Abstract
Juvenile ciliopathy syndromes that are associated with renal cysts and premature renal failure are commonly the result of mutations in the gene encoding centrosomal protein CEP290. In addition to centrosomes and the transition zone at the base of the primary cilium, CEP290 also localizes to the nucleus; however, the nuclear function of CEP290 is unknown. Here, we demonstrate that reduction of cellular CEP290 in primary human and mouse kidney cells as well as in zebrafish embryos leads to enhanced DNA damage signaling and accumulation of DNA breaks ex vivo and in vivo. Compared with those from WT mice, primary kidney cells from Cep290-deficient mice exhibited supernumerary centrioles, decreased replication fork velocity, fork asymmetry, and increased levels of cyclin-dependent kinases (CDKs). Treatment of Cep290-deficient cells with CDK inhibitors rescued DNA damage and centriole number. Moreover, the loss of primary cilia that results from CEP290 dysfunction was rescued in 3D cell culture spheroids of primary murine kidney cells after exposure to CDK inhibitors. Together, our results provide a link between CEP290 and DNA replication stress and suggest CDK inhibition as a potential treatment strategy for a wide range of ciliopathy syndromes.
Introduction
Mutations in CEP290 are known to cause the entire spectrum of ciliopathies, including childhood renal failure due to nephronophthisis (NPHP) (1), Joubert syndrome (JS; MIM 610188), the lethal Meckel Grüber syndrome (MKS; MIM 611134), Bardet-Biedl syndrome (BBS) (MIM 209900), Senior-Løken syndrome (SLS) (MIM 610189), and up to 25% of Leber congenital amaurosis (LCA; MIM 611755) cases, although no precise genotype-phenotype correlations have been found (2). Around 50% of patients with JS in which there is a cerebello-oculo-renal phenotype have mutations in CEP290 (3). Progressive kidney damage, secondary to NPHP, leads to end-stage renal disease (ESRD) in affected patients and occurs at a mean age of 14 years (4). NPHP is the most frequent monogenic cause of ESRD in the first 3 decades of life (5) and accounts for 5% to 10% of all children with ESRD. Disease mechanisms underlying NPHP, which is characterized by a tubulointerstitial fibrosis, tubular atrophy, and corticomedullary cyst formation, implicate abnormal ciliary and centrosomal proteins (5).
CEP290 is a large multidomain centrosomal protein (6); identified binding partners of CEP290 include centrosomal proteins CEP131 and CCDC13; scaffold proteins pericentrin and PCM1; transcription factors, including ATF4; and proteins that are implicated in the DNA damage response (DDR), for example, ataxia telangiectasia and RAD3-related (ATR) (1, 7–9). The localization of CEP290 to the centrosome is dynamic, depending on the stage of cell cycle and expression of the primary cilium (1). The primary cilium is expressed in G0 after exit from the cell cycle, when the mother centriole is docked to the plasma membrane. Cells disassemble their cilium at the end of G1 in order to duplicate their centrosome for mitotic spindle formation (10). CEP290 localizes to the transition zone at the base of the primary cilium as well as at the centrosomes in a complex with other centrosomal proteins: NPHP1, INVS (also known as NPHP2), NPHP4, IQCB1 (also known as NPHP5), RPGRIP1L (also known as NPHP8), and NEK8 (also known as NPHP9) (11–13); mutations in any of these proteins can cause one or more ciliopathy syndromes as well. CEP290 also localizes to the nucleus, although its function there is entirely unknown (1). One possibility is that CEP290 acts in a manner similar that of the other ciliary proteins mutated in renal ciliopathies (CEP164, ZNF423, SDCCAG8, NEK8), which have been associated with enhanced DDR signaling (14–16). A single study examining the events leading to DNA damage in this setting recently established a role for NEK8 in the ATR-regulated replication stress response and in the regulation S-phase cyclin-dependent kinase (CDK) activity (16). However, only three families with mutations in NEK8 have been described (17), making these data less clinically relevant.
We set out to extend this correlation to a broader clinical base and investigate the role of CEP290 loss in DDR signaling and replication stress. To confirm that defects in DDR signaling underlie progressive renal disease seen in NPHP would allow a novel rationale for therapeutic interventions in these patients. Our findings support the overall hypothesis that NPHP-related ciliopathies (NPHP-RC) are initially caused by DNA damage and replication stress during early stages of development (18). Here, we used primary cells isolated from kidneys of Cep290LacZ/LacZ mice with JS symptoms and their WT littermates (19) to investigate DNA damage signaling and the replication stress response. We found enhanced DNA damage signaling and concomitant DNA breaks in Cep290LacZ/LacZ cells, in addition to supernumerary centrioles. Decreased replication fork velocity and fork asymmetry underlie the DNA damage. Additionally, application of CDK inhibitors (CDKi) rescues the DNA damage phenotypes and restores the ability of Cep290LacZ/LacZ cells to ciliate. These findings provide insight into the disease mechanism of NPHP-RC and will help to refine treatment strategies.
Results
CEP290 depletion causes DNA damage ex vivo, in vitro, and in vivo.
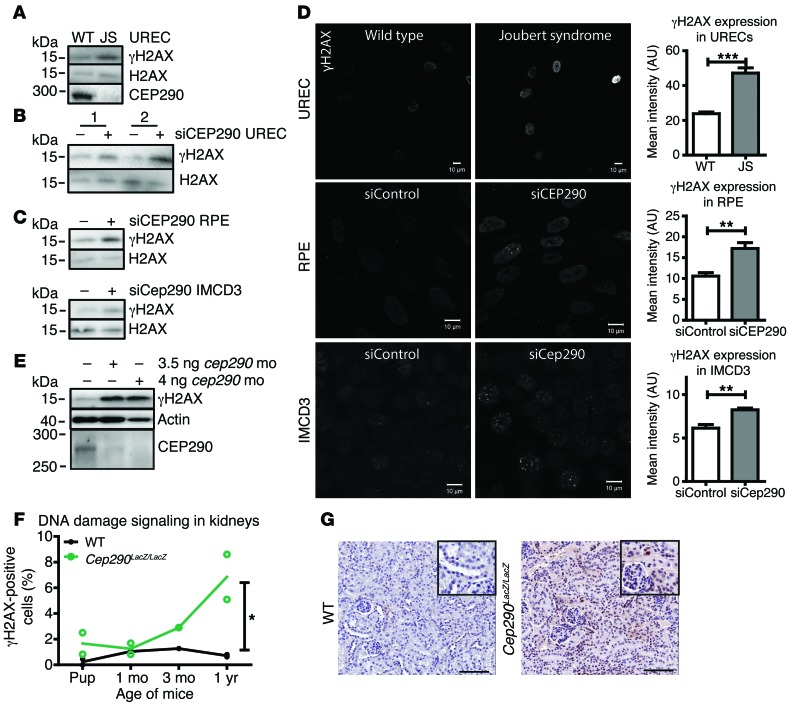
Recent studies have implicated DDR and sensitivity to replication stress in the development of ciliopathies. To explore the breadth of molecular effects in different human ciliopathies, we tested siRNA targeting of Ofd1 and Mks1, which is characteristic of orofaciodigital syndrome (MIM 311200) and MKS, respectively. Increased levels of DDR signaling were observed for all siRNAs tested (Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI80657DS1). To test the effect of CEP290 mutations in patient material, we isolated urine-derived renal epithelial cells (URECs) from a patient with JS with compound heterozygous mutations in CEP290, p.Q950Pfs*6, and p.K939N and observed increased γH2AX levels and decreased CEP290 levels compared with those of a healthy donor by Western blot (Figure 1A and Supplemental Figure 1C). We next lowered endogenous levels of CEP290 in URECs from two healthy donors and determined siRNA efficiency to be about 70% in the single donor tested (due to limited material, n = 1; P < 0.01; Supplemental Figure 1A). Western blots from both donors revealed increased DNA damage signaling levels (γH2AX) after 48 hours of siCEP290 transfection (Figure 1B and Supplemental Figure 1B). Similar results were observed upon siRNA knockdown of CEP290 in mIMCD3 and hTERT-RPE cell lines, which suggests that increased DNA damage signaling is not restricted to renal cells alone (n = 3; Figure 1C and Supplemental Figure 1, A and B). Immunofluorescence showed increased γH2AX foci in URECs from the patient with JS as well as in RPE and mIMCD3 cells after siRNA knockdown of CEP290, compared with that in controls (n = 3; P < 0.01 and P < 0.001 Figure 1D). Next, we examined whether DDR signaling would be induced after a cep290 ATG-targeting morpholino (mo) was injected in zebrafish (1). Western blot of 3.5 ng or 4 ng mo–injected fish targeting cep290 showed increased γH2AX 48 hours after fertilization, compared with that in controls (n = 3; Figure 1E and Supplemental Figure 1, B and D). Furthermore, γH2AX levels increased with the age of mouse Cep290LacZ/LacZ kidneys as compared with those in WT littermates, suggesting a role for DNA damage or DNA damage signaling in the pathophysiology of NPHP-RC (n = 13, P < 0.05; Figure 1, F and G).
Figure 1. DDR signaling is enhanced ex vivo, in vitro, and in vivo.
(A) Western blot of UREC lysates of a healthy donor (WT) and a patient with JS with compound heterozygous mutations in CEP290, p.Q950Pfs*6, and p.K939N. CEP290 protein expression loss and increased γH2AX levels are detected in the JS protein lysate. H2AX was used as loading control; samples were run on parallel gels contemporaneously. (B) Increased γH2AX levels were detected in siCEP290-transfected WT URECs, 48 hours after siCEP290 transfection. H2AX was used as loading control. (C) RPE and mIMCD3 cells depleted of CEP290 by siRNA show increased γH2AX levels compared with control siRNA transfection (56 hours). H2AX was used as loading control (n = 3). (D) Immunofluorescent staining of γH2AX in WT and JS URECs and RPE and mIMCD3 cells depleted of CEP290 by siRNA and quantification of staining intensity per nucleus (scale bar: 10 μm; n = 3; 100 cells scored per condition; t test, **P < 0.01, ***P < 0.001). (E) Western blot of zebrafish injected with 3.5 or 4 ng cep290 ATG-targeting mo. Loss of CEP290 protein expression and increased γH2AX levels were detected in cep290 mo-injected zebrafish lysates 48 hours after fertilization. Actin was used as loading control (n = 3); samples were run on parallel gels contemporaneously. (F) More γH2AX (brown) staining in kidneys of homozygote Cep290LacZ/LacZ gene trap mice compared with WT mice, which increased with age (n = 13; between 5,000–7,000 cells scored per animal) (linear model, goodness-of-fit test R2 = 0.86; *P < 0.05). (G) Example of WT mouse kidney and Cep290LacZ/LacZ gene trap mouse kidney at age 3 months stained for γH2AX (brown). Insets show high-magnification images of kidneys. Scale bars: 100 μm; original magnification ×2 (inset). Quantification of Western blots is shown in Supplemental Figure 1.
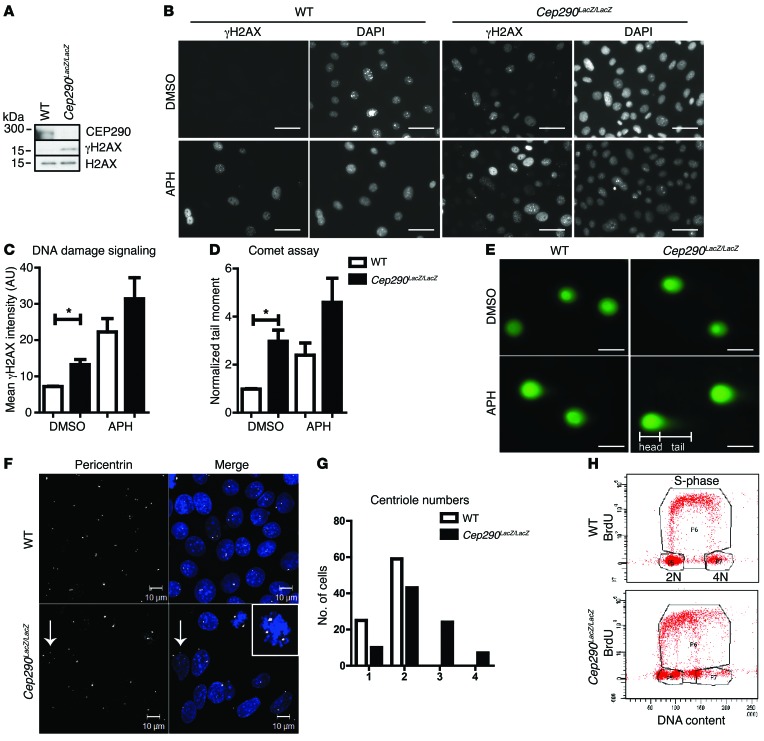
Results from our Western blot analyses support the presence of enhanced DNA damage signaling (γH2AX) in primary mouse collecting duct cells isolated from Cep290LacZ/LacZ kidneys (19) as opposed to their WT littermates (Figure 2A). Immunofluorescence confirmed higher intensities of γH2AX expression per nucleus in the Cep290LacZ/LacZ cells, which was further enhanced by incubation with the replication stress-inducing agent aphidicolin (APH) (20) (400 nM, 18 hours; n = 3; P < 0.05; Figure 2, B and C).
Figure 2. DNA damage is enhanced in CEP290-depleted cells.
(A) Western blot of Cep290LacZ/LacZ and WT primary mouse kidney cell lysates. Increased H2AX phosphorylation was detected in the CEP290-depleted cells. H2AX was used as loading control. (B) Immunofluorescent staining of γH2AX in control cells treated with DMSO and in cells treated with 400 nM APH for 18 hours. Scale bar: 10 μm. (C) Quantification of γH2AX staining intensity per nucleus in control cells treated with DMSO and in cells treated with 400 nM APH for 18 hours (n = 3; 100 cells scored per condition; 2-way ANOVA, *P < 0.05). (D) Quantification of comet tail moments of cells treated with 400 nM APH for 18 hours normalized to control DMSO-treated WT cells (n = 3; 50 cells scored per condition; 2-way ANOVA, *P < 0.05). (E) Images of SYBR Gold–stained DNA in comet tail assays. Scale bar: 10 μm. (F) Immunofluorescent staining of WT and Cep290LacZ/LacZ cells for pericentrin (white) reveals supernumerary centrioles (arrows) in Cep290LacZ/LacZ cells. The inset shows multipolar (>2) spindles. Scale bars: 10 μm; original magnification ×2 (inset). (G) Quantification of centriole number in WT and Cep290LacZ/LacZ cells (84 cells scored per cell line; n = 3). (H) BrdU FACS of WT and Cep290LacZ/LacZ cells shows normal DNA content and cell cycle for WT cells; however, Cep290LacZ/LacZ cells have irregular DNA content (10,000 events measured; n = 3).
To differentiate between actual DNA fragmentation and simple increased DDR signaling, we assessed whether DNA double-strand breaks were formed using neutral comet assays. Increased comet tail moments were observed in Cep290LacZ/LacZ cells and exacerbated by APH incubation (400 nM, 18 hour; n = 5; P < 0.05; Figure 2, D and E). We concluded that loss of CEP290 generates double-stranded breaks in DNA. Because CEP290 is a centrosomal protein, we examined centriole numbers. Supernumerary centrioles were observed in the Cep290LacZ/LacZ cells (Figure 2, F and G), which might contribute to genome instability. We therefore performed DNA content analysis by FACS (DAPI and BrdU); Cep290LacZ/LacZ cells demonstrated changes in DNA content as compared with the WT control cells. WT cells had a normal G1 (2 N DNA) and G2 (4 N DNA) cell cycle profile, whereas Cep290LacZ/LacZ cells had an aberrant DNA content profile that did not match 2 N or 4 N DNA (Figure 2H). To ensure that this result was not confounded by contamination or clonal drift, we grew Cep290LacZ/LacZ cells clonally and observed similar data in all 7 clones tested (Supplemental Figure 2, B and C). Our data support a role for CEP290 in genome stability.
CEP290 protects cells from replication stress.
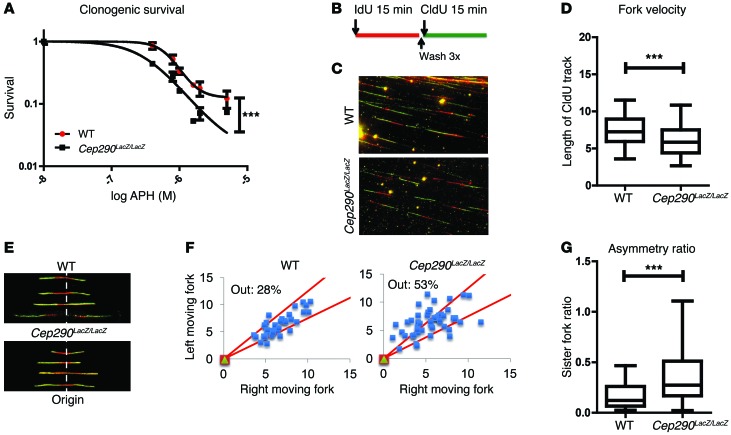
The formation of DNA breaks is frequently a consequence of perturbed DNA replication, a phenomenon referred to as replication stress (21). Clonogenic survival assays using the WT and Cep290LacZ/LacZ mouse primary kidney cells incubated with different doses of APH for 18 hours suggested that Cep290LacZ/LacZ cells are more sensitive to replication stress (n = 3; P < 0.001; Figure 3A). To investigate the underlying molecular mechanism, we next performed single-molecule DNA fiber analysis of replication fork progression in the WT and Cep290LacZ/LacZ mouse primary kidney cells. To label newly synthesized DNA, cells were pulsed with iododeoxyuridine (IdU) for 15 minutes and then with chlorodeoxyuridine (CldU) for 15 minutes. The double labeling with IdU and CldU allows the assessment of fork symmetry and velocity as well as origin firing (ref. 22; Figure 3, B and E). We first asked whether CEP290 modulated the fork velocity, also referred to as the rate of DNA synthesis, by measuring the lengths of the CldU tracts. We found that the replication fork velocity was decreased (n = 4; P < 0.001; Figure 3, B–D) in the Cep290LacZ/LacZ cells. After origin firing, forks moved bidirectionally in a relatively synchronous manner, unless there was a problem with fork progression or restart. Thus, we also analyzed the symmetry of sister forks emanating from a common origin, as fork asymmetry is an indicator of fork stalling. To do so, we measured the lengths of the CldU tracts for each pair of sister forks and plotted each pair as the right length versus the left length. From this analysis, we found that Cep290LacZ/LacZ cells had an increase in asymmetric forks, indicative of an inherent loss of fork stability in the absence of CEP290 (n = 2; P < 0.001; Figure 3, E–G). Importantly, sister fork asymmetry and reduced fork velocity are both indicators of replication stress and suggest that DNA break formation in Cep290LacZ/LacZ cells occurs as a result of increased replication stress. To examine cell cycle S-phase progression defects, which are potentially induced by replication stress, we quantified cells in early S-phase 18 hours after BrdU pulse labeling of cells in S-phase. IMCD3 cells had a higher proportion of cells in early S-phase after CEP290 siRNA treatment compared with controls (n = 3; P < 0.01; Supplemental Figure 3A), suggesting that S-phase progression is slower due to replication fork defects. As these results mirror those seen in Nek8–/– cells, it seems likely that defective DNA replication dynamics are a common feature of NPHP-RC.
Figure 3. Loss of CEP290 leads to replication stress.
(A) Clonogenic survival of control cells treated with DMSO and of cells treated with 500 to 5000 nM APH for 18 hours, showing sensitivity to replication stress of Cep290LacZ/LacZ cells. Log IC50 curves are compared with nonlinear regression (curve fit; n = 3; ***P < 0.001). (B) Schematic overview of pulse labeling of cells with IdU and CldU for 15 minutes. (C) Immunofluorescent staining of DNA fibers after pulse labeling with IdU (red) and CldU (green), respectively. Original magnification, x40. (D) Quantification of replication fork velocity by measuring length of the CldU track. Cep290LacZ/LacZ cells demonstrate decreased fork velocity (700 tracks scored per condition; n = 4; t test, ***P < 0.001). Horizontal bars indicate the median values, boxes indicate 25th to 75th percentiles, and whiskers indicate 5th and 95 percentiles. (E) Immunofluorescent staining of DNA fibers after pulse labeling with IdU (red) and CldU (green), respectively. Asymmetric replication at origins in Cep290LacZ/LacZ cells is present. Original magnification, x40. (F) Quantification of fork asymmetry at origins in Cep290LacZ/LacZ cells. When the replication track differs more than 25% between the right and left moving fork, the measurement falls below the cutoff (50 tracks scored per condition). (G) Fork asymmetry ratio, as calculated by the ratio of the right and left moving forks (50 tracks scored per condition; n = 2; t test, ***P < 0.001). Horizontal bars indicate the median values, boxes indicate 25th to 75th percentiles, and whiskers indicate 5th and 95 percentiles.
CEP290 is stabilized upon DNA damage induction.
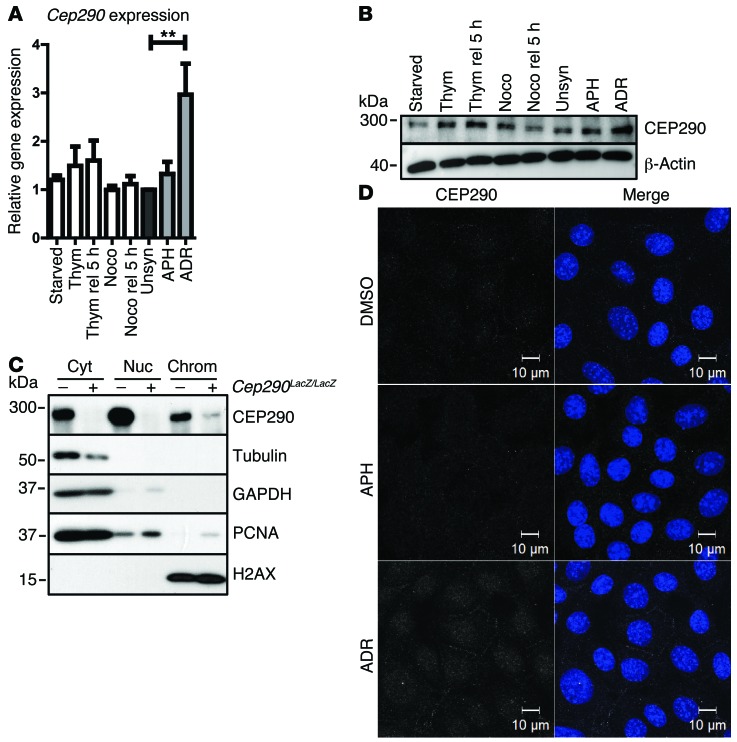
To examine CEP290 expression throughout the cell cycle, WT primary kidney cells were synchronized with thymidine or nocodazole and released for 5 hours. We found that CEP290 levels varied, and the highest levels of CEP290 were observed in S-phase (n = 4; Figure 4, A and B, and Supplemental Figure 2A). Since loss of CEP290 causes replication stress, we examined whether CEP290 precipitates with chromatin. Standard chromatin fractionation assays convincingly showed endogenous CEP290 to be enriched in the chromatin cell fraction (n = 2; Figure 4C). Immunofluorescence revealed expression at the base of the primary cilium (Supplemental Figure 2D) and nuclear expression as well (n = 3; Figure 4D). Endogenous CEP290 protein levels did not increase or relocalize after treatment with APH (400 nM, 24 hours). However, the DNA-damaging agent adriamycin (ADR) (1 μM, 24 hours) upregulated CEP290 expression (n = 4; P < 0.01; Figure 4, A, B, and D), indicating a CEP290 response to DNA damage independent of DNA replication progression.
Figure 4. Expression and localization of CEP290.
(A) Normalized Cep290 gene expression levels were measured by RT-qPCR of WT cells synchronized by serum starvation (G1), thymidine (S), nocodazole (G2) and release for 5 hours of these synchronizations, and unsynchronized, APH (400 nM) and ADR (1 μM) (24 hours) treated cells (n = 4; 1-way ANOVA, **P < 0.01). (B) Concomitantly, protein expression was detected by Western blot, using β-actin as loading control. (C) Western blot of chromatin fractionation of WT (–) and Cep290LacZ/LacZ (+) cells. CEP290 is expressed in the cytosol (GAPDH, tubulin), nucleus (PCNA), and chromatin-enriched fraction (H2AX). Samples were run on parallel gels contemporaneously. (D) Immunofluorescence staining of CEP290 expression (white) of WT cells treated with DMSO, APH (400 nM), and ADR (1 μM) (18 hours). ADR exposure caused increased CEP290 levels. Scale bar: 10 μm.
CDKi rescue DNA damage and cilia.
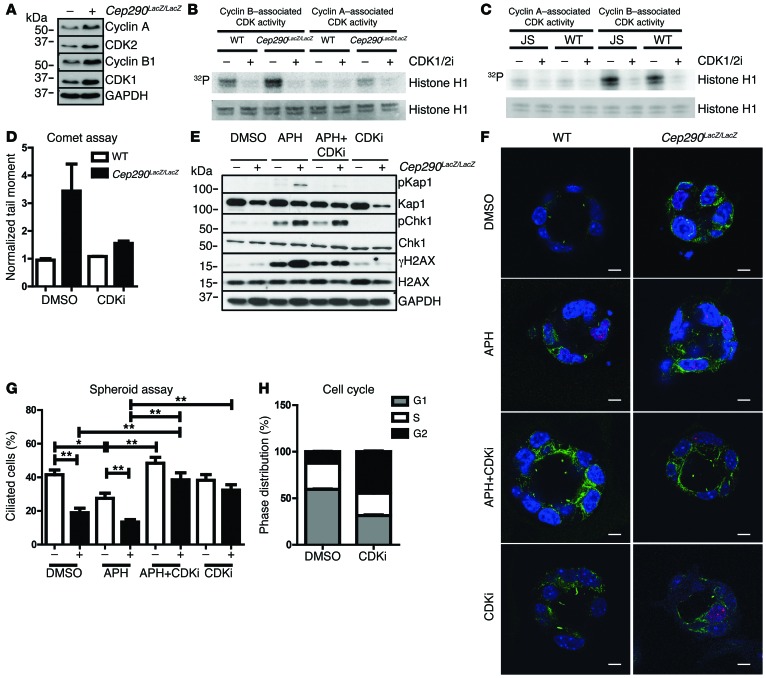
The DNA fiber results suggest that CEP290 has an important function during DNA replication to ensure efficient fork progression and stability. It is known that elevated cyclin A–associated CDK1/2 activity can cause similar defects on replication fork dynamics. Furthermore, we have previously shown that partial inhibition of CDK1/2 activity can largely suppress the DNA damage resulting from NEK8 loss (16). We therefore hypothesized that enhanced CDK1/2 activity in Cep290LacZ/LacZ primary kidney cells could be responsible for the increased DNA damage signaling and DNA breaks during disturbed DNA replication (23, 24). Indeed, total protein levels of CDK1 and CDK2 as well as cyclin A and cyclin B levels were higher in Cep290LacZ/LacZ cells (Figure 5A). In vitro CDK1 and CDK2 kinase assays revealed a trend toward enhanced cyclin A–associated CDK activity (+26%) and cyclin B–associated CDK activity (+7%) in Cep290LacZ/LacZ cells (n = 4; Figure 5B and Supplemental Figure 4A), although the intraexperimental variation was high. In addition, the single kinase assay of WT URECs and URECs from a patient with JS revealed modestly increased cyclin A–associated CDK activity (+7%) and cyclin B–associated CDK activity (+14%) as well (n = 1; Figure 5C and Supplemental Figure 4B). We followed up by testing whether the increase in DNA damage signaling (using phosphorylated KAP1, γH2AX, and phosphorylated CHK1 as markers; n = 3) and the increase in DNA breaks, as measured by neutral comet assays (n = 2), could be rescued in Cep290LacZ/LacZ cells by treatment with a CDK1/2 inhibitor (CDK1/2i) (200 nM, 18 hours; Figure 5, D and E). Both phenotypes could be rescued by CDK1/2 inhibition. To investigate whether reduced cilia frequency could be similarly rescued by CDK1/2 inhibition, we used a 3D spheroid culture assay and compared Cep290LacZ/LacZ mouse primary kidney cells to WT littermate cells. We observed a rescue of ciliation by CDK1/2 inhibition (n = 2; P < 0.05 and P < 0.01; Figure 5, F and G). Because the effects of CDK1/2 inhibition on the cell cycle might bias our ciliary frequency data, we measured cell cycle distribution; after CDK1/2 inhibition the G1 cell population decreased from 60% to 31%, whereas the G2 cell population increased from 12% to 45% (200 nM, 18 hours, n = 3; Figure 5H). This control experiment indicates that the gain of cilia was not caused by accumulation of cells in G1. Likewise, we wanted to exclude the idea that rescue of the DNA damage and cilia phenotypes by CDK1/2 inhibition was caused by apoptosis. FACS revealed no enhanced apoptosis after CDK1/2 inhibition (18 hours, 200 nM; n = 3; Supplemental Figure 3C). Furthermore, centriole numbers in Cep290LacZ/LacZ mouse primary kidney cells were normalized after treatment with CDK1/2i (18 hours, 200 nM; Supplemental Figure 3B).
Figure 5. CDK1/2i rescues DNA damage and primary cilia.
(A) Western blot of Cep290LacZ/LacZ and WT cell lysates showing increased levels of CDK1, CKD2, cyclin A, and cyclin B1 in CEP290-depleted cells. GAPDH was used as loading control; samples were run on parallel gels contemporaneously (B) In vitro CDK kinase assay of Cep290LacZ/LacZ and WT cell lysates after precipitation of cyclin A–CDK and cyclin B1–CDK complexes (n = 4). (C) CDK kinase assay of WT URECs and URECs from a patient with JS (n = 1). Histone 1 represents substrate level control. (D) DNA breaks are rescued by CDKi in comet tail moments of cells treated with 200 nM CDK1/2i (18 hours) normalized to control DMSO-treated WT cells (n = 3; 50 cells scored per condition). (E) Western blot of Cep290LacZ/LacZ and WT cells treated with 400 nM APH and/or 200 nM CDKi (18 hours). Higher phosphorylation levels of KAP1, H2AX, and CHK1 in Cep290LacZ/LacZ cells with or without APH treatment, rescued by CDK1/2i. Unphosphorylated protein and GAPDH are loading controls; samples were run on parallel gels contemporaneously. (F) 3D spheroids of WT and Cep290LacZ/LacZ cells, stained for γH2AX (red) and ciliation (acetylated tubulin; green), treated with 400 nM APH and/or 200 nM CDKi (18 hours). Scale bars: 5 μm. (G) CDKi rescues ciliation in Cep290LacZ/LacZ and APH-treated spheroids. (35 spheroids scored per condition) (n = 3; data shown for single experiment; 3-way ANOVA, *P < 0.05, **P < 0.01). (H) Cell cycle distribution of WT cells treated with DMSO or 200 nM CDKi (18 hours) reveals increased cell number in G2 (black) and decreased cell number in G1 (gray) after CDKi treatment (10,000 events measured; n = 3).
Discussion
CEP290 loss causes enhanced DNA damage signaling, DNA breaks, replication stress, and supernumerary centrioles. These data describe the first function to our knowledge for CEP290 in the nucleus, in which the protein cofractionates with chromatin. We show that CEP290 levels increase after DNA damage. Our data indicate a causal relationship of enhanced DDR to a broad clinical range of CEP290-related ciliopathies. Accumulation of DNA damage can be attributed to disturbed replication forks. These insights into the pathogenesis of CEP290 loss are reminiscent of the molecular functions of another ciliopathy protein, NEK8 (16). Furthermore, increased DNA damage signaling was seen in CEP164-, ZNF423-, and SDCCAG8-associated NPHP (14, 15) and hints at a general disease mechanism. It is interesting to note that CEP290 loss affects centriole duplication outside the nucleus. The centriolar satellite complex, including CEP290, has been previously reported to function in maintaining genome stability (7, 8). A systematic analysis of all ciliopathy genes is required to confirm the breadth and depth of replication stress and centriole duplication defects. Furthermore, we demonstrate the use of disease modeling by siRNA-mediated gene depletion in URECs. Potential interventions as well as modeling molecular/cellular temporal-spatial events leading to the pathophysiology of renal ciliopathies from URECs will help tailor personalized treatments.
It is interesting to note that both NEK8- and CEP290-depleted cells demonstrate strikingly similar sensitivity to replication stress, decreased fork velocity, and fork asymmetry (16). In Nek8–/– cells, enhanced cyclin A–associated CDK activity was observed and cyclin B–associated CDK activity was unchanged (16). Both Nek8–/– and Cep290LacZ/LacZ phenotypes are equally rescued by the pan CDK1/2i, suggesting that the pathophysiology is conserved.
We believe that the functional consequences of CEP290 depletion beyond cilia and ciliary signaling are novel. The hallmarks of the ciliopathy-associated NPHP are corticomedullary cyst formation and interstitial fibrosis. The renal ciliopathy field has been dominated by studies of ciliary dysfunction and renal cysts; however, the more clinically relevant aspect of renal fibrosis has been largely ignored. We have obtained functional insight into the NPHP-RC pathogenesis and investigated the effect of CDK1/2i on loss of function of CEP290. Use of CDKi as a potential treatment strategy has already shown some success in vivo (25, 26), and our data confirm that changes in CDK protein levels may be widespread in patients with NPHP, justifying this approach. We would like to extend this knowledge to other forms of chronic kidney disease (27) and renal fibrosis, which are associated with DNA damage signaling.
Methods
Cell culture.
mIMCD3 and hTERT-RPE cells (ATCC) were grown in DMEM/F12 (1:1) medium with 10% FCS, 2 mM l-glutamine, and penicillin/streptomycin in 5% CO2 at 37°C. Cep290LacZ/LacZ H-2Kb-tsA58+/− and Cep290+/+ H-2Kb-tsA58+/− collecting duct cells were isolated from kidneys of 1-month-old transgenic mice (19). Cells were incubated at 33°C in 5% CO2. CCD media was used for cell culture as previously described (28). URECs were obtained from a patient with JS and a healthy gender- and age-matched control and from additional healthy individuals. URECs were derived as previously described (19, 29). All cells were tested for mycoplasma every 2 weeks and were negative.
Antibodies and reagents.
Antibodies to phospho S139-H2AX and phospho S345-CHK1 (Cell signaling, 9718S and 2348L, respectively); phospho S824-KAP1 (Bethyl Laboratory, A300-767A); CHK1 (G-4), cyclin A (C-19), cyclin B1 (H433), CDK1 (Cdc2 p34; ref. 17), CDK2 (D-12), and PCNA (Santa Cruz Biotechnologies, 8408, 596, 752, 54, 6248, and 56, respectively); KAP1 (Transduction Lab, K57620); pericentrin, H2AX, and GAPDH (Abcam, Ab4448, Ab11175, and Ab8245, respectively); α-tubulin, acetylated α-tubulin, and β-actin (Sigma-Aldrich, T6199, T6793, and A5441, respectively); actin (C4, ImmunO, 691001); CEP290 (Novus Biologicals, NB100-86991) are all commercially available. The ON-TARGETplus siRNA SMARTpools (Thermo Scientific Dharmacon) for nontargeting pool UGGUUUACAUGUCGACUAA/UGGUUUACAUGUUGUGUGA/UGGUUUACAUGUUUUCUGA/UGGUUUACAUGUUUUCCUA (D-001810-10) and the siRNAs targeting human CEP290, GGAUUCGGAUGAAAUGAAA/GGAAUUGACUUACCUGAUG/AAAGUUAAUGAGCAAUUG/GAAGUAGAGUCCCUCAGAA (L-014590-00), and mouse Cep290, GCCAAUAAUCAGAUGGAUA/AGACUUACCCGAUGGGAUA/CGUCCAAACUCCAGAAGAU/ACUUAGAGCUGGUGAACGA (L-052034-00), were purchased from ThermoFisher. All cell transfections were performed using 20 nM siRNA with Lipofectamine RNAimax (Invitrogen, 13778-150) following the manufacturer’s protocols. The CDK1/2i (217714) was purchased from Millipore, and APH was purchased from Serva (13696).
Immunofluorescence.
For immunostaining, cells were grown on coverslips and fixed for 15 minutes in 4 % paraformaldehyde or 5 minutes in ice-cold methanol, followed by a 15-minute permeabilization step in 0.1% Triton X-100/1% BSA/PBS. Primary antibody incubations were performed overnight at 4°C in 1% BSA/PBS blocking buffer. Alexa Fluor 488, 568, and 647 secondary antibodies (Invitrogen, dilution 1:500) and DAPI incubations were performed for 1 hour at room temperature. Coverslips were mounted in Fluormount G (Cell Lab, Beckman Coulter). Confocal imaging was performed using a Zeiss confocal laser microscope, and images were processed with the ZEN 2011 software. Mean γH2AX fluorescence intensity per nucleus was quantified using ImageJ. Approximately 150 events per condition were scored.
Spheroids.
Ten thousand collecting duct cells from Cep290LacZ/LacZ or Cep290+/+ mice were mixed 1:1 with growth factor–depleted Matrigel (BD Biosciences) in Labtech chamber slides. After polymerization for 30 minutes at 33°C, warm medium was dripped over the matrix until just covered, and cells were cultured at 33°C, 5% CO2. The cells formed spheroids with cleared lumens 3 to 4 days later (19). During the last 18 hours, spheroids were cultured with medium without serum containing 400 nM APH, with or without 200 nM CDK1/2i for 18 hours. Medium was removed by pipetting, and the gels were washed 3 times for 10 minutes with warm PBS supplemented with calcium and magnesium. General fixation and staining protocols were used (30). Images and z-stacks were taken with a Zeiss LSM700 confocal microscope, and approximately 35 spheroids per condition were scored. Three-way ANOVA with Bonferroni post-hoc test was used for statistical analysis.
Western blot.
Protein lysates were prepared using RIPA lysis buffer and sonicated. To correct for protein content, BCA protein assay (Pierce) was performed. After blotting, the PVDF membranes were blocked in 5% BSA or 5% dried skim milk in TBS with 0.5% Tween. The primary antibodies were incubated overnight at 4°C. Coomassie blue staining of the gel was performed as additional loading control. The secondary HRP-conjugated swine anti-rabbit and rabbit anti-mouse antibodies (DAKO, dilution 1:2,000) were incubated for 1 hour at room temperature. The ECL Chemiluminescent Peroxidase Substrate Kit (Sigma-Aldrich, CPS1120-1KT) was used for development. Scans of the blots were made with the Bio-Rad ChemiDoc XRS+ device, and Image Lab software 4.0 was used for quantification.
Comet tail assay.
Comet assays were performed with the Electrophoresis Kit (Trevigen) according to the manufacturer’s protocols. DNA was stained with SYBR Gold (Invitrogen, S11494, 1:30,000), and comet tail moments were calculated by counting 50 cells for each sample and analyzed with CometScore software (TriTek Corp).
In vitro kinase assay.
Cells were lysed in high-salt buffer (300 mM NaCl, 50 mM HEPES, pH 7.5, 0.8% Triton X-100, 8% glycerol, 1 mM EDTA, 1 mM DTT, protease inhibitors, and phosphatase inhibitors). Cell lysates were clarified by centrifugation and were immunoprecipitated with cyclin A (Santa Cruz Biotechnologies) or cyclin B1 (Santa Cruz Biotechnologies) antibody-bound protein A-Sepharose beads (GE Healthcare) for 2 hours at 4°C. Precipitates were washed 3 times with high-salt buffer and once with kinase buffer (20 mM HEPES, pH 7.5, 5 mM MnCl2, 5 mM MgCl2, 5 mM β-glycerophosphate, 1 mM NaF, 1 mM DTT, 1 mM Na3VO4). Kinase reactions were carried out by incubating immunoprecipitated cyclin A/B1 with 50 μl of kinase buffer with 10 μM ATP, 5 μM γ32P-ATP, and 5 μg of substrate histone H1 for 30 minutes at 30°C with and without 2 μM CDKi and stopped by adding SDS sample buffer. Samples were boiled in Leammli buffer and analyzed by SDS-PAGE. Ponceau S or Coomassie blue staining was used to validate equal substrate loading.
Chromatin fractionation.
Chromatin fractionation was performed as previously described (16, 31). Approximately 107 cells were used, and fractions were resuspended in Laemmli sample buffer and analyzed by Western blotting. The chromatin fraction was sheared by sonication. Equal loading of fractions was verified by Ponceau S staining and immunoblotting against α-tubulin and GAPDH (cytosolic fraction), PCNA (nuclear and chromatin fractions), and H2AX (nuclear fraction).
Clonogenic survival.
Cep290LacZ/LacZ and Cep290+/+ cells were treated with APH at the concentrations indicated in figure legends for 18 hours. Subsequently, cells were seeded onto 6-cm plates at a density of 1,000 cells. After 11 days, cells were fixed in 1% formaldehyde in PBS, stained with 0.5% Crystal Violet (Sigma-Aldrich) for 10 minutes, and rinsed with tap water. Resulting colonies were manually counted. Each each point represents three experimental replicates.
DNA fiber analysis.
Fiber analysis was performed, and data were processed, as previously described (22). Briefly, Cep290LacZ/LacZ and WT cells were pulsed labeled with IdU (Sigma-Aldrich) for 30 minutes, quickly washed with PBS, and pulse labeled with CldU (Sigma-Aldrich) for 30 minutes. DNA fibers were visualized using primary antibodies specific for IdU and CldU (BD Biosciences [347580, 1:50] and Abcam [Ab6326, 1:100], respectively) and stained with Alexa Fluor 488– or 594–conjugated (1:500) secondary antibodies (Molecular Probes). The DNA fibers were captured with fluorescence microscopy using a 40x objective with a QICAM 12bits CCD camera, such that 1 pixel was equivalent to 4.65 μm. ImageJ was used for pulse labeling analysis.
RT-qPCR.
Cells were lysed, and total RNA was isolated (RNeasy Mini Kit, Qiagen, 74106) and measured (NanoDrop Spectrophotometer ND-1000, Thermo Fischer Scientific Inc.). cDNA was synthesized from a 1,000-ng RNA template using the iScript cDNA Synthesis Kit (Bio-Rad, 170-8891) according to the supplier’s protocol. Dilutions were made for RT-qPCR analysis to determine mRNA expression levels, which were normalized against a reference gene. The iQ SYBR Green Supermix (Bio-Rad, 170-8880) was used to multiply and measure the cDNA with a CFX96 Touch Real-Time PCR Detection System (Bio-Rad). All samples were run in triplicate in 20 μl reactions. The following PCR program was used: 95°C for 3 minutes; followed by 40 cycles of 10 seconds at 95°C, 30 seconds at the annealing temperature indicated with the primer sequence, and 30 seconds at 72°C; and then 10 seconds at 95°C, followed by a melt of the product from 65°C to 95°C. The primer sequences (Sigma-Aldrich) used and concomitant annealing temperatures are as follows: RPLP0 forward 5′-TGCACAATGGCAGCATCTAC, RPLP0 reverse 5′-ATCCGTCTCCACAGACAAGG, 58°C, CEp290 forward 5′-TGACTGCTAAGTACAGGGACATCTTG, CEP290 reverse 5′-AGGAGATGTTTTCACACTCCAGGT, 65°C, Rpl27 forward 5′-CGCCCTCCTTTCCTTTCTGC, Rpl27 reverse 5′-GGTGCCATCGTCAATGTTCTTC, 53°C, Cep290 forward 5′-GACAGTGACTACCGATCACAGT, Cep290 reverse 5′-CGCTTTCATCCTGTTGTACTCA, 64°C. The ΔΔCT method was used for statistical analysis to determine gene expression levels.
Histology.
Immunocytochemistry was performed as previously described (27). Mouse kidney tissue sections (n = 13) embedded in paraffin were deparaffinized, treated with peroxidase block for 15 minutes, and incubated at 100°C in citrate-HCl buffer (pH 6) for 20 minutes. The sections were stained with rabbit antibody to γH2AX (1:200) overnight at 4°C. Samples were incubated with BrightVision Anti-Rabbit Poly HRP for 1 hour at room temperature. The Nova RED Substrate Kit for Peroxidase (Vector, SK-4800) was used, and samples were counterstained with hematoxylin. Analysis was performed using Aperio ImageScope software. Five random tubular fields (approximately 1,000 cells per field) were analyzed for positively stained nuclei using an in-house ImageJ macro. Linear model analysis was used for statistical analysis.
FACS.
To quantify cell cycle phase distribution, WT and Cep290LacZ/LacZ cells were incubated with 10 μM BrdU for 30 minutes and fixed in ice-cold 70% EtOH. To quantify S-phase progression, IMCD3 cells (48 hours after siRNA) were incubated with 10 μM BrdU for 30 minutes, cultured for 18 hours, and fixed in ice-cold 70% EtOH. Samples were stained for FACS analysis with BrdU mouse mAb Alexa Fluor 647 conjugate (1:200; Invitrogen) in 0.1% BSA-PBS-T for 1 hour on ice and stained with DAPI in PBS. To quantify apoptosis, unfixed cells were stained with 7-AAD and Hoechst as described previously (32). 10,000 events were measured with a BD FACSCanto II Flow Cytometer and analyzed using BD FACSDiva Software.
Zebrafish.
WT embryos at the 1- to 2-cell stage were injected with 3.5 ng or 4 ng of a mo oligonucleotide targeting cep290 ATG codon sequence (Gene-Tools) (1) in pure water with 0.1% phenol red using a Nanoject2000 Microinjector (World Precision Instruments). The sequence of the standard control mo was as follows: 5′-CCTCTTACCTCAGTTACAATTTATA-3′. For Western blot, 18 embryos were pooled in 36 μl Triton X-100 lysis buffer and were sonicated.
Statistics.
P values were calculated for normally distributed data sets using a 2-tailed Student’s t test, 1-way ANOVA with Dunnett’s post-hoc test, 2-way ANOVA and 3-way ANOVA with Bonferroni post-hoc tests, or as otherwise indicated. Statistical analyses represent the mean of at least 3 independent experiments or as otherwise indicated. Error bars represent SEM. P values of less than 0.05 were considered significant.
Study approval.
URECs were obtained from a 3-year-old boy with clinical JS with a retinal, renal, and cerebellar phenotype, with compound heterozygous mutations in CEP290, p.Q950Pfs*6 het, and p.K939N het. Ethical approval was obtained from the National Research Ethics Service (NRES) Committee North East (14/NE/1076). URECs were obtained from healthy controls that have been included in the Aetiologic research into Genetic and Occupational/environmental Risk factors for Anomalies in children (AGORA) biobank project. The Regional Committee on Research involving Human Subjects (Commissie Mensgebonden Onderzoek [CMO] Arnhem/Nijmegen) approved the study protocol. Written informed consent was obtained from the parents. All mouse work was performed under licenses granted from the Home Office (United Kingdom) in accordance with the guidelines and regulations for the care and use of laboratory animals outlined by the Animals (Scientific Procedures) Act 1986 and with the approval of the Newcastle University Ethical Review Committee. Mice with a Cep290LacZ/LacZ hypomorphic mutation bred on a pure 129/Ola genetic background (F1 heterozygotes were backcrossed with C57/b6 for 6 generations) were used as previously described (19). Male and female mice were sacrificed at various ages and genotyped. All zebrafish experiments were conducted in accordance with the Dutch guidelines for the care and use of laboratory animals, with the approval of the Animal Experimentation Committee (DEC) of the Royal Netherlands Academy of Arts and Sciences (KNAW) and were approved by the Animal Care Committee of the University Medical Center Utrecht in the Netherlands.
Supplementary Material
Acknowledgments
We thank the patients and the controls for their participation in this study. We thank the Flow Cytometry Core Facility and the Cell Microscopy Center at the University Medical Center Utrecht for providing expert services. We thank Ive Logister and Henry Ajzenberg for culturing urine cells, Hendrik Gremmels for help with statistics, and all Cimprich and Giles lab members for scientific input. We thank Sophie Saunier for help with the molecular genetic diagnosis of patients with JS. We acknowledge generous support from Kids Kidney Research (07/05/13; to A.M. Hynes), Kidney Research UK (RP40/2009 and TF1/2013; to J.A. Sayer, C. Miles, A.M. Hynes, and S. Srivastava), the Medical Research Council (MR/M012212/1 to J.A. Sayer and C. Miles) and Northern Counties Kidney Research Fund (07.2013; J.A. Sayer), the NIH (grant ES016486; to K.A. Cimprich), the American Cancer Society Postdoctoral Fellowship (123391-PF-12-156-01-DMC; to A.C. Kile), the Postdoctoral Enrichment Program Award from the Burroughs Wellcome Fund and the NIH (grant T32CA09151; to J.C. Saldivar), the European Community’s Seventh Framework Programme FP7/2009 (under grant agreement no. 241955, SYSCILIA), and the Dutch Kidney Foundation Consortium CP11.18 “KOUNCIL” and 13A3D103 (to R.H. Giles). G.G. Slaats further acknowledges support from Foundation “De Drie Lichten” in the Netherlands (14/13), the Dutch Kidney Foundation (14OKK01), the Simons Foundation Fund (1059), and the EMBO fellowship program (554-2013). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2015;125(9):3657–3666. doi:10.1172/JCI80657.
References
- 1.Sayer JA, et al. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat Genet. 2006;38(6):674–681. doi: 10.1038/ng1786. [DOI] [PubMed] [Google Scholar]
- 2.Coppieters F, Lefever S, Leroy BP, De Baere E. CEP290, a gene with many faces: mutation overview and presentation of CEP290base. Hum Mutat. 2010;31(10):1097–1108. doi: 10.1002/humu.21337. [DOI] [PubMed] [Google Scholar]
- 3.Valente EM, Brancati F, Boltshauser E, Dallapiccola B. Clinical utility gene card for: Joubert syndrome — update 2013. Eur J Hum Genet. 2013;21(10) doi: 10.1038/ejhg.2013.10. ). doi: 10.1038/ejhg.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halbritter J, et al. High-throughput mutation analysis in patients with a nephronophthisis-associated ciliopathy applying multiplexed barcoded array-based PCR amplification and next-generation sequencing. J Med Genet. 2012;49(12):756–767. doi: 10.1136/jmedgenet-2012-100973. [DOI] [PubMed] [Google Scholar]
- 5.Hildebrandt F, Attanasio M, Otto E. Nephronophthisis: disease mechanisms of a ciliopathy. J Am Soc Nephrol. 2009;20(1):23–35. doi: 10.1681/ASN.2008050456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moradi P, Davies WL, Mackay DS, Cheetham ME, Moore AT. Focus on molecules: centrosomal protein 290 (CEP290) Exp Eye Res. 2011;92(5):316–317. doi: 10.1016/j.exer.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 7.Staples CJ, et al. The centriolar satellite protein Cep131 is important for genome stability. J Cell Sci. 2012;125(pt 20):4770–4779. doi: 10.1242/jcs.104059. [DOI] [PubMed] [Google Scholar]
- 8.Staples CJ, et al. Ccdc13 is a novel human centriolar satellite protein required for ciliogenesis and genome stability. J Cell Sci. 2014;127(pt 13):2910–2919. doi: 10.1242/jcs.147785. [DOI] [PubMed] [Google Scholar]
- 9.Valdes-Sanchez L, et al. ATR localizes to the photoreceptor connecting cilium and deficiency leads to severe photoreceptor degeneration in mice. Hum Mol Genet. 2013;22(8):1507–1515. doi: 10.1093/hmg/dds563. [DOI] [PubMed] [Google Scholar]
- 10.Basten SG, Giles RH. Functional aspects of primary cilia in signaling, cell cycle and tumorigenesis. Cilia. 2013;2(1):6. doi: 10.1186/2046-2530-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sang L, et al. Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell. 2011;145(4):513–528. doi: 10.1016/j.cell.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolf MT, Hildebrandt F. Nephronophthisis. Pediatr Nephrol. 2011;26(2):181–194. doi: 10.1007/s00467-010-1585-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Omran H. NPHP proteins: gatekeepers of the ciliary compartment. J Cell Biol. 2010;190(5):715–717. doi: 10.1083/jcb.201008080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaki M, et al. Exome capture reveals ZNF423 and CEP164 mutations, linking renal ciliopathies to DNA damage response signaling. Cell. 2012;150(3):533–548. doi: 10.1016/j.cell.2012.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Airik R, et al. Renal-retinal ciliopathy gene Sdccag8 regulates DNA damage response signaling. J Am Soc Nephrol. 2014;25(11):2573–2583. doi: 10.1681/ASN.2013050565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi HJ, et al. NEK8 links the ATR-regulated replication stress response and S phase CDK activity to renal ciliopathies. Mol Cell. 2013;51(4):423–439. doi: 10.1016/j.molcel.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otto EA, Trapp ML, Schultheiss UT, Helou J, Quarmby LM, Hildebrandt F. NEK8 mutations affect ciliary and centrosomal localization and may cause nephronophthisis. J Am Soc Nephrol. 2008;19(3):587–592. doi: 10.1681/ASN.2007040490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slaats GG, Giles RH. Are renal ciliopathies (replication) stressed out? Trends Cell Biol. 2015;25(6):317–319. doi: 10.1016/j.tcb.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 19.Hynes AM, et al. Murine Joubert syndrome reveals Hedgehog signaling defects as a potential therapeutic target for nephronophthisis. Proc Natl Acad Sci U S A. 2014;111(27):9893–9898. doi: 10.1073/pnas.1322373111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baranovskiy AG, Babayeva ND, Suwa Y, Gu J, Pavlov YI, Tahirov TH. Structural basis for inhibition of DNA replication by aphidicolin. Nucleic Acids Res. 2014;42(22):14013–14021. doi: 10.1093/nar/gku1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16(1):2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140(6):1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanchez Y, et al. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science. 1997;277(5331):1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- 24.Beck H, et al. Regulators of cyclin-dependent kinases are crucial for maintaining genome integrity in S phase. J Cell Biol. 2010;188(5):629–638. doi: 10.1083/jcb.200905059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bukanov NO, et al. CDK inhibitors R-roscovitine and S-CR8 effectively block renal and hepatic cystogenesis in an orthologous model of ADPKD. Cell Cycle. 2012;11(21):4040–4046. doi: 10.4161/cc.22375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bukanov NO, Smith LA, Klinger KW, Ledbetter SR, Ibraghimov-Beskrovnaya O. Long-lasting arrest of murine polycystic kidney disease with CDK inhibitor roscovitine. Nature. 2006;444(7121):949–952. doi: 10.1038/nature05348. [DOI] [PubMed] [Google Scholar]
- 27.Zhou W, et al. FAN1 mutations cause karyomegalic interstitial nephritis, linking chronic kidney failure to defective DNA damage repair. Nat Genet. 2012;44(8):910–915. doi: 10.1038/ng.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bens M, et al. Corticosteroid-dependent sodium transport in a novel immortalized mouse collecting duct principal cell line. J Am Soc Nephrol. 1999;10(5):923–934. doi: 10.1681/ASN.V105923. [DOI] [PubMed] [Google Scholar]
- 29.Zhou T, et al. Generation of human induced pluripotent stem cells from urine samples. Nat Protoc. 2012;7(12):2080–2089. doi: 10.1038/nprot.2012.115. [DOI] [PubMed] [Google Scholar]
- 30.Giles RH, Ajzenberg H, Jackson PK. 3D spheroid model of mIMCD3 cells for studying ciliopathies and renal epithelial disorders. Nat Protoc. 2014;9(12):2725–2731. doi: 10.1038/nprot.2014.181. [DOI] [PubMed] [Google Scholar]
- 31.Smits VA, Reaper PM, Jackson SP. Rapid PIKK-dependent release of Chk1 from chromatin promotes the DNA-damage checkpoint response. Curr Biol. 2006;16(2):150–159. doi: 10.1016/j.cub.2005.11.066. [DOI] [PubMed] [Google Scholar]
- 32.Schmid I, Uittenbogaart C, Jamieson BD. Live-cell assay for detection of apoptosis by dual-laser flow cytometry using Hoechst 33342 and 7-amino-actinomycin D. Nat Protoc. 2007;2(1):187–190. doi: 10.1038/nprot.2006.458. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.