Abstract
It was long assumed that eukaryotic precursor mRNAs (pre-mRNAs) are almost always spliced to generate a linear mRNA that is subsequently translated to produce a protein. However, it is now clear that thousands of protein-coding genes can be non-canonically spliced to produce circular noncoding RNAs, some of which are expressed at much higher levels than their associated linear mRNAs. How then does the splicing machinery decide whether to generate a linear mRNA or a circular RNA? Recent work has revealed that intronic repetitive elements, including sequences derived from transposons, are critical regulators of this decision. In most cases, circular RNA biogenesis appears to be initiated when complementary sequences from 2 different introns base pair to one another. This brings the splice sites from the intervening exon(s) into close proximity and facilitates the backsplicing event that generates the circular RNA. As many pre-mRNAs contain multiple intronic repeats, distinct circular transcripts can be produced depending on which repeats base pair to one another. Intronic repeats are thus critical regulatory sequences that control the functional output of their host genes, and potentially cause the functions of protein-coding genes to be highly divergent across species.
Keywords: ADAR, Alu, backsplicing, base pairing, circRNA, circular RNA, LINE1, noncoding RNA, pre-mRNA splicing, retrotransposition
Although most genetic information is thought to be expressed as proteins, less than 2% of the human genome actually codes for protein. Much (∼70%) of our genome is instead derived from repetitive elements with LINE-1 (L1) retrotransposons and Alu elements being particularly abundant, accounting for ∼17% and 10% of the total genomic sequence, respectively.1,2 During evolution, these transposable elements took advantage of the autonomous retrotransposition activity of L1 to spread as “selfish” DNA and expand the size of our genome.3-5 In addition, they facilitated genomic rearrangements, insertional mutagenesis events, and the formation of new exons and genes. Nevertheless, only ∼80–100 L1s are thought to still be capable of retrotransposition today.6 The other >99.9% of transposon-derived sequences have been rendered inactive, which has led some to suggest they may simply represent remnants of our evolutionary past, or so-called “junk” DNA. It is, however, becoming increasingly clear that repetitive elements perform multiple critical functions, including regulating transcription,7 mRNA localization,8 mRNA degradation,9 and mRNA editing.10 Recent work from multiple groups,11-14 including our own, has now revealed a novel function for the many repetitive elements that are present in introns of protein-coding genes.15 Fascinatingly, these intronic repeats regulate the functional output of their host genes by facilitating the formation of circular RNAs.
As their name suggests, circular RNAs have covalently linked ends and were originally identified in pathogens, including viroids (virus-like infectious particles) and hepatitis δ virus.16,17 A handful of circular RNAs generated via splicing of eukaryotic genes were subsequently identified in the 1990s, but these transcripts were generally expressed at low levels (∼0.01% the level of their associated linear mRNA).18–20 Because eukaryotic genes contain introns, their precursor mRNAs (pre-mRNAs) must be modified such that introns are removed and exons joined together. Depending on how a pre-mRNA is spliced, different linear or circular RNAs can be produced, each potentially with its own unique function (Fig. 1). If a pre-mRNA is spliced in the standard way (namely, exon 1 is joined to exon 2, which is joined to exon 3, etc.), a linear mRNA is generated that can subsequently be translated to produce a protein. If “backsplicing” instead occurs and a splice donor is joined to an upstream splice acceptor (e.g. the end of exon 3 is joined to the beginning of exon 2), that pre-mRNA now produces a circular RNA. In most cases, these transcripts are noncoding as the start and/or stop codons have been removed, although artificial circles containing an open reading frame and an IRES (internal ribosome entry site) can be translated.21,22 Considering that canonical splicing and backsplicing are in direct competition with one another,23 the pre-mRNA splicing machinery must be tightly regulated so that the appropriate mature RNAs are produced. In some cases, backsplicing is combined with exon skipping so that a linear mRNA as well as a circular RNA comprised of the skipped exon(s) can be generated from a single pre-mRNA.20,24,25
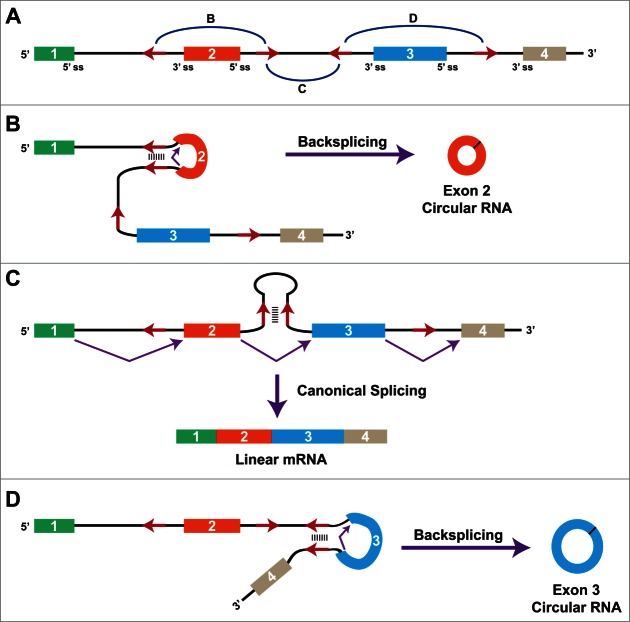
Figure 1.
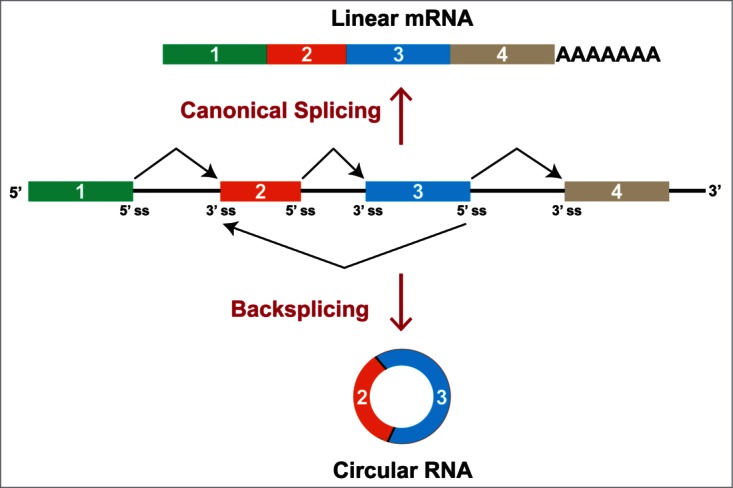
Pre-mRNA splicing can generate linear or circular RNAs. If the pre-mRNA splice sites (ss) are joined in the canonical order, a mature linear mRNA is generated that is subsequently polyadenylated (top). Alternatively, the splicing machinery can backsplice and join a splice donor to an upstream splice acceptor, generating a circular RNA whose ends are covalently linked (bottom). Here, a circular RNA composed of 2 exons is generated, although backsplicing can result in the production of circular RNAs that comprise one or many exons.
With the exception of the first and last exons of genes, every other exon in the genome has splicing signals at its 5′ and 3′ ends and theoretically can circularize. However, most exons do not generate circular RNAs, in part because pre-mRNA splicing generally occurs co-transcriptionally.26,27 Simply put, most introns are rapidly spliced as soon as they are transcribed, thereby removing the upstream splice acceptors that are needed for backsplicing to occur. Nevertheless, it is now clear that thousands of human genes produce circular RNAs, sometimes at a level that exceeds the associated linear mRNA by a factor of 10.13,28-32 Over 25,000 circular RNAs, derived from ∼15% of actively transcribed genes, were recently identified in a single human cell type.13 These highly stable transcripts contain almost exclusively exonic sequences and accumulate in the cytoplasm, but do not generally associate with ribosomes.30 In at least 2 cases, circular RNAs function as efficient sponges for microRNAs,29,33,34 but most others contain few microRNA binding sites and likely have a different function.30 Regardless of their ultimate fate, what inputs initially determine whether a pre-mRNA is spliced to generate a linear mRNA or any of its possible circular RNAs?
A connection between repetitive elements and circular RNA biogenesis was first suggested at the mouse Sry locus, which efficiently produces a ∼1.2-kb circular RNA comprised of a single exon.35 Remarkably, this exon is flanked by ∼50-kb of near perfectly complementary sequences (>99.7% identity), and complete removal of either of the repeats eliminated Sry circular RNA production from an expression plasmid.36 Smaller deletions revealed that 400-nt of complementary intronic repeats were sufficient for Sry circle biogenesis. This suggested a model in which the intronic repeats may base pair to one another, bringing the splice sites into close proximity to facilitate backsplicing. In support of this idea, the addition of complementary intronic repeats to in vitro RNA splicing substrates was found to promote the formation of circular RNAs.37
However, very few exons are naturally flanked by repeats as long as those at the Sry locus. Computational analysis instead revealed that pairs of Alu elements, which are ∼300-nt in length, are enriched in the introns flanking human exons that generate circular RNAs.12,13 In fact, almost 90% of circular RNAs appear to have complementary Alu elements in their flanking introns.14 To directly determine if these repeats or other nearby sequences regulate the production of circular RNAs, we and others recently mutagenized plasmids that express various human circular RNAs.11,12,38,39 This was most extensively done with the human ZKSCAN1 locus, which produces an abundant 668-nt circular RNA containing exons 2 and 3 in human brain and liver.11 Surprisingly, miniature (<90-nt) introns containing only the splice sites along with short (∼30 to 40-nt) inverted repeats were sufficient to allow the intervening ZKSCAN1 exons to efficiently circularize in cells (Fig. 2). As expected, mutating the splice sites completely eliminated ZKSCAN1 circular RNA production. Likewise, disrupting base pairing between the intronic repeats by mutating several nucleotides in one of the Alu elements also prevented circularization. Introducing compensatory mutations into the other repeat did, however, rescue circular RNA biogenesis. Base pairing between the intronic repeats is thus necessary for efficient ZKSCAN1 circularization and similar results were obtained with the human HIPK3 and EPHB4 genes.11 Notably, several other circular RNA expression plasmids can weakly generate circles when no complementary sequences are present in the flanking introns.38,39 However, even in these cases, the presence of inverted repeats drastically increases (>10-fold) the efficiency of circular RNA production, indicating that interactions between flanking introns can strongly promote circularization. In total, these data suggest that the Sry circular RNA biogenesis model is likely applicable at thousands of human genes, with a short stretch of base pairing between intronic repeats appearing to often be sufficient.11 Importantly, this mechanism appears to also be commonly used across eukaryotes as inverted repeats generally flank circular RNAs in mice and C. elegans.14
Figure 2.
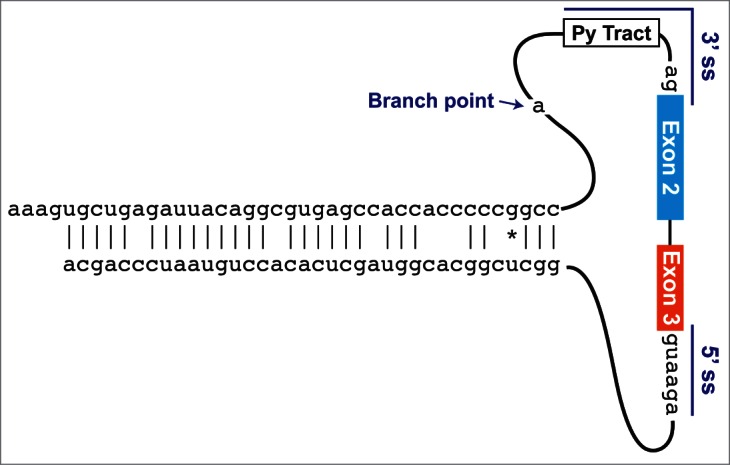
Minimal intronic elements that are sufficient for ZKSCAN1 circular RNA production. Using extensive mutagenesis, minimal sequences that are sufficient for generating the circular RNA from exons 2 and 3 of human ZKSCAN1 were defined.11 In the upstream intron, 87 nt are sufficient, which include 40 nt of an Alu element as well as the 3′ splice site (comprised of a polypyrimidine (Py) tract followed by AG) and branch point sequences. In the downstream intron, 59 nt are sufficient, which include the 5′ splice site and 36 nt of an Alu element. As the 2 Alu elements are highly complementary to one another, the repeats can base pair and form a hairpin, which promotes backsplicing.
Complicating the regulation of circular RNA biogenesis is the fact that introns commonly contain multiple repetitive elements. Depending on how the complementary sequences base pair to one another, very different splicing patterns can result (Fig. 3A).12 When base pairing occurs across different introns (as discussed above), backsplicing is induced and the intervening exon(s) form a circular RNA (Fig. 3B). If base pairing instead occurs between repeats within a single intron, canonical splicing occurs and a linear mRNA is produced (Fig. 3C). The expression of a given circular RNA can thus be largely controlled by competition for base pairing between the various complementary sequences. The number of repeats, their degree of homology, and the distance between them are likely all key determining factors.40 Furthermore, this competition allows multiple circular RNAs to be generated from a single protein-coding gene (Fig. 3D).12,30,41,42
Figure 3.
The presence of multiple intronic repeats allows a variety of mature RNAs to be generated from a single gene. (A) Schematic of a 4-exon gene, with intronic repeat elements depicted as red arrows. Depending on which repeats base pair to one another (denoted by blue arcs), distinct mature RNAs are produced. (B) If the repeats flanking exon 2 base pair to one another, backsplicing (denoted in purple) is induced and a circular RNA composed of exon 2 is generated. (C) If, however, the repeats in the second intron base pair to one another, canonical splicing occurs and a linear mRNA is produced. (D) If the repeats flanking exon 3 base pair to one another, a circular RNA composed of exon 3 is generated. Finally, base pairing between the first and last repeats would yield a circular RNA composed of exons 2 and 3 (not shown).
Although base pairing plays a critical role in circular RNA regulation, it is clear that more than simple thermodynamics is at play. We notably found that not all repeat sequences can support circular RNA production from plasmids.11 For example, strengthening the hairpin between the ZKSCAN1 minimal repeat elements sometimes actually inhibited circularization, especially when low complexity sequences (such as poly(A) tracts) or multiple G-U wobble base pairs were included. These data suggest that the cell is somehow able to sense subtle distortions in the hairpin formed between the intronic repeats, likely in part via double-stranded RNA binding proteins. Supporting this idea, ADAR (adenosine deaminase acting on RNA) enzymes, which convert adenosines in double-stranded regions to inosines, were recently shown to inhibit the production of over 80 different human circular RNAs.14 By modifying nucleotides within the intronic repeats, ADAR activity likely causes the hairpin between the repeats to become unwound, thereby directly disrupting the interactions that promote backsplicing.
As the genomic repeat landscape varies significantly among species (e.g., Alu elements are specific to primates),43 one might expect that distinct circular RNAs are expressed in different eukaryotes. The ultimate function of a gene could thus be quite different in each organism, even if the protein-coding exons are perfectly conserved. This has interesting evolutionary implications and suggests that insertions of transposable elements into introns may have played a critical role in modulating the functions and/or expression of their host genes. Surprisingly, an analysis of human and mouse genes with clear one-to-one orthologs revealed that if a given human gene produces a circular RNA, then its mouse ortholog also does so in 66% of cases.30 The splice sites used for backsplicing were often found to be orthologous (∼37% of cases) or partially overlapping (∼32%), indicating that human and mouse circular RNAs are commonly generated from the exact same exons.13,30 As the functions for nearly all of these circular RNAs are not yet known, it is unclear if and why evolution may have convergently selected for the production of these particular circular transcripts.
L1 and Alu elements contribute to the genetic variation among individuals and some continue to retrotranspose, with L1 being active in the brain44 and new Alu insertions occurring every ∼20 births.45 It is thus possible that new retrotransposition insertions into introns could change the output of a gene from a protein-coding mRNA to a circular RNA (or vice versa), thereby dysregulating gene function. Retrotransposition insertions have previously been shown to cause various diseases, including hemophilia, cystic fibrosis, and cancer,46 and it will be insightful to determine if some intronic insertions may affect circular RNAs to likewise drive various human diseases.
Although repetitive elements appear to be necessary for the biogenesis of many circular RNAs, there may be some notable exceptions. In particular, it appears that the introns that flank many Drosophila circular RNAs do not contain complementary sequences.47 This suggests that circular RNA biogenesis in flies may often occur via a distinct mechanism, e.g. via the binding of splicing factors to both flanking introns, as has been proposed at the Drosophila mbl locus.23 Analogous to how base pairing between intronic repeats can bring the intervening splice sites into close proximity of one another, interactions between proteins that bind 2 separate introns could likewise promote backsplicing. Although not yet definitively demonstrated, recent work suggests that the splicing factor Quaking may bind flanking introns and promote the production of some circular RNAs via such a mechanism.48
Considering that repeat sequences appear to be critical players in the generation of most circular RNAs in human, mice, and C. elegans, it would be quite surprising if Drosophila used a completely distinct mechanism. Nevertheless, cells do appear to generate a variety of other circles via unique strategies. For example, a completely distinct class of circular RNAs are now known to be generated from the introns of some protein-coding genes when these introns fail to be debranched.49 In addition, various noncoding RNAs in archaea are processed to yield circular RNAs through poorly understood mechanisms.50
In summary, thousands of protein-coding genes generate circular RNAs13,28-32 and we are beginning to understand the key elements that facilitate their biogenesis. Both computational analyses and plasmid-based experiments have pointed to a key role for base pairing between intronic complementary sequences in promoting most backsplicing events. Therefore, repetitive elements in introns are not simply “junk” DNA but critical regulatory sequences that modulate the functional output of protein-coding genes. With the development of efficient genome editing techniques using the CRISPR/Cas9 system,51,52 it will now be important to directly modify endogenous gene loci and test the effect of specifically adding or removing repeat elements. An analogous approach can be used to identify additional cis-acting elements, especially those that may act to slow the splicing of upstream introns so that backsplicing can occur. Trans-acting factors that collaborate with the repeats are also beginning to be identified, and further work will reveal detailed mechanisms that regulate circular RNA expression levels, especially across different tissues.
The expression of circular RNAs has recently been reported to generally decrease in human cancers53 and increase as flies age,47 suggesting that their expression can be drastically altered and potentially contribute to pathological states. However, the biological functions of most circular RNAs are still completely unclear. Recently described11 circular RNA expression plasmids will serve as valuable tools for identifying their cellular roles as well as their interacting proteins. Analogous to how bacterial operons function, it may be that RNA circles often work in the same pathway as the protein produced from its parental gene, thereby integrating noncoding RNAs and proteins into the same regulatory paradigms. Alternatively, circular RNAs may allow the formation of large RNA-protein complexes or even be translated in some cases. In total, exon circularization represents a new underappreciated way that the transcriptome is expanded, and future research will likely continue to provide many more unexpected insights into their biogenesis, regulation, and functions.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
I thank Jeff Wilusz, Deirdre Tatomer, and the other members of my laboratory for suggestions and discussions.
Funding
This work was supported by NIH R00-GM104166.
References
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al.. Initial sequencing and analysis of the human genome. Nature 2001; 409:860-921; PMID:11237011; http://dx.doi.org/ 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- 2.de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD.. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet 2011; 7:e1002384; PMID:22144907; http://dx.doi.org/ 10.1371/journal.pgen.1002384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orgel LE, Crick FH.. Selfish DNA: the ultimate parasite. Nature 1980; 284:604-7; PMID:7366731; http://dx.doi.org/ 10.1038/284604a0 [DOI] [PubMed] [Google Scholar]
- 4.Beck CR, Garcia-Perez JL, Badge RM, Moran JV.. LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet 2011; 12:187-215; PMID:21801021; http://dx.doi.org/ 10.1146/annurev-genom-082509-141802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cordaux R, Batzer MA.. The impact of retrotransposons on human genome evolution. Nat Rev Genet 2009; 10:691-703; PMID:19763152; http://dx.doi.org/ 10.1038/nrg2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, Badge RM, Moran JV.. LINE-1 retrotransposition activity in human genomes. Cell 2010; 141:1159-70; PMID:20602998; http://dx.doi.org/ 10.1016/j.cell.2010.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA.. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell 2008; 29:499-509; PMID:18313387; http://dx.doi.org/ 10.1016/j.molcel.2007.12.013 [DOI] [PubMed] [Google Scholar]
- 8.Chen LL, DeCerbo JN, Carmichael GG.. Alu element-mediated gene silencing. EMBO J 2008; 27:1694-705; PMID:18497743; http://dx.doi.org/ 10.1038/emboj.2008.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong C, Maquat LE.. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 2011; 470:284-8; PMID:21307942; http://dx.doi.org/ 10.1038/nature09701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniel C, Silberberg G, Behm M, Ohman M.. Alu elements shape the primate transcriptome by cis-regulation of RNA editing. Genome Biol 2014; 15:R28; PMID:24485196; http://dx.doi.org/ 10.1186/gb-2014-15-2-r28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang D, Wilusz JE.. Short intronic repeat sequences facilitate circular RNA production. Genes Dev 2014; 28:2233-47; PMID:25281217; http://dx.doi.org/ 10.1101/gad.251926.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L.. Complementary sequence-mediated exon circularization. Cell 2014; 159:134-47; PMID:25242744; http://dx.doi.org/ 10.1016/j.cell.2014.09.001 [DOI] [PubMed] [Google Scholar]
- 13.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE.. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013; 19:141-57; PMID:23249747; http://dx.doi.org/ 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C, et al.. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep 2015; 10:170-7; PMID:25558066; http://dx.doi.org/ 10.1016/j.celrep.2014.12.019 [DOI] [PubMed] [Google Scholar]
- 15.Sela N, Mersch B, Gal-Mark N, Lev-Maor G, Hotz-Wagenblatt A, Ast G.. Comparative analysis of transposed element insertion within human and mouse genomes reveals Alu's unique role in shaping the human transcriptome. Genome Biol 2007; 8:R127; PMID:17594509; http://dx.doi.org/ 10.1186/gb-2007-8-6-r127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK.. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A 1976; 73:3852-6; PMID:1069269; http://dx.doi.org/ 10.1073/pnas.73.11.3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H.. The hepatitis delta (delta) virus possesses a circular RNA. Nature 1986; 323:558-60; PMID:2429192; http://dx.doi.org/ 10.1038/323558a0 [DOI] [PubMed] [Google Scholar]
- 18.Cocquerelle C, Mascrez B, Hetuin D, Bailleul B.. Mis-splicing yields circular RNA molecules. FASEB J 1993; 7:155-60; PMID:7678559 [DOI] [PubMed] [Google Scholar]
- 19.Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B.. Scrambled exons. Cell 1991; 64:607-13; PMID:1991322; http://dx.doi.org/ 10.1016/0092-8674(91)90244-S [DOI] [PubMed] [Google Scholar]
- 20.Zaphiropoulos PG. Exon skipping and circular RNA formation in transcripts of the human cytochrome P-450 2C18 gene in epidermis and of the rat androgen binding protein gene in testis. Mol Cell Biol 1997; 17:2985-93; PMID:9154796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen CY, Sarnow P.. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 1995; 268:415-7; PMID:7536344; http://dx.doi.org/ 10.1126/science.7536344 [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Wang Z.. Efficient backsplicing produces translatable circular mRNAs. RNA 2015; 21:172-9; PMID:25449546; http://dx.doi.org/ 10.1261/rna.048272.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S.. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 2014; 56:55-66; PMID:25242144; http://dx.doi.org/ 10.1016/j.molcel.2014.08.019 [DOI] [PubMed] [Google Scholar]
- 24.Surono A, Takeshima Y, Wibawa T, Ikezawa M, Nonaka I, Matsuo M.. Circular dystrophin RNAs consisting of exons that were skipped by alternative splicing. Hum Mol Genet 1999; 8:493-500; PMID:9949208; http://dx.doi.org/ 10.1093/hmg/8.3.493 [DOI] [PubMed] [Google Scholar]
- 25.Kelly S, Greenman C, Cook PR, Papantonis A.. Exon Skipping Is Correlated with Exon Circularization. J Mol Biol 2015; Epub ahead of print; PMID:25728652; http://dx.doi.org/ 10.1016/j.jmb.2015.02.018 [DOI] [PubMed] [Google Scholar]
- 26.Brugiolo M, Herzel L, Neugebauer KM.. Counting on co-transcriptional splicing. F1000Prime Rep 2013; 5:9; PMID:23638305; http://dx.doi.org/ 10.12703/P5-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pandya-Jones A, Black DL.. Co-transcriptional splicing of constitutive and alternative exons. RNA 2009; 15:1896-908; PMID:19656867; http://dx.doi.org/ 10.1261/rna.1714509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO.. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012; 7:e30733; PMID:22319583; http://dx.doi.org/ 10.1371/journal.pone.0030733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al.. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013; 495:333-8; PMID:23446348; http://dx.doi.org/ 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 30.Guo JU, Agarwal V, Guo H, Bartel DP.. Expanded identification and characterization of mammalian circular RNAs. Genome Biol 2014; 15:409; PMID:25070500; http://dx.doi.org/ 10.1186/s13059-014-0409-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, Dinneny JR, Brown PO, Salzman J.. Circular RNA is expressed across the eukaryotic tree of life. PLoS One 2014; 9:e90859; PMID:24609083; http://dx.doi.org/ 10.1371/journal.pone.0090859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al.. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nature Neurosci 2015; 18:603-10; PMID:25714049; http://dx.doi.org/ 10.1038/nn.3975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J.. Natural RNA circles function as efficient microRNA sponges. Nature 2013; 495:384-8; PMID:23446346; http://dx.doi.org/ 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 34.Wilusz JE, Sharp PA.. Molecular biology. A circuitous route to noncoding RNA. Science 2013; 340:440-1; PMID:23620042; http://dx.doi.org/ 10.1126/science.1238522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell-Badge R.. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 1993; 73:1019-30; PMID:7684656; http://dx.doi.org/ 10.1016/0092-8674(93)90279-Y [DOI] [PubMed] [Google Scholar]
- 36.Dubin RA, Kazmi MA, Ostrer H.. Inverted repeats are necessary for circularization of the mouse testis Sry transcript. Gene 1995; 167:245-8; PMID:8566785; http://dx.doi.org/ 10.1016/0378-1119(95)00639-7 [DOI] [PubMed] [Google Scholar]
- 37.Pasman Z, Been MD, Garcia-Blanco MA.. Exon circularization in mammalian nuclear extracts. RNA 1996; 2:603-10; PMID:8718689 [PMC free article] [PubMed] [Google Scholar]
- 38.Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, Bindereif A.. Exon circularization requires canonical splice signals. Cell Rep 2015; 10:103-11; PMID:25543144; http://dx.doi.org/ 10.1016/j.celrep.2014.12.002 [DOI] [PubMed] [Google Scholar]
- 39.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al.. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015; 22:256-64; PMID:25664725; http://dx.doi.org/ 10.1038/nsmb.2959 [DOI] [PubMed] [Google Scholar]
- 40.Athanasiadis A, Rich A, Maas S.. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol 2004; 2:e391; PMID:15534692; http://dx.doi.org/ 10.1371/journal.pbio.0020391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO.. Cell-type specific features of circular RNA expression. PLoS Genet 2013; 9:e1003777; PMID:24039610; http://dx.doi.org/ 10.1371/journal.pgen.1003777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE.. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet 2010; 6:e1001233; PMID:21151960; http://dx.doi.org/ 10.1371/journal.pgen.1001233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shedlock AM, Okada N.. SINE insertions: powerful tools for molecular systematics. BioEssays 2000; 22:148-60; PMID:10655034; http://dx.doi.org/ 10.1002/(SICI)1521-1878(200002)22:2%3c148::AID-BIES6%3e3.0.CO;2-Z [DOI] [PubMed] [Google Scholar]
- 44.Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O'Shea KS, Moran JV, Gage FH.. L1 retrotransposition in human neural progenitor cells. Nature 2009; 460:1127-31; PMID:19657334; http://dx.doi.org/ 10.1038/nature08248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cordaux R, Hedges DJ, Herke SW, Batzer MA.. Estimating the retrotransposition rate of human Alu elements. Gene 2006; 373:134-7; PMID:16522357; http://dx.doi.org/ 10.1016/j.gene.2006.01.019 [DOI] [PubMed] [Google Scholar]
- 46.Babatz TD, Burns KH.. Functional impact of the human mobilome. Curr Opin Genet Dev 2013; 23:264-70; PMID:23523050; http://dx.doi.org/ 10.1016/j.gde.2013.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC.. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep 2014; 9:1966-80; PMID:25544350; http://dx.doi.org/ 10.1016/j.celrep.2014.10.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ.. The RNA Binding Protein Quaking Regulates Formation of circRNAs. Cell 2015; 160:1125-34; PMID:25768908; http://dx.doi.org/ 10.1016/j.cell.2015.02.014 [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL.. Circular intronic long noncoding RNAs. Mol Cell 2013; 51:792-806; PMID:24035497; http://dx.doi.org/ 10.1016/j.molcel.2013.08.017 [DOI] [PubMed] [Google Scholar]
- 50.Danan M, Schwartz S, Edelheit S, Sorek R.. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res 2012; 40:3131-42; PMID:22140119; http://dx.doi.org/ 10.1093/nar/gkr1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al.. Multiplex genome engineering using CRISPR/Cas systems. Science 2013; 339:819-23; PMID:23287718; http://dx.doi.org/ 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM.. RNA-guided human genome engineering via Cas9. Science 2013; 339:823-6; PMID:23287722; http://dx.doi.org/ 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW, Zeillinger R, Pils D.. Correlation of circular RNA abundance with proliferation – exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep 2015; 5:8057; PMID:25624062; http://dx.doi.org/ 10.1038/srep08057 [DOI] [PMC free article] [PubMed] [Google Scholar]