SUMMARY
To accommodate genomes in the limited space of the cell nucleus and ensure the correct execution of gene expression programs, genomes are packaged in complex fashion in the three-dimensional cell nucleus. As a consequence of the extensive higher-order organization of chromosomes, distantly located genomic regions on the same or distinct chromosomes undergo long-range interactions. This article discusses the nature of long interactions, mechanisms of their formation, and their emerging functional roles in gene regulation and genome maintenance.
Physical interactions between distant genomic regions are important for the regulation of genes, imprinting, X inactivation, and other processes.
Overview
The higher-order packing of DNA in the cell nucleus is both driven by and leading to physical interactions between genomic regions. The formation of physical interactions between loci, even distantly located ones, is not surprising in itself. The question of whether these interactions are functionally meaningful or merely a by-product of the tight packing of DNA in a limited space is what needs deciphering. Although some interactions are likely purely because of the random proximity of the genome regions in the nucleus, two facts argue very strongly against mere haphazardness and for the existence of recurrent and functionally relevant interactions. The first is that if interactions were functionally irrelevant, then one would predict that they would occur largely randomly in a population. Based on unbiased mapping studies this is not the case—specific, high-frequency interactions can be mapped in a large range of organisms (Splinter and de Laat 2011). The second argument stems from the numerous examples of long-range interactions, both intra- and interchromosomal, that have been molecularly dissected and shown to have significant functional consequences. Among the best characterized are the globin genes, the Hox genes, imprinted loci, and interaction involved in X-inactivation (see discussion below).
Long-range interactions can occur intrachromosomally between regions located on the same chromosome or interchromosomally between regions on distinct chromosomes (Deng and Blobel 2010; Dean 2011). Intrachromosomal interactions have been reported between promoters and terminators over several kilobases, as have interactions between promoters and enhancers located tens of kilobases up to megabases away (Deng and Blobel 2010; Dean 2011). Another type of intrachromosomal interaction is insulator-mediated contacts, which appear to contribute to the organization of the genome into functionally distinct regions by separating differentially regulated regions from each other (Phillips and Corces 2009).
Compared with intrachromosomal interactions, many fewer interchromosomal interactions are known. Interchromosomal interactions are mostly involved in promoting the formation of chromatin domains such as centromere clusters. There are some intriguing examples suggesting that these types of interactions may also be involved in gene regulation, for example, in the control of olfactory receptors, interferon-responsive genes, and also more globally in the establishment of X-inactivation (discussed below and also the topic of Brockdorff and Turner 2014; Busslinger and Tarakhovsky 2014; Lomvardas and Maniatis 2014). Beyond gene regulation, interchromosomal interactions are also now appreciated as playing a particularly critical role in the formation of chromosomal translocations because the rejoining of broken chromosomes requires their physical interaction (Misteli and Soutoglou 2009).
Traditionally, genomic interactions have been detected using fluorescence in situ hybridization (FISH). In this approach, cells are chemically fixed, chromatin denatured, and fluorescently labeled probes hybridized to their targets. This enables the visualization of specific genes, genomic regions, or entire chromosomes by fluorescence microscopy. In these approaches genomic regions are detected as bright fluorescent signals and use of multicolor labels allows detection of a few loci simultaneously. Distances between fluorescent spots can be measured to provide information regarding the proximity of the investigated regions. For example, this approach was used to investigate the organization of the immune IgH gene during VDJ recombination (discussed in Busslinger and Tarakhovsky 2014). The advantage of FISH is the ability to interrogate interactions at the single cell level, thus allowing determination of the fraction of cells in a population that harbors a particular interaction. However, FISH methods are limited in the resolution with which they can determine spatial proximity of genomic regions and they do not provide information about actual physical association of two loci. Furthermore, FISH can only be applied in candidate approaches and is not easily amenable to the unbiased discovery of novel genome interactions.
More recently, biochemical approaches that map physical genome interactions have been developed (Sanyal et al. 2011; Splinter and de Laat 2011). These so-called “C” techniques (3C, 4C, 5C, and Hi-C) involve chemical cross-linking of the genomes, fragmenting chromatin (e.g., by digestion with restriction enzymes), religating cross-linked regions, and identifying the interacting regions either by polymerase chain reaction (PCR) or DNA sequencing (Sanyal et al. 2011; Splinter and de Laat 2011; Hakim and Misteli 2012). Most of these methods allow the unbiased and genome-wide mapping of interactions without prior knowledge of partners, thus permitting the discovery of new interactions. Their downside is that these are population-based approaches that only generate averaged data and cannot interrogate the behavior of single cells or subpopulations. A combination of the two methods is a powerful approach to solving the question of how genomes are spatially organized.
1. Introduction: The challenge of storing DNA in vivo
Genomes are organized in complex ways in the 3D space of the cell nucleus (Misteli 2007; Rajapakse and Groudine 2011; Cavalli and Misteli 2013). A typical higher eukaryotic cell contains ∼2 m of DNA that must be packed into a nucleus ∼10 µm in diameter. Assuming approximately five trillion cells in the human body, the total amount of DNA in an individual is on the order of 10 trillion meters—the equivalent of spanning the distance from Earth to Sun more than 100 times! Clearly, cells face a tremendous challenge to ensure the safekeeping and accurate propagation of DNA during replication and cell division, at the same time allowing accessibility of regulatory factors to genes at the right time and in the right place.
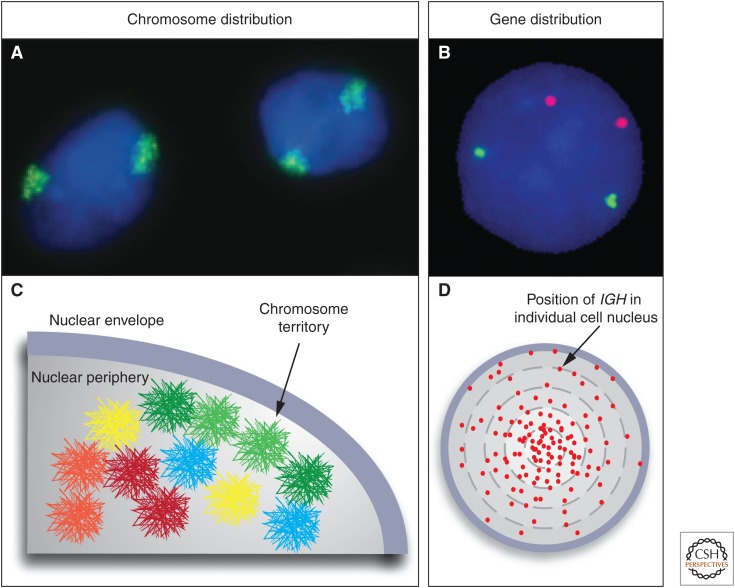
To accommodate the immense length of DNA in the nucleus and to ensure functionality, the genetic material is wrapped into higher-order chromatin fibers culminating in the organization of chromatin into chromosome territories (Fig. 1A,C). A chromosome territory is the physical space taken up by a given chromosome in the interphase nucleus (Cremer et al. 1982; Lanctot et al. 2007). Despite the extraordinary length of DNA in a cell, it is noteworthy that DNA-containing chromatin only takes up an estimated 15% of the nuclear volume.
Figure 1.
Chromosome territories and genes in three-dimensional (3D) space. Fluorescence in situ hybridization (FISH) visualizes the spatial organization of the genome. (A) Chromosomes exist in the form of chromosome territories in the interphase nucleus. The DNA of each chromosome occupies a spatially well-defined fraction of the nuclear volume, typically about 1–2 μm in diameter. Chromosome 11 (green) in the nucleus (blue) of MCF10A breast cancer cells is shown. (B) Individual genes appear as distinct spots. MYC (red) and TGFBR2 (green) in MCF10A cells are shown. (C) The position of a chromosome or a gene can be expressed as the distance from the center of the nucleus or relative to other genes. The distribution of chromosomes and genes is nonrandom with some chromosomes (red) preferentially occupying internal positions and others (green) occupying peripheral positions. The nonrandom radial positioning also gives rise to nonrandom genome neighborhoods. (D) The distribution of a gene is probabilistic. Mapping of the position of the IGH gene in human lymphocytes in several hundred individual cells shows that its distribution is distinct from a random distribution; however, the IGH locus can be found in variable locations in individual cells. Each red dot represents the position of an IGH allele in a single cell. (C, Modified from Meaburn and Misteli 2007.)
Obviously, to achieve the necessary compaction of DNA, the chromatin fiber must be looped onto itself and condensed. How precisely this occurs in vivo is one of the enigmas in modern cell and molecular biology. The traditional view holds that DNA is hierarchically folded into higher-order fibers of 10 nm, 30 nm, 100 nm, and beyond (Felsenfeld and Groudine 2003; Li and Reinberg 2011). Although there is compelling evidence for the 10-nm fiber, several recent studies have questioned the existence under physiological conditions of the regular structure beyond the 30-nm fiber (Fussner et al. 2010). Higher-order regimes of organization are even less well defined and the physical properties of higher-order chromatin are currently largely unknown. The reason for this considerable uncertainty is the inability to accurately visualize higher-order chromatin fibers at the required level of resolution, thus limiting our ability to ascertain its structure. Despite the difficulty in assessing the path of the chromatin fiber in single cells, important insights into the higher-order spatial organization of chromatin have been obtained by improved imaging approaches and 3C-based technologies.
2. Long-range interactions in the context of nuclear architecture
2.1. Genomes Are Nonrandomly Organized in the Cell Nucleus
A fundamental property of genomes in higher eukaryotes is their nonrandom spatial organization in 3D space (Fig. 1) (Misteli and Soutoglou 2009; Rajapakse and Groudine 2011; Cavalli and Misteli 2013). The nonrandomness of a genome’s spatial arrangement is easily shown in FISH experiments by determining the position of a chromosome or a gene relative to the center of the nucleus (Fig. 1C,D). This is referred to as its radial position. These experiments reveal that each chromosome, and many genes, have a characteristic distribution of positioning within the cell nucleus. The prototypical examples of radial chromosome positioning are human chromosomes 18 and 19, which localize preferentially to the center (chromosome 19) and the periphery (chromosome 18) of the nucleus in human lymphocytes (Croft et al. 1999). Similar preferential location patterns have been documented for all chromosomes in the human genome (Boyle et al. 2001). The radial position of genes and chromosomes is cell-type- and tissue-specific. For example, X chromosomes localize more peripherally in liver cells compared to kidney cells (Parada et al. 2004). Along the same lines, chromosome position often differs in disease cells. For example, in pancreatic cancer, chromosome 8 shifts to a more peripheral location (Wiech et al. 2005). Similarly, chromosomes 18 and 19 change nuclear location in several cancers types, including cervical and colon cancer (Cremer et al. 2003).
It is important to point out that the spatial position of a chromosome or a gene is a statistical property, and not an absolute indicator of its location (Parada et al. 2003; Rajapakse and Groudine 2011). A gene’s location may be highly variable between individual cells (Fig. 1D). In other words, although a gene may, for example, preferentially be located toward the center of the nucleus, any given allele may be found at the nuclear periphery in a subpopulation of cells. The position of a gene is thus most accurately reflected by the statistical distribution of its positions in a population of cells.
The radial arrangement of chromosomes and genes in the nuclear space has an important consequence: Distinct genome neighborhoods arise through the preference for a genomic region to either be located toward the center or the periphery of the nucleus (Fig. 1C) (Meaburn and Misteli 2007). For example, more internal chromosomes or genes have a low probability of associating with genomic regions that are enriched at the nuclear periphery, thus physically segregating genomic regions within the nuclear space. Prominent examples of nonrandom spatial genome neighborhoods are the nucleolus, which harbors ribosomal RNA gene clusters located on human chromosomes 13, 14, 15, 21, and 22 or the congregation of chromosomes 12, 14, 15, which associate with each other with high frequency in mouse lymphocytes and are involved in chromosomal translocations in lymphoma (Parada et al. 2002).
For many genes the precise radial positioning is generally not sufficient to determine gene activity, nor is it predictive of gene activity. For instance, in a systematic study of 20 genes in a breast cancer model, no consistent correlation between gene activity and radial position was found (Meaburn and Misteli 2008). Active genes were found both internally and peripherally, as were inactive genes. Along the same lines, visualization of active transcription sites in the cell nucleus reveals a relatively homogenous distribution of active genes without any preferential accumulations. It can be concluded that the radial position of a gene alone is not sufficient to determine its activity.
2.2. Physical Interaction with the Nuclear Periphery as a Regulatory Mechanism
Although the precise location of a gene within the cell nucleus is not likely to be a key determinant of its activity, its positioning with respect to the nuclear envelope can have regulatory function (Kind and van Steensel 2010; Egecioglu and Brickner 2011). In most higher eukaryotes, the nuclear periphery is enriched in condensed heterochromatin, generally associated with the transcriptional repression. Furthermore, in budding yeast Saccharomyces cerevisiae, silent telomeric regions are preferentially found at the nuclear periphery (Egecioglu and Brickner 2011).
Large-scale mapping (e.g., using targeted DNA methylation approaches [DamID] and antibody pull-down [chromatin immunoprecipitation, ChIP]) has identified genomic regions that preferentially associate with the nuclear lamina (Pickersgill et al. 2006; Guelen et al. 2008; McCord et al. 2013), a meshwork of the intermediate filament proteins lamin A, B, and C and associated proteins (Dechat et al. 2010). In humans, there are more than 1000 lamina-associated domains (LADs) and these regions are typically 0.1–1 Mb in size and generally gene poor, and the contained genes are either silent or expressed at low levels (Pickersgill et al. 2006; Guelen et al. 2008; Kind and van Steensel 2010). LADs are likely organized into higher-order structures as their boundaries are frequent binding sites for the insulator protein CTCF. Not surprisingly, because they are enriched in inactive genes, histone marks associated with active genes (e.g., H3K4me3) are depleted in LADs (Kind and van Steensel 2010).
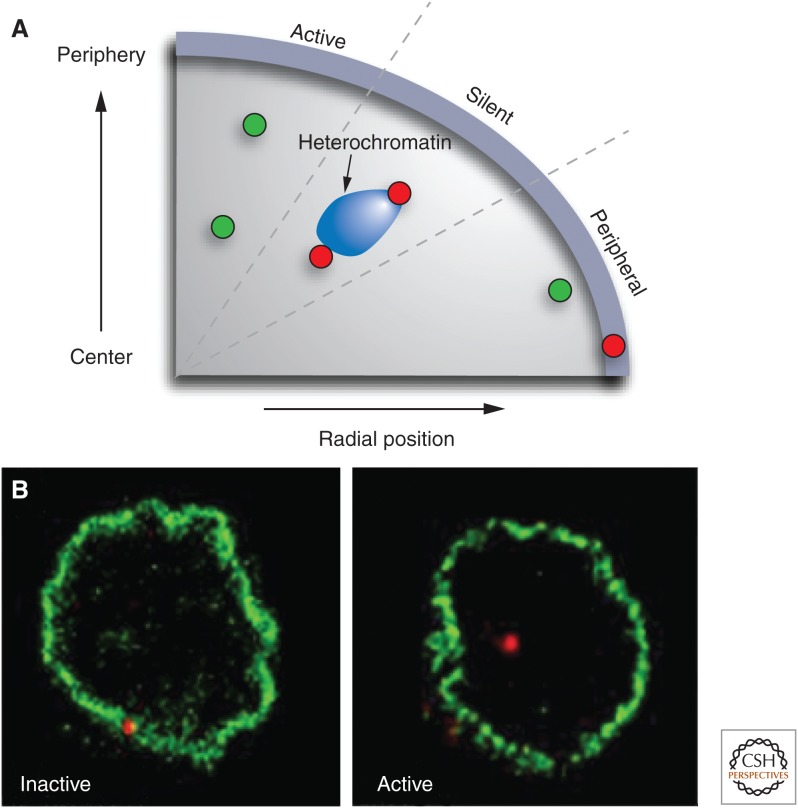
The presence of heterochromatin and inactive genes suggests that the periphery is a transcriptionally repressive environment (Egecioglu and Brickner 2011). But is interaction with the nuclear periphery a gene regulatory mechanism? A functional role is suggested by the observation of widespread gene misregulation in cells lacking major components of the nuclear lamina such as lamin A or lamin C (Dechat et al. 2010). Similar effects are seen upon overexpression of lamins or dominant mutants of lamin A, many of which are associated with human diseases (Scaffidi and Misteli 2008). Furthermore, experimental repositioning of active genes to the nuclear periphery by artificial tethering to a component of the nuclear lamina results in silencing of at least some genes, suggesting that interaction of genes with the nuclear lamina can be sufficient to alter gene activity (Fig. 2A) (Finlan et al. 2008; Kumaran and Spector 2008; Reddy et al. 2008).
Figure 2.
The role of the nuclear periphery in gene regulation. (A) Active genes (green) show a large range of radial positions; the precise radial position of a locus does not correlate with its activity level. Inactive genes (red) may associate with heterochromatin blocks at various radial positions. In contrast, physical association with the nuclear periphery is often linked to silencing. Genes that are in close proximity to the nuclear envelope but do not physically interact with it may be active. (B) Association of a gene locus with the nuclear edge often correlates with activity. The IgH locus (red) associates with the nuclear lamina when it is inactive in hematopoietic progenitor cells, but dissociates on activation in activated pro B cells. (A, Redrawn from Takizawa et al. 2008; B, reproduced, with permission, from Kosak et al. 2002, © AAAS.)
Gene repositioning relative to the nuclear envelope is likely a physiologically relevant regulatory mechanism because physical relocation away from the nuclear envelope has been implicated in several gene activation events during differentiation. For instance, the IgH and IgK loci dissociate from their location at the nuclear periphery in hematopoietic progenitor cells and assume a more internal location upon stimulation into pro-B-cells (Fig. 2B) (Kosak et al. 2002). Similarly, the CFTR gene detaches from the nuclear envelope upon its activation (Zink et al. 2004). These types of relocation events correlated with activity changes appear to be universal and can also be seen in diverse organisms such as Caenorhabditis elegans (Meister et al. 2010).
The mechanisms by which mammalian genes are regulated through interaction with the nuclear periphery are poorly understood, but likely involve specific sets of histone modifications (Egecioglu and Brickner 2011). A hint as to the molecular mechanisms involved in silencing peripheral genes comes from analysis of their histone modification patterns. Nuclear envelope–associated genes are generally hypoacetylated and treatment with histone deacetylase inhibitors reverses silencing of genes tethered to the periphery (Finlan et al. 2008; Guelen et al. 2008). In addition, peripherally located genes are enriched in H3K9me2 (Yokochi et al. 2009). Interestingly, dimethylation of H3K9 (H3K9me2) is mediated by the G9A histone methyltransferase, which physically interacts with BAF, one of the major lamina-interacting proteins (Montes de Oca et al. 2011). Elimination of G9A results in derepression of predominantly late-replicating genes typically associated with heterochromatin (Yokochi et al. 2009). A model therefore emerges in which genes may be marked as silent by specific histone modifications and tethered to the periphery by virtue of these modifications.
Although the nuclear periphery, particularly in higher eukaryotes, has predominantly a repressive effect on gene activity, in S. cerevisiae it is often the site of gene activity (Brickner and Walter 2004; Egecioglu and Brickner 2011). The prototypical examples of gene activation at the yeast nuclear periphery are the galactose-inducible GAL genes (Brickner and Walter 2004; Casolari et al. 2004). These genes localize to the nuclear interior when repressed, but rapidly associate with the nuclear pore complex (NPC) upon stimulation. The nuclear pore interaction is likely to be a key step in their activation because several NPC components and NPC-associated factors are required for activation of these genes. Targeting of these genes to the NPC requires their promoter region, but is independent of sequences in the gene body or the 3′ untranslated region (Ahmed et al. 2010). Importantly, mutational analysis of the promoter region of INO1, an NPC-targeted gene in which transcription is ablated, indicates that targeting to the pore can be uncoupled from transcription (Light et al. 2010). The INO1 promoter regions that target the gene to the pore are also distinct from the regions that control its activity. These so-called gene recruitment sequences are also sufficient to reposition an adjacent reporter gene to the pore (Ahmed et al. 2010; Light et al. 2010). It remains to be seen to what extent active genes are also found at the nuclear periphery in higher eukaryotes. In genome-wide mapping studies in Drosophila, hundreds of genomic sites containing active genes have been identified that physically interact with the nuclear pore complex (Kalverda et al. 2010).
2.3. Spatial Gene Clustering
The nonrandom positioning of genes in the nucleus raises the possibility that functionally related genes are spatially clustered in 3D space. Clustering of genes may be caused by sharing of overlapping activating or repressing protein complexes, and may provide a mechanism to coregulate groups of genes through their association with subnuclear compartments enriched in specific regulatory proteins and increase efficiency of transcription and RNA processing. There are several examples of spatial gene clustering.
2.3.1. The Nucleolus
The nucleolus is the site of ribosomal RNA (rRNA) synthesis and processing (Pederson 2010). The structure contains three morphologically distinct regions: the fibrillar center, which contains the rRNA genes; the dense fibrillar component, which contains newly synthesized rRNA transcripts at early stages of processing; and the granular component, which contains mature rRNA and early ribosomal assembly intermediates. Eukaryotic cells contain multiple copies of rDNA genes, typically several hundred in number, organized as tandem repeats located on multiple chromosomes. The rDNA repeat regions of the genome are referred to as nucleolar organizer regions (NORs). In humans, five chromosomes (chromosomes 13, 14, 15, 21, 22) contain NORs, each ∼3 Mb in size, made up of approximately 80 copies of the 43-kb rDNA repeat. In contrast to the five NOR clusters, typically only two to three nucleolar foci are observed in human cells. FISH analysis shows that the reason for the numerical discrepancy between NORs and nucleoli is that rDNA regions from multiple chromosomes physically aggregate to form an individual nucleolus. A typical nucleolus occupies up to 25% of the nuclear volume and represents the most dramatic example of gene clustering (Pederson 2010).
The formation and maintenance of the nucleolus is driven by the transcriptional activity of the ribosomal genes (Dundr and Misteli 2010). Nucleoli form in late M phase/early G1 as transcription of rDNA genes resumes. Importantly, nucleoli only form around active, not inactive, NORs. Initially, small prenucleolar bodies (PNBs) form in late M phase, which serve as precursors of mature nucleoli. PNBs likely contain partially processed rRNA molecules and, upon resumption of full rDNA transcription, mature into nucleoli (McStay and Grummt 2008). Evidence for a requirement for transcription as a driving force in formation of nucleoli is the observation that inhibition of rDNA transcription leads to disassembly of the nucleolar structure (Karpen et al. 1988). On the other hand, introduction into Drosophila cells of mini-NORs on plasmids is sufficient to nucleate structures, which, by morphological criteria, are indistinguishable from endogenous nucleoli (Karpen et al. 1988).
Although the formation of the nucleolus per se is strictly dependent on transcriptional activity, the association of rDNA genes is not. This was indicated in elegant hybrid experiments exploiting the fact that rDNA transcription is species specific. When a human chromosome containing a NOR is introduced into mouse cells, the human rDNA genes are not transcribed (Sullivan et al. 2001). However, the human chromosome still physically associates with preexisting mouse nucleoli, demonstrating that physical interaction of rDNA gene clusters is independent of their transcription (Sullivan et al. 2001).
Why do rDNA genes cluster? They are unusual in that they are expressed at exceedingly high levels. Although most RNA Pol II–transcribed genes contain a few polymerases at any given time, there are typically hundreds of RNA Pol I complexes associated with an rDNA gene. Live-cell imaging experiments have estimated that individual polymerases are separated by <100 nt and a new initiation event occurs every few seconds (Beyer et al. 1980; Dundr et al. 2002). As a result, thousands of rRNA transcripts are generated from the several hundred rDNA genes every minute, all of which need to undergo extensive processing. The spatial clustering of the rDNA genes and formation of a dedicated nuclear subcompartment, in which all necessary processing factors are enriched, may serve to facilitate efficient rRNA processing and promote ribosome assembly.
2.3.2. tRNA Gene Clustering
The transfer RNAs (tRNA), which act as adaptor molecules during translation, are encoded by multiple tRNA genes; humans contain ∼500, S. cerevisiae 274, and C. elegans 620 tRNA genes, distributed over multiple chromosomes (Phizicky and Hopper 2010). Analysis of S. cerevisiae tRNA genes has provided a compelling case for gene clustering. When localized by FISH analysis, the tRNA genes, which are located on all 16 S. cerevisiae chromosomes, are found in a large spatial gene cluster near the nucleolus (Thompson et al. 2003). The proximity of the tRNA cluster to the nucleolus may be of functional relevance because tRNAs and rRNAs are often coregulated in response to environmental conditions. Clustering of tRNA genes persists through all phases of the cell cycle, including cell division when transcription ceases, further supporting the notion that gene clustering can be independent of transcriptional activity (Thompson et al. 2003). Clustering of tRNA genes has to date only been observed in S. cerevisiae and it remains to be seen whether higher eukaryotes organize their tRNA genes in a similar fashion.
The formation of tRNA clusters near the nucleolus appears to be a two-step process (Haeusler et al. 2008). The clustering of individual tRNA genes requires condensin subunits. In temperature-sensitive mutants of the condensin subunits, smc2-8, smc4-1, ycg1-2, ycs4-1, and brn1-9 tRNA genes lose their clustered appearance and are dispersed throughout the nucleus. The condensin effect may reflect a global role of the complex on higher-order genome organization. But, interestingly, the fact that condensin specifically interacts with tRNA genes via the TFIIIB/TFIIIC complex and does not require the RNA Pol III complex that transcribes tRNAs, points to a direct role of condensin in the spatial clustering of tRNA genes. The second step in the clustering process is the position of the cluster near the nucleolus. This step is dependent on microtubules as indicated by the fact that disruption of microtubules results in displacement of the tRNA cluster from the nucleolus (Haeusler et al. 2008).
2.3.3. Transcription Factories
The notion of gene clustering, as observed in specialized classes of genes such as rRNA and tRNA genes, has been generalized in the concept of transcription factories (Fig. 3) (Jackson et al. 1993; Eskiw et al. 2011). These are defined as nuclear structures containing multiple active genes and multiple copies of the RNA Pol II transcription complex. In particular, they are characterized by the presence of hyperphosphorylated RNA Pol II, representing the actively elongating form of the polymerase. The existence of transcriptional hot spots, which are shared by multiple genes, is suggested by quantitative analysis of transcription sites. Although expression profiling indicates that typically several thousand genes are expressed in a given cell type, visualization of transcription using imaging methods reveals a much lower number of sites of nuclear transcription. Quantitative analysis in HeLa cells suggests that on average approximately eight polymerases and eight active genes cluster in a transcription factory (Jackson et al. 1993; Schoenfelder et al. 2010a).
Figure 3.
A view into the nuclear interior. Genomes exist in vivo in the form of chromosome territories made up of chromatin folded into still poorly understood higher-order fibers (multicolored areas). Chromatin fibers occupy the entire nuclear space, yet despite the presence of up to 2 m of DNA in a mammalian nucleus, there is considerable open nucleoplasmic space. Active genes (red) may loop out from chromosome territories and aggregate in centers of transcription (shaded areas), which contain appropriate transcription factors (blue, green). It has been suggested that coregulated genes cluster in shared transcription sites. (Reproduced from Misteli 2011; illustration by Anatomy Blue.)
In contrast to the traditional view of the transcription machinery being recruited to a gene, in the transcription factory model, genes are recruited to these centralized hot spots of transcription. Association of genes does not involve their physical interaction with each other, but their association with the protein components of the transcription factory or with adjacent splicing factor compartments (Brown et al. 2006). A key question then is whether transcription factories are specialized—that is, whether the genes they contain are coregulated or whether association of genes is largely random. Several observations raise the possibility of preferential clustering of coregulated genes. For example, in erythrocytes, the cell-type-specific Hba, Hbb, and Xpo7 genes can be found in a triplet cluster in a transcription factory at frequencies higher than expected based on random association (Schoenfelder et al. 2010b). Furthermore, pairwise clustering of Hba and Hbb with several other erythroid-specific genes has been observed at frequencies higher than expected. These genes are regulated by the transcription factor Krüppel-like factor 1, and their association with the transcription factory appears to be mediated by Klf1 because its loss interferes with clustering (Schoenfelder et al. 2010b). Another example of recruitment of coordinately regulated genes to a shared transcription factory is TNF-responsive genes (Papantonis et al. 2011). Upon stimulation, TNF-responsive genes, located on both the same and different chromosomes, spatially associate with each other in a coordinated fashion closely following their temporal expression pattern, suggesting that their physical clustering is directly related to their activity (Papantonis and Cook 2010). Further examples include the juxtaposition of estrogen-responsive genes (Fullwood et al. 2009) and the physical association of androgen-responsive genes (Lin et al. 2009). However, these studies also show that expressed genes are not exclusively clustering with coordinately regulated genes. For instance, the hbb locus not only interacts with a number of Klf4-regulated genes, but also with hundreds of apparently unrelated but coexpressed genes (Schoenfelder et al. 2010b). Clearly, many parameters, ranging from general transcription status to binding of specific sets of transcriptions factors, contribute to determining the composition of clusters of active genes.
These observations suggest that spatial clustering of genes is not limited to high copy number genes such as rRNA and tRNA genes, but may also play a role in expression of RNA Pol II transcribed genes. An unanswered question is how globally applicable functional clustering of genes is. Even in the most prominent examples, such as the clustering of erythroid-specific genes, coalescence of multiple genes was only observed in 5%–10% of cells in a population, suggesting that clustering is not a prerequisite for proper gene expression. An additional related question is how many distinct transcription factories a nucleus contains. There is currently no strong evidence to suggest that a transcription factory caters exclusively to a specific set of genes and it seems more likely that most transcription factories contain genes that are regulated by multiple, distinct mechanisms.
2.4. Proximity in the Formation of Chromosomal Translocations
Translocations are a common hallmark of cancer cells. These genomic rearrangements are formed by the illegitimate joining of DNA ends on distinct or the same chromosomes. Translocations may represent a downstream bystander effect due to enhanced genomic instability and DNA repair defects in cancer cells. Translocations can be causal in carcinogenesis, as in the case of fusion between the BCR-ABL genes in chronic myelogenous leukemia or the PML-RAR genes in promyelocytic leukemia. The nonrandom arrangement of the genome in 3D space and the physical interaction of genomic regions located on different chromosomes can have a critical impact on the frequency of formation of chromosomal translocations (Fig. 4) (Misteli and Soutoglou 2009).
Figure 4.
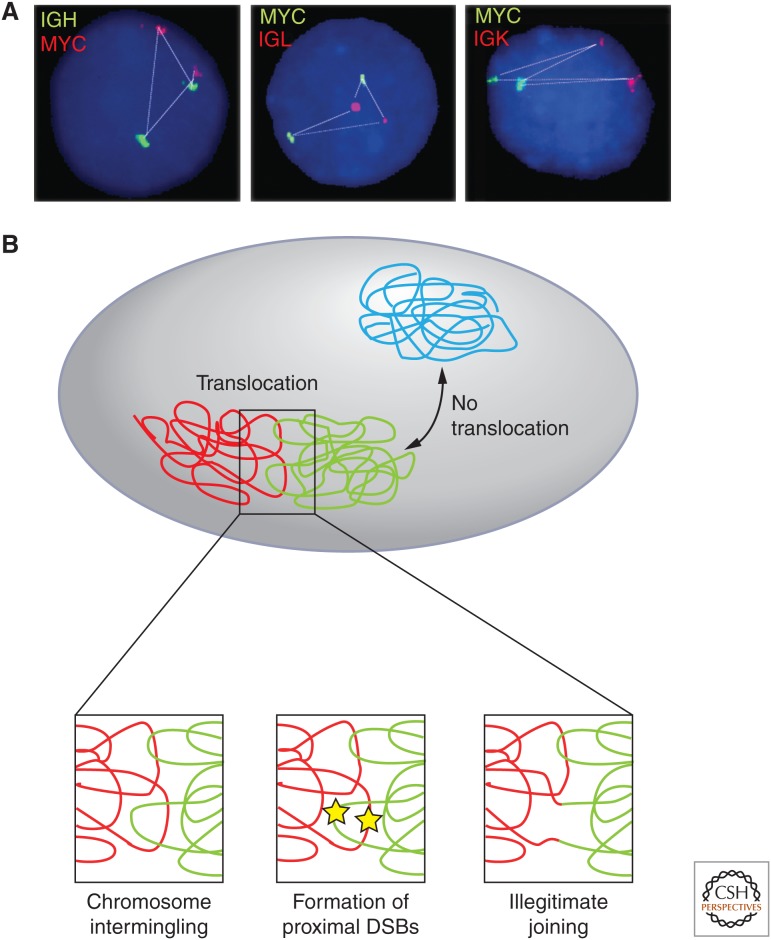
Roles of genome organization in determining chromosome translocations. The nonrandom organization of genes and chromosomes contributes to the formation of cancer translocations. (A) The physical distance of MYC to its translocation partners IGH, IGL, and IGK correlates with their translocations frequency (MYC-IGH > MYC-IGL > MYC-IGK). (B) Translocations preferentially occur between proximally positioned chromosomes (red, green), and only rarely between distally located chromosomes (blue). Closely juxtaposed double-strand breaks (yellow stars) occurring at the interface between chromosomes create free chromosome ends that may recombine to form a chromosome translocation by illegitimate joining. (A, Adapted from Roix et al. 2003; B, reproduced, with permission, from Misteli 2010, © Cold Spring Harbor Laboratory Press.)
A prerequisite for the formation of a chromosome translocation is the physical association of the two translocating chromosomes in the nuclear space. Live-cell imaging experiments have shown that in mammalian cells the free chromosome ends generated by double-strand breaks are largely immobile in the cell nucleus (Soutoglou et al. 2007). As a consequence, translocations occur preferentially between genomic regions that are often in close spatial proximity because of the nonrandom arrangement of the genome in the interphase nucleus. In support, FISH experiments and, more recently, biochemical mapping approaches of frequent translocation partners have revealed juxtaposition of translocating regions in the intact cell nucleus (Neves et al. 1999; Roix et al. 2003; Osborne et al. 2007; Kind and van Steensel 2010; Chiarle et al. 2011). For example, the MYC and IGH genes, whose translocation gives rise to Burkitt’s lymphoma, are frequently found interacting or located in close proximity in B-cells (Roix et al. 2003; Osborne et al. 2007). Similar correlations between translocation frequency and physical proximity have been found for numerous translocation partners in mouse and human cells (Mani et al. 2009; Mathas et al. 2009). The notion that spatial interactions between translocating regions are a key determinant in translocation formation is also supported by observations on prostate cancer translocations. In prostate cancer, translocations between the TMPRSS2 gene and either ETV1 or ERG are frequent (Lin et al. 2009; Mani et al. 2009). In normal prostate cells, these genes do not physically associate, yet stimulation with androgen leads to their physical association and, when challenged with UV-irradiation, their rapid translocation. The importance of spatial positioning in the formation of chromosomal translocations is further shown by unbiased approaches that are able to map physical interactions of chromatin fibers at a genome-wide scale (see below). Clearly, analyses of naturally occurring and experimentally inducible random translocations have revealed a strong correlation between translocation frequency and spatial proximity (Chiarle et al. 2011; Klein et al. 2011; Hakim et al. 2012; Zhang et al. 2012).
Further support for the notion that the nonrandom spatial proximity of chromosomes is key in determining translocation frequency is the finding that adjacent chromosomes intermingle their DNA (Branco and Pombo 2006). Careful visualization and mapping indicates that chromosomes invade the body of their adjacent neighbors, thus creating a zone of chromatin intermingling. It appears likely that translocations preferentially occur when DNA damage occurs in this region creating double-strand breaks, which are immobile and prone to undergo illegitimate joining with proximal breaks. This view is supported by the finding that translocation frequencies correspond closely to the degree of chromosome overlap (Branco and Pombo 2006) (Fig. 4).
The finding that translocations occur preferentially between proximally positioned regions means that the nonrandom spatial organization of the genome is a key factor in determining translocation partners (Misteli and Soutoglou 2009). The tissue-specific arrangement of chromosomes is a significant contributor to the tissue-specific appearance of translocations because we know genomes are arranged differently in different cell types and tissues. In agreement with this notion, chromosomes 5 and 6, which are frequently translocated in mouse hepatoma, are often neighbors in normal liver cells, whereas in mouse lymphocytes it is translocation partners chromosomes 12 and 15 that are frequent neighbors (Parada et al. 2004). These observations support a role for the nonrandom spatial genome interactions in the formation of chromosomal translocations.
3. Analysis of chromatin interactions and gene regulation
3.1. Introduction: Gene Elements and Methodologies
Perhaps the most direct and specific role of long-range chromatin interactions and spatial chromatin organization is in controlling gene expression. In the large genomes of metazoans, only a small portion of the genome is protein coding. The enormous noncoding fraction of the genome, including both intragenic and intergenic DNA, is thought to represent a large reservoir of gene regulatory elements. A wide range of genome-wide approaches are being applied to identify the full set of functional elements in the human genome and initial estimates indicate the presence of hundreds of thousands of regulatory elements scattered throughout the genome (ENCODE Project Consortium 2011, 2012). Gene regulatory elements include enhancers, repressors, insulators, and possibly other classes of yet-to-be-discovered elements. These elements may be located at significant genomic distances from the nearest gene, suggesting that they are able to communicate over considerable distances in the linear genome. Consistently, studies using transgenes and reporter constructs have revealed that significant stretches of flanking DNA (e.g., hundreds of kilobases) are required to recapitulate the normal regulation of a gene.
One obvious mechanism by which long-range gene control can be achieved is through direct physical associations between regulatory elements and gene promoters (e.g., through the 3D folding and looping of the chromatin fiber). Technological innovations have facilitated detection of chromatin loops and indicate that they indeed play critical roles in gene regulation by bringing widely spaced genes and regulatory elements in direct physical contact.
Microscopic approaches can be used for the analysis of the relative spatial positioning of loci within individual nuclei, as illustrated above, but they generally lack in resolution and comprehensiveness. On the other hand, crystallographic studies can be used to determine chromatin structure at the level of individual nucleosomes. However, both these techniques do not determine the folding and structure of the chromatin fiber. This resolution gap has, for a long time, been an impediment to studying the role of chromatin structure in mediating long-range gene regulation often involving regulatory elements located tens to hundreds of kilobases from their target genes.
Over the last several years, an expanding set of related molecular methodologies based on the chromosome conformation capture (3C) technique have been developed to allow the quantification of chromatin interactions at increasing resolution (kb) and scale (whole genome; Dekker et al. 2002; van Steensel and Dekker 2011; Hakim and Misteli 2012). These 3C-based approaches are particularly powerful for the identification of chromatin looping interactions, and increasingly for determining whole-chromosome and whole-genome folding at the resolution of several kilobases.
3.2. 3C
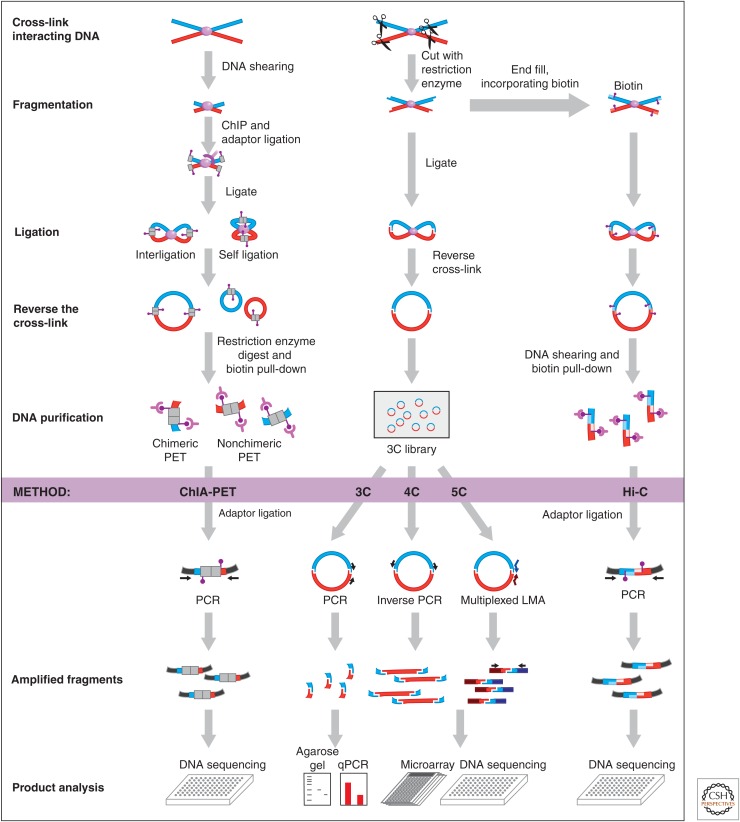
The basic concept of the 3C method is that loci in close spatial proximity (e.g., they directly interact to form a chromatin loop or a contact between two chromosomes) can be chemically cross-linked (Fig. 5). The most frequently used cross-linking agent is formaldehyde; it easily permeates cells and induces cross-links between proteins, and between proteins and DNA. Thus, physically interacting segments of chromatin fibers throughout the genome become cross-linked with protein complexes that associate with them. Next, chromatin is fragmented using a restriction enzyme. Cross-linked chromatin segments are then ligated under dilute conditions to strongly favor intramolecular ligations over random intermolecular ligation events. Finally, cross-links are broken by simply heating the chromatin to 95°C, and DNA is purified to obtain a library of 3C ligation products. Effectively, 3C converts pairs of loci that are spatially proximal into unique hybrid DNA molecules that can then be detected and quantified using any of a wide range of standard DNA detection methods (Fig. 5).
Figure 5.
Overview of 3C-based methods. All 3C-based methods rely on covalently linking spatially proximal chromatin segments. Through a series of steps chromatin is then fragmented and religated and ligation products are detected using PCR, microarrays, or deep sequencing. In ChIA-PET (chromatin interaction analysis with paired-end tag sequencing), chromatin fragmentation is achieved by shearing and ligation junctions are marked by adaptors that contain recognition sites for type I restriction enzymes. Redigestion of ligation products with such enzymes yields small ligation, junction-containing molecules that can be analyzed by deep sequencing. 3C, 4C, and 5C use restriction enzyme digestion to fragment cross-linked chromatin. Religation of DNA then produces ligation products that can be directly analyzed by PCR (3C), inverse PCR (4C), or ligation-mediated amplification (LMA; 5C). Hi-C is as 3C, but includes a step to incorporate biotinylated nucleotides before religation. This facilitates purification of ligation junctions that are then analyzed by deep sequencing. (Adapted from Sanyal et al. 2011.)
In a classical 3C experiment, a genome-wide 3C library is generated that can be used as DNA template in PCR assays using locus-specific primers to determine the presence and relative abundance of specific ligation products of interest (Dekker et al. 2002). Thus, interaction frequencies are determined one at a time using a candidate approach, which puts practical limits on the number of interactions that can reasonably be tested. Classical 3C is often used to determine interaction frequencies among up to several dozen individual genomic restriction fragments (e.g., to test whether a given gene promoter of interest is frequently interacting with a particular regulatory element). One of the first applications of 3C was to detect chromatin looping in the β-globin locus between the active globin genes and the upstream locus control region (LCR; Tolhuis et al. 2002). Since then, many other studies have used 3C to identify similar looping interactions in other loci.
The 3C technology has revolutionized the study of chromatin folding, particularly because it has spawned the development of a number of 3C-based technologies with increased resolution, sensitivity, and, most importantly, throughput. It has achieved this by changing the way 3C ligation product libraries are interrogated (van Steensel and Dekker 2011). In Sections 3.3 and 3.4, we will outline these detection methods in more detail and then describe how their application is starting to uncover the principles that determine the spatial organization of individual gene loci, chromosomes, and whole genomes, and how these different layers of genome organization impact gene expression.
3.3. Global Analysis of Complete 3C Libraries
The recent development of high-throughput DNA sequencing platforms has greatly facilitated comprehensive analysis of chromatin interactions. Improvements in 3C technology have culminated in the development of the Hi-C method, the first genome-wide 3C method, which includes a step to label the ends of restriction fragments with biotinylated nucleotides before DNA ligation, thereby labeling ligation junctions with biotin (Fig. 5) (Lieberman-Aiden et al. 2009). This facilitates purification and sequencing of DNA molecules only containing ligation junctions, greatly improving the number of informative DNA sequence reads. Moreover, more recently, it has become feasible, and no longer cost-prohibitive, to directly sequence a 3C library to obtain an unbiased genome-wide chromatin interaction map. Indeed, direct sequencing of 3C libraries of Drosophila embryos has yielded genome-wide maps of long-range interactions at a resolution of several kilobases (Sexton et al. 2012).
Genome-wide interaction maps provide a global overview of the spatial organization of genomes. As we will discuss in more detail in Section 4, these maps have revealed several novel features of higher-order chromosome structure and nuclear organization (Cavalli and Misteli 2013).
3.4. Comprehensive Analysis of Parts of the 3C Library
When relatively small genomes are analyzed (e.g., that of yeast), comprehensive interaction detection methods such as Hi-C can provide structural information at a resolution of a few kilobases. However, for large metazoan genomes, these global methods generally yield much lower resolution, typically 0.1–1 Mb. The reason for this is that the complexity of 3C libraries obtained for large genomes is truly enormous. For instance, for the human genome a 3C library generated with the HindIII enzyme will contain as many as 1012 unique ligation products, and the abundance of these products can vary over several orders of magnitude. Thus, to obtain a reliable chromatin interaction data set at a resolution of single restriction fragments, one needs to sequence far more molecules than is currently feasible.
To overcome the resolution limit of unbiased global detection methods, several targeted detection methods have been developed. These methods use different strategies to comprehensively sequence and analyze only a subset of the 3C library (Fig. 5). By targeting only a selected portion of the 3C library, one increases the sequencing coverage per interaction and thereby the resolution with which long-range interactions can be identified and quantified. This is particularly relevant for the analysis of long-range gene regulation that involves specific functional elements that are usually smaller than a kilobase.
3.4.1. 4C Technology
In many studies, one is interested in identifying all genomic loci that interact with a particular gene or element (e.g., to find putative long-range regulatory elements that loop to and regulate a gene of interest). Thus, instead of probing the entire 3C library in an unbiased fashion, one would like to analyze a subfraction of it at high resolution. To comprehensively identify all fragments that are ligated to a single restriction fragment of interest (the “bait” or “anchor” fragment), the 4C technology combines 3C with inverse PCR detection of 3C ligation products (Fig. 5) (Simonis et al. 2006; Zhao et al. 2006). Briefly, many of the 3C ligation products are circular, or can be circularized by redigestion and re-ligation of 3C ligation products. Inverse PCR with primers pointing outward from the bait fragment can therefore amplify all DNA fragments ligated to the bait. 4C PCR products can then be detected and quantified by hybridization to a genome-wide DNA tiling array or direct DNA sequencing. A 4C experiment provides a genome-wide interaction profile of a bait locus to reveal the positions and identities of other loci at kilobase resolution on the same chromosome or on other chromosomes that the locus frequently interacts with.
3.4.2. 5C Technology
The 5C (abbreviation for 3C-carbon copy) technology is used specifically to determine interaction frequencies between two selected sets of loci (e.g., between a set of promoters and a set of enhancers [Fig. 5] [Dostie et al. 2006]). 5C differs from 3C only by the way ligation products are detected. In 5C, LMA is used with pairs of locus-specific forward and reverse primers, termed 5C primers. These are designed to anneal directly across predicted 3C ligation junctions. Only when a ligation product is present in the 3C library can the two primers anneal and become ligated by a nick-specific DNA ligase. Ligated 5C primer pairs are subsequently PCR amplified using primers that recognize their universal tails. The advantage of 5C is that LMA can be performed at high levels of multiplexing, in contrast to PCR. For instance, one can design 5C reverse primers for thousands of genes and 5C forward primers for thousands of putative distal regulatory elements, and interrogate the millions of pairwise gene-element interactions in a single reaction. The resulting 5C library represents a “carbon copy” of a selected fraction of the 3C library and can be analyzed by direct DNA sequencing. In many respects, 5C is similar to other enrichment methods such as hybrid capture approaches for selectively purifying parts of the genome (e.g., all exons) before targeted sequencing (Mamanova et al. 2010).
3.4.3. ChIP-Loop and ChIA-PET Technologies
Another approach to selectively enrich for specific interactions is to combine 3C with ChIP to isolate cross-linked complexes that contain a protein of interest (Fig. 5). For instance, by immunoprecipitating chromatin complexes one can enrich for interactions that involve specific proteins of interest. The ChIP-loop assay and ChIA-PET assays use PCR and deep sequencing, respectively, to analyze the purified ligation junctions (Cai et al. 2006; Fullwood et al. 2009; Li et al. 2010).
3.5. 3C-Based Data Reveal Cellular Heterogeneity in Genome Folding
The 3C procedure and all its high-throughput variants described in Sections 3.3 and 3.4 generate a highly complex mixture of hybrid DNA molecules in which each individual DNA molecule represents a single chromatin interaction event in a single cell. It is critical to appreciate that the 3C procedure captures interactions across a large population of cells, and thus the abundance of a single ligation product in the library represents the fraction of cells in which the two corresponding loci are sufficiently proximal to become cross-linked. When sufficient numbers of cells are studied, a given genomic locus can be found contacting almost any other locus in the genome, but with contact probabilities ranging over several orders of magnitude. Thus, 3C-based data can only be understood in terms of enormous cell-to-cell variability in chromosome folding and nuclear organization (Kalhor et al. 2011). Of course, this is exactly what has been observed by direct imaging of loci, revealing preferences in subnuclear localizations, but no determined and reproducible positions. 3C libraries thus reflect the contact probability landscapes of loci, revealing trends in spatial organization of the genome in a large cell population. These data sets must be carefully analyzed to determine patterns in contact probabilities of sets for loci, indicating the presence of looping interactions and other chromatin structural features (Bulger and Groudine 1999, 2011; Dekker 2006; Cavalli and Misteli 2013). Similarly, any model indicating the role of higher-order chromatin structure in gene regulation must take into consideration the significant cell-to-cell variability in chromatin folding.
4. Multiple categories of chromatin looping interactions
Studies of local and global patterns of chromatin organization suggest that looping and long-range interactions, in general, reflect different levels of chromatin organization with distinct roles in gene regulation. At the finest scale, we can discern frequent, precise, and specific looping interactions between gene promoters and their distal regulatory elements. At a more global scale, specific sets of loci that can be located on different chromosomes associate with each other. Finally, zones of interactions between and within large megabase-sized chromosomal regions indicate the presence of distinct higher-order structural domains, reflecting nuclear organization and functional compartmentalization, in general. This last category involves infrequent, broad, and rather nonspecific interactions. These different categories of chromatin associations are described in Sections 4.1–4.6 in more detail in relation to their impact on gene regulation (Cavalli and Misteli 2013).
4.1. Looping between Genes and Their Regulatory Elements
Enhancers are elements that can control expression over very large genomic distances. One of the best studied enhancers is the LCR in the β-globin locus, located 40–80 kb upstream of its cluster of target genes (Fig. 6). An even more dramatic example is the sonic hedgehog gene regulated by an enhancer located more than 1 Mb away. Many genetic experiments have shown that long-range regulation of gene expression is a general phenomenon in many complex genomes. Over the years, several mechanisms have been proposed by which these elements can activate their target genes. Most of these models propose the formation of a direct physical contact between the element and the target promoter, but they differ in the mechanisms by which these contacts are established (Bulger and Groudine 1999, 2011).
Figure 6.
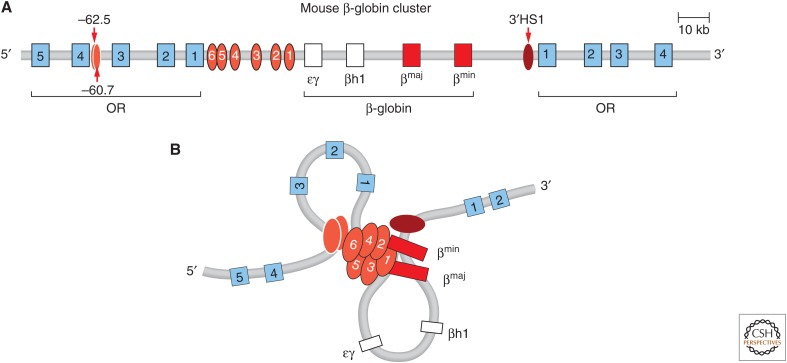
The mouse β-globin locus. (A) Schematic representation of the β-globin locus. (B) Looping between the LCR, the globin genes, and up- and downstream elements bound by CTCF (HS-62.5/-60.7 and 3′HS1) is observed in globin-expressing erythroid cells. (Redrawn from de Laat and Grosveld 2003, with permission from Springer Science and Business Media.)
The application of 3C-based assays has led to the identification of long-range interactions between gene regulatory elements and genes. This has confirmed the idea that chromatin looping plays a major role in gene regulation. We will describe some of the best-characterized examples of long-range looping interactions.
4.2. The α- and β-Globin Loci
The α- and β-globin loci express high levels of α- and β-type globin proteins that combine to form hemoglobin. Expression of these loci has been studied in great detail and they provide a paradigm for long-range gene regulation by looping (Fig. 6).
Both loci are composed of clusters of related genes that are developmentally regulated. The order of the genes in the cluster corresponds to the order in which they are expressed during development. The β-globin gene cluster is activated by an extraordinarily strong composite element, the LCR. The α-globin genes are activated by a strong enhancer located 40–60 kb upstream of the gene cluster. 3C studies of the β-globin locus were among the first to show that the LCR directly touches the globin genes, resulting in the formation of a chromatin loop that is 40–80 kb in size (Tolhuis et al. 2002). This looping interaction is strictly correlated with gene expression; it is not observed in cells that do not express the locus, but appears in cells that express it. Further, during development, the LCR interacts sequentially with the appropriate gene in the cluster that is expressed at the corresponding stage (Palstra et al. 2003). 3C studies of the α-globin locus show, similarly, that the α-globin genes directly interact with their enhancer located 40 kb upstream of the genes, and again this interaction is only observed in cells that express the genes (Vernimmen et al. 2007; Bau et al. 2010). Since these initial studies, many additional examples of looping between genes and enhancers have been found, including in other complex gene loci such as the HoxD locus (Montavon et al. 2011), the immunoglobulin heavy chain locus (Guo et al. 2011), and the Th2 Interleukin cluster (Spilianakis et al. 2005).
Looping interactions are not limited to very highly expressed gene clusters or developmentally controlled genes, but are also observed in single gene loci encoding proteins involved in a large variety of biological processes. For instance, looping interactions with distal enhancers have been observed for CFTR (Ott et al. 2009; Gheldof et al. 2010), c-MYC (Wright et al. 2010), FoxL2 (D’Haene et al. 2009), and many other genes. A recent comprehensive analysis of more than 600 gene promoters in the human genome identified several looping interactions per gene with elements resembling enhancers, promoters, and insulator elements (bound by the CTCF protein; see Sec. 5.1) located within up to hundreds of kilobases from the promoter (Sanyal et al. 2012).
Long-range interactions are not only involved in gene activation, but can also lead to gene silencing. In particular, Polycomb complexes repress genes by compacting chromatin (discussed in Grossniklaus and Paro 2014) and mediating looping between silencing elements and genes. For example, in the Drosophila bithorax complex looping interactions have been identified between Polycomb response elements and silenced hox genes (Lanzuolo et al. 2007). Extrapolating from these studies, one can infer that most genes are engaged in highly specific long-range interactions with regulatory elements located up to hundreds of kilobases around the promoter, leading to either activation or repression.
4.3. Looping between Regulatory Elements
Many genes are regulated by more than one enhancer. For instance, the HoxD locus is regulated by numerous elements spread out over a large gene desert that act in concert to ensure proper control of the gene cluster (Montavon et al. 2011). 3C and 4C studies show that the hoxD genes not only interact with each of these elements at the appropriate time and location during development, but that these elements also associate with each other. One interpretation is that a highly complex multilooped structure is formed. Similar observations have been made in the β-globin locus in which the globin genes interact with the LCR, but both also associate with elements bound by the CTCF protein located upstream of and downstream from the locus (Tolhuis et al. 2002). These observations have led to models in which complex higher-order structures are formed that may act as singular units to integrate regulatory input from multiple elements. Although appealing, there is currently no firm evidence that all pairwise interactions detected using 3C-based methods occur simultaneously in a cell to form stable and reproducible structures, leaving open the possibility that these interactions and the assembly of multilooped structures are dynamic and/or vary among cells in the population.
4.4. Interchromosomal Interactions between Specific Loci
It has long been known from genetic studies in Drosophila that regulatory elements on one chromosome can affect gene expression on the homologous chromosome. This phenomenon, called transvection, is thought to be made possible by close pairing of homologs chromosomes (e.g., in polytene chromosomes), which leads to close spatial proximity of homologous regions located on the two chromosomes. In that context, a regulatory element can activate or repress the target gene it is linked to in cis, as well as the target gene located on the nearby homolog in trans. One well-studied example is the yellow locus, in which the presence of an enhancer on one homolog can activate the yellow target gene on the homologous chromosome lacking the enhancer, in a process that depends on close interactions between the homologs (Morris et al. 1999; Ou et al. 2009). Thus, the mechanism of transvection may be related to other long-range gene regulation phenomena by also involving direct physical interactions.
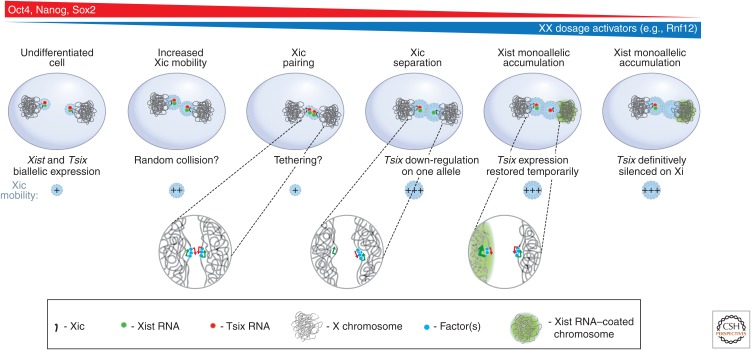
Several tantalizing examples have been described in mammalian cells suggesting that interchromosomal interactions between regulatory elements on one chromosome can affect gene activity on other chromosomes including nonhomologous chromosomes. The best-characterized case involves the pair of X chromosomes in female mouse cells during the initiation of the X-chromosome inactivation process, which is the topic of Brockdorff and Turner (2014) (Fig. 7). In early embryonic development one of the two X chromosomes is chosen to become inactivated. This process is directed by a large and complex locus called the X inactivation center (Xic). This locus encodes the Xist noncoding RNA, which is activated and spreads in cis along the X chromosome from the site of transcription from the chromosome that becomes inactive. The Xist RNA recruits silencing factors such as Polycomb complexes that will then silence expression of most genes all along the X chromosome. The process of X inactivation ensures that one and only one of the two X chromosomes will express Xist and become silenced. Both DNA FISH and 3C studies show that during X inactivation the two Xic loci on the two X chromosomes briefly pair and directly interact (Bacher et al. 2006; Xu et al. 2006). The Xic pairing process requires specific CTCF-bound loci around the Xist gene, as well as a larger region located several hundred kilobases from the Xic (Augui et al. 2007; Donohoe et al. 2009). It is thought that this encounter somehow establishes that one of the two X chromosomes will activate the Xist gene, whereas the other will not. Recently, this was studied by live-cell imaging of tagged Xic loci, followed by RNA FISH analysis, demonstrating that pairing is followed by asymmetric expression of Tsix, the Xist antisense transcript and regulator, leading ultimately to monoallelic Xist up-regulation (Masui et al. 2011). Thus, the physical association between the Xics may directly coordinate the establishment of opposite epigenetic states on the X chromosomes. Indeed, mutants that affect pairing of Xics affect appropriate regulation of X-chromosome inactivation.
Figure 7.
Interchromosomal interactions between Xics during early embryonic development. During early stages of embryonic stem (ES) cell differentiation, the two X chromosomes are highly mobile, possibly to allow pairing of the two Xics. During this pairing stage, through a process still largely unknown, one of the two X chromosomes will down-regulate expression of Tsix, a negative regulator of the Xist RNA gene. On that chromosome, Xist will be expressed and Xist RNA will accumulate resulting in X-inactivation. The other X chromosome will continue to express Tsix and the chromosome will remain active. (Redrawn, with permission, from Masui et al. 2011, © Elsevier.)
Other examples of interchromosomal interactions have been described that may be involved in similar coordinated events, but their relevance is highly debated. In olfactory neurons, only one of hundreds of olfactory receptor genes is expressed. 3C studies and imaging have shown that the mouse genome contains a single enhancer (the H-enhancer) that associates with a single olfactory gene either in cis or in trans in a given cell, leading to the activation of that gene, whereas the other gene copies remain silent (Lomvardas et al. 2006). Although this certainly is a very intriguing finding, subsequent deletion of the H-enhancer found no effect on olfactory genes located on other chromosomes that do not have the H-enhancer, putting in doubt the functional relevance of the interchromosomal interactions (Fuss et al. 2007). Another example of interchromosomal interaction is that of the Th2 interleukin cluster on mouse chromosome 11 and the interferon γ locus on chromosome 10 in naïve T cells (Spilianakis et al. 2005; Noordermeer et al. 2011). This interaction is lost when the cells differentiate into either Th1 or Th2 cells that express the interleukin cluster or interferon-γ gene, respectively. The interaction between the loci before commitment may keep the loci in a poised state, ready to coordinate activation and repression of the loci during subsequent differentiation. However, whether these interactions cause the poised state, or simply reflect it, remains an open question.
A direct test of the occurrence and relevance of interchromosomal interactions was performed by analyzing the effect of an ectopically inserted LCR on the expression of the endogenous β-globin genes (Noordermeer et al. 2011). 4C analyses showed that an ectopic LCR can directly associate with the endogenous globin locus, but only in a very small minority of cells. Interestingly, combined RNA and DNA FISH experiments showed that in the rare cases in which the ectopic LCR interacted with the globin locus, the globin genes were up-regulated. This important study shows that interchromosomal interactions occur very infrequently, even for a very strong enhancer, but that these rare events can have functional consequences for the interacting genes in the few cells in which the interaction occurs.
Despite these examples of potential functional interchromosomal interactions between specific loci and regulatory elements, the relevance of such interactions for gene regulation is currently far from established. In fact, many of these interchromosomal interactions may be of a different nature than interactions typically involved in intrachromosomal looping for gene regulation via bona fide gene elements. Interchromosomal associations may be related to overall nuclear organization of gene expression in which groups of genes are found gathered at subnuclear sites enriched in transcription machinery or silencing factors. These less-specific and typically low-frequency associations may be the result of gene activity, rather than the cause, and are discussed next.
4.5. Associations between Groups of Active and Inactive Loci
Although active genes can be found throughout the genome, within the nucleus transcription is highly nonhomogeneous and occurs most prominently at multiple foci or subnuclear regions enriched in transcribing RNA polymerases and splicing machineries. These sites can be visualized microscopically in fixed cells using antibodies against active RNA Pol II, splicing proteins, or by BrU labeling of nascent RNA. One interpretation is that groups of active genes, including those located on different chromosomes, cluster together in the nucleus at these transcription foci that are sometimes referred to as transcription factories. Indeed, both imaging and 3C-based studies show that colocalization and direct physical contacts between active genes are readily detected (Osborne et al. 2004; Simonis et al. 2006; Lieberman-Aiden et al. 2009). In general, there appears to be limited specificity in these interactions; 4C experiments in erythroid cells detect interactions between the β-globin locus and many other active genes throughout the genome (Simonis et al. 2006). These interactions involve larger chromatin segments (e.g., entire genes instead of precise gene regulatory elements) and occur significantly less frequently in the cell population as compared with the chromatin looping interactions described in Sections 4.1–4.3 that drive gene regulation.
Hi-C analysis has shown that active chromosomal domains, typically hundreds of kilobases up to several megabases in size, preferentially associate with any other active chromosomal domains throughout the genome (Lieberman-Aiden et al. 2009). Similarly, inactive domains associate with any other inactive domain. There seems to be limited specificity in these interactions, although there are some exceptions. In Drosophila, loci repressed by the Polycomb complex are found clustered at a limited number of Polycomb bodies. In one case, it was found that disrupting the association of a locus with other Polycomb-repressed loci can affect gene regulation of that locus, albeit weakly and only after several generations, although still suggesting that clustering of loci directly impacts on gene expression (Bantignies et al. 2011).
These experiments highlight a recurring theme for these types of chromosomal interactions; active genes are found with other active genes, and inactive or silenced genes are found at other subnuclear sites (Cavalli and Misteli 2013) as indicated by direct imaging, although particular interactions are highly variable and nonspecific—that is, the precise subset of interacting genes and loci differs between cells, even in an otherwise homogeneous cell population. This does not rule out that some preferential clustering of loci regulated by overlapping transcriptional regulators may occur over and above the general tendency of colocalization.
4.6. Topologically Associating Domains: Building Blocks of Chromosomes
Both high-resolution, genome-wide Hi-C analysis and targeted 5C analysis of the X-chromosome inactivation center has led to the discovery that chromosomes are composed of series of chromosomal domains that are several tens of kilobases (in flies), or several hundreds of kilobases (mouse and human) in size (Dixon et al. 2012; Nora et al. 2012; Sexton et al. 2012; Cavalli and Misteli 2013). These domains are characterized by relatively high interaction frequencies between loci located within one domain, but lower interaction frequencies between loci located in different domains. Therefore, these domains have been dubbed “topologically associating domains” (TADs; Nora et al. 2012), or “topological domains” (Dixon et al. 2012). TADs are separated by genetically defined boundary elements (Nora et al. 2012), but the precise mechanism of their formation is currently unknown. The much larger active and inactive chromatin domains described above that interact with other large active and inactive domains genome-wide, respectively, are often composed of multiple smaller TADs.
Initial analyses have indicated that groups of genes located within the same TAD are correlated in their expression during differentiation (Nora et al. 2012), but how general this is across the genome is still an open question. Chromatin looping interactions between genes and their regulatory elements occurs mostly within TADs, which is perhaps not surprising given that chromatin interactions between TADs, in general, are of lower frequency (Sanyal et al. 2012; Shen et al. 2012). TADs may therefore represent not only structural building blocks of chromosomes, but also functional units of gene regulation. It has been proposed that TADs are a critical structural and functional level in the hierarchy of higher-order chromosome folding (Gibcus and Dekker 2013), but much is still unknown about this level of chromosome organization (Cavalli and Misteli 2013).
5. Building chromatin loops
Up to now, we have described the various types of chromatin interactions that can be observed in cells. But what mediates these interactions? We will focus on the class of precise and specific looping interactions between gene promoters and long-range acting gene regulatory elements because these have been characterized in most detail, although many details driving these phenomena remain to be discovered.
Enhancer and promoter elements are bound by large protein complexes containing a variety of different types of proteins, ranging from specific DNA binding transcription factors to cofactors with a variety of enzymatic activities such as histone acetylation and methylation and noncoding RNAs. Long-range interactions between gene regulatory elements and promoters likely involve direct protein–protein interactions and may also require specific bridging complexes. The role of specific proteins in mediating looping has been confirmed for several cases. For example, the looping between the LCR and β-globin genes requires binding of the GATA1 and EKLF1 transcription factor to both the enhancer and the target gene (Drissen et al. 2004; Vakoc et al. 2005). Besides transcription factors that directly bind DNA, other complexes have been implicated in chromatin loop formation. For instance, the chromatin-remodeling enzyme Brg1 is involved in looping in the α- and β-globin loci (Kim et al. 2009a; 2009b). Brg1 directly binds GATA1 and EKLF complexes and may be required for remodeling chromatin to facilitate binding of other factors.
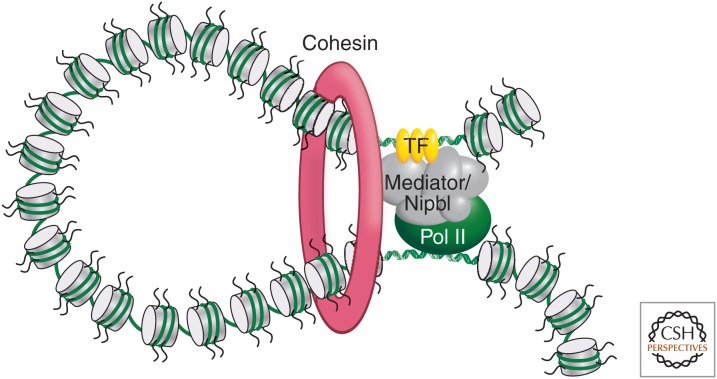
The presence of specific combinations of proteins on pairs of interacting elements may explain, at least in part, the specificity of looping interactions between promoters and their regulatory elements. Interestingly, there are also general factors that may contribute to long-range interactions. The Mediator complex is a transcriptional regulator that interacts with both the basal transcription machinery on the promoter and transcriptional activators, making this an ideal bridging factor between complexes bound at promoters and distal regulatory elements. Indeed, 3C analysis in mouse ES cells identified Mediator-dependent looping between promoters of pluripotency gene promoters and their enhancers (Fig. 8). These interactions also required the presence of the cohesin complex at these sites (Kagey et al. 2010). The cohesin complex mediates interactions between sister chromatids, but is increasingly implicated in mediating other types of long-range interactions, including looping between gene regulatory elements (Hadjur et al. 2009) and often in collaboration with the CTCF protein.
Figure 8.
Cohesin and Mediator complexes mediate long-range interactions between promoters and enhancers. The Cohesin and Mediator have been shown to act together to form and stabilize chromatin looping interactions between gene promoters and distal enhancers (Kagey et al. 2010). A model is shown of how these complexes might mediate chromatin looping, although molecular details are currently unknown. (Redrawn, with permission, from Young 2011, © Elsevier.)
5.1. The General Role of the CTCF Protein in Looping and Chromosome Organization
The CTCF protein binds tens of thousands of sites throughout the mouse and human genomes. The protein is particularly enriched near genes (Kim et al. 2007). Initial studies found that the protein acts as an insulator or boundary complex to block long-range gene regulation by enhancers, at least in reporter constructs. Recently, it has become clear that the protein plays a particularly prominent role in building higher-order chromatin architectures by facilitating intra- and interchromosomal looping interactions (Phillips and Corces 2009).
As described in Sections 4.2–4.4, active gene promoters often interact with distal sites bound by the CTCF protein (e.g., for example, in the α- and β-globin loci) but also in many other cases. CTCF-bound elements also interact with each other, which has led to a model that these loci play general roles in organizing chromosomes (Phillips and Corces 2009). CTCF-bound sites appear to cluster in the nucleus, which will greatly constrain the folding of chromosomes (Yusufzai et al. 2004). How assembly of these higher-order chromatin structures plays roles in regulating genes is far from clear. CTCF-binding to the H19 imprinting control region affects the pattern of looping on the imprinted Igf2 locus, elaborated in Barlow and Bartolomei (2014), consistent with a role in controlling other looping interactions in a chromosomal locus. However, other studies are not consistent with such an insulator-related role. Deletion of single CTCF-binding elements in the β-globin locus does not affect globin gene regulation or promoter–enhancer looping, possibly because they are redundant (Bender et al. 2006; Splinter et al. 2006). This observation is not consistent with a simple enhancer-blocking model in which CTCF-bound sites determine which enhancer–promoter looping interactions can occur. One additional complicating factor, however, is the possibility that CTCF-bound sites include a variety of different types of elements depending on additional protein and RNA factors that are recruited to these sites (Yao et al. 2010).
6. Looping interactions and gene regulation
Although the occurrence of looping interactions between promoters and regulatory elements is now well established, how these interactions contribute to modulating expression levels of the target genes is less well understood. Several observations indicate that direct interactions between promoters and distal enhancers facilitate the recruitment of critical transcription regulators to the gene. For instance, the α-globin enhancer has been shown to recruit RNA Pol II first, which may then be transferred to the target promoter through looping (Vernimmen et al. 2007). Other protein complexes can be recruited in a similar manner. Also, the LCR influences the transition of RNA polymerase from its initiating phase to transcriptional elongation (Sawado et al. 2003), suggesting that the LCR-promoter interaction can help recruit elongation factors to the promoter.
7. Conclusions
The statement, “genomes exist in space and time in the cell nucleus” is a trivial one, but one that has long been ignored in our studies of gene function. During the decades of pioneering work that have led to a now detailed molecular understanding of transcription, the consequences of spatial and temporal aspects of genome organization were largely ignored. This was necessary to reduce the complexity and to make the question amenable to experimental interrogation. The last decade, however, has brought the realization that gene expression is not merely controlled by the information contained in the DNA sequence, but that higher-order chromatin structure features such as nucleosome positioning, chromatin fiber formation, intra- and interchromosomal chromatin interactions, and even chromosome and gene localization contribute to genome function. Not only have we realized that these aspects of genome organization are critical, we are in the fortunate position to have experimental tools available to test their contribution. Although imaging-based methods have long been used for these studies, the emergence of biochemical methods to test physical genome interactions, coupled with affordable next-generation sequencing, has revolutionized our ability to now test their functional relevance. Using these methods, we can now fairly routinely generate complete maps of genome interactions.
Mapping of genome interactions is only the very first step in understanding the functional relevance of spatial and temporal genome organization. What needs to be done? A key step will be the generation of a large number of genome-wide interaction maps for various cell types, tissues, developmental stages, and diseases. Several international consortia, such as ENCODE and IHEC, are aiming to generate such data. Once this information is gathered, physical interaction profiles must be compared with gene expression profiles, and histone modification and DNA methylation patterns. These correlative studies should provide some hints as to the functional relationship between genome organization and function. To truly test their relevance, genome organization patterns will need to be manipulated experimentally. This can be achieved by knockdown and overexpression approaches. This should reveal, on the one hand, how genome organization affects function and, on the other hand, what cellular factors determine genome organization.
Mapping and understanding genome organization has important implications for disease. For instance, we know that the nonrandom genome organization plays a key role in determining cancer translocations and 3C-based methods are being used to detect translocations. It is intriguing, and not utopian anymore, to speculate that we will be able to use genome organization for diagnostic and prognostic purposes. Aberrant chromatin organization is increasingly recognized as a hallmark of various diseases ranging from common cancer to rare diseases. Of particular interest is the possibility of using genome organization as a marker for early stage detection of disease, taking advantage of the fact that changes in chromatin and genome organization often precede genetic changes.
In hindsight, it seems obvious that considering the spatial and temporal organization of the genome would be an integral part of epigenetic and genetic function, and needs to be taken into account when attempting to unravel the mysteries of the genome. For the first time, we are now in a position to explore this key aspect of genome function experimentally; there is no doubt that what we will find will enrich our understanding and appreciation of the complexity and intricacy of genome function.
Footnotes
Editors: C. David Allis, Marie-Laure Caparros, Thomas Jenuwein, Danny Reinberg, and Monika Lachner
Additional Perspectives on Epigenetics available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Ahmed S, Brickner DG, Light WH, Cajigas I, McDonough M, Froyshteter AB, Volpe T, Brickner JH. 2010. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat Cell Biol 12: 111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augui S, Filion GJ, Huart S, Nora E, Guggiari M, Maresca M, Stewart AF, Heard E. 2007. Sensing X chromosome pairs before X inactivation via a novel X-pairing region of the Xic. Science 318: 1632–1636. [DOI] [PubMed] [Google Scholar]
- Bacher CP, Guggiari M, Brors B, Augui S, Clerc P, Avner P, Eils R, Heard E. 2006. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat Cell Biol 8: 293–299. [DOI] [PubMed] [Google Scholar]
- Bantignies F, Roure V, Comet I, Leblanc B, Schuettengruber B, Bonnet J, Tixier V, Mas A, Cavalli G. 2011. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell 144: 214–226. [DOI] [PubMed] [Google Scholar]
- *.Barlow DP, Bartolomei MS. 2014. Genomic imprinting in mammals. Cold Spring Harb Perspect Biol 6: a018382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bau D, Sanyal A, Lajoie BR, Capriotti E, Byron M, Lawrence JB, Dekker J, Marti-Renom MA. 2010. The three-dimensional folding of the α-globin gene domain reveals formation of chromatin globules. Nat Struct Mol Biol 18: 107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender MA, Byron R, Ragoczy T, Telling A, Bulger M, Groudine M. 2006. Flanking HS-62.5 and 3′ HS1, and regions upstream of the LCR, are not required for β-globin transcription. Blood 108: 1395–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer AL, Miller OL, McKnight SL. 1980. Ribonucleoprotein structure in nascent hnRNA is nonrandom and sequence-dependent. Cell 20: 75–84. [DOI] [PubMed] [Google Scholar]
- Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, Bickmore WA. 2001. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet 10: 211–219. [DOI] [PubMed] [Google Scholar]
- Branco MR, Pombo A. 2006. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol 4: e138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner JH, Walter P. 2004. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol 2: e342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Brockdorff N, Turner BM. 2014. Dosage compensation in mammals. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Leach J, Reittie JE, Atzberger A, Lee-Prudhoe J, Wood WG, Higgs DR, Iborra FJ, Buckle VJ. 2006. Coregulated human globin genes are frequently in spatial proximity when active. J Cell Biol 172: 177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Groudine M. 1999. Looping versus linking: Toward a model for long-distance gene activation. Genes Dev 13: 2465–2477. [DOI] [PubMed] [Google Scholar]
- Bulger M, Groudine M. 2011. Functional and mechanistic diversity of distal transcription enhancers. Cell 144: 327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Busslinger M, Tarakhovsky A. 2014. Epigenetic control of immunity. Cold Spring Harb Perspect Biol 6: a019307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Lee CC, Kohwi-Shigematsu T. 2006. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet 38: 1278–1288. [DOI] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117: 427–439. [DOI] [PubMed] [Google Scholar]
- Cavalli G, Misteli T. 2013. Functional implications of genome topology. Nat Struct Mol Biol 20: 290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarle R, Zhang Y, Frock RL, Lewis SM, Molinie B, Ho YJ, Myers DR, Choi VW, Compagno M, Malkin DJ, et al. 2011. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell 147: 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer C, Schneider T, Baumann H, Hens L, Kirsch-Volders M. 1982. Analysis of chromosome positions in the interphase nucleus of Chinese hamster cells by laser-UV-microirradiation experiments. Hum Genet 62: 201–209. [DOI] [PubMed] [Google Scholar]
- Cremer M, Kupper K, Wagler B, Wizelman L, von Hase J J, Weiland Y, Kreja L, Diebold J, Speicher MR, Cremer T. 2003. Inheritance of gene density-related higher order chromatin arrangements in normal and tumor cell nuclei. J Cell Biol 162: 809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. 1999. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol 145: 1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean A. 2011. In the loop: Long range chromatin interactions and gene regulation. Brief Funct Genomics 10: 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T, Adam SA, Taimen P, Shimi T, Goldman RD. 2010. Nuclear lamins. Cold Spring Harb Perspect Biol 2: a000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J. 2006. The three ‘C’s of chromosome conformation capture: Controls, controls, controls. Nat Methods 3: 17–21. [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. 2002. Capturing chromosome conformation. Science 295: 1306–1311. [DOI] [PubMed] [Google Scholar]
- de Laat W, Grosveld F. 2003. Spatial organization of gene expression: The active chromatin hub. Chromosome Res 11: 447–459. [DOI] [PubMed] [Google Scholar]
- Deng W, Blobel GA. 2010. Do chromatin loops provide epigenetic gene expression states? Curr Opin Genet Dev 20: 548–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Haene B, Attanasio C, Beysen D, Dostie J, Lemire E, Bouchard P, Field M, Jones K, Lorenz B, Menten B, et al. 2009. Disease-causing 7.4 kb cis-regulatory deletion disrupting conserved non-coding sequences and their interaction with the FOXL2 promotor: Implications for mutation screening. PLoS Genet 5: e1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe ME, Silva SS, Pinter SF, Xu N, Lee JT. 2009. The pluripotency factor Oct4 interacts with Ctcf and also controls X-chromosome pairing and counting. Nature 460: 128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, et al. 2006. Chromosome Conformation Capture Carbon Copy (5C): A massively parallel solution for mapping interactions between genomic elements. Genome Res 16: 1299–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W. 2004. The active spatial organization of the β-globin locus requires the transcription factor EKLF. Genes Dev 18: 2485–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Misteli T. 2010. Biogenesis of nuclear bodies. Cold Spring Harb Perspect Biol 2: a000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Hoffmann-Rohrer U, Hu Q, Grummt I, Rothblum LI, Phair RD, Misteli T. 2002. A kinetic framework for a mammalian RNA polymerase in vivo. Science 298: 1623–1626. [DOI] [PubMed] [Google Scholar]
- Egecioglu D, Brickner JH. 2011. Gene positioning and expression. Curr Opin Cell Biol 23: 338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. 2011. A user’s guide to the Encyclopedia of DNA Elements. PLoS Biol 9: e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium, Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskiw CH, Cope NF, Clay I, Schoenfelder S, Nagano T, Fraser P. 2011. Transcription factories and nuclear organization of the genome. Cold Spring Harb Symp Quant Biol 75: 501–506. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G, Groudine M. 2003. Controlling the double helix. Nature 421: 448–453. [DOI] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA. 2008. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet 4: e1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, et al. 2009. An oestrogen-receptor-α-bound human chromatin interactome. Nature 462: 58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuss SH, Omura M, Mombaerts P. 2007. Local and cis effects of the H element on expression of odorant receptor genes in mouse. Cell 130: 373–384. [DOI] [PubMed] [Google Scholar]
- Fussner E, Ching RW, Bazett-Jones DP. 2010. Living without 30 nm chromatin fibers. Trends Biochem Sci 36: 1–6. [DOI] [PubMed] [Google Scholar]
- Gheldof N, Smith EM, Tabuchi TM, Koch CM, Dunham I, Stamatoyannopoulos JA, Dekker J. 2010. Cell-type-specific long-range looping interactions identify distant regulatory elements of the CFTR gene. Nucleic Acids Res 38: 4325–4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibcus JH, Dekker J. 2013. The hierarchy of the 3D genome. Mol Cell 49: 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Grossniklaus U, Paro R. 2014. Transcriptional silencing by Polycomb-group proteins. Cold Spring Harb Perspect Biol 6: a019331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453: 948–951. [DOI] [PubMed] [Google Scholar]
- Guo C, Gerasimova T, Hao H, Ivanova I, Chakraborty T, Selimyan R, Oltz EM, Sen R. 2011. Two forms of loops generate the chromatin conformation of the immunoglobulin heavy-chain gene locus. Cell 147: 332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. 2009. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460: 410–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR. 2008. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev 22: 2204–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim O, Misteli T. 2012. Chromosome conformation techniques. Cell 148: 1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim O, Resch W, Yamane A, Klein I, Kieffer-Kwon KR, Jankovic M, Oliveira T, Bothmer A, Voss TC, Ansarah-Sobrinho C, et al. 2012. DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature 484: 69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Hassan AB, Errington RJ, Cook PR. 1993. Visualization of focal sites of transcription within human nuclei. EMBO J 12: 1059–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhor R, Tjong H, Jayahilaka N, Alber F, Chen L. 2011. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat Biotechnol 30: 90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalverda B, Pickersgill H, Shloma VV, Fornerod M. 2010. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell 140: 360–371. [DOI] [PubMed] [Google Scholar]
- Karpen GH, Schaefer JE, Laird CD. 1988. A Drosophila rRNA gene located in euchromatin is active in transcription and nucleolus formation. Genes Dev 2: 1745–1763. [DOI] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. 2007. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128: 1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SI, Bresnick EH, Bultman SJ. 2009a. BRG1 directly regulates nucleosome structure and chromatin looping of the α globin locus to activate transcription. Nucleic Acids Res 37: 6019–6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SI, Bultman SJ, Kiefer CM, Dean A, Bresnick EH. 2009b. BRG1 requirement for long-range interaction of a locus control region with a downstream promoter. Proc Natl Acad Sci 106: 2259–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, van Steensel B. 2010. Genome-nuclear lamina interactions and gene regulation. Curr Opin Cell Biol 22: 320–325. [DOI] [PubMed] [Google Scholar]
- Klein IA, Resch W, Jankovic M, Oliveira T, Yamane A, Nakahashi H, Di Virgilio M, Bothmer A, Nussenzweig A, Robbiani DF, et al. 2011. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell 147: 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. 2002. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science 296: 158–162. [DOI] [PubMed] [Google Scholar]
- Kumaran RI, Spector DL. 2008. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol 180: 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. 2007. Dynamic genome architecture in the nuclear space: Regulation of gene expression in three dimensions. Nat Rev Genet 8: 104–115. [DOI] [PubMed] [Google Scholar]
- Lanzuolo C, Roure V, Dekker J, Bantignies F, Orlando V. 2007. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat Cell Biol 9: 1167–1174. [DOI] [PubMed] [Google Scholar]
- Li G, Reinberg D. 2011. Chromatin higher-order structures and gene regulation. Curr Opin Genet Dev 21: 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Fullwood MJ, Xu H, Mulawadi FH, Velkov S, Vega V, Ariyaratne PN, Mohamed YB, Ooi HS, Tennakoon C, et al. 2010. ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing. Genome Biol 11: R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light WH, Brickner DG, Brand VR, Brickner JH. 2010. Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Mol Cell 40: 112–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, Zhang J, Rose DW, Fu XD, Glass CK, et al. 2009. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell 139: 1069–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Lomvardas S, Maniatis T. 2014. Epigenetics regulation in neuronal development. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. 2006. Interchromosomal interactions and olfactory receptor choice. Cell 126: 403–413. [DOI] [PubMed] [Google Scholar]
- Mamanova L, Coffey AJ, Scott CE, Kozarewa I, Turner EH, Kumar A, Howard E, Shendure J, Turner DJ. 2010. Target-enrichment strategies for next-generation sequencing. Nat Methods 7: 111–118. [DOI] [PubMed] [Google Scholar]
- Mani RS, Tomlins SA, Callahan K, Ghosh A, Nyati MK, Varambally S, Palanisamy N, Chinnaiyan AM. 2009. Induced chromosomal proximity and gene fusions in prostate cancer. Science 326: 1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui O, Bonnet I, Le Baccon P, Brito I, Pollex T, Murphy N, Hupe P, Barillot E, Belmont AS, Heard E. 2011. Live-cell chromosome dynamics and outcome of X chromosome pairing events during ES cell differentiation. Cell 145: 447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathas S, Kreher S, Meaburn KJ, Johrens K, Lamprecht B, Assaf C, Sterry W, Kadin ME, Daibata M, Joos S, et al. 2009. Gene deregulation and spatial genome reorganization near breakpoints prior to formation of translocations in anaplastic large cell lymphoma. Proc Natl Acad Sci 106: 5831–5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCord RP, Nazario-Toole A, Zhang H, Chines PS, Zhan Y, Erdos MR, Collins FS, Dekker J, Cao K. 2013. Correlated alterations in genome organization, histone methylation, and DNA-lamin A/C interactions in Hutchinson-Gilford progeria syndrome. Genome Res 23: 260–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay B, Grummt I. 2008. The epigenetics of rRNA genes: From molecular to chromosome biology. Annu Rev Cell Dev Biol 24: 131–157. [DOI] [PubMed] [Google Scholar]
- Meaburn KJ, Misteli T. 2007. Cell biology: Chromosome territories. Nature 445: 379–781. [DOI] [PubMed] [Google Scholar]
- Meaburn KJ, Misteli T. 2008. Locus-specific and activity-independent gene repositioning during early tumorigenesis. J Cell Biol 180: 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister P, Towbin BD, Pike BL, Ponti A, Gasser SM. 2010. The spatial dynamics of tissue-specific promoters during C. elegans development. Genes Dev 24: 766–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. 2007. Beyond the sequence: Cellular organization of genome function. Cell 128: 787–800. [DOI] [PubMed] [Google Scholar]
- Misteli T. 2010. Higher-order genome organization in human disease. Cold Spring Harb Perspect Biol 2: a000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. 2011. The inner life of the genome. Sci Am 304: 66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Soutoglou E. 2009. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol 10: 243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, de Laat W, Spitz F, Duboule D. 2011. A regulatory archipelago controls Hox genes transcription in digits. Cell 147: 1132–1145. [DOI] [PubMed] [Google Scholar]
- Montes de Oca R, Andreassen PR, Wilson KL. 2011. Barrier-to-autointegration factor influences specific histone modifications. Nucleus 2: 580–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JR, Geyer PK, Wu CT. 1999. Core promoter elements can regulate transcription on a separate chromosome in trans. Genes Dev 13: 253–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves H, Ramos C, da Silva MG, Parreira A, Parreira L. 1999. The nuclear topography of ABL, BCR, PML, and RARalpha genes: Evidence for gene proximity in specific phases of the cell cycle and stages of hematopoietic differentiation. Blood 93: 1197–1207. [PubMed] [Google Scholar]
- Noordermeer D, de Wit E, Klous P, van de Werken H, Simonis M, Lopez-Jones M, Eussen B, de Klein A, Singer RH, de Laat W. 2011. Variegated gene expression caused by cell-specific long-range DNA interactions. Nat Cell Biol 13: 944–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485: 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, et al. 2004. Active genes dynamcially colocalize to shared sites of ongoing transcription. Nat Genet 36: 1065–1071. [DOI] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Mitchell JA, Horton A, Wood AL, Bolland DJ, Corcoran AE, Fraser P. 2007. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol 5: e192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott CJ, Blackledge NP, Kerschner JL, Leir SH, Crawford GE, Cotton CU, Harris A. 2009. Intronic enhancers coordinate epithelial-specific looping of the active CFTR locus. Proc Natl Acad Sci 106: 19934–19939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou SA, Chang E, Lee S, So K, Wu CT, Morris JR. 2009. Effects of chromosomal rearrangements on transvection at the yellow gene of Drosophila melanogaster. Genetics 183: 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W. 2003. The β-globin nuclear compartment in development and erythroid differentiation. Nat Genet 35: 190–194. [DOI] [PubMed] [Google Scholar]
- Papantonis A, Cook PR. 2010. Genome architecture and the role of transcription. Curr Opin Cell Biol 22: 271–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papantonis A, Larkin JD, Wada Y, Ohta Y, Ihara S, Kodama T, Cook PR. 2011. Active RNA polymerases: Mobile or immobile molecular machines? PLoS Biol 8: e1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada L, McQueen P, Munson P, Misteli T. 2002. Conservation of relative chromosome positioning in normal and cancer cells. Curr Biol 12: 1692. [DOI] [PubMed] [Google Scholar]
- Parada LA, Roix JJ, Misteli T. 2003. An uncertainty principle in chromosome positioning. Trends Cell Biol 13: 393–396. [DOI] [PubMed] [Google Scholar]
- Parada L, McQueen P, Misteli T. 2004. Tissue-specific spatial organization of genomes. Genome Biol 7: R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. 2010. The nucleolus. Cold Spring Harb Perspect Biol 3: a000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. 2009. CTCF: Master weaver of the genome. Cell 137: 1194–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky EM, Hopper AK. 2010. tRNA biology charges to the front. Genes Dev 24: 1832–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B. 2006. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet 38: 1005–1014. [DOI] [PubMed] [Google Scholar]
- Rajapakse I, Groudine M. 2011. On emerging nuclear order. J Cell Biol 192: 711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H. 2008. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 452: 243–247. [DOI] [PubMed] [Google Scholar]
- Roix JJ, McQueen PG, Munson PJ, Parada LA, Misteli T. 2003. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat Genet 34: 287–291. [DOI] [PubMed] [Google Scholar]
- Sanyal A, Bau D, Marti-Renom MA, Dekker J. 2011. Chromatin globules: A common motif of higher order chromosome structure? Curr Opin Cell Biol 23: 325–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J. 2012. The long-range interaction landscape of gene promoters. Nature 489: 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawado T, Halow J, Bender MA, Groudine M. 2003. The β-globin locus control region (LCR) functions primarily by enhancing the transition from transcription initiation to elongation. Genes Dev 17: 1009–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T. 2008. Lamin A-dependent misregulation of adult stem cells associated with accelerated ageing. Nat Cell Biol 10: 452–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Clay I, Fraser P. 2010a. The transcriptional interactome: Gene expression in 3D. Curr Opin Genet Dev 20: 127–133. [DOI] [PubMed] [Google Scholar]
- Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, et al. 2010b. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet 42: 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. 2012. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148: 458–472. [DOI] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. 2012. A map of the cis-regulatory sequences in the mouse genome. Nature 488: 116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. 2006. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat Genet 38: 1348–1354. [DOI] [PubMed] [Google Scholar]
- Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, Danuser G, Misteli T. 2007. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol 9: 675–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. 2005. Interchromosomal associations between alternatively expressed loci. Nature 435: 637–645. [DOI] [PubMed] [Google Scholar]
- Splinter E, de Laat W. 2011. The complex transcription regulatory landscape of our genome: Control in three dimensions. EMBO J 30: 4345–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W. 2006. CTCF mediates long-range chromatin looping and local histone modification in the β-globin locus. Genes Dev 20: 2349–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan GJ, Bridger JM, Cuthbert AP, Newbold RF, Bickmore WA, McStay B. 2001. Human acrocentric chromosomes with transcriptionally silent nucleolar organizer regions associate with nucleoli. EMBO J 20: 2867–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa T, Meaburn KJ, Misteli T. 2008. The meaning of gene positioning. Cell 135: 9–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M, Haeusler RA, Good PD, Engelke DR. 2003. Nucleolar clustering of dispersed tRNA genes. Science 302: 1399–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. 2002. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol Cell 10: 1453–1465. [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. 2005. Proximity among distant regulatory elements at the β-globin locus requires GATA-1 and FOG-1. Mol Cell 17: 453–462. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Dekker J. 2011. Genomics tools for unraveling chromosome architecture. Nat Biotechnol 28: 1089–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernimmen D, De Gobbi M, Sloane-Stanley JA, Wood WG, Higgs DR. 2007. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. EMBO J 26: 2041–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiech T, Timme S, Riede F, Stein S, Schuricke M, Cremer C, Werner M, Hausmann M, Walch A. 2005. Human archival tissues provide a valuable source for the analysis of spatial genome organization. Histochem Cell Biol 123: 229–238. [DOI] [PubMed] [Google Scholar]
- Wright JB, Brown SJ, Cole MD. 2010. Upregulation of c-MYC in cis through a large chromatin loop linked to a cancer risk-associated single-nucleotide polymorphism in colorectal cancer cells. Mol Cell Biol 30: 1411–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Tsai CL, Lee JT. 2006. Transient homologous chromosome pairing marks the onset of X inactivation. Science 311: 1149–1152. [DOI] [PubMed] [Google Scholar]
- Yao H, Brick K, Evrard Y, Xiao T, Camerini-Otero RD, Felsenfeld G. 2010. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev 24: 2543–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokochi T, Poduch K, Ryba T, Lu J, Hiratani I, Tachibana M, Shinkai Y, Gilbert DM. 2009. G9a selectively represses a class of late-replicating genes at the nuclear periphery. Proc Natl Acad Sci 106: 19363–19368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA. 2011. Control of the embryonic stem cell state. Cell 144: 940–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. 2004. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell 13: 291–298. [DOI] [PubMed] [Google Scholar]
- Zhang Y, McCord RP, Ho YJ, Lajoie BR, Hildebrand DG, Simon AC, Becker MS, Alt FW, Dekker J. 2012. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell 148: 908–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, et al. 2006. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet 38: 1341–1347. [DOI] [PubMed] [Google Scholar]
- Zink D, Amaral MD, Englmann A, Land S, Clarke LA, Rudolph C, Alt F, Luther K, Braz C, Sadoni N, et al. 2004. Transcription-dependent spatial arrangement of CFTR and adjacent genes in human cell nuclei. J Cell Biol 166: 815–825. [DOI] [PMC free article] [PubMed] [Google Scholar]