Abstract
The coelacanth has long been regarded as a “living fossil,” with extant specimens looking very similar to fossils dating back to the Cretaceous period. The hypothesis of a slowly or even not evolving genome has been proposed to account for this apparent morphological stasis. While this assumption seems to be sustained by different evolutionary analyses on protein-coding genes, recent studies on transposable elements have provided more conflicting results. Indeed, the coelacanth genome contains many transposable elements and has been shaped by several major bursts of transposition during evolution. In addition, comparison of orthologous genomic regions from the genomes of the 2 extant coelacanth species L. chalumnae and L. menadoensis revealed multiple species-specific insertions, indicating transposable element recent activity and contribution to post-speciation genome divergence. These observations, which do not support the genome stasis hypothesis, challenge either the impact of transposable elements on organismal evolution or the status of the coelacanth as a “living fossil.” Closer inspection of fossil and molecular data indicate that, even if coelacanths might evolve more slowly than some other lineages due to demographic and/or ecological factors, this variation is still in the range of a “non-fossil” vertebrate species.
Keywords: coelacanth, evolution, fish, genome stasis, living fossil, transposable elements, tetrapods
The Coelacanth: An Assumed “Living Fossil”
Coelacanths are lobe-finned fish that were thought to be extinct for 70 million years (Mega-annum, Ma), until a first living specimen was discovered fortuitously in South Africa in 1938 by Marjorie Courtenay-Latimer1 (Fig. 1). Two extant coelacanth species have been described to date, Latimeria chalumnae in Africa and L. menadoensis in Indonesia.2 Almost 80 years later, this fish continues to puzzle scientists. Coelacanths indeed present several unique and intriguing features such as unpaired lobbed-fins looking much like paired fins and highly modified lungs/swim bladder. Together with lungfish, they are the closest relatives to tetrapods, and are therefore essential to study the emergence of terrestrial vertebrates through comparative studies (Fig. 2).3,4 Accordingly, coelacanths share with tetrapods several morpho-anatomical features that are not found in more distantly related vertebrates such as ray-finned fishes. For example, the study of the structure of their paired lobbed-fins is instrumental to reconstruct the ancestral organization of the paired appendages of the tetrapod ancestry before the water-to-land transition that required the evolution of fins to limbs.5
Figure 1.

The African coelacanth Latimeria chalumnae (photo credit: Aquamarine Fukushima).
Figure 2.
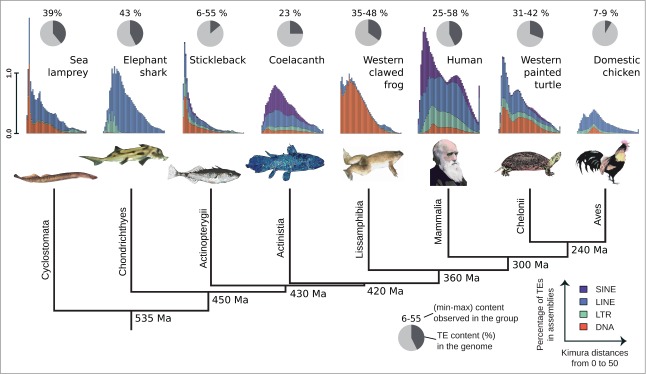
A phylogram of vertebrates with TE content and landscape in genomes (black: % of TEs, gray: % of non-TE DNA). TE content in coelacanth is from reference 3, a higher value has been reporter in another study.4 Landscapes highlight transposon activity during evolution: TE copies are clustered according to Kimura distances to their consensus sequence (X-axis), with more recent copies on the left of the graph and older more divergent copies on the right. Actinistia: Latimeria chalumnae (African coelacanth); Actinopterygii: Gasterosteus aculeatus (three-spined stickleback); Aves: Gallus gallus (chicken); Chelonii: Chrysemys picta (Western painted turtle); Chondrichthyes: Callorhinchus milii (elephant shark); Cyclostomata: Petromyzon marinus (sea lamprey); Lissamphibia: Xenopus tropicalis (Western clawed frog); Mammalia: Homo sapiens (human). Coelacanth, stickleback, elephant shark and sea lamprey landscape data are from reference 29. Chicken, Western painted turtle, human and Western clawed frog landscape values were retrieved from RepeatMasker Genomic datasets (http://www.repeatmasker.org/genomicDatasets/RMGenomicDatasets.html).
In addition, when the first extant coelacanth was discovered, it reminded so much fossil forms from the Cretaceous period (150 to 65 Ma ago) that it was designated as a “living fossil”, i.e. a species with a morphology that did not evolve much over a long period of time.1,6 Notably, as no other species belonging to the subclass Actinistia survived up to now but the 2 extant Latimeria species, it is expected that the species the most similar to coelacanths are fossils. To account for this apparent morphological stasis, it has often been suggested that coelacanths possess a slowly or even not evolving genome.7,8 The recent sequencing of the genomes of the 2 extant coelacanth species allowed testing this assumption.3,4
Do Coelacanth Protein-Coding Genes Evolve Slowly?
A first insight into genome dynamics in coelacanths has been obtained from the study of protein-coding genes. Before the sequencing of the complete genome, a number of analyses have been devoted to the study of specific loci of interest, such as the Hox and protocadherin gene clusters of L. menadoensis. Interestingly, the analysis of protein-coding sequences suggested a significantly reduced rate of evolution compared to other vertebrates.9-11 These observations were further supported by the genome-wide data obtained through the sequencing of the genomes of L. chalumnae3 and L. menadoensis.4 In the former study, 251 orthologous protein-coding genes of L. chalumnae and tetrapods were concatenated and analyzed. An average amino-acid substitution rate/site of 0.89 was observed in the Latimeria lineage. This is substantially lower than the values found in some tetrapods, representing 73% and 81% of the substitution rates in the human (1.21) and chicken lineages (1.09), respectively. This result was refined in the latter study by analyzing a set of 5,247 orthologous genes. Depending on the method of estimation, the average amino-acid substitution rate in Latimeria was 57% and 72% of the rate observed in human.4 In this analysis, genetic diversity in coelacanth was found to be extremely low, this being likely to be due to both a small population size and a reduced mutation rate.4
The alignment of genomic sequences from both coelacanth species revealed a very high percentage of interspecific nucleotide sequence identity: 98.7% globally and 99.7% for exonic sequences only.3,12 Nikaido and colleagues even proposed a nucleotide divergence of only 0.18% between the complete nuclear genomes of both species.4 Considering these nuclear sequences, the divergence time between both Latimeria species has been estimated to be slightly older than that of human and chimp (6–8 Ma),3 which show a similar degree of nucleotide sequence identity.13 However, other – and variable – divergence times have been obtained using mitochondrial (mt) DNA: from 4.7–6.3 Ma using only 7 mt genes (cytb, 12SrRN, 16SRNA and 4 tRNA genes)2 up to 30–40 Ma using the whole mtDNA genome.14 These major differences might be linked to a number of technical issues, and a more precise divergence estimate would probably require further investigations. Using a divergence time of 20–30 Ma, which might be a gross overestimation, it has been proposed that the nucleotide substitution rate is 30–40 times lower between coelacanths than between human and chimp.4 Taken together, all these observations suggest that the evolutionary rate of protein-coding genes is lower in coelacanths than in human and some other tetrapod lineages.
However, reduction of evolutionary rate in coelacanth is not supported by all studies, and might even not be specific to the coelacanth lineage in vertebrates. Indeed, a recent study based on a whole genome alignment of several vertebrate species did not confirm the finding of a low mutation rate in protein-coding regions of Latimeria. The analysis of fourfold degenerate sites (4D sites) revealed that the coelacanth displays a mutation rate that equals that of mammals and is about twice higher than those of sauropsids.15 However, this estimation might be biased by the fact that the coelacanth is at the end of a long branch and that the saturation at 4D sites is likely to be very high. Interestingly, it was also found that the non-coding DNA flanking ultra-conserved elements evolved at similar rates in coelacanth and mammals.15
Of note, amino-acid substitution rate and morphological evolution do not correlate easily: results gathered during decades of developmental genetics and “evo-devo” studies have established that morphological evolution is rather due to changes in regulatory sequences than to amino-acid substitutions.16 This is particularly well documented in the case of the Hox genes, which encode a transcription factor family that plays a key role in morphogenesis. On the one hand, the Hox complexes are very similar in human and mouse, whereas these species display quite different morphologies (for a recent review, see ref.17). On the other hand, extant sharks resemble Cladoselache, a fossil from Devonian, and were therefore often considered as “living fossils,” but undeservedly.18 However, sharks and rays have lost their entire HoxC cluster after their divergence from chimerae.19,20 Therefore the Hox complexes of Cladoselache are thought to be much more closely related to those of chimerae than to those of modern sharks and rays. The tuatara and the Western painted turtle are additional examples of contrasting aspects of morphological and molecular evolution. The tuatara displays very ancestral morphological traits but a rapid rate of genome evolution.21 On the contrary, the Western painted turtle shows very derived traits and adaptation to anoxia and tissue freezing but has a slow rate of genome evolution.15,22
Transposable Elements have Shaped the Genome of Coelacanth
Transposable elements (TEs) are powerful drivers of evolution. These mobile sequences are able to insert into new locations within genomes. Since they are generally repeated, they can also serve as substrates for homologous recombination and induce the formation of rearrangements such as deletions, duplications, inversions and translocations. TEs can provide new coding and regulatory sequences to the genes of their hosts, and can even be recruited as novel RNA and protein-coding genes in a process called molecular domestication.23,24 According to the “living fossil” hypothesis, TE activity should be strongly reduced and even absent in coelacanths.25
Several lines of evidence indicate that TEs have not been particularly “quiet” in the coelacanth lineage since its divergence from tetrapods. After firsts reports of the presence of Short Interspersed Nuclear Elements (SINEs) and DNA transposons in Latimeria,26,27 many different types of TEs have been identified in coelacanths after genome-wide analysis.28 Depending on the study, TEs have been reported to cover 23%–50% of the coelacanth genome.3,4 This fits well the range of the values found in other vertebrate genomes (from 6 to 55%,29 Fig. 2). Hence, coelacanth genomes are not particularly poor in transposable elements.
The most prominent types of TEs in the coelacanth genome belong to retrotransposons, particularly Long Interspersed Nuclear Elements (LINEs) and SINEs, but also a specific family of DNA transposons called LatiHarb1.3,4,27,28,30 From the point of view of TE diversity, the number of TE families is lower in coelacanth than in teleost fish, but higher than in chicken and mammals. This is consistent with the phylogenetic position of coelacanth in vertebrates, and with the sequential loss of TE families observed in the lineage leading to humans and birds after divergence from ray-finned fish.29
Clues on the evolutionary dynamics of each type of TEs during the evolution of a specific lineage can be obtained from a «landscape» analysis, which represents the distribution of TE copies according to their degree of sequence identity with their family consensus sequence31 (Fig. 2). In such an analysis, copies very similar to their consensus, probably resulting from more recent transposition events, are found on the left, while older copies are on the right of the plot. While the most ancient transposition burst in the coelacanth genome might have resulted from the expansion of LINE1 retrotransposons, 3 subsequent waves were detected, involving CR1, LINE2 and Deu retrotransposons as well as several DNA transposons such as Harbinger.4,28 This suggested transposition activity in the past and more recent evolutionary history of the coelacanth lineage.
Transposable Elements Contributed to Post-Speciation Genome Divergence in Coelacanths
An important direct proof of recent TE transposition is based on the identification of polymorphic insertions through the comparison of orthologous genomic sequences from different related species or individuals. The close nucleotide sequence similarity between L. chalumnae and L. menadoensis genomes allowed searching for TE insertions present at a particular locus in one species but absent from the other.
By comparing 36 Bacterial Artificial Chromosomes from L. menadoensis with their orthologous regions in L. chalumnae covering about 0.2% of the genome, 27 species-specific TE insertions were identified (13 in L. chalumnae and 14 in L. menadoensis, respectively).32 Notably, 15 of them (55%) were CR1 retrotransposons. Several insertions presented different hallmarks of recent transposition such as an important number of highly similar copies in the genome and the presence of identical target site duplications flanking the insertion. By extrapolating the results obtained to the whole genome, we would expect at least 6,500 TE insertions in one of the Latimeria species that are not found in the other. This number is probably underestimated, since only gene-rich regions were considered, which are expected to be more refractory to TE insertions than gene-poor regions. Interestingly, this value is of the same order of magnitude as the number of specific TE insertions detected between human and chimp,33 suggesting that coelacanth genomes do not evolve much more slowly than primate genomes in terms of TE activity.
Consistent with current activity, search for TEs in transcriptomes from liver, testis and muscle revealed transcription of a significant proportion of TE families, with a particular contribution of CR1 LINE, LF-SINE and LINE2 retrotransposons.30 For CR1 notably, several elements were found to have a high level of expression but low copy numbers in the genome, suggesting true activity rather than basal background transcription. While presence of TE sequences in transcriptome is not a definitive proof of current activity, the detected high expression of CR1 elements is in line with their observed recent transposition in the 2 coelacanth species. Hence, the data obtained indicate that TEs have been and are probably still active in coelacanth genomes. Since TEs are assumed to be powerful drivers of evolution, there is therefore a discrepancy between the apparent morphological stasis in coelacanths and the transposition activity detected in their genomes.
The Coelacanth: Living but not Fossil?
If we assume that coelacanths did not significantly change in terms of morphological appearance over millions of years, such an observation would strongly challenge the postulated impact of transposable elements on organismal evolution. Importantly, coelacanths certainly do not display the often assumed evolutionary-frozen morphology and are far more diverse than alleged.34 Indeed, the term coelacanth is loosely used to designate a family of more than 70 species regrouped in about 30 genera. Actually, no coelacanth fossil is available since the end of the Cretaceous period; we therefore have no information at all concerning the evolution of coelacanth morphology during the last 70 million years of evolution.
In addition, there is no representative of the Latimeria genera in the fossil record, and differences between Latimeria and its closest relative genus Macropoma are obvious. They not only present many differences in length, vertebral column regionalization, fin location and skull anatomy, but they are also adapted to very different environments. Indeed, the extant Latimeria species have an oil-filled swim bladder adapted to deep sea, whereas Macropoma have an ossified swim bladder/lung, suggesting that they were shallow water dwellers.5 Strikingly, the studies assuming a frozen morphology for coelacanths refer to original descriptions published in the first half of the past century, and/or inappropriately cite articles from Peter Forey that actually challenge the long held view of coelacanths as morphologically static from a cladistic point of view.5
Recently, a phylogenetic analysis of the pace of morphological changes in the lineages of coelacanths, teleosts and tetrapods came as a support for the slowly evolving coelacanth hypothesis. Even if the comparison of paces of evolution between lineages on the basis of non-homologous characters can be challenged, the rate of morphological evolution for the Latimeria lineage was estimated to be 3 times lower than in teleosts, and 6 times lower than in birds.35 Indeed, it is not surprising that the bird ancestry, which adapted first to terrestrial and then to aerial life, displays more dramatic osteological changes than lineages that stayed in water. In addition, even if it was claimed that the coelacanth skeleton has accumulated synapomorphies at a rate equal to one third of the rate found in teleosts, it can hardly be considered as having a frozen morphology, since many changes in shape and size are not identified as synapomorphies.
Conclusions
It is not surprising to observe that the Latimeria genome has been shaped by and still contains active transposable elements: the coelacanth has evolved, and therefore does not deserve its status of “living fossil.” Clearly, coelacanth morphology has been less constant during evolution than usually alleged. Moreover, nothing is known about the evolution of metabolism, immunity, development and behavior in this lineage: the quite stable organization of its skeleton does not imply that other traits did not evolve. At the genomic level, several studies converged to the conclusion that the protein-coding sequences of the coelacanth evolved significantly more slowly than the sequences of other vertebrates including humans and other mammals. However, only 30%–50% reductions in evolutionary rates have been reported, which is far from no evolution at all. Moreover, a low substitution rate was not found in an independent study on 4D sites15 and the Ka/Ks is higher in coelacanths than in human/chimp comparison.4 There is thus no strong evidence supporting that a lower mutation rate and/or a stronger negative selection could be at the origin of a lower substitution rate at the amino-acid level. Finally, the protein-coding sequences of some other tetrapod lineages such as sauropsids might evolve more slowly than those of coelacanths.15
It is well established that both the mutation and the amino-acid substitution rates are not constant across lineages. Even if the coelacanth might in some aspects evolve more slowly than other lineages due to demographic and/or ecological factors, we assume that this variation is in the range of “non-fossil” vertebrates. This is supported by the fact that coelacanth genomes contain, like those of other vertebrates, active transposable elements. More work is required to assess the impact of these sequences as a source of mutations and new regulatory and coding sequences on the genomic and organismal diversity of this fascinating fish.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Smith JLB. A living fish of mesozoic type. Nature 1939; 143:455-6 [Google Scholar]
- 2.Holder MT, Erdmann MV, Wilcox TP, Caldwell RL, Hillis DM. Two living species of coelacanths? Proc Natl Acad Sci 1999; 96:12616-20; PMID:10535971; http://dx.doi.org/ 10.1073/pnas.96.22.12616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amemiya CT, Alföldi J, Lee AP, Fan S, Philippe H, MacCallum I, Braasch I, Manousaki T, Schneider I, Rohner N, et al.. The African coelacanth genome provides insights into tetrapod evolution. Nature 2013; 496:311-6; PMID:23598338; http://dx.doi.org/ 10.1038/nature12027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nikaido M, Noguchi H, Nishihara H, Toyoda A, Suzuki Y, Kajitani R, Suzuki H, Okuno M, Aibara M, Ngatunga BP, et al.. Coelacanth genomes reveal signatures for evolutionary transition from water to land. Genome Res 2013; 23:1740-8; PMID:23878157; http://dx.doi.org/ 10.1101/gr.158105.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forey PL, Gardiner BG, Patterson C. Origins of the higher groups of tetrapods, controversy and consensus Chapter 5, The lungfish, the coelacanth, and the cow revisited. Schultze H-P, Trueb L Eds., Ithaca: Cornell University Press; 1991; p:145-72. [Google Scholar]
- 6.Smith JLB. Old Fourlegs, the story of the coelacanth. London, New York: Longmans; 1956:260 p. [Google Scholar]
- 7.Thomson KS. Living fossil: The story of the Coelacanth. W.W. Norton; 1992:252 p. [Google Scholar]
- 8.Friedman M, Coates MI. A newly recognized fossil coelacanth highlights the early morphological diversification of the clade. Proc Biol Sci 2006; 273:245-50; PMID:16555794; http://dx.doi.org/ 10.1098/rspb.2005.3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noonan JP, Grimwood J, Danke J, Schmutz J, Dickson M, Amemiya CT, Myers RM. Coelacanth genome sequence reveals the evolutionary history of vertebrate genes. Genome Res 2004; 14:2397-405; PMID:15545497; http://dx.doi.org/ 10.1101/gr.2972804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amemiya CT, Powers TP, Prohaska SJ, Grimwood J, Schmutz J, Dickson M, Miyake T, Schoenborn MA, Myers RM, Ruddle FH, et al.. Complete HOX cluster characterization of the coelacanth provides further evidence for slow evolution of its genome. Proc Natl Acad Sci 2010; 107:3622-7; PMID:20139301; http://dx.doi.org/ 10.1073/pnas.0914312107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higasa K, Nikaido M, Saito TL, Yoshimura J, Suzuki Y, Suzuki H, Nishihara H, Aibara M, Ngatunga BP, Kalombo HWJ, et al.. Extremely slow rate of evolution in the HOX cluster revealed by comparison between Tanzanian and Indonesian coelacanths. Gene 2012; 505:324-32; PMID:22698790; http://dx.doi.org/ 10.1016/j.gene.2012.05.047 [DOI] [PubMed] [Google Scholar]
- 12.Pallavicini A, Canapa A, Barucca M, Alfőldi J, Biscotti M, Buonocore F, De Moro G, Di Palma F, Fausto A, Forconi M, et al.. Analysis of the transcriptome of the Indonesian coelacanth Latimeria menadoensis. BMC Genomics 2013; 14:538; PMID:23927401; http://dx.doi.org/ 10.1186/1471-2164-14-538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The Chimpanzee Sequencing and Analysis Consortium . Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 2005; 437:69-87; PMID:16136131; http://dx.doi.org/ 10.1038/nature04072 [DOI] [PubMed] [Google Scholar]
- 14.Inoue JG, Miya M, Venkatesh B, Nishida M. The mitochondrial genome of Indonesian coelacanth Latimeria menadoensis (Sarcopterygii: Coelacanthiformes) and divergence time estimation between the two coelacanths. Gene 2005; 349:227-35; PMID:15777665; http://dx.doi.org/ 10.1016/j.gene.2005.01.008 [DOI] [PubMed] [Google Scholar]
- 15.Green RE, Braun EL, Armstrong J, Earl D, Nguyen N, Hickey G, Vandewege MW, John JAS, Capella-Gutiérrez S, Castoe TA, et al.. Three crocodilian genomes reveal ancestral patterns of evolution among archosaurs. Science 2014; 346:1254449; PMID:25504731; http://dx.doi.org/ 10.1126/science.1254449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carroll SB, Grenier JK, Weatherbee SD. From DNA to diversity: molecular genetics and the evolution of animal design. 2nd ed. Malden, MA: Blackwell Scientific; 2005:214 p. [Google Scholar]
- 17.Liang D, Wu R, Geng J, Wang C, Zhang P. A general scenario of Hox gene inventory variation among major sarcopterygian lineages. BMC Evol Biol 2011; 11:25; PMID:21266090; http://dx.doi.org/ 10.1186/1471-2148-11-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cuny G. Les requins sont-ils des fossiles vivants? EDP Sciences; 2002:205 p. [Google Scholar]
- 19.Oulion S, Debiais-Thibaud M, d' Aubenton-Carafa Y, Thermes C, Da Silva C, Bernard-Samain S, Gavory F, Wincker P, Mazan S, Casane D. Evolution of Hox gene clusters in gnathostomes: insights from a survey of a shark (Scyliorhinus canicula) transcriptome. Mol Biol Evol 2010; 27:2829-38; PMID:20616144; http://dx.doi.org/ 10.1093/molbev/msq172 [DOI] [PubMed] [Google Scholar]
- 20.King BL, Gillis JA, Carlisle HR, Dahn RD. A natural deletion of the HoxC cluster in elasmobranch fishes. Science 2011; 334:1517; PMID:22174244; http://dx.doi.org/ 10.1126/science.1210912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hay JM, Subramanian S, Millar CD, Mohandesan E, Lambert DM. Rapid molecular evolution in a living fossil. Trends Genet 2008; 24:106-9; PMID:18255186; http://dx.doi.org/ 10.1016/j.tig.2007.12.002 [DOI] [PubMed] [Google Scholar]
- 22.Shaffer HB, Minx P, Warren DE, Shedlock AM, Thomson RC, Valenzuela N, Abramyan J, Amemiya CT, Badenhorst D, Biggar KK, et al.. The western painted turtle genome, a model for the evolution of extreme physiological adaptations in a slowly evolving lineage. Genome Biol 2013; 14:R28; PMID:23537068; http://dx.doi.org/ 10.1186/gb-2013-14-3-r28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Volff J-N. Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. Bioessays 2006; 28:913-22; PMID:16937363; http://dx.doi.org/ 10.1002/bies.20452 [DOI] [PubMed] [Google Scholar]
- 24.Rebollo R, Romanish MT, Mager DL. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu Rev Genet 2012; 46:21-42; PMID:22905872; http://dx.doi.org/ 10.1146/annurev-genet-110711-155621 [DOI] [PubMed] [Google Scholar]
- 25.Oliver KR, Greene WK. Transposable elements: powerful facilitators of evolution. Bioessays 2009; 31:703-14; PMID:19415638; http://dx.doi.org/ 10.1002/bies.200800219 [DOI] [PubMed] [Google Scholar]
- 26.Bejerano G, Lowe CB, Ahituv N, King B, Siepel A, Salama SR, Rubin EM, Kent WJ, Haussler D. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature 2006; 441:87-90; PMID:16625209; http://dx.doi.org/ 10.1038/nature04696 [DOI] [PubMed] [Google Scholar]
- 27.Smith JJ, Sumiyama K, Amemiya CT. A living fossil in the genome of a living fossil: Harbinger transposons in the coelacanth genome. Mol Biol Evol 2012; 29:985-93; PMID:22045999; http://dx.doi.org/ 10.1093/molbev/msr267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chalopin D, Fan S, Simakov O, Meyer A, Schartl M, Volff J-N. Evolutionary active transposable elements in the genome of the coelacanth. J Exp Zool B Mol Dev Evol 2013; 332:322-33; PMID:23908136 [DOI] [PubMed] [Google Scholar]
- 29.Chalopin D, Naville M, Plard F, Galiana D, Volff J-N. Comparative analysis of transposable elements highlights mobilome diversity and evolution in vertebrates. Genome Biol Evol 2015; 7:567-80; PMID:25577199; http://dx.doi.org/ 10.1093/gbe/evv005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forconi M, Chalopin D, Barucca M, Biscotti MA, De Moro G, Galiana D, Gerdol M, Pallavicini A, Canapa A, Olmo E, et al.. Transcriptional activity of transposable elements in coelacanth. J Exp Zool B Mol Dev Evol 2014; 322:379-89; PMID:24038780; http://dx.doi.org/ 10.1002/jez.b.22527 [DOI] [PubMed] [Google Scholar]
- 31.Smit A, Hubley R, Green P. RepeatMasker Open-3.0. c1996-2010. Available from: http://www.repeatmasker.org [Google Scholar]
- 32.Naville M, Chalopin D, Volff J-N. Interspecies insertion polymorphism analysis reveals recent activity of transposable elements in extant coelacanths. PloS One 2014; 9:e114382; PMID:25470617; http://dx.doi.org/ 10.1371/journal.pone.0114382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mills RE, Bennett EA, Iskow RC, Devine SE. Which transposable elements are active in the human genome? Trends Genet 2007; 23:183-91; PMID:17331616; http://dx.doi.org/ 10.1016/j.tig.2007.02.006 [DOI] [PubMed] [Google Scholar]
- 34.Casane D, Laurenti P. Why coelacanths are not “living fossils”. Bioessays 2013; 35:332-8; PMID:23382020; http://dx.doi.org/ 10.1002/bies.201200145 [DOI] [PubMed] [Google Scholar]
- 35.Cavin L, Guinot G. Coelacanths as “almost living fossils”. Paleontology 2014; 2:49 [Google Scholar]


