Abstract
The trans-splicing of a spliced-leader RNA to a subset of mRNAs is a phenomenon that occurs in many species, including Caenorhabditis elegans, and yet the driving force for its evolution in disparate groups of animals remains unclear. Polycistronic mRNA resulting from the transcription of operons is resolved via trans-splicing, but operons comprise only a sub-set of trans-spliced genes. Using the marine chordate, Oikopleura dioica, we recently tested the hypothesis that metazoan operons accelerate recovery from growth arrest. We found no supporting evidence for this in O. dioica. Instead we found a striking relationship between trans-splicing and maternal mRNA in O. dioica, C. elegans and the ascidian, Ciona intestinalis. Furthermore, in O. dioica and C. elegans, we found evidence to suggest a role for mTOR signaling in the translational control of growth-related, trans-spliced maternal mRNAs. We propose that this may be a mechanism for adjusting egg number in response to nutrient levels in these species.
Keywords: Endocycle, growth arrest, maternal RNA, operon, oogenesis, TOP motif, TOR signaling
Introduction
The organization of genes into operons is a genomic feature that is found in disparate branches of the tree of life. Transcription from the single promoter of an operon results in a polycistronic transcript encoding multiple protein products. Trans-splicing of a separately transcribed spliced leader (SL) RNA at unpaired splice acceptor sites on a polycistron results in translatable monocistronic transcripts. These share a 5′ leader sequence and a modified, hyper-methylated cap structure. The occurrence of trans-splicing is also prevalent in monocistronic transcripts that have unpaired 5′ acceptor sites. In this case the function and evolutionary advantage of trans-splicing is unclear. In C. elegans, 70% of mRNAs are trans-spliced and 17% of mRNAs are found in operons.1,2 There are 2 main SL RNAs in C. elegans: SL1 is trans-spliced to monocistrons and to the first gene in an operon; SL2 (which has several variants) is trans-spliced to downstream operon genes and assists in 3′ end-formation. In the ascidian C. intestinalis, 58% of mRNAs are trans-spliced with a single SL species and 20% of genes are organized into operons.3 In the appendicularian Oikopleura dioica, 39% of mRNAs are trans-spliced to a single SL and 28% are found in operons.4-6
Several hypotheses for the functions and evolutionary advantages of both operons and trans-splicing (Fig. 1) have been proposed.7 Genes encoding proteins with related functions, or proteins that require the same stoichiometry, organized into an operon can be co-regulated at the transcriptional level, since they share a promoter. There are, however, many cases where the mRNA abundances transcribed from genes within operons are not correlated, indicating differing regulation at the post-transcriptional level. One study has suggested that germline expression drives operon evolution in C. elegans,8 whereas another has proposed that metazoan operons provide an advantage in recovery from growth arrest.9 Since this latter hypothesis was based mainly on data from C. elegans and C. intestinalis we sought to determine if this was also the case in another metazoan; the marine chordate Oikopleura dioica.5,6 Our work revealed a distinctive role for trans-splicing in the regulation of maternal mRNA.4
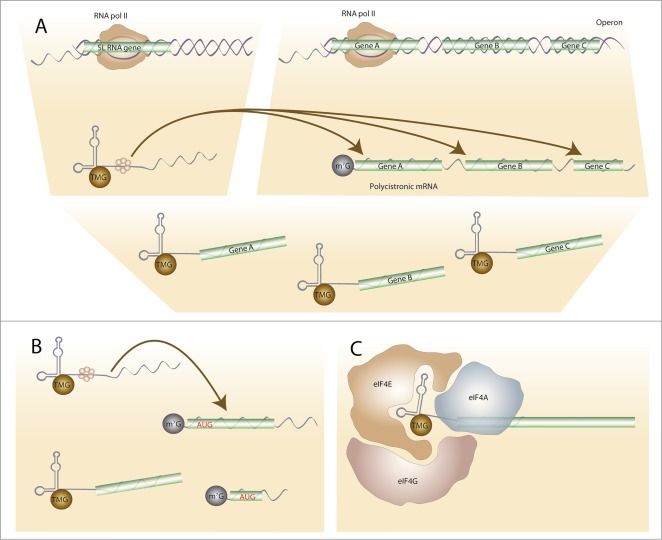
Figure 1.
Functions of spliced-leader trans-splicing. (A) The best-known function of SL trans-splicing is resolving polycistronic mRNAs (the products of operon transcription) into capped, monocistronic, mRNAs. The SL RNA is transcribed separately (by RNA Pol II). It contains a binding site for Sm proteins and one or more stem-loop secondary structures. Its exon sequence is trans-spliced at unpaired acceptor sites upstream of the 5′ ends of polycistronic coding regions. Its intron-like moiety together with the 5′ ‘outron’ from the region upstream of a trans-splice site is degraded. This process is also coupled to 3′ end formation and leads to translatable monocistrons. Since mRNAs from non-operon genes are also trans-spliced, other functions of trans-splicing have been proposed including (B) ‘sanitising’ the 5′ UTRs (e.g. removing premature or out of frame start codons) and increasing the efficiency of translation through the modified TMG cap structure together with the 5′ SL sequence (C).
Operons and Recovery from Growth Arrest
C. elegans larvae that hatch in the absence of food enter a growth-arrested state (L1 diapause). They may also enter a growth arrested state (dauer) later, at the second molt, in response to temperature, food availability and, importantly, population density. O. dioica can also enter a growth arrested state, during a post-metamorphosis and pre-meiotic-entry window, in response to food availability and population density.10 Similar to C. elegans, growth arrest in O. dioica is reversible; when conditions become favorable the animal resumes normal growth (primarily due to the expansion of endocycling somatic cells) and germline progression (germ cells enter meiosis).
We induced growth arrest in O. dioica by culturing the animals in crowded conditions, initiated recovery by diluting them to normal population density and measured the transcriptional response of polycistronic operon genes and monocistronic non-operon genes. In C. elegans operon genes are preferentially up-regulated during recovery from both L1 and dauer arrested states.9 In contrast, we found that in O. dioica non-operon genes are preferentially upregulated during recovery from growth arrest.4 Thus, the idea that operons have evolved to accelerate recovery from growth arrested states cannot be generalized to all metazoans.
A Shift at the Maternal to Zygotic Transition
Maternal RNAs and proteins that are stocked in the oocyte during vitellogenesis provide initial resources to the fertilized egg and control early mitotic cell divisions, cell fates and patterning. During the maternal to zygotic transition, maternal products are eliminated and the zygotic genome takes control over development. In O. dioica, as in C. elegans, the maternal to zygotic transition is rapid; most maternal mRNAs are degraded by the 2–8 cell stage, which occurs less than 1 hour post-fertilization. We found a striking shift in the transcription of trans-spliced mRNAs at the maternal to zygotic transition in O. dioica: most maternal mRNA is trans-spliced; most zygotic mRNA is not trans-spliced. Further examination of existing data from C. elegans and C. intestinalis revealed a common association between maternal mRNA and trans-spliced mRNA in all 3 species. We hypothesize that trans-splicing a common leader sequence to a subset of mRNAs allows for post-transcriptional co-regulation of maternal mRNAs.
TOP of the Trans-Splicing Class
A characteristic of growth is an increase in the production of the translational machinery to allow for an increase in protein synthesis. mRNAs that encode the translational machinery (in particular ribosomal proteins) are characterized by the presence of a 5′ Terminial OligoPyrimidine tract (a TOP motif) and are known as TOP mRNAs. When energy resources are limited the translation of TOP mRNAs is suppressed to reduce catabolic processes. The energy that is saved can then be redirected to the transcription and translation of stress-response genes. The mTOR pathway mediates this translational response to changes in nutrient levels via the TOP motif.11,12
From our work and that of others, we know that in both O. dioica and C. elegans TOP mRNAs are predominantly trans-spliced. This raises the interesting possibility that the SL sequence, which is trans-spliced onto these mRNAs, replaces the cis-regulatory role of the TOP motif in these species. Indeed, SL sequences tend to be pyrimidine-enriched.
After the onset of meiosis, O. dioica no longer enters a growth-arrested state in unfavorable conditions.10 The number of eggs produced is, however, dependent on nutrient levels.13 Eggs in O. dioica are produced via the partitioning of the common cytoplasm of a single-celled multinucleate germline.14-17 Based on our data showing that trans-splicing is associated with maternal mRNAs and TOP mRNAs,4 we propose that the production of maternal proteins during vitellogenesis is controlled in a nutrient-dependent manner via the mTOR pathway and cis-regulatory sequences in the spliced-leader (Fig. 2). If nutrient levels are low then the translation of trans-spliced maternal mRNA is suppressed. This results in a reduced cytoplasmic volume in the ovary that is partitioned into consequently fewer eggs.
Figure 2.
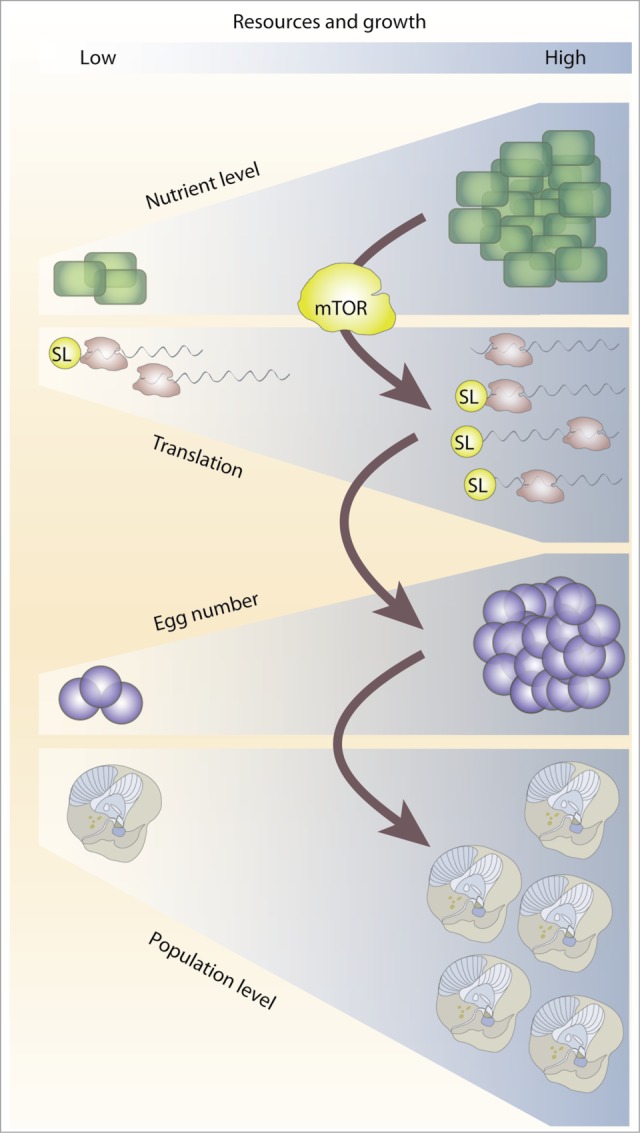
Model linking trans-splicing to nutrient dependent translational control of maternal mRNA. Growth is characterized by an increase in the production of the protein synthesis machinery (via the translation of TOP mRNAs) and is limited by resource availability. The mTOR pathway mediates the response to nutrient levels by controlling the translation of TOP mRNAs. Egg number in O. dioica is dependent on nutrient levels and the majority of maternal mRNAs and TOP mRNAs are trans-spliced. We propose a model whereby the spliced-leader fulfills the function of the TOP motif and allows the translational control of trans-spliced mRNAs. This allows the animal to adjust reproductive output, and therefore population level, in accordance with dynamic resource levels (including algal blooms) in its environment. Control at the translational level allows a more rapid response to changes in nutrient levels compared to transcriptional control, and this may be of particular importance for organisms with short life cycles.
Growth Arrest and Translational Control
Operons in C. elegans are up-regulated during recovery from growth arrest. The reduction in number of transcriptional machinery units required for operon transcription may be advantageous for conserving energy, as previously proposed.9 However, in light of our data, we propose that this up-regulation of operons is primarily a response to the availability of food. In order to facilitate rapid growth the transcription of growth-related genes is increased. In addition, we suggest that these same genes are regulated at the translational level in a nutrient-dependent manner. Indeed, a recent study found that the translation of ribosomal protein mRNA was strongly suppressed during L1 arrest and resumed upon recovery in 4 different worm species.18 It is possible that we do not observe the same transcriptional upregulation of operons in O. dioica during recovery from growth arrest since, unlike C. elegans, O. dioica continues to feed during growth arrest. Post-metamorphic growth in O. dioica occurs primarily via the expansion of endocycling somatic cells.19 Regulating translation is a rapid and direct means of adjusting cytoplasmic volume. It is possible therefore that growth in O. dioica may rely more on translational control, rather than transcriptional control. Indeed, we found that the transcription of mRNAs encoding the translational machinery is strongly upregulated during stages of post-metamorphic growth in O. dioica with little change in the transcription of mRNAs encoding the transcriptional machinery.4 We speculate that translational control may predominate the recovery from growth arrest in O. dioica.
Translational Control as a Unifying Function of Trans-Splicing
The ability to control protein abundance at the translational level is advantageous when a rapid response to environmental or physiological cues is critical for survival and/or reproductive success. One important environmental parameter is nutrient availability, which determines the balance of resource allocation between reproduction and somatic maintenance. We hypothesize that the spliced-leader (via its sequence, secondary structure and/or modified cap structure) plays an important role in the translational control of mRNAs in order to adjust reproductive output in response to environmental conditions. Given the TOP-like characteristics of the SL in C. elegans and O. dioica and the trans-splicing of known TOP mRNAs, it may be that this control, at least for a subset of trans-spliced mRNAs, is mediated by TOR signaling (Fig. 1). There may also be other environmental parameters or processes that require translational control via the SL without the involvement of TOR signaling. Trans-spliced mRNAs in C. intestinalis do not include TOP mRNAs but instead are enriched for genes related to plasma and endomembrane systems, Ca2+ homeostasis/regulation, cell-cell signaling and cortical cytoskeleton.3 These are all related to important processes in the maturation of oocytes: the cytoskeleton is critical in the reorganization of organelles; Ca2+ signaling pathways are remodeled, with accompanying changes in membrane permeability and ion currents, in preparation for fertilization. Further studies of translational control in these and other species that utilize trans-splicing are needed to increase our understanding of this phenomenon and the forces that drive its appearance in various, disparately related, lineages.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Blumenthal T, Gleason KS. Caenorhabditis elegans operons: form and function. Nat Rev Genet 2003; 4:110-8; http://dx.doi.org/ 10.1038/nrg995 [DOI] [PubMed] [Google Scholar]
- 2.Allen MA, Hillier LW, Waterston RH, Blumenthal T. A global analysis of C. elegans trans-splicing. Genom Res 2011; 21:255-64; PMID:21177958; http://dx.doi.org/ 10.1101/gr.113811.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsumoto J, Dewar K, Wasserscheid J, Wiley GB, Macmil SL, Roe BA, Zeller RW, Satou Y, Hastings KEM. High-throughput sequence analysis of Ciona intestinalis SL trans-spliced mRNAs: Alternative expression modes and gene function correlates. Genom Res 2010; 20:636-45; PMID:20212022; http://dx.doi.org/ 10.1101/gr.100271.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Danks GB, Raasholm M, Campsteijn C, Long AM, Manak JR, Lenhard B, Thompson EM. Trans-splicing and operons in metazoans: translational control in maternally regulated development and recovery from growth arrest. Mol Biol Evol 2015; 32:585-99; PMID:25525214; http://dx.doi.org/ 10.1093/molbev/msu336 [DOI] [PubMed] [Google Scholar]
- 5.Ganot P, Kallesøe T, Reinhardt R, Chourrout D, Thompson EM. Spliced-leader RNA trans splicing in a chordate, Oikopleura dioica, with a compact genome. Mol Cell Biol 2004; 24:7795-805; PMID:15314184; http://dx.doi.org/ 10.1128/MCB.24.17.7795-7805.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denoeud F, Henriet S, Mungpakdee S, Aury J-M, Da Silva C, Brinkmann H, Mikhaleva J, Olsen LC, Jubin C, Cañestro C, et al.. Plasticity of animal genome architecture unmasked by rapid evolution of a pelagic tunicate. Science 2010; 330:1381-5; PMID:21097902; http://dx.doi.org/ 10.1126/science.1194167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hastings KEM. SL trans-splicing: easy come or easy go? Trend Genet 2005; 21:240-7. [DOI] [PubMed] [Google Scholar]
- 8.Reinke V, Cutter AD. Germline expression influences operon organization in the Caenorhabditis elegans genome. Genetics 2009; 181:1219-28; PMID:19204375; http://dx.doi.org/ 10.1534/genetics.108.099283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaslaver A, Baugh LR, Sternberg PW. Metazoan operons accelerate recovery from growth-arrested states. Cell 2011; 145:981-92; PMID:21663799; http://dx.doi.org/ 10.1016/j.cell.2011.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Subramaniam G, Campsteijn C, Thompson EM. Lifespan extension in a semelparous chordate occurs via developmental growth arrest just prior to meiotic entry. PLoS ONE 2014; 9:e93787; PMID:24695788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meyuhas O. Synthesis of the translational apparatus is regulated at the translational level. Eur J Biochem 2001; 267:6321-30; PMID:11029573 [DOI] [PubMed] [Google Scholar]
- 12.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 2012; 485:109-13; PMID:22552098; http://dx.doi.org/ 10.1038/nature11083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Troedsson C, Bouquet JM, Aksnes D, Thompson EM. Resource allocation between somatic growth and reproductive output in the pelagic chordate Oikopleura dioica allows opportunistic response to nutritional variation. Mar Ecol Progr Ser 2002; 243:83-91. [Google Scholar]
- 14.Ganot P, Bouquet JM, Thompson EM. Comparative organization of follicle, accessory cells and spawning anlagen in dynamic semelparous clutch manipulators, the urochordate Oikopleuridae. Biol Cell 2006; 98: 389-401; PMID:16478443; http://dx.doi.org/ 10.1042/BC20060005 [DOI] [PubMed] [Google Scholar]
- 15.Ganot P, Bouquet JM, Kallesøe T, Thompson EM. The Oikopleura coenocyst, a unique chordate germ cell permitting rapid, extensive modulation of oocyte production. Dev Biol 2007; 302:591-600; PMID:17126826; http://dx.doi.org/ 10.1016/j.ydbio.2006.10.021 [DOI] [PubMed] [Google Scholar]
- 16.Ganot P, Kallesoe T, Thompson EM. The cytoskeleton organizes germ nuclei with divergent fates and asynchronous cycles in a common cytoplasm during oogenesis in the chordate Oikopleura. Dev Biol 2007; 302: 577-90; PMID:17123503; http://dx.doi.org/ 10.1016/j.ydbio.2006.10.022 [DOI] [PubMed] [Google Scholar]
- 17.Ganot P, Moosmann-Schulmeister A, Thompson EM. Oocyte selection is concurrent with meiosis resumption in the coenocystic oogenesis of Oikopleura. Dev Biol 2008; 324:266-76; PMID:18845138; http://dx.doi.org/ 10.1016/j.ydbio.2008.09.016 [DOI] [PubMed] [Google Scholar]
- 18.Stadler M, Fire A. Conserved translatome remodeling in nematode species executing a shared developmental transition. PLoS Genet 2013; 9:e1003739; PMID:24098135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganot P, Thompson EM. Patterning through differential endoreduplication in epithelial organogenesis of the chordate, Oikopleura dioica. Dev Biol 2002; 252:59-71; PMID:12453460; http://dx.doi.org/ 10.1006/dbio.2002.0834 [DOI] [PubMed] [Google Scholar]