubules. Proc Natl Acad Sci 109: 20029–20034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai TYC, Theriot JA, Ferrell JE. 2014. Changes in oscillatory dynamics in the cell cycle of early Xenopus laevis embryos. PLoS Biol 12: e1001788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga V, Leduc C, Bormuth V, Diez S, Howard J. 2009. Kinesin-8 motors act cooperatively to mediate length-dependent microtubule depolymerization. Cell 138: 1174–1183. [DOI] [PubMed] [Google Scholar]
- Verde F, Dogterom M, Stelzer E, Karsenti E, Leibler S. 1992. Control of microtubule dynamics and length by cyclin A- and cyclin B-dependent kinases in Xenopus egg extracts. J Cell Biol 118: 1097–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur JD, Heald R. 2013. Mitotic spindle scaling during Xenopus development by kif2a and importin α. eLife 2: e00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wühr M, Chen Y, Dumont S, Groen AC, Needleman DJ, Salic A, Mitchison TJ. 2008. Evidence for an upper limit to mitotic spindle length. Curr Biol 18: 1256–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wühr M, Tan ES, Parker SK, Detrich HW III, Mitchison TJ. 2010. A model for cleavage plane determination in early amphibian and fish embryos. Curr Biol 20: 2040–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]