Abstract
Microtubules (MTs) are cytoskeletal filaments essential for many processes in eukaryotic cells. Assembled of tubulin subunits, MTs are dynamic structures that undergo successive and stochastic phases of polymerization and depolymerization, a behavior called dynamic instability. Dynamic instability has been extensively studied in cultured cells and in vitro using cytoplasmic extracts or reconstituted MTs. However, how MTs behave in intact tissues and how their dynamics are affected by or affect tissue function are poorly understood. Recent advances in high-resolution live imaging have helped overcome technical limitation in order to visualize MTs in intact living organisms including Drosophila or Caenorhabditis elegans. We recently took advantage of the well-characterized development, small size and transparency of C. elegans to monitor MT dynamics throughout tissue biogenesis with high spatial and temporal resolution. Using the sex myoblast lineage that generates the egg-laying muscles from 2 mitotic precursors, we identified selective dynamics in precursor versus differentiated cells, and molecular regulation of MT dynamics changes that occur during cell differentiation. We discuss here how this approach led to novel insights into the regulation of MTs dynamics and organization in vivo.
Keywords: cell differentiation, cytoskeleton, high-resolution live imaging, microtubule dynamics, microtubule differentiation, tissue biogenesis
Abbreviations
- MT
microtubules
- MAP
microtubule associated protein
- SM
sex myoblast
- UMC
uterine muscle cell
High-Resolution Imaging Using C. elegans
Even before the discovery of MT dynamic instability in 1984,1,2 MTs inspired curiosity in cell biologists and investigators in other disciplines by their ability to self assemble, the conservation of MT-based processes among eukaryotes, and MTs’ important role in mitosis that makes them an important target of anti-cancer drugs. Despite extensive study, many aspects of MT dynamics remain enigmatic, including how MTs behave in differentiated somatic cells, and how their dynamics relate to cell and tissue function. Recently, some aspects of MT dynamics have been described during development in the fruit fly3 and zebrafish.4,5 MT organization was followed during neuronal morphogenesis in the embryonic cerebrum in mouse using extravital imaging6 and MT dynamics have been observed in vivo using intravital imaging of mouse muscle fibers.7 The latter study revealed that MTs display different dynamic properties whether the same muscle fibers were observed in culture or in vivo. These observations motivate minimally invasive study of MT dynamics and its regulation in a tissue context in an intact animal.
One of our aims was to understand how MT dynamics change during cell differentiation and tissue biogenesis. One obstacle to develop such an approach is to achieve the high spatial and temporal resolution required to measure MT dynamics while preserving animal integrity. We found C. elegans to be ideal model system for fluorescence live imaging of MT dynamics since it is a small transparent animal that develops rapidly and is amenable to the use of powerful genetic tools. One common concern for any fluorescent imaging is phototoxicity. While oxygen scavengers can be used with in vitro systems to overcome this issue,8 the only way to prevent the stress generated by imaging an intact animal is to minimize the illumination of the specimen. Doing so compromises the signal to noise ratio, thus necessitating the use of sensitive detectors and optical or arithmetic techniques that reduce or remove out-of-focus light. To obtain the desired temporal and spatial resolution, as well as signal to noise ratio, we used swept-field confocal microscopy, which allows essentially simultaneous illumination of the focal plane via a sweeping array of pinholes or slits.9,10 Swept-field confocal microscopy also achieved desirable axial resolution in thicker samples such as gravid adult worms (100–120 μm diameter). To verify that this technique is a non-invasive way of measuring MT dynamics in intact C. elegans, we tested the effects of imaging, and presumably some phototoxicity, on growth rate and catastrophe frequency. We imaged our cell of interest, the uterine muscle cells (UMCs), in situ for an extended period of time and measured MT dynamics in control worms. We found that MT dynamics did not significantly differ between the beginning and end of the imaging period (unpublished data). To further ensure that MT dynamics are robust to our imaging conditions, we then used 4-fold less illumination intensity for our experiments. In addition, even after 2 hours of imaging, our C. elegans worms displayed normal behavior including egg laying after being recovered from the imaging chamber to a culture plate. Thus, our in situ system not only revealed MT dynamics in a tissue context but also provided a means to assess the physiological relevance of our experimental conditions.
Microtubule Dynamics Regulation During Cell Differentiation
In order to follow MT dynamics over the course of differentiation and development in situ, we generated a strain that expresses GFP::tubulin specifically in several cell lineages, including the sex myoblasts (SM) that give rise to the egg-laying apparatus. This lineage comprises 2 precursors (SM) that give rise to the 16 egg-laying muscles including UMCs (Fig. 1). Divisions and differentiation of the SMs and their descendants occur within 24 hours of growth at 25°C, beginning at the third larval stage (L3) and ending during the adult stage. To describe MT behavior in the SM lineage, we focused on the 4 main parameters that describe MT dynamics11 i.e., the rates of assembly and disassembly (growth and shrinkage rates) and frequencies of the transitions between these events (catastrophe: growth to shrinkage, and rescue: shrinkage to growth). We found that all 4 parameters significantly change during differentiation of SM into UMCs. Growth and shrinkage rates increase while the frequencies of catastrophe and rescue are reduced over the course of differentiation.
Figure 1.
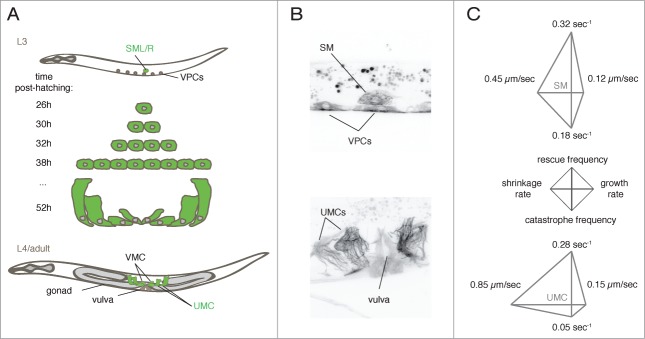
MTs dynamics followed during differentiation in situ. (A) SM lineage. The left and right sex myoblasts (SML/R), divide and differentiated to generate 16 egg-laying muscles. UMCs = uterine muscle cells, VMCs = vulva muscle cells, VPCs = vulval precursor cells. (B) Maximal intensity projections of confocal z-series images of worms expressing GFP-tubulin under the control of the unc-62 promoter, at the L3 (top) and adult stages (bottom). (C) Diamond graphs representing dynamics data of MTs in the SM and UMCs. Data from Lacroix et al.12
The multidimensional nature of MT dynamics data can confound visualization and comparison among different conditions. To overcome this complexity, we created 4 axis plots, which we call diamond graphs (Fig. 1) and that simultaneously represent the 4 main parameters of MT dynamics (see details in Lacroix et al.12). The shapes and sizes of diamond graphs thus allow simple and instantaneous comparison of multiple data sets in a manner far more accessible than conventional tables. We predict that diamond graphs (or other shapes with 3 or more struts) will be adopted for other multidimensional data analyses.
We combined our in situ imaging regime with RNAi-based protein depletion to test how MT dynamics change with different sub-processes of differentiation. We depleted LIN-12Notch and HLH-8Twist to compromise UMCs differentiation. We found that each dynamic parameter is distinctly regulated during differentiation. Specifically, rescue frequency changes to post-differentiation levels as cells exit the mitotic state, whereas the other parameters change only when morphogenesis occurs later during differentiation. Thus, we identified the selective regulation of MT dynamics during different steps of the differentiation process and the uncoupling of the dynamics parameters.
Molecular and Dynamics Signatures of Mts During Differentiation
To understand how changes in dynamics were triggered during differentiation and how MTs are required for differentiated cell function, we depleted conserved and predicted tubulin isoforms, tubulin-modifying enzymes, and MT-associated proteins (MAPs). Targets were depleted either from the first larval stage, during tissue biogenesis, or starting from the fourth larval stage to affect only tissue function. The UMCs are essential for egg-laying apparatus development and function, so any defect in SM cell division or differentiation will perturb egg laying and manifest as lower brood size with eggs accumulating in the uterine cavity.12,13 For approximately 100 targets, we assessed the gross morphology of the animal and of the vulva, as well as brood size. We verified which MAPs’ depletion perturbed egg laying due to a role specifically in the UMCs by performing tissue specific RNAi and by stimulating egg laying with drugs to confidently attribute the defects to a dysfunction of the muscle cells and not to other tissues including the neurons that control egg laying.
The screen enabled us to identify conserved MAPs required for SM division and differentiation, or for function of differentiated cells. MAPs were classified based on phenotypes and on their ability to perturb either early developmental stages or tissue function. For example, we identified a set of proteins including CLS-1CLASP, EBP-2EB1 SAS-3γ-tubulin, SAS-4, SAS-6 and TPXL-1TPX2 essential for divisions and expansion of SM but not for the function of differentiated cells. Interestingly, some MAPs with redundant functions appeared in the same categories. For example, KLP-7MCAK and KLP-13Kip3 are 2 kinesins with similar MT-depolymerizing activity and were both found to be essential for the function of differentiated cells only. This result suggests that in differentiated cells, the regulation of MT depolymerization is tightly controlled by at least these 2 kinesins and that their functions are not redundant. Since differentiated cells are larger and have multiple distinct subcellular regions, it is possible that local regulation via local enrichment of specific MAPs generates different MT populations that co-exist in the same cell and are essential for proper cell function. Defining MT-related proteins that act specifically in proliferating cells is of interest for the development of drugs that selectively target cancer cells without affecting differentiated cell function.
We further correlated tissue-level phenotype with MT dynamics in UMCs to test whether MAP depletions causing muscle dysfunction shared a MT dynamics signature. Among the perturbations of MT dynamics in UMCs that caused egg-laying defects, they all had in common a global reduction of MT dynamics. Conversely, increased dynamics did not perturb function, suggesting that high dynamicity, and not just polymer level, is essential for proper function of these differentiated cells.
Features of MT Differentiation
Comparing various static conditions such as cell types and species demonstrate that a range of MT dynamics parameters exists throughout nature. This diversity is achieved via the combinatorial effects of tubulin isoforms and post-translational modifications, MAPs and MT dynamics. The molecular and dynamic changes that MTs undergo during cell differentiation could collectively be thought of as “MT differentiation.” MTs in differentiated cells are commonly thought to be more stable that in proliferating cells. Thus it was unexpected that our study revealed high dynamicity of MTs in differentiated cells. A single expression of dynamicity is the average distance that a MT can cover per unit time. Interestingly, dynamicity was not significantly different (p = 0.2416) between SM (3.3 ± 0.3 μm/min) and UMC (2.9 ± 0.2 μm/min), meaning that MTs in differentiated cells are as dynamic as those in the precursor cell of the same lineage. Our work is substantiated by the observation of highly dynamic MTs in mouse muscles in vivo.7
The general idea that MTs are more stable in differentiated cells was in part speculated based on in vitro experiments that highlighted the presence of MTs resistant to depolymerization in differentiated cells in culture.14–18 These so-called stable MTs were also decorated by several post-translational modifications of tubulin, which therefore became a marker of MT stability. These post-translational modifications of tubulin are indeed often enriched in differentiated cells as compared to proliferative cells. Recently, tubulin post-translational modifications have come to be associated not necessarily with stable MTs, but rather with specific populations of MTs within the cells.19
In support of the idea that tubulin post-translational modifications are important for the differences in MT dynamics between SM and UMCs, our screen revealed that several enzymes responsible for these modifications are essential for differentiated cell function and not for precursors. Future work will be aimed at developing tools to detect tubulin post-translational modifications in C. elegans, assessing their presence during differentiation, and determining if this specific requirement of modifying enzymes is correlated with the presence of the modifications.
An emerging concept is that differentiated cells have different populations of MTs with distinct dynamics, rather than a global stabilization of the MT network. Based on our observations and on what others have also reported in C. elegans neurons20 and in mouse muscle fibers,7 differentiated cells appear to share the common feature of fast growing MTs. Our work revealed that the main differences between differentiated and mitotic cells are the length of MTs and the persistence of polymer elongation (higher in differentiated cells). Thus, in differentiated cells, MT tracks appear to be longer and to grow at higher rate. As suggested by Ghosh-Roy et al.20 and by our work, the regulation of this process might involve a “selection” of robust fast growing MTs and elimination of short MTs with a rapid turnover (Fig. 2).
Figure 2.
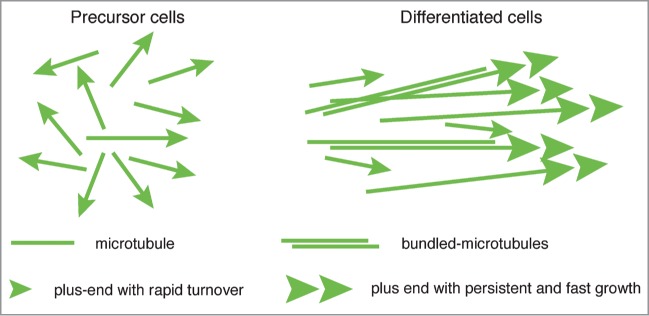
Model of MT differentiation. Generic model of MT adaptation to cell differentiation. MTs organization and dynamic properties change as cells differentiate, forming a more parallel MT array with fast and persistent growth excursions. Differentiated cells also contain different MT populations such as bundles, with distinct properties.
Another general characteristic of differentiated cells is also the loss of centrosomal organization.21 As a consequence, MTs in differentiated cells adopt a more parallel organization, as we observed in UMCs (Figs. 1B and 2). This spatial reorganization polarizes the location of dynamic MT-ends within the cell, and thus could influence MT dynamics, cell organization and function. While our work began to define several molecular and dynamic aspects of MT differentiation, additional studies of different cell types or species will be useful to understand the evolution of MTs during the different steps of cell differentiation.
How do Tissue Context and Cell Environment Affect MT Dynamics?
Measuring MT dynamics in an intact tissue allowed us to directly compare cellular and subcellular organelle function with tissue- and animal level phenotypes. Another advantage of such an approach that we did not investigate is, conversely, the influence of the neighboring environment on an intracellular behavior.
Our approach combined with tissue specific RNAi could be used to determine if altering the development or function of neighboring tissues affects MT dynamics in the UMCs in a cell non-autonomous manner. For example, we have used LIN-12Notch depletion to block the differentiation of the SM daughter cells. Depletion of LIN-12Notch is also known to affect the development of the uterine cells that directly contact the UMCs.22-24 To test how differentiation in neighboring tissues impacts MT dynamics in UMCs, one could use a strain background in which all cells were RNAi-incompetent, and rescuing RNAi with specific expression of RNAi machinery in the uterus or other tissues in contact with the UMCs.
Concluding Remarks
The C. elegans zygote has proven to be a powerful cell biological system for the study of conserved cytoskeletal components. We have extended the use of this model species to investigate MT functions and dynamics with high spatial and temporal resolution, at different stages of development, in the intact animal. Furthermore, since our cells of interest are embedded in a tissue, sustaining functions, we were able to correlate the cell biology of MT dynamics with cellular behavior. Given that MT related proteins and processes are highly conserved between nematodes and mammals, we expect that discoveries made with C. elegans in situ somatic cell biology will help us understand MT regulation in general. Conversely, since our system allows combining cell biology and developmental biology with affordable genetic tools such as efficient RNAi mediated protein depletions, we expect that it will be employed to follow up on the in vivo roles of MT related proteins and processes characterized via in vitro reconstitution with purified brain MTs. We are confident that such a dialog between in vitro and in situ approaches will strengthen the use of C. elegans for MT biology.
Disclosure of Potential Conflicts of Interest
There were no potential conflicts of interest to disclose.
Acknowledgments
We thank Julien Dumont (Institut Jacques Monod, France) for critical reading of the manuscript.
Funding
BL was supported by Cancer Research Society (Canada) F2011–16307 and the Fondation pour la Recherche Médicale (France) ARF2014012955. ASM is supported by the National Institutes of Health (GM102390).
References
- 1. Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature 1984; 312:237-42; PMID:6504138; http://dx.doi.org/ 10.1038/312237a0 [DOI] [PubMed] [Google Scholar]
- 2. Mitchison T, Kirschner M. Microtubule assembly nucleated by isolated centrosomes. Nature 1984; 312:232-7; PMID:6504137; http://dx.doi.org/ 10.1038/312232a0 [DOI] [PubMed] [Google Scholar]
- 3. Brodu V, Baffet AD, Le Droguen PM, Casanova J, Guichet A. A developmentally regulated two-step process generates a noncentrosomal microtubule network in Drosophila tracheal cells. Dev Cell 2010; 18:790-801; PMID:20493812; http://dx.doi.org/ 10.1016/j.devcel.2010.03.015 [DOI] [PubMed] [Google Scholar]
- 4. Distel M, Hocking JC, Volkmann K, Koster RW. The centrosome neither persistently leads migration nor determines the site of axonogenesis in migrating neurons in vivo. J Cell Biol 2010; 191:875-90; PMID:21059852; http://dx.doi.org/ 10.1083/jcb.201004154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yoo SK, Lam PY, Eichelberg MR, Zasadil L, Bement WM, Huttenlocher A. The role of microtubules in neutrophil polarity and migration in live zebrafish. J Cell Sci 2012; 125:5702-10; PMID:22992461; http://dx.doi.org/ 10.1242/jcs.108324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sakakibara A, Sato T, Ando R, Noguchi N, Masaoka M, Miyata T. Dynamics of Centrosome Translocation and Microtubule Organization in Neocortical Neurons during Distinct Modes of Polarization. Cereb Cortex 2013; 24:1301-10; PMID:23307632; http://dx.doi.org/ 10.1093/cercor/bhs411 [DOI] [PubMed] [Google Scholar]
- 7. Oddoux S, Zaal KJ, Tate V, Kenea A, Nandkeolyar SA, Reid E, Liu W, Ralston E. Microtubules that form the stationary lattice of muscle fibers are dynamic and nucleated at Golgi elements. J Cell Biol 2013; 203:205-13; PMID:24145165; http://dx.doi.org/ 10.1083/jcb.201304063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gell C, Bormuth V, Brouhard GJ, Cohen DN, Diez S, Friel CT, Helenius J, Nitzsche B, Petzold H, Ribbe J, et al. . Microtubule dynamics reconstituted in vitro and imaged by single-molecule fluorescence microscopy. Methods Cell Biol 2010; 95:221-45; PMID:20466138; http://dx.doi.org/ 10.1016/S0091-679X(10)95013-9 [DOI] [PubMed] [Google Scholar]
- 9. Castellano-Munoz M, Peng AW, Salles FT, Ricci AJ. Swept field laser confocal microscopy for enhanced spatial and temporal resolution in live-cell imaging. Microsc Microanal 2012; 18:753-60; PMID:22831554; http://dx.doi.org/ 10.1017/S1431927612000542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maddox AS, Maddox PS. High-resolution imaging of cellular processes in Caenorhabditis elegans. Methods Cell Biol 2012; 107:1-34; PMID:22226519; http://dx.doi.org/ 10.1016/B978-0-12-394620-1.00001-1 [DOI] [PubMed] [Google Scholar]
- 11. Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol 1997; 13:83-117; PMID:9442869; http://dx.doi.org/ 10.1146/annurev.cellbio.13.1.83 [DOI] [PubMed] [Google Scholar]
- 12. Lacroix B, Bourdages KG, Dorn JF, Ihara S, Sherwood DR, Maddox PS, Maddox AS. In situ imaging in C. elegans reveals developmental regulation of microtubule dynamics. Dev Cell 2014; 29:203-16; PMID:24780738; http://dx.doi.org/ 10.1016/j.devcel.2014.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. . Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 2003; 421:231-7; PMID:12529635; http://dx.doi.org/ 10.1038/nature01278 [DOI] [PubMed] [Google Scholar]
- 14. Baas PW, Slaughter T, Brown A, Black MM. Microtubule dynamics in axons and dendrites. J Neurosci Res 1991; 30:134-53; PMID:1795398; http://dx.doi.org/ 10.1002/jnr.490300115 [DOI] [PubMed] [Google Scholar]
- 15. Bre MH, Pepperkok R, Kreis TE, Karsenti E. Cellular interactions and tubulin detyrosination in fibroblastic and epithelial cells. Biol Cell 1991; 71:149-60; PMID:1912941; http://dx.doi.org/ 10.1016/0248-4900(91)90061-Q [DOI] [PubMed] [Google Scholar]
- 16. Bulinski JC, Gundersen GG. Stabilization of post-translational modification of microtubules during cellular morphogenesis. Bioessays 1991; 13:285-93; PMID:1892478; http://dx.doi.org/ 10.1002/bies.950130605 [DOI] [PubMed] [Google Scholar]
- 17. Lim SS, Sammak PJ, Borisy GG. Progressive and spatially differentiated stability of microtubules in developing neuronal cells. J Cell Biol 1989; 109:253-63; PMID:2745551; http://dx.doi.org/ 10.1083/jcb.109.1.253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schulze E, Kirschner M. Dynamic and stable populations of microtubules in cells. J Cell Biol 1987; 104:277-88; PMID:3543024; http://dx.doi.org/ 10.1083/jcb.104.2.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Janke C, Bulinski JC. Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat Rev Mol Cell Biol 2011; 12:773-86; PMID:22086369; http://dx.doi.org/ 10.1038/nrm3227 [DOI] [PubMed] [Google Scholar]
- 20. Ghosh-Roy A, Goncharov A, Jin Y, Chisholm AD. Kinesin-13 and tubulin posttranslational modifications regulate microtubule growth in axon regeneration. Dev Cell 2012; 23:716-28; PMID:23000142; http://dx.doi.org/ 10.1016/j.devcel.2012.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bartolini F, Gundersen GG. Generation of noncentrosomal microtubule arrays. J Cell Sci 2006; 119:4155-63; PMID:17038542; http://dx.doi.org/ 10.1242/jcs.03227 [DOI] [PubMed] [Google Scholar]
- 22. Newman AP, Sternberg PW. Coordinated morphogenesis of epithelia during development of the Caenorhabditis elegans uterine-vulval connection. Proc Natl Acad Sci U S A 1996; 93:9329-33; PMID:8790329; http://dx.doi.org/ 10.1073/pnas.93.18.9329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Newman AP, White JG, Sternberg PW. The Caenorhabditis elegans lin-12 gene mediates induction of ventral uterine specialization by the anchor cell. Development 1995; 121:263-71; PMID:7768171 [DOI] [PubMed] [Google Scholar]
- 24. Oommen KS, Newman AP. Co-regulation by Notch and Fos is required for cell fate specification of intermediate precursors during C. elegans uterine development. Development 2007; 134:3999-4009; PMID:17942488; http://dx.doi.org/ 10.1242/dev.002741 [DOI] [PubMed] [Google Scholar]


