Abstract
Aging and increased amyloid burden are major risk factors for cognitive diseases such as Alzheimer’s disease (AD). Effective therapies for these diseases are lacking. Here, we evaluated mouse models of age-associated memory impairment and amyloid deposition to study transcriptome and cell type–specific epigenome plasticity in the brain and peripheral organs. We determined that aging and amyloid pathology are associated with inflammation and impaired synaptic function in the hippocampal CA1 region as the result of epigenetic-dependent alterations in gene expression. In both amyloid and aging models, inflammation was associated with increased gene expression linked to a subset of transcription factors, while plasticity gene deregulation was differentially mediated. Amyloid pathology impaired histone acetylation and decreased expression of plasticity genes, while aging altered H4K12 acetylation–linked differential splicing at the intron-exon junction in neurons, but not nonneuronal cells. Furthermore, oral administration of the clinically approved histone deacetylase inhibitor vorinostat not only restored spatial memory, but also exerted antiinflammatory action and reinstated epigenetic balance and transcriptional homeostasis at the level of gene expression and exon usage. This study provides a systems-level investigation of transcriptome plasticity in the hippocampal CA1 region in aging and AD models and suggests that histone deacetylase inhibitors should be further explored as a cost-effective therapeutic strategy against age-associated cognitive decline.
Introduction
Epigenetic processes, including posttranslational modifications of histones, regulate gene-expression programs and play a key role in genome-environment interactions (1, 2). Recent data suggest that histone acetylation regulates memory consolidation and may contribute to cognitive decline associated with neurodegenerative diseases (2–5). Histone acetylation is regulated by the counteracting activity of histone acetyltransferases (HATs) and histone deacetylases (HDACs). Recent can improve memory function in rodents (2). Various HDACi have been developed that show different activity toward the 11 zinc-dependent HDACs encoded in the mammalian genome (6). Studies using genetic mouse models (7–10) suggest that specifically the inhibition of class I HDACs as well as HDAC6 could be most beneficial in treating cognitive decline in neurodegenerative diseases such as Alzheimer’s disease (AD) (2, 3). Importantly, the HDACi suberoylanilide hydroxamic acid (SAHA, vorinostat, Zolinza) displays such a profile (11). SAHA is an FDA-approved drug for the treatment of subcutaneous T cell lymphoma (12, 13), suggesting that administration of SAHA could be a suitable strategy to test the effect of HDACi in AD patients. This step is currently hindered by the fact that the molecular effects of HDACi in models for cognitive decline are not well understood. Therefore, the available data on the effect of HDACi in the brain are not cell-type specific and are so far limited to the analysis of selected genes via quantitative reverse-transcriptase PCR (qRT-PCR), bulk changes in histone acetylation, or acetylation for selected genes (2). One particular argument we and others have been facing in our efforts to drive HDACi into clinical trials treating AD patients is that such drugs would rather unspecifically induce massive changes in gene expression. Thus, a systems biology approach for studying the effect of memory impairment and HDACi administration at the genome-wide level is needed. Moreover, oral administration of SAHA has not been tested in preclinical experiments in relation to memory impairment, which is of utmost importance for translational research.
Thus, we decided to test the efficacy of oral SAHA treatment in mouse models for age-associated memory impairment that had already developed severe pathology to closely mimic the clinical situation. To this end, we used 20-month-old mice as a model for age-associated memory impairment and 10-month-old APP/PS1-21 mice (where APP indicates amyloid precursor protein) (14, 15) as a model for severe amyloid deposition. We combined behavioral characterization with an in-depth analysis of epigenome and transcriptome plasticity performing RNA-sequencing (RNA-seq) and ChIP-sequencing (ChIP-seq) of the neuronal versus nonneuronal population of the hippocampal CA1 region. We also tested the effect of SAHA administration on the liver as a major target for most orally applied drugs. Our data reveal a number of important findings. First, we observed that, in cognitively impaired mice, neuronal but not nonneuronal acetylation of histone 4 at lysine 12 (H4K12) was decreased in the hippocampal CA1 region. Oral SAHA treatment rescued this histone acetylation deficit in neurons, but had no effect on nonneuronal cells. At the transcript levels, we observed that age-associated memory impairment correlated with an inflammatory response mediated by elevated gene expression. Notably, SAHA treatment reduced the expression of genes linked to inflammation, which is at least in part due to effects linked to nonhistone acetylation. In aged mice, deregulation of genes linked to synaptic plasticity did not manifest at the level of gene expression, but was almost exclusively linked to differential splicing that was associated with decreased H4K12 acetylation (H4K12ac) at the intron-exon junction. Notably, SAHA treatment completely reinstated physiological exon usage and expression of plasticity genes. This effect was linked to increased H4K12ac at the intron-exon junction of deregulated genes. We furthermore provide evidence that deregulation of RNA splicing impairs hippocampal synaptic plasticity and memory function in mice and that these phenotypes can only be partially rescued by SAHA administration, suggesting a mechanism by which SAHA regulates neuronal plasticity. While amyloid pathology also increased inflammatory processes, deregulation of plasticity genes was not linked to altered exon usage but to differences in gene-expression levels, which were partially reverted by SAHA treatment.
In conclusion, our data not only provide molecular insight into the mechanisms underlying cognitive decline and the action of the HDACi SAHA, but also strongly suggest that SAHA should be tested in clinical trials for its potential to improve memory function.
Results
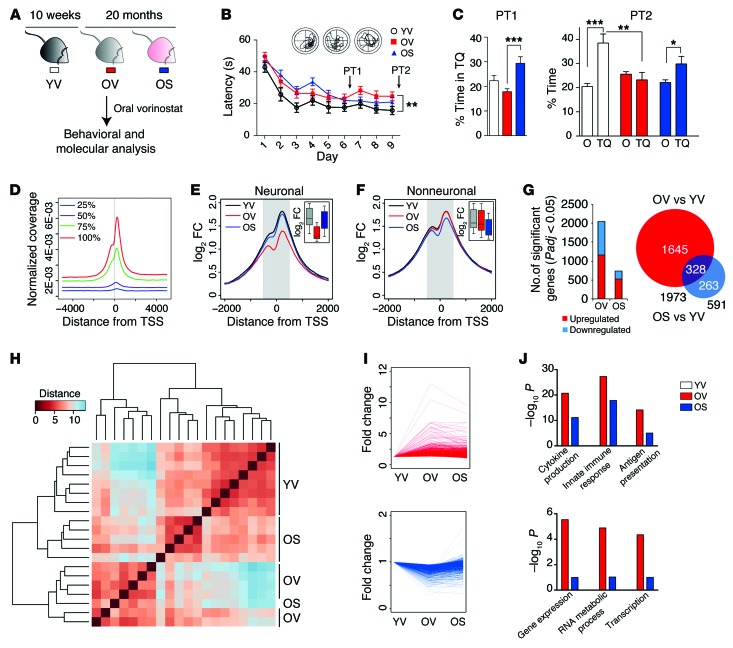
Since aging is the most important risk factor for dementia, we investigated age-associated memory impairment and the response of aged mice to oral SAHA administration. To this end, we employed 20-month-old mice that received SAHA for 4 weeks. We used 20-month-old animals that received vehicle solution as the control group. An additional control group consisted of 3-month-old mice treated with vehicle (Figure 1A). SAHA treatment did not influence basal activity (Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI79942DS1) or anxiety, as measured in the elevated plus maze test (Supplemental Figure 1B). To test hippocampus-dependent spatial memory function, mice were subjected to the Morris water maze (MWM) test. In line with previous findings (16–18), memory was impaired in vehicle-treated 20-month-old mice when compared with the 3-month group (Figure 1, B and C). Spatial reference memory was significantly improved in SAHA-treated animals when compared with the corresponding vehicle group (Figure 1, B and C), showing that oral administration of SAHA can ameliorate age-associated memory impairment. As a first step toward elucidating the underlying mechanisms, we characterized histone acetylation in the CA1 region of the hippocampus, the key brain structure required for spatial learning (19, 20). We performed ChIP-seq analysis for H4K12ac, since this mark has been linked to age-associated memory impairment (7, 16, 21). To be more specific on the molecular mechanisms, we sorted neuronal and nonneuronal nuclei and examined these populations separately. In agreement with previous reports (22), H4K12ac correlated well with the absolute level of gene expression in CA1 neurons (Figure 1D). We found that, in comparison with young mice, 20-month-old animals treated with vehicle displayed decreased H4K12ac that was specific to the neuronal cell population, since nonneuronal cells were not affected (Figure 1, E and F). SAHA reinstated physiological levels of H4K12ac in neurons, while no effect was seen in nonneuronal cells (Figure 1, E and F). These data demonstrate that oral administration of SAHA has quantifiable effects on neuronal histone acetylation in the aged brain.
Figure 1. Oral administration of SAHA improves memory function in an aging model and normalizes epigenetic and transcriptional activity.
(A) Experimental design. Old SAHA (OS), 20-month-old mice treated with SAHA for 4 weeks; old vehicle (OV), 20-month-old animals treated with vehicle; young vehicle (YV), 3-month-old mice treated with vehicle. (B) Escape latency during MWM training. Aged animals display significantly impaired escape latency in the MWM task. Repeated measures ANOVA, F(1,30) = 8.961, P < 0.01. (C) Probe test (PT) performance expressed as percentage of time spent in target quadrant (TQ) vs. average of other quadrants (O) for early (PT1) and later (PT2) probe test performed at indicated time points. *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t test after Holm-Sidak correction. n = 14 (YV), 18 (OV), 16 (OS) (A–C). (D) Profile plot of H4K12ac around TSS across whole genome for genes in 25th, 50th, 75th and 100th expression percentiles. Plot refers to CA1 neurons from YV-treated animals. (E) Profile plot of H4K12ac around TSS across genome for neuronal population. (F) Profile plot of H4K12ac around TSS across e genome for nonneuronal population. For E and F, inset shows box plot of the difference in coverage within shaded area (± 500 bp around TSS). Coverage is expressed as log2 fold-change over corresponding input. FC, fold change. (G) Left: number of genes at indicated significance cutoff up- or downregulated in the YV vs. OV and YV vs. OS comparisons. Right: Venn diagram illustrating overlap between genes in comparisons described above. Padj, P value adjusted. (H) Distance heat map with hierarchical clustering of YV, OV, and OS samples. Note that OS animals tend to cluster together with YV animals. (I) For every significant age-regulated gene, fold changes in OV and OS condition are plotted for upregulations (top) and downregulations (bottom) separately. (J) Sample functional categories overrepresented within upregulated (top) and downregulated (bottom) genes and their significance in animals treated with vehicle or SAHA. Error bars indicate SEM.
Next, we studied the effect of aging and oral SAHA administration on gene expression in the hippocampal CA1 region using RNA-seq. We found a very robust age-associated gene-expression signature consisting of 1,973 differentially expressed genes (Figure 1G, Supplemental Figure 2A, and Supplemental Table 1). Of note, these age-associated changes in gene expression were partially (83%) reversed after SAHA treatment (Figure 1G, Supplemental Figure 2A, and Supplemental Tables 2 and 3). In line with these data, hierarchical clustering and principal component analysis (Figure 1H and Supplemental Figure 2B) showed that aged mice treated with SAHA clustered more closely with young mice, while young and aged mice treated with vehicle clearly formed separate clusters (Figure 1H). These data further support the view that SAHA treatment partially normalizes age-associated changes in gene expression. Interestingly, this effect was seen for both up- and downregulated genes (Figure 1I). Functional enrichment analysis showed that age-associated increases in gene expression were linked to inflammatory processes, while decreases represented pathways linked to metabolism and transcriptional response (Figure 1J and Supplemental Figure 2, C and D). Of note, SAHA treatment had a clear dampening effect on the significance of all of these pathways (Figure 1J and Supplemental Figure 2, C and D).
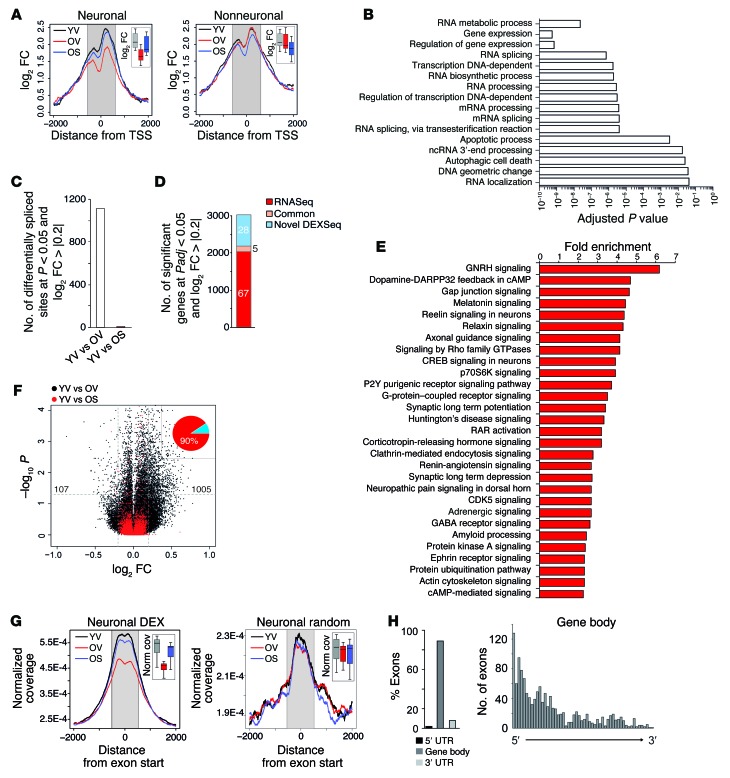
Interestingly, we did not find any pathways linked to synaptic plasticity, which is known to decrease in the aging brain (23). These data suggest that the impact of aging on gene expression and the effect of SAHA are likely to be complex. In order to address this issue further, we first performed a more detailed analysis concentrating on genes that were significantly (false discovery rate < 0.05) downregulated in the aged CA1 region and fully restored to physiological expression after SAHA treatment. When we analyzed H4K12ac for the 328 genes that fulfilled these criteria, we found a strong decrease of acetylation levels in the aging hippocampus that was almost fully restored by SAHA treatment when we analyzed neuronal cells. This effect was not seen in nonneuronal cells (Figure 2A). Pathway analysis showed that these genes were functionally linked to RNA splicing and noncoding RNA processing, suggesting that RNA processing and splicing are affected in CA1 neurons during aging (Figure 2B). To validate this finding, we measured protein levels of the dual specificity kinase CLK1, which is a critical regulator of the spliceosome and has been linked to alternative splicing (24–26). Consistent with our RNA-seq data, the levels of CLK1 were decreased in aged animals (Supplemental Figure 3).
Figure 2. SAHA reinstates physiological exon usage and H4K12ac levels at intron-exon junctions.
(A) Profile plot for H4K12ac for genes downregulated during aging and whose expression is fully reversed by SAHA in the neuronal (left) and nonneuronal (right) population. Inset shows box plots of the signal within the shaded area. (B) Functional categories and their significance associated with aging-downregulated genes whose expression is fully reversed by SAHA. (C) Number of splice sites significantly different in each of the indicated comparisons. (D) Of all genes found to be differentially expressed at the gene or exon level, the number and percentage (numbers inside each segment) of genes found in the RNA-seq only, in the DEX-seq analysis, or in common are represented. (E) Sample functional categories enriched within genes that contain 1 or more differential splicing events. (F) Volcano plot illustrating the magnitude and significance of changes at the exonic level that occur during aging after treatment with vehicle (black) or SAHA (red). Note that, whereas the distribution is clearly skewed toward the right (upregulations) in aged animals, this is reversed after SAHA treatment. Numbers within the graph denote the specific count of up- and downregulated exons at the indicated threshold for aged animals. Inset: percentage of exons with a higher inclusion rate (red) and a lower inclusion rate (blue) during aging. (G) H4K12ac signal around differentially spliced exons (left) and around a random set of exons (right) in the neuronal population. Insets represent box plots of the signal within the shaded area. DEX, differentially expressed exon. (H) Distribution of differentially spliced exons in aging across genes (left) and within the gene body (right).
This prompted us to specifically analyze alternative splicing in vehicle- and SAHA-treated animals. Our data show that splicing is massively affected during aging and demonstrate a quantitatively similar deregulation of differential gene expression and exon usage in the CA1 region (Figure 2C and Supplemental Table 4, compare to Figure 1D). SAHA was able to reinstate the exon-expression pattern in 20-month-old animals to levels almost equivalent to those of 3-month-old animals (Figure 2C and Supplemental Table 5). There was minimal overlap among the genes affected by splicing and those affected at the total expression level (Figure 2D). Functional analysis revealed that genes affected by alternative splicing were almost exclusively linked to synapse function (Figure 2E). We validated differential exon usage for selected genes via qRT-PCR (Supplemental Figure 4). Individual examples of genes affected by alternative exon usage are depicted in Supplemental Figures 5, 6, 7, and 8 and include a likely case of differential promoter usage (neuroplastin, Supplemental Figure 5), a case of alternative transcript usage (Sptbn1, Supplemental Figure 6), and a case of nonsense-mediated decay expression (Usp21, Supplemental Figure 7). Notably, the microtubule-associated protein Tau was also affected at the level of exon usage. We observed increased expression of the first exon that contains microtubule repeats (Supplemental Figure 8). Since this exon contains Tau phosphorylation sites that affect Tau solubility and microtubule stability (27), the increased inclusion of this exon might be particularly critical in age-associated dementia. SAHA fully restituted the physiological pattern of exon usage (Supplemental Figure 8).
Of note, age-associated memory impairment was mainly linked to the aberrant inclusion of exons (90%) rather than to exon skipping (Figure 2F). Recent reports indicate that histone acetylation can influence transcriptional elongation rate and thereby affect exon inclusion/exclusion (28–30). Thus, we examined H4K12ac in neurons at the intron-exon junctions of differentially expressed exons. There was a significant decrease in neuronal H4K12ac at the intron-exon junction in aged animals that was almost fully restituted after SAHA treatment. This was not the case when we analyzed a random set of exons (Figure 2G), indicating a potential causal relation between histone acetylation and exon usage at these particular sites. In agreement with the transcriptional rate hypothesis, closer examination of the distribution of differentially expressed exons indicated that these accumulate in the gene body, showing a 5′-to-3′ gradient (Figure 2H).
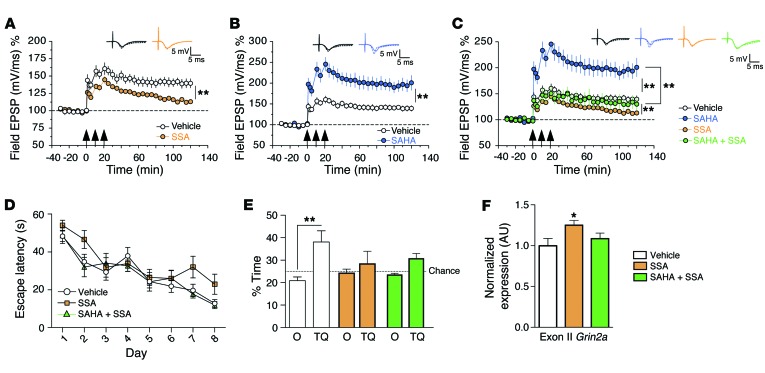
These data suggest that differential splicing contributes to age-associated memory decline and point to a mechanism by which SAHA improves cognitive function. Thus, we decided to further investigate the role of RNA splicing on synaptic plasticity and memory function employing spliceostatin A (SSA), a compound that binds the spliceosome component SF3b, thereby deregulating spliceosome function (31, 32). When administered to hippocampal slices, SSA significantly impaired long-term potentiation (LTP), which is believed to be a molecular correlate of learning and memory (Figure 3A).Next, we analyzed whether the administration of SAHA could rescue SSA-mediated impairment of hippocampal LTP. We first confirmed that SAHA significantly enhanced LTP (Figure 3B). This effect was dramatically reduced when SAHA was coadministered with SSA (Figure 3C). On the basis of these data, we decided to test the effect of SSA on hippocampus-dependent memory function. Microcannulae were implanted into the dorsal CA1 region, and SSA was injected after each training session of the MWM test. A probe test to assay spatial reference memory was performed after 8 days of training. While administration of SSA did not dramatically alter the escape latency during training, it significantly impaired performance during the probe test (Figure 3D). Thus, mice that had received SSA did not show any preference for the target quadrant, indicating that SSA impairs hippocampus-dependent spatial memory (Figure 3E). In an additional experiment, SSA and SAHA were coadministered into the hippocampal CA1 region during water maze learning. In line with the electrophysiological data, SAHA only partially rescued SSA-induced memory impairment during the probe test (Figure 3, D and E). Next, we employed qRT-PCR to study differential splicing of Grin2a, which encodes the ionotropic glutamate receptor N-methyl-D-aspartate subunit 2A, and was among the genes affected in the aging CA1 region. To this end, spliceostatin was injected into the CA1 region of mice and tissue was isolated for qRT-PCR analysis. Similar to the deregulation seen in the aging CA1 region, administration of SSA significantly increased the inclusion of Grin2a exon II (Figure 3F). This effect was partially rescued when SSA was coinjected with SAHA (Figure 3F). These findings suggest that (a) proper RNA splicing is essential for hippocampal synaptic plasticity and memory formation and (b) SAHA improves synaptic function and memory formation at least in part via mechanisms that involve differential exon usage.
Figure 3. Impairment of hippocampal LTP and in spatial memory by deregulation of RNA splicing is only partially recovered via SAHA administration.
(A) When SSA (190 nM) was applied 1 hour prior to LTP induction, a significant reduction of LTP was observed. Repeated measures ANOVA, F(1,10) = 10.31, P < 0.01. (B) Administration of SAHA (10 μM) robustly enhances hippocampal LTP. Repeated measures ANOVA, F(1,10) = 15.21, P < 0.01. (C) Coadministration of SAHA with SSA could only partially recover the effect of SAHA. While LTP was significantly increased when comparing SSA alone to SSA plus SAHA (repeated measures ANOVA, F[1,10] = 14.37, P < 0.01), this effect barely reached the LTP level seen in the vehicle group. For each group, n = 6. Arrows indicate high-frequency stimulation to induce LTP. (D) Escape latency for vehicle-, SSA-, and SSA plus SAHA–treated animals. (E) Percentage of time spent in target quadrant vs. other quadrants during the probe test performed at day 9. **P < 0.01, t test after Holm-Sidak correction, n = 9 (vehicle), 8 (SSA), 7 (SSA plus SAHA). (F) qRT-PCR demonstrating increased expression of Grin2a’s exon II in SSA-treated animals. *P < 0.05, t test after Holm-Sidak correction, n = 7 (vehicle), 8 (SSA), 7 (SSA plus SAHA). Error bars indicate SEM.
So far, we have shown that administration of SAHA can partially reinstate physiological gene expression and exon usage in aged mice at the genome-wide level. Since little is know of the effect of SAHA on the expression of microRNAs (miRNAs), which act as important regulators of synaptic plasticity and memory function and are also linked to cognitive impairment (33), we also analyzed the miRNAome of the CA1 region via small RNA-seq. We detected age- and SAHA-associated changes in the expression of miRNAs (Supplemental Table 6 and Supplemental Figure 9A). Based on target prediction, changes in these miRNAs could account for up to 13% and 7% of the aging-associated up- and downregulated genes, respectively (Supplemental Figure 9B and Supplemental Table 7). Unlike the expression of genes, age-associated miRNA changes were not reinstated in the presence of SAHA (Supplemental Figure 9A and Supplemental Table 6). Rather, we found that there was a subset of miRNAs that was specifically associated with SAHA administration (Supplemental Figure 9C).
We also analyzed the expression of long, noncoding RNAs (lncRNAs) (34). In line with a recent study showing aging-associated changes in lncRNAs in the cortex (35), we found 10 lncRNAs that were significantly altered in the aging CA1 region (Supplemental Table 8), including Neat1, which can act as a scaffolding molecule for nuclear paraspeckles and has been recently reported to be altered in Huntington’s disease (36). Of note, SAHA reversed or partially restituted physiological expression levels for 6 of these lncRNAs (Supplemental Table 8).
Since histone acetylation has been generally associated with facilitated gene expression, we have so far focused our analyses on aging-downregulated genes whose expression is reinstated by SAHA. Our data, however, also show that SAHA could partially reinstate physiological expression of aging-upregulated genes that were linked to inflammation (Figure 1, I and J, and Supplemental Figure 2). Since this effect cannot easily be explained by changes in histone acetylation, we speculate that processes related to the acetylation of nonhistone proteins, such as transcription factors, might be involved (37–39). To address this issue, we first examined transcription factor–binding sites linked to aging-upregulated genes. We found an enrichment for transcription factors that act as key regulators of inflammatory processes (Supplemental Figure 10A), most prominently STAT proteins. STAT1 is known to be regulated by acetylation (40, 41), suggesting a link between aging and SAHA on the activity of proteins like STAT1. In line with this hypothesis, we found hippocampal STAT1 activity to be increased in the CA1 region of aged mice, while SAHA ameliorated this effect (Supplemental Figure 10B). These data suggest that part of the antiinflammatory effect of SAHA may be mediated through the STAT pathway.
While our data provide clear evidence that oral SAHA administration affects brain function in a therapeutic manner, determining the effect of SAHA treatment on other organs would be clinically relevant. Thus, we also studied transcriptome plasticity in the liver via RNA-seq, since the liver represents a primary target region for oral drug administration. Our data show that aging affects the liver transcriptome and is associated with inflammatory gene expression, albeit through the regulation of a transcriptional program fundamentally different from that observed in the hippocampal CA1 region (Supplemental Figure 11 and Supplemental Tables 9–12). Also, in contrast with the results obtained in the brain, SAHA did not appear to normalize the age-associated gene-expression response observed in the liver. Most importantly, our analysis did not provide any evidence that SAHA would have detrimental effects on liver function (Supplemental Figure 11).
We have thus shown that aging is associated with a neuron-specific decrease in H4K12ac, which is linked to impaired gene expression and aberrant exon usage leading to deregulation of RNA processing and synaptic plasticity programs, respectively. All of these mechanisms are partially reinstated by oral administration of SAHA, the effect on exon usage being particularly robust.
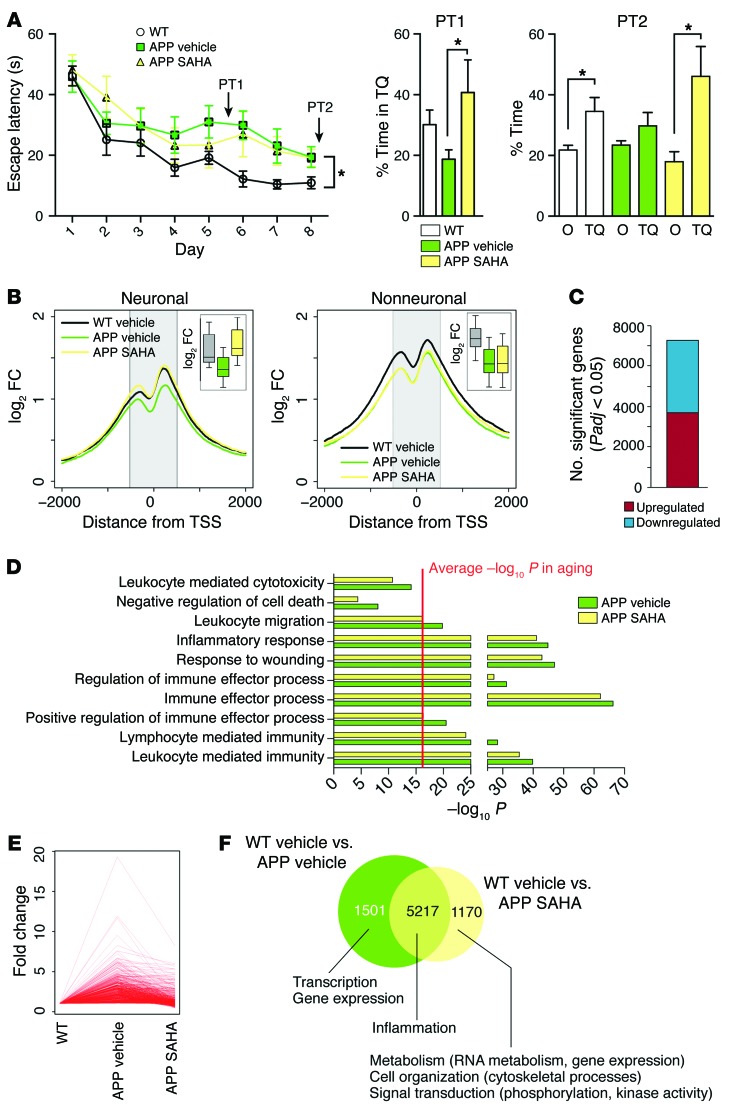
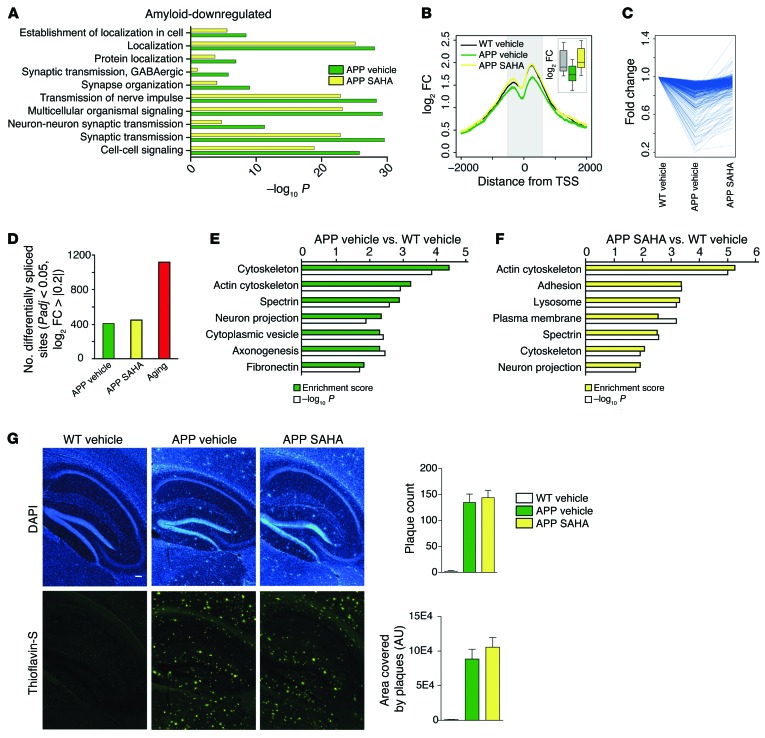
While aging is the most important risk factor for dementia and AD, other factors also contribute to this pathogenesis. A major hallmark of AD pathogenesis in humans is amyloid deposition, which can be investigated in mouse models overexpressing mutant proteins linked to familial AD and altered APP processing. A mouse model with particularly severe amyloid deposition is APP/PS1-21, which overexpresses mutant forms of both App and Psen1 (14). In order to test how robust and extendable the efficacy of oral SAHA administration is, we sought to determine whether oral administration of SAHA would also rescue memory function in 10-month-old APP/PS1-21 mice that had already developed severe amyloid pathology and hippocampus-dependent learning impairments (14, 15), thus representing a relevant preclinical model for drug testing. Ten-month old APP/PS1-21 mice received oral SAHA administration for 4 weeks (APP SAHA group) and were subsequently subjected to behavior testing. APP/PS1-21 mice and control littermates that received vehicle solution served as control groups. Similar to the data obtained in aged animals, oral administration of SAHA improved spatial reference memory in APP/PS1-21 mice (Figure 4A). Cell-type–specific ChIP-seq analysis of H4K12ac in the hippocampal CA1 region showed a global decrease in the signal for this mark around the transcription start site (TSS) in neuronal and nonneuronal cells, and SAHA reinstated H4K12ac specifically in neurons (Figure 4B). Next, we analyzed the transcriptome of the hippocampal CA1 region using RNA-seq. When comparing APP/PS1-21 mice with the corresponding control group, we found over 6,000 differentially expressed genes (Figure 4C and Supplemental Table 13). Pathway analysis indicated that the upregulated genes were almost exclusively linked to inflammatory processes (Figure 5D). Despite the comparatively larger magnitude of these changes with respect to the data obtained in our aging model, SAHA exerted a normalizing effect on gene expression, namely, it dampened the inflammatory transcriptional response in APP/PS1-21 mice (Figure 4, D–F, Supplemental Figure 12, and Supplemental Table 14). We furthermore found that the core of the inflammatory response observed in aged animals was not only concordant with that seen in APP/PS1-21 mice representing a stage of advanced amyloid pathology, but was even more pronounced at the single gene level (Supplemental Figure 12B). This effect was also observed at the level of noncoding RNAs (Supplemental Table 15). Thus, we found 5 lncRNAs commonly regulated between aging and APP/PS1-21 animals, most prominently Neat1, which was more strongly induced in APP/PS1-21 mice. Interestingly, the number of significant lncRNAs was larger in APP/PS1-21 animals (compare Supplemental Tables 8 and 15), suggesting a distinct and potentially relevant role of these molecules specifically associated to amyloid pathology.
Figure 4. SAHA recapitulates the effects seen on aging in a model for amyloid deposition.
(A) Escape latency (left) and result from an early (PT1, middle) and later (PT2, right) probe test performed in APP/PS1-21 animals the received SAHA from 10 months of age. APP/PS1-21 animals treated with vehicle displayed significant learning impairment, as indicated by the escape latency (left panel). Repeated measures ANOVA, F(1,18) = 5.174. *P < 0.05, Student’s t test after Holm-Sidak correction, n = 11 (WT vehicle), 9 (APP vehicle), 7 (APP SAHA). (B) H4K12 profile plot across the genome around the TSS in the neuronal (left) and nonneuronal (right) population. Insets show the differences in signal within the shaded area. (C) Number of significantly up- and downregulated genes in APP/PS1-21 animals. Note the large difference in magnitude compared with aged WT animals. (D) Main functional categories associated with amyloid-induced upregulated genes and their significance. The red line indicates the average significance level for similar categories in aged animals for comparison purposes. (E) Fold change of genes upregulated in APP/PS1-21 mice that received vehicle solution (APP vehicle) and APP SAHA animals. Every line represents an individual gene. (F) Overlap of genes found to be significant in the control littermates that received vehicle solution (WT vehicle) vs. APP vehicle and WT vehicle vs. APP SAHA comparisons. The main functional categories associated with each set of genes are indicated. Error bars indicate SEM.
Figure 5. SAHA can partially reinstate amyloid-induced downregulation of transcriptional programs, but does not affect plaque load.
(A) Main functional categories associated with genes downregulated in APP vehicle animals and their significance. (B) Profile plot for H4K12ac signal for downregulated genes around the TSS in the neuronal population. The inset shows a box plot of the coverage within the shaded area. (C) Fold change for downregulated genes in APP vehicle and APP SAHA animals. Each line represents an individual gene. (D) Number of significant splice events under the indicated conditions. Note that aging is associated with almost a 3-fold greater deregulation of exon usage. (E and F) Functional categories associated with genes that contain 1 or more differentially expressed exons in APP vehicle (E) and APP SAHA (F) animals. (G) Representative images and quantification of plaque load in APP/PS1-21 animals treated with vehicle or SAHA. n = 4 (WT vehicle), 4 (APP vehicle), 5 (APP SAHA). Scale bar: 100 μm.
In agreement with the data obtained in our aging model, amyloid-mediated increased gene expression was strongly associated with STAT transcription factors and SAHA moderately but systematically decreased the significance of these predictions (Supplemental Figure 10D), further endorsing the interpretation that amyloid pathology represents a very augmented aging response, at least regarding inflammatory processes.
In contrast to the data obtained in our aging model, we found that the genes downregulated in the CA1 region of APP/PS1-21 mice were associated with functional pathways mainly linked to synaptic plasticity (Figure 5A). Downregulation of such genes was linked to decreased neuronal H4K12ac and was rescued by SAHA administration (Figure 5B). In line with these data, SAHA treatment partially normalized this transcriptional program and its associated pathways (Figure 5, A and C). Since, in the aging model, we observed that genes linked to synaptic plasticity were deregulated via differential exon usage, we also analyzed splicing events in APP/PS1-21 mice. We observed that exon usage was affected in APP/PS1-21 mice, but to a much lower degree when compared with the aging model (Figure 4D). Interestingly, differential splicing in APP/PS1-21 mice was predominantly associated with genes linked to cytoskeleton function and neuronal projections (Figure 5E). SAHA treatment did not reinstate physiological exon usage in APP/PS1-21 mice as observed in the aging model. Nevertheless, despite a modest approximately 30% overlap of differentially spliced genes between vehicle- and SAHA-treated mice (Supplemental Tables 16 and 17), functional analysis revealed that the same biological processes, namely cytoskeleton function and neuronal projections, were affected in vehicle- and SAHA-treated APP/PS1-21 mice (Figure 5, E and F).
Of note, SAHA treatment did not affect plaque load in APP/PS1-21 mice (Figure 5G), suggesting that reinstated memory function is at least in part due to the normalization of gene-expression levels affecting synaptic plasticity and inflammatory responses.
These data show that oral administration of SAHA also ameliorates hippocampus-dependent memory decline in a mouse model of amyloid deposition. As in the aging model, SAHA impinges upon inflammatory processes in APP/PS1-21. While aging affects the expression of genes linked to synaptic plasticity at the level of differential splicing, such genes are downregulated in response to amyloid pathology. This effect is partially reversed by SAHA administration. Generally, amyloid pathology does not seem to affect differential exon usage to the same degree as observed in our aging model (Figure 6).
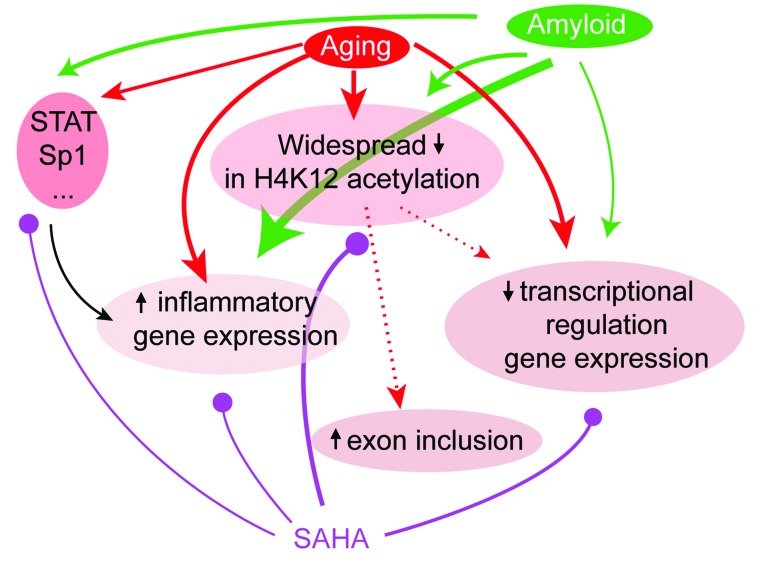
Figure 6. A graphical summary of the effects of aging and amyloid at the epigenetic, gene, and splicing level and the points of action of SAHA.
Both aging and amyloid pathology have indirect (epigenetic alterations [H2K12ac], regulation of transcription factors) and possibly direct effects on gene expression. We found that in aging these effects are primarily related to inflammation and splicing regulation. The effect on histone acetylation may also directly influence exon inclusion. Amyloid primarily targets inflammatory gene expression and, to a lesser extent, H4K12ac and nonhistone targets. SAHA acts on all of these molecular processes at all levels, though the exact mechanisms of action remain to be further investigated. Note that line thickness represents the extent to which a particular process is involved in aging/amyloid pathology or is targeted by SAHA according to our observations.
In conclusion, our data provide what we believe is the first genome-wide approach investigating the transcriptional events associated with aging and amyloid deposition in hippocampal neurons after oral treatment with an HDACi. We show that oral administration of SAHA improves memory function in models for AD and provide mechanistic insight into the action of HDACi. Our data strongly support the idea that SAHA — an FDA-approved drug (vorinostat/Zolinza) — should be tested in clinical settings treating patients with AD.
Discussion
Our study is the first, to our knowledge, to investigate the transcriptome and epigenome of the hippocampal CA1 region in response to aging, amyloid pathology, and oral SAHA treatment using genome-wide and cell-type–specific approaches. Our data show that the HDACi SAHA partially reinstates spatial memory function when administered orally to 20-month-old mice and 10-month-old APP/PS1-21 mice representing models for age-associated memory decline and amyloid deposition. This effect is paralleled by reinstatement of H4K12ac in neurons of the hippocampal CA1 region, demonstrating that oral delivery of SAHA affects histone acetylation in the brain. These data further support previous findings from imaging studies suggesting that SAHA can cross the blood-brain barrier (42–45) and are generally in line with studies showing that administration of HDACi via i.p. injection or direct injection into the brain can improve memory function in mice (11, 16, 46–55). However, none of these studies used genome-wide approaches to investigate the molecular underpinning of HDACi treatment. While it has been suggested that HDACi affects memory function via the regulation of histone acetylation and gene expression (56, 57), genome-wide evidence to support this view is very limited (58–63) and there are no data available related to age-associated memory decline or AD.
Interestingly, we observed that aging was associated with decreased H4K12ac in hippocampal CA1 neurons. A particularly interesting finding of our study is that nonneuronal cells of the CA1 region showed normal H4K12ac that was also not affected by the administration of SAHA. A similar pattern was observed in the CA1 region of APP/PS1-21 mice, and although the effect of amyloid deposition on nonneuronal cells was more pronounced than in the aging model, SAHA did not have a major impact on nonneuronal cells. These findings may indicate that — at least in vivo — nonproliferating cells such as neurons are more sensitive to homeostatic changes that affect histone acetylation. Importantly, while SAHA reinstated physiological H4K12ac in neuronal cells, we did not find evidence for an unspecific genome-wide increase in neuronal or nonneuronal H4K12ac, suggesting that HDACi treatment can ameliorate existing deficits, but does not act on cells in which histone acetylation is in homeostasis. The fact that neuron-specific impairment in age-associated H4K12ac was restored — but not further increased — by SAHA treatment correlated with increased expression of genes that were downregulated during aging. We did not find evidence that SAHA would cause massive and unselective changes in hippocampal gene expression. Genes downregulated in the aging CA1 region were functionally linked to the regulation of gene expression, metabolic function, and RNA processing, including RNA splicing. Expression of these genes was completely or partially restored in response to SAHA treatment. In line with the fact that RNA processing was affected in the aging CA1 region, we found massive changes in differential exon usage. While exon usage is known to be subject to intense regulation during brain development (64, 65), there is so far no genome-wide evidence that RNA splicing is linked to cognitive decline and the action of HDACi. This is likely due to the fact that previous studies investigating gene expression in age-associated cognitive impairment did not use RNA-seq. Our data are, however, in line with recent reports showing massive changes in RNA splicing when comparing the cortex from embryos to that of adult mice (66) or the prefrontal cortex and cerebellum from humans and primates at various ages (65). Furthermore, previous studies report age-associated splicing changes at the level of selected genes (64, 67). For 90% of the genes affected by altered splicing in the aging hippocampus, we observed aberrant inclusion of exons at the 5′ end of the coding region. These exons were characterized by decreased H4K12ac that was reinstated to physiological levels after SAHA treatment. These data are in line with previous reports linking histone acetylation to splicing regulation (29, 30, 68). In particular, it was shown that both H3 acetylation (at lysine 9) and H4 acetylation are linked to exon selection through regulation of the transcriptional elongation rate. Therefore, increased histone acetylation was found to increase RNA polymerase II (RNAPol-II) processivity and exon skipping in 3 different genes and cellular systems (29, 30, 68). Taking also into account that H4K12ac has been linked to transcriptional elongation (16), these data suggest that aberrant inclusion of exons in the aging CA1 regions is — at least in part — linked to reduced H4K12ac at the intron-exon junctions. The almost complete reinstatement of physiological splicing after SAHA treatment is most likely also linked to the restoration of genes implicated with the splicing machinery, such as Clk1, Srsf10, Srsf7, Tra2a, or Srsf3, which are downregulated during aging in the CA1 region and fully restored after SAHA treatment. Clk1 is a protein kinase essential for phosphorylation of Serine/Arginine-rich (SR) proteins (69), which in turn are fundamental components of the spliceosome and can form nuclear speckles (70). This view is further supported by recent data implicating CLK1 with AD pathogenesis (71).
In conclusion, our data indicate that RNA splicing plays a key role in the regulation of synaptic plasticity and the therapeutic action of SAHA. Furthermore, we show that SSA-mediated deregulation of RNA splicing impairs hippocampal LTP and hippocampus-dependent memory function in mice. To the best of our knowledge, such an effect has never been demonstrated before, but is in line with targeted approaches showing, for example, that splicing of neuronal surface receptors such as neurexins is linked to synaptic function (72). Most importantly, administration of SAHA could only partially rescue SSA-mediated impairment of hippocampal LTP and memory formation. These data suggest that the regulation of RNA splicing is critical, but of course, is not the only mechanism by which SAHA affects neuronal plasticity.
In contrast to the aging CA1 region, where genes linked to synaptic plasticity were affected solely at the level of differential exon usage, in APP/PS1-21 mice, functional pathways linked to synaptic plasticity were affected via reduced gene expression. The comparatively few genes affected by splicing in APP/PS1-21 were functionally linked to cytoskeleton-related processes. Interestingly, SAHA treatment did not reinstate physiological exon usage in APP/PS1-21 animals, but it altered the splicing of genes linked to the same molecular pathways. While differential splicing of selected genes has been observed in postmortem brain tissue from AD patients (73, 74) and in models for amyloid deposition (75), our data suggest that aging and amyloid deposition affect expression of synaptic plasticity genes via fundamentally different mechanisms. This is of utmost importance, taking into account that in human AD patients, both risk factors coincide. One potential explanation might be altered metabolic processes. Thus, histone acetylation is intimately linked to citrate metabolism (76), which is known to decrease in the aging brain (16, 77, 78). While energy metabolism is affected in brains of AD patients (79), there is no evidence so far that citrate levels decrease early in response to amyloid pathology. It has to be reiterated that in our study, as well as in most preclinical settings, age-matched APP/PS1-21 mice are used for analysis, thus neglecting age-associated changes. Thus, our data suggest that at least at the level of gene expression, therapeutic interventions directed toward amyloid pathology would fail to improve deregulation of plasticity genes linked to age-associated changes in exon usage. Of note, SAHA affects gene expression and RNA splicing and thus was able to reinstate physiological expression of plasticity genes in the aging and amyloid model, further highlighting its therapeutic potential.
Both aging and amyloid deposition were associated with a transcriptional program predominately linked to inflammation, which is in line with previous findings (80–83). Since aging is not associated with massive changes in the number of glial cells (37, 84), our data indicate that the gene-expression program linked to inflammation reflects activity changes. Given that the same pathways were affected in APP/PS1-21 mice, albeit much more strongly, our data suggest that, also in response to amyloid pathology, inflammation is initially mediated by changes in cell activity, but later also dominated by increased cell number. This is in line with previous studies on the role of astro- and microglia (85). Surprisingly, SAHA treatment was able to decrease the expression of genes linked to inflammation in our aging model and, to some extent, also in the amyloid model. These data are in line with previous reports showing antiinflammatory actions of HDACi. More specifically, SAHA was found to mediate antiinflammatory actions in various models of inflammation, such as corneal neovascularization (86), sepsis (87), intrapulmonary inflammation (88), and encephalomyelitis (89). Of note, neither in our aging nor in the amyloid model was increased expression of inflammatory genes associated with elevated H4K12ac in nonneuronal cells. These data suggest that H4K12ac does not simply reflect transcriptional activity, but plays a rather specific role in transcriptional regulation of cells in the adult brain. Thus, antiinflammatory effects of SAHA are likely to be mediated via processes that involve nonhistone protein acetylation, which is supported by recent findings (90, 91). Consistent with these data, we found that at least part of the inflammatory response might be mediated by the action of transcription factors such as STAT1, which are regulated by acetylation (41, 92, 93) and have been shown to control inflammatory processes (94, 95). Our data suggest that SAHA treatment indirectly ameliorates the age-associated increase in STAT1 activity. This is of particular importance, since STAT1 has recently been linked to cognitive function. Mice lacking STAT1 show enhanced memory performance and are resistant to memory impairment induced by Aβ peptides injected into the hippocampus (96).
Although we focused our analysis on the brain, oral administration of an HDACi such as SAHA certainly targets other organs, foremost the liver. Very few reports have examined aging in this organ before, and those that have have shown mixed results. While some have found none or very modest changes (83, 97, 98), others report hundreds or even thousands of gene-expression changes associated with age (99, 100). Our results are consistent with the more recent reports and show extensive age-associated transcriptional changes in the liver related to inflammatory and metabolic processes (100). The differences in magnitude with respect to previous studies likely reflect the increases of sequencing methods over microarrays. The gene-expression changes observed in the liver were not reversed by administration of SAHA, although SAHA had an overall affect on gene expression in the liver. Of note, the analysis of liver-specific gene expression did not yield any evidence for detrimental effects of chronic SAHA administration. In contrast to the widespread gene-expression changes, aging was associated with comparatively few alterations in differential splicing. These data are in agreement with reports showing that differential exon usage in the liver is generally less complex when compared with that in the brain (101, 102).
In conclusion, our data show for the first time, to our knowledge, that oral administration of SAHA can ameliorate memory impairment in mouse models of age-associated cognitive decline and amyloid deposition. Both aging and amyloid pathology are associated with inflammation and impaired synaptic function at the gene-expression level. However, during aging, deregulation of synaptic plasticity genes manifests mainly at the level of differential splicing, which is not further affected by amyloid pathology. On top of age-associated changes in splicing, amyloid deposition causes downregulation of plasticity genes, underscoring the importance of carefully interpreting results from animal models that only represent one aspect of complex humans diseases, such as AD. Thus, our data are of the utmost importance for the development of therapeutic strategies for treating AD patients in whom amyloid pathology occurs on the background of an aged brain. Notably, SAHA was able to impinge on all of these pathological changes and we did not find evidence for massive and unspecific changes in transcriptome plasticity after oral administration of SAHA. The later argument and the fact that mechanistic insight on the action of HDACi in the brain is limited is currently hindering the use of HDACi in clinical trials treating AD patients. Thus, our study provides important insight that will inspire clinical studies aiming to treat dementia. Since SAHA is an approved drug for human therapy, our data strongly advocate testing SAHA in a clinical setting treating AD patients, and we are currently preparing to perform such a study.
Methods
Animals.
Both 3-month-old and 20-month-old animals from a C57BL/6JRj background were obtained from Janvier. APP/PS1-21 mice belong to the Tg(Thy1-APPSw,Thy1-PSEN1*L166P)21Jckr colony. All animals were housed in standard cages with food and water ad libitum. They were allowed 2 to 4 weeks of habituation to the holding room before being subjected to any manipulation or experiment. Mice were single caged prior to behavioral testing.
SAHA administration.
SAHA was dissolved in drinking water following a previously published formula (42): 0.67 g of SAHA (Cayman Chemical Co.) was dissolved in 1 l of drinking water containing 18 g of β-cyclodextrin (Sigma-Aldrich, Ref 332607). The solution was heated to 95°C and stirred until no SAHA particles were visible (approximately 20 minutes) and then allowed to cool down to room temperature before making it available to animals. Animals drank on average 3 ml/d, which equals 2 mg SAHA intake per day. The maximum tolerable dose in humans is 400 mg/d. We used the following formula to covert the dose used in mice into human terms: human dose (mg/kg) = rodent dose (mg[kg] × rodent KM/rodent KM/human KM; KM rodent = 3 and KM human = 37, where KM = ratio of body weight [kg] to surface area [m2]) (103). Thus, the dose applied to mice in this study approximates 5 mg/kg in humans. Taking into account an average weight of 75 kg, the dose used would equal 375 mg/day and is thus within the maximum tolerable dose.
NGS data access.
All original microarray data were deposited in the NCBI’s Gene Expression Omnibus (RNA-seq: GEO GSE63943; small RNA–seq: GEO GSE63944; ChIP-seq: GEO GSE63942).
Detailed descriptions of all molecular and bioinformatics procedures are given in the Supplemental Methods.
Statistics.
All statistical analyses and graphical representations except those specifically described for RNA-seq and ChIP-seq were done with Prism 6 (GraphPad). All data are represented as mean ± SEM. Statistical tests employed for data analysis are described in greater detail within the figure legends and depended on experimental design. All t tests performed were 2 tailed. A P value of less than 0.05 was considered significant.
Study approval.
All procedures were performed by experienced experimenters and according to protocols approved by the Lower Saxony State Office for Consumer Protection and Food Safety.
Supplementary Material
Acknowledgments
This project was partly supported by the following sources to A. Fischer: the Euryi Award (DFG grant FI 981), the ERA NET Neuron Project EPITHERAPY, the Schram Foundation, the Hans and Ilse Breuer Foundation, and DZNE Göttingen.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2015;125(9):3572–3584. doi:10.1172/JCI79942.
References
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Fischer A. Epigenetic memory: the Lamarckian brain. EMBO J. 2014;33(9):945–967. doi: 10.1002/embj.201387637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fischer A, Sananbenesi F, Mungenast A, Tsai LH. Targeting the right HDAC(s) to treat congitive diseases. Trends Pharmacol Sci. 2010;31(12):605–617. doi: 10.1016/j.tips.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Gräff J, Tsai LH. Histone acetylation: molecular mnemonics on the chromatin. Nat Rev Neurosci. 2013;14(2):97–111. doi: 10.1038/nrn3427. [DOI] [PubMed] [Google Scholar]
- 5.Mahgoub M, Monteggia L. Epigenetics and psychiatry. Neurotherapeutics. 2014;10(4):734–741. doi: 10.1007/s13311-013-0213-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta P, Reid RC, Iyer A, Sweet MJ, Fairlie DP. Towards isozyme-selective HDAC inhibitors for interrogating disease. Curr Top Med Chem. 2012;12(14):1479–1499. doi: 10.2174/156802612802652420. [DOI] [PubMed] [Google Scholar]
- 7.Guan JS, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459(7243):55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McQuown SC, et al. HDAC3 is a critical negative regulator of long-term memory formation. J Neurosci. 2011;31(2):764–774. doi: 10.1523/JNEUROSCI.5052-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gräff J, et al. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature. 2012;483(7388):222–226. doi: 10.1038/nature10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Govindarajan N, et al. Reducing HDAC6 ameliorates cognitive deficits in a mouse model for Alzheimer’s disease. EMBO Mol Med. 2013;5(1):52–63. doi: 10.1002/emmm.201201923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kilgore M, et al. Inhibitors of class 1 histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer’s disease. Neuropsychopharmacology. 2010;35(4):870–880. doi: 10.1038/npp.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grant S, Dent P. Simultaneous interruption of signal transduction and cell cycle regulatory pathways: implications for new approaches to the treatment of childhood leukemias. Curr Drug Targets. 2007;8(6):751–759. doi: 10.2174/138945007780830764. [DOI] [PubMed] [Google Scholar]
- 13.Thaler F, Mercurio C. Towards selective inhibition of histone deacetylase isoforms: what has been achieved, where we are and what will be next. ChemMedChem. 2014;9(3):523–526. doi: 10.1002/cmdc.201300413. [DOI] [PubMed] [Google Scholar]
- 14.Radde R, et al. Abeta42-driven cerebral amyloidosis in transgenic mice reveals early and robust pathology. EMBO Rep. 2006;7(9):940–946. doi: 10.1038/sj.embor.7400784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Govindarajan N, Agis-Balboa C, Walter J, Sananbenesi F, Fischer A. Sodium butyrate improves memory function in an Alzheimer’s disease mouse model when administered at an advanced stage of disease progression. J Alzheimers Dis. 2011;24(1):1–11. doi: 10.3233/JAD-2011-110080. [DOI] [PubMed] [Google Scholar]
- 16.Peleg S, et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328(5979):753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- 17.Fordyce DE, Wehner JM. Effects of aging on spatial learning and hippocampal protein kinase C in mice. Neurobiol Aging. 1993;14(4):309–317. doi: 10.1016/0197-4580(93)90116-S. [DOI] [PubMed] [Google Scholar]
- 18.Lamberty Y, Gower AJ. Simplifying environmental cues in a Morris-type water maze improves place learning in old NMRI mice. Behav Neurosci. 1991;117(3):485–495. doi: 10.1016/0163-1047(91)90315-h. [DOI] [PubMed] [Google Scholar]
- 19.Morris RG, Garrud P, Rawlins JN, O’Keefe J. Place navigation impaired in rats with hippocampal lesions. Nature. 1982;297(5868):618–683. doi: 10.1038/297681a0. [DOI] [PubMed] [Google Scholar]
- 20.Moser MB, Moser EI. Functional differentiation in the hippocampus. Hippocampus. 1998;8(6):608–619. doi: 10.1002/(SICI)1098-1063(1998)8:6<608::AID-HIPO3>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 21.Stilling RM, Fischer A. The role of histone acetylation in age-associated memory impairment and Alzheimer’s disease. Neurobiol Learn Mem. 2011;96(1):19–26. doi: 10.1016/j.nlm.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 22.Lopez-Atalaya JP, Ito S, Valor LM, Benito E, Barco A. Genomic targets, and histone acetylation and gene expression profiling of neural HDAC inhibition. Nucleic Acids Res. 2013;41(17):8072–8084. doi: 10.1093/nar/gkt590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar A. Long-term potentiation at CA3-CA1 hippocampal synapses with special emphasis on aging, disease, and stress. Front Aging Neurosci. 2011;20:7. doi: 10.3389/fnagi.2011.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duncan PI, Stojdl DF, Marius RM, Bell JC. In vivo regulation of alternative pre-mRNA splicing by the Clk1 protein kinase. Mol Cell Biol. 1997;17(10):5996–6001. doi: 10.1128/mcb.17.10.5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duncan PI, Stojdl DF, Marius RM, Scheit KH, Bell JC. The Clk2 and Clk3 dual-specificity protein kinases regulate the intranuclear distribution of SR proteins and influence pre-mRNA splicing. Exp Cell Res. 1998;241(2):300–308. doi: 10.1006/excr.1998.4083. [DOI] [PubMed] [Google Scholar]
- 26.Hartmann AM, et al. Regulation of alternative splicing of human tau exon 10 by phosphorylation of splicing factors. Mol Cell Neurosci. 2001;18(1):80–90. doi: 10.1006/mcne.2001.1000. [DOI] [PubMed] [Google Scholar]
- 27.Wang JZ, Xia YY, Grundke-Iqbal I, Iqbal K. Abnormal hyperphosphorylation of tau: sites, regulation, and molecular mechanism of neurofibrillary degeneration. J Alzheimers Dis. 2013;33(suppl 1):123–139. doi: 10.3233/JAD-2012-129031. [DOI] [PubMed] [Google Scholar]
- 28.Schor IE, Rascovan N, Pelisch F, Alló M. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc Natl Acad Sci U S A. 2009;106(11):4325–4330. doi: 10.1073/pnas.0810666106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schor IE, Kornblihtt AR. Playing inside the genes: Intragenic histone acetylation after membrane depolarization of neural cells opens a path for alternative splicing regulation. Commun Integr Biol. 2009;2(4):341–343. doi: 10.4161/cib.2.4.8550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou HL, et al. Hu proteins regulate alternative splicing by inducing localized histone hyperacetylation in an RNA-dependent manner. Proc Natl Acad Sci U S A. 2011;108(36):E627–E635. doi: 10.1073/pnas.1103344108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaida D, et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol. 2007;3(9):576–583. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
- 32.Roybal GA, Jurica MS. Spliceostatin A inhibits spliceosome assembly subsequent to prespliceosome formation. Nucleic Acids Res. 2010;38(19):6664–6672. doi: 10.1093/nar/gkq494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fiore R, Khudayberdiev S, Saba R, Schratt G. MicroRNA function in the nervous system. Prog Mol Biol Transl Sci. 2011;102:47–100. doi: 10.1016/B978-0-12-415795-8.00004-0. [DOI] [PubMed] [Google Scholar]
- 34.Roberts TC, Morris KV, Wood MJ. The role of long non-coding RNAs in neurodevelopment, brain function and neurological disease. Philos Trans R Soc Lond B Biol Sci. 2014;369(1652):20130507. doi: 10.1098/rstb.2013.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wood SH, Craig T, Li Y, Merry B, de Magalhães JP. Whole transcriptome sequencing of the aging rat brain reveals dynamic RNA changes in the dark matter of the genome. Age. 2013;35(3):763–776. doi: 10.1007/s11357-012-9410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson R. Long non-coding RNAs in Huntington’s disease neurodegeneration. Neurobiol Dis. 2012;46(2):245–254. doi: 10.1016/j.nbd.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 37.Stilling R, et al. K-Lysine acetlytransferase 2A regualtes a hippocampal gene-expression network linked to memory formation. EMBO J. 2014;33(17):1912–1927. doi: 10.15252/embj.201487870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fischer A. Targeting histone-modifications in Alzheimer’s disease. What is the evidence that this is a promising therapeutic avenue? Neuropsychopharmacology. 2014;80:95–102. doi: 10.1016/j.neuropharm.2014.01.038. [DOI] [PubMed] [Google Scholar]
- 39.Bose P, Dai Y, Grant SG. Histone deacetylase inhibitor (HDACI) mechanisms of action: emerging insights. Pharmacol Ther. 2014;143(3):323–336. doi: 10.1016/j.pharmthera.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim CP, Cao X. Structure, function, and regulation of STAT proteins. Mol Biosyst. 2006;2(11):536–550. doi: 10.1039/b606246f. [DOI] [PubMed] [Google Scholar]
- 41.Wieczorek M, Ginter T, Brand P, Heinzel T, Krämer OH. Acetylation modulates the STAT signaling code. Cytokine Growth Factor Rev. 2012;23(6):293–305. doi: 10.1016/j.cytogfr.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 42.Mielcarek M, et al. SAHA decreases HDAC 2 and 4 levels in vivo and improves molecular phenotypes in the R6/2 mouse model of Huntington’s disease. PLos One. 2011;6(11):e27746. doi: 10.1371/journal.pone.0027746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hockly E, et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington’s disease. Proc Natl Acad Sci U S A. 2003;100(4):2041–2046. doi: 10.1073/pnas.0437870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reid AE, et al. Evaluation of 6-([(18)F]fluoroacetamido)-1-hexanoicanilide for PET imaging of histone deacetylase in the baboon brain. Nucl Med Biol. 2009;36(3):247–258. doi: 10.1016/j.nucmedbio.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yeh HH, et al. Imaging epigenetic regulation by histone deacetylases in the brain using PET/MRI with 18F-FAHA. Neuroimage. 2013;64:630–639. doi: 10.1016/j.neuroimage.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ricobaraza A, Cuadrado-Tejedor M, Pérez-Mediavilla A, Frechilla D, Del Río J, García-Osta A. Phenylbutyrate ameliorates cognitive deficit and reduces tau pathology in an Alzheimer’s disease mouse model. Neuropsychopharmacology. 2009;34(7):1721–1732. doi: 10.1038/npp.2008.229. [DOI] [PubMed] [Google Scholar]
- 47.Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447(7141):178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- 48.Fontan-Lozano A, Romero-Granados R, Troncoso J, Munera A, Delgado-Garcia JM, Carrion AM. Histone deacetylase inhibitors improve learning consolidation in young and in KA-induced-neurodegeneration and SAMP-8-mutant mice. Mol Cell Neurosci. 2008;39(2):193–201. doi: 10.1016/j.mcn.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 49.Francis YI, et al. Dysregulation of histone acetylation in the APP/PS1 mouse model of Alzheimer’s disease. J Alzheimers Dis. 2009;18(1):131–139. doi: 10.3233/JAD-2009-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Govindarajan N, Agis-Balboa RC, Walter J, Sananbenesi F, Fischer A. Sodium butyrate improves memory function in an Alzheimer’s disease mouse model when administered at an advanced stage of disease progression. J Alzheimers Dis. 2011;26(1):187–197. doi: 10.3233/JAD-2011-110080. [DOI] [PubMed] [Google Scholar]
- 51.Kilgore M, et al. Inhibitors of class 1 histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer’s disease. Neuropsychopharmacology. 2010;35(4):870–880. doi: 10.1038/npp.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peleg S, et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328(5979):753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- 53.Reolon GK, et al. Posttraining systemic administration of the histone deacetylase inhibitor sodium butyrate ameliorates aging-related memory decline in rats. Behav Brain Res. 2011;221(1):329–332. doi: 10.1016/j.bbr.2011.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ricobaraza A, Cuadrado-Tejedor M, Garcia-Osta A. Long-term phenylbutyrate administration prevents memory deficits in Tg2576 mice by decreasing Aβ. Front Biosci (Elite Ed) 2011;3:1375–1384. doi: 10.2741/e340. [DOI] [PubMed] [Google Scholar]
- 55.Ricobaraza A, Cuadrado-Tejedor M, Perez-Mediavilla A, Frechilla D, Del Rio J, Garcia-Osta A. Phenylbutyrate ameliorates cognitive deficit and reduces tau pathology in an Alzheimer’s disease mouse model. Neuropsychopharmacology. 2009;34(7):1721–1732. doi: 10.1038/npp.2008.229. [DOI] [PubMed] [Google Scholar]
- 56.Fischer A. Epigenetic memory: the Lamarckian brain. EMBO J. 2014;33(9):945–967. doi: 10.1002/embj.201387637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Graff J, Tsai LH. Histone acetylation: molecular mnemonics on the chromatin. Nat Rev Neurosci. 2013;14(2):97–111. doi: 10.1038/nrn3427. [DOI] [PubMed] [Google Scholar]
- 58.Graff J, et al. Epigenetic priming of memory updating during reconsolidation to attenuate remote fear memories. Cell. 2014;156(1–2):261–276. doi: 10.1016/j.cell.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Host L, et al. Inhibition of histone deacetylases in rats self-administering cocaine regulates lissencephaly gene-1 and reelin gene expression, as revealed by microarray technique. J Neurochem. 2010;113(1):236–247. doi: 10.1111/j.1471-4159.2010.06591.x. [DOI] [PubMed] [Google Scholar]
- 60.Sanchis-Segura C, Lopez-Atalaya JP, Barco A. Selective boosting of transcriptional and behavioral responses to drugs of abuse by histone deacetylase inhibition. Neuropsychopharmacology. 2009;34(13):2642–2654. doi: 10.1038/npp.2009.125. [DOI] [PubMed] [Google Scholar]
- 61.Schroeder FA, et al. A selective HDAC 1/2 inhibitor modulates chromatin and gene expression in brain and alters mouse behavior in two mood-related tests. PLoS One. 2013;8(8):e71323. doi: 10.1371/journal.pone.0071323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomas EA, et al. The HDAC inhibitor 4b ameliorates the disease phenotype and transcriptional abnormalities in Huntington’s disease transgenic mice. Proc Natl Acad Sci U S A. 2008;105(40):15564–15569. doi: 10.1073/pnas.0804249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lopez-Atalaya JP, Ito S, Valor LM, Benito E, Barco A. Genomic targets, and histone acetylation and gene expression profiling of neural HDAC inhibition. Nucleic Acids Res. 2013;41(17):8072–8084. doi: 10.1093/nar/gkt590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tollervey JR, et al. Analysis of alternative splicing associated with aging and neurodegeneration in the human brain. Genome Res. 2011;21(10):1572–1582. doi: 10.1101/gr.122226.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mazin P, et al. Widespread splicing changes in human brain development and aging. Mol Syst Biol. 2013;9:633. doi: 10.1038/msb.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dillman AA, et al. mRNA expression, splicing and editing in the embryonic and adult mouse cerebral cortex. Nat Neurosci. 2013;16(4):499–506. doi: 10.1038/nn.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Erickson KI, et al. Exercise training increases size of hippocampus and improves memory. Proc Natl Acad Sci U S A. 2011;108(7):3017–3022. doi: 10.1073/pnas.1015950108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hnilicová J, et al. Histone deacetylase activity modulates alternative splicing. PLoS One. 2011;6(2):e167272. doi: 10.1371/journal.pone.0016727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Duncan P, Stojdl DF, Marius RM, Bell JC. In vivo regulation of alternative pre-mRNA splicing by the Clk1 protein kinase. Mol Cell Biol. 1997;17(10):5996–6001. doi: 10.1128/mcb.17.10.5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou Z, Fu XD. Regulation of splicing by SR proteins and SR protein-specific kinase. Chromosoma. 2013;122(3):191–207. doi: 10.1007/s00412-013-0407-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jain P, Karthikeyan C, Moorthy NS, Waiker DK, Jain AK, Trivedi P. Human CDC2-like kinase 1 (CLK1): a novel target for Alzheimer’s disease. Curr Drug Targets. 2014;15(5):539–550. doi: 10.2174/1389450115666140226112321. [DOI] [PubMed] [Google Scholar]
- 72.Schreiner D, et al. Targeted combinatorial alternative splicing generates brain region-specific repertoires of neurexins. Neuron. 2014;84(2):386–398. doi: 10.1016/j.neuron.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 73.Twine NA, Janitz K, Wilkins MR, Janitz M. Whole transcriptome sequencing reveals gene expression and splicing differences in brain regions affected by Alzheimer’s disease. PLoS One. 2011;6(1):e16266. doi: 10.1371/journal.pone.0016266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mills JD, et al. RNA-seq analysis of the parietal cortex in Alzheimer’s disease reveals alternatively spliced isoforms related to lipid metabolism. Neurosci Lett. 2013;536:90–95. doi: 10.1016/j.neulet.2012.12.042. [DOI] [PubMed] [Google Scholar]
- 75.Kim KH, Moon M, Yu SB, Mook-Jung I, Kim JI. RNA-seq analysis of frontal cortex and cerebellum from 5XFAD mice at early stage of disease pathology. J Alzheimers Dis. 2012;29(4):793–808. doi: 10.3233/JAD-2012-111793. [DOI] [PubMed] [Google Scholar]
- 76.Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324(5930):1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jiang N, et al. NMR-based metabonomic investigations into the metabolic profile of the senescence-accelerated mouse. J Proteome Res. 2008;7(9):3678–3686. doi: 10.1021/pr800439b. [DOI] [PubMed] [Google Scholar]
- 78.Wang H, et al. Age-related alterations in the metabolic profile in the hippocampus of the senescence-accelerated mouse prone 8: a spontaneous Alzheimer’s disease mouse model. J Alzheimers Dis. 2014;39(4):841–848. doi: 10.3233/JAD-131463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jové M, Portero-Otín M, Naudí A, Ferrer I, Pamplona R. Metabolomics of human brain aging and age-related neurodegenerative diseases. J Neuropathol Exp Neurol. 2014;73(7):640–657. doi: 10.1097/NEN.0000000000000091. [DOI] [PubMed] [Google Scholar]
- 80.Blalock EM, et al. Gene microarrays in hippocampal aging: statistical profiling identifies novel processes correlated with cognitive impairment. J Neurosci. 2003;23(9):3807–3819. doi: 10.1523/JNEUROSCI.23-09-03807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Erraji-Benchekroun L, et al. Overexpression of β2-adrenergic receptors in mouse liver alters the expression of gluconeogenic and glycolytic enzymes. Am J Physiol Endocrinol Metab. 2005;288(4):15–22. doi: 10.1152/ajpendo.00113.2004. [DOI] [PubMed] [Google Scholar]
- 82.Lee CK, Weindruch R, Prolla TA. Gene-expression profile of the ageing brain in mice. Nature Genetics. 2000;25(3):294–297. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- 83.Zahn JM, et al. AGEMAP: a gene expression database for aging in mice. PLoS Genet. 2007;3(11):e201. doi: 10.1371/journal.pgen.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Long JM, Mouton PR, Jucker M, Ingram DK. What counts in brain aging? Design-based stereological analysis of cell number. J Gerontol A Biol Sci Med Sci. 1999;54(10):B407–B417. doi: 10.1093/gerona/54.10.B407. [DOI] [PubMed] [Google Scholar]
- 85.Hosokawa M, Klegeris A, Maguire J, McGeer PL. Expression of complement messenger RNAs and proteins by human oligodendroglial cells. Glia. 2003;42(4):417–423. doi: 10.1002/glia.10234. [DOI] [PubMed] [Google Scholar]
- 86.Zhou H, Jiang S, Chen J, Su SB. Suberoylanilide hydroxamic acid suppresses inflammation-induced neovascularization. Can J Physiol Pharmacol. 2014;92(10):879–885. doi: 10.1139/cjpp-2014-0117. [DOI] [PubMed] [Google Scholar]
- 87.Zhao T, et al. Histone deacetylase inhibitor treatment attenuates coagulation imbalance in a lethal murine model of sepsis. Surgery. 2014;156(2):214–220. doi: 10.1016/j.surg.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen HY, Li L, Fu ZJ. Histone deacetylase inhibitors trichostatin A and suberoylanilide hydroxamic acid attenuate ventilator-induced lung injury. Pharmazie. 2014;69(1):55–59. [PubMed] [Google Scholar]
- 89.Ge Z, et al. Vorinostat, a histone deacetylase inhibitor, suppresses dendritic cell function and ameliorates experimental autoimmune encephalomyelitis. Exp Neurol. 2013;241:56–66. doi: 10.1016/j.expneurol.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 90.Grabiec AM, Tak PP, Reedquist KA. Targeting histone deacetylase activity in rheumatoid arthritis and asthma as prototypes of inflammatory disease: should we keep our HATs on? Arthritis Res Ther. 2008;10(5):226. doi: 10.1186/ar2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shakespear MR, Halili MA, Irvine KM, Fairlie DP, Sweet MJ. Histone deacetylases as regulators of inflammation and immunity. Trends Immuncol. 2011;32(7):325–343. doi: 10.1016/j.it.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 92.Kramer OH, et al. A phosphorylation-acetylation switch regulates STAT1 signaling. Genes Dev. 2009;23(2):223–235. doi: 10.1101/gad.479209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhuang S. Regulation of STAT signaling by acetylation. Cell Signal. 2013;25(9):1924–1931. doi: 10.1016/j.cellsig.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.de Prati AC, et al. STAT1 as a new molecular target of anti-inflammatory treatment. Curr Med Chem. 2005;12(16):1819–1828. doi: 10.2174/0929867054546645. [DOI] [PubMed] [Google Scholar]
- 95.Sikorski K, Czerwoniec A, Bujnicki JM, Wesoly J, Bluyssen HA. STAT1 as a novel therapeutical target in pro-atherogenic signal integration of IFNγ, TLR4, and IL-6 in vascular disease. Cytokine Growth Factor Rev. 2011;22(4):211–219. doi: 10.1016/j.cytogfr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 96.Hsu WL, Ma YL, Hsieh DY, Liu YC, Lee EH. STAT1 negatively regulates spatial memory formation and mediates the memory-impairing effect of Aβ. Neuropsychopharmacology. 2014;39(3):746–758. doi: 10.1038/npp.2013.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cao SX, Dhahbi JM, Mote PL, Spindler SR. Genomic profiling of short- and long-term caloric restriction effects in the liver of aging mice. Proc Natl Acad Sci U S A. 2001;98(19):10630–10635. doi: 10.1073/pnas.191313598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tollet-Egnell P, Flores-Morales A, Stahlberg N, Malek RL, Lee N, Norstedt G. Gene expression profile of the aging process in rat liver: normalizing effects of growth hormone replacement. Mol Endocrinol. 2001;15(2):308–318. doi: 10.1210/mend.15.2.0594. [DOI] [PubMed] [Google Scholar]
- 99.Thomas RP, Guigneaux M, Wood T, Evers BM. Age-associated changes in gene expression patterns in the liver. J Gastrointest Surg. 2002;6(3):445–453. doi: 10.1016/S1091-255X(01)00010-5. [DOI] [PubMed] [Google Scholar]
- 100.Yu Y, et al. A rat RNA-seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nat Commun. 2014;5:3230. doi: 10.1038/ncomms4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ramskold D, Wang ET, Burge CB, Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PloS Comput Biol. 2009;5(12):e1000598. doi: 10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yeo G, Holste D, Kreiman G, Burge CB. Variation in alternative splicing across human tissues. Genom Biol. 2004;5(10):R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Reagan-Shaw S, Nihal M, Ahmad N. Dose translation from animal to human studies revisited. FASEB J. 2007;22(3):659–661. doi: 10.1096/fj.07-9574LSF. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.