Abstract
It is now well established that the immune system can recognize developing cancers and that therapeutic manipulation of immunity can induce tumor regression. The capacity to manifest remarkably durable responses in some patients has been ascribed in part to T cells that can (a) kill tumor cells directly, (b) orchestrate diverse antitumor immune responses, (c) manifest long-lasting memory, and (d) display remarkable specificity for tumor-derived proteins. This specificity stems from fundamental differences between cancer cells and their normal counterparts in that the former develop protein-altering mutations and undergo epigenetic and genetic alterations, resulting in aberrant protein expression. These events can result in formation of tumor antigens. The identification of mutated and aberrantly expressed self-tumor antigens has historically been time consuming and laborious. While mutant antigens are usually expressed in a tumor-specific manner, aberrantly expressed antigens are often shared between cancers and, therefore, in the past, have been the major focus of therapeutic cancer vaccines. However, advances in next-generation sequencing and epitope prediction now permit the rapid identification of mutant tumor neoantigens. This review focuses on a discussion of mutant tumor neoantigens and their use in personalizing cancer immunotherapies.
Introduction
The idea that tumors of nonviral origin possess unique, tumor-specific antigens (TSAs) arose from work reported in the first half of the twentieth century by several groups, particularly those led by Gross, Foley, Prehn, and Old (1–4). These studies showed that when inbred mice bearing carcinogen-induced tumors were cured of their cancers by surgical resection, they were immune to subsequent rechallenge with the same tumor cells, but not with other distinct tumor cells, even those derived in the same manner from different hosts. In the ensuing next quarter century, the roles of the MHC proteins in antigen presentation were discovered (5, 6), methods were developed to propagate antigen-specific cytolytic T lymphocytes (CTL) in culture (7, 8), and it became possible to clone and express gene products using molecular biology techniques. Together, these developments provided the elements needed for the first molecular identification of TSAs. Using mutagenized forms of preexisting P815 mastocytoma tumor cells, Boon and colleagues isolated a highly immunogenic, nontumorigenic tumor cell variant (tum–) and used CTL-based expression cloning approaches to show that a point mutation in a ubiquitously expressed protein (P91A) was responsible for the immune rejection of these tumor cells in naive, syngeneic WT mice (9). Shortly thereafter, Hans Schreiber and colleagues showed that tumor-specific mutant proteins could also function as tumor neoantigens in a set of naturally arising, highly immunogenic, primary UV–induced mouse tumors (10, 11).
During this time, independent efforts by Knuth and Old and the Rosenberg group identified T cells in the peripheral blood and tumors of melanoma patients that predominately reacted with melanoma cells but not normal cells, suggesting that human cancers also possessed tumor antigens whose expression was either tumor specific or showed limited expression in normal cells (12, 13). In 1991, Boon and colleagues used their T cell–based approach to clone the first human tumor antigen (MAGEA1) (14), then subsequently cloned a range of different human tumor antigens that included, among others, those derived from tumor-specific mutant genes, alternatively initiated proteins, normal proteins that displayed aberrant quantitative or qualitative expression in tumor cells, and proteins expressed only in germ cells and tumor cells (15). At about the same time, Sahin et al. used an autologous antibody-based cloning approach (SEREX) and also identified different types of human tumor antigens (16). Together, these pioneering efforts led to the acknowledgement that the tumor antigenome comprised both tumor-specific and tumor-associated antigens (TAAs).
Tumor antigen classification
Today we recognize three broad classifications of tumor antigens: TSAs, TAAs, and cancer-germline/cancer testis antigens (CTAs) (15, 17–19). TSAs are antigens that are not encoded in the normal host genome and may represent either oncogenic viral proteins or abnormal proteins that arise as a consequence of somatic mutations (i.e., neoantigens). During cancer initiation and progression, tumor cells acquire protein-altering mutations that are either responsible for transformation (driver mutations) or are a byproduct of the genomic instability that accompanies cellular transformation (passenger mutations) (20–23). Some of these alterations may result in expression of mutant proteins that are perceived as foreign proteins by the immune system. This class of antigens is likely to be less susceptible to mechanisms of immunological tolerance and therefore may represent more visible targets for immune-mediated tumor control (19, 24). TAAs include proteins encoded in the normal genome and may be either normal differentiation antigens or aberrantly expressed normal proteins. Overexpressed normal proteins that possess growth/survival-promoting functions, such as Wilms tumor 1 (WT1) (25) or Her2/neu (26), represent TAAs that directly participate in the oncogenic process. Posttranslational modifications of proteins such as phosphorylation may also lead to formation of TAAs (27, 28). Because TAAs are normal proteins, their antigenicity depends on abnormal expression levels or context to circumvent naturally occurring mechanisms of immunological tolerance (29, 30). Along these lines, TAAs usually have lower T cell receptor (TCR) affinity compared with TSAs or foreign antigens (31). The third category comprises CTAs, which are normally expressed in testis, fetal ovaries, and trophoblasts, but can also be expressed in cancer cells (17). Because they are encoded in the normal genome but display highly restricted tissue expression, CTAs have received considerable attention as attractive targets for immunotherapy (32).
Paving the way for TSA-based cancer immunotherapy
In 2005, two important human studies stimulated increased interest in tumor neoantigens as therapeutic targets for cancer immunotherapy. First, using expression-cloning approaches, T. Wölfel et al. showed that the naturally occurring antitumor T cell response in a melanoma patient was directed toward neoantigens formed by somatic point mutations in five distinct genes and that T cell responses against these TSAs prevailed over responses to TAAs (33). Second, Rosenberg and Robbins showed that ex vivo–expanded tumor-infiltrating lymphocytes (TILs), when adoptively transferred into a melanoma patient who subsequently underwent a complete tumor regression, contained T cells that were specific for two mutant tumor antigens (34). T cells specific for these neoantigens persisted in the blood and tumor of the patient after adoptive transfer. Together, these data provided support for the concept that T cells recognizing tumor neoantigens could indeed provide substantial therapeutic benefit to human cancer patients.
These results came at a time when the majority of efforts in the field were focused on identifying TAAs and CTAs for use in cancer immunotherapy. During this period, limited effort was expended on TSAs because their identification was so difficult and represented a case of too much effort for too little gain for too few individuals. However, by 2008, cancer genome sequencing had clearly established that all cancers express somatic mutations (35, 36). These observations led Allison and Vogelstein to conduct in silico analyses of exome-sequencing data from breast and colorectal cancers, where they found several mutations that were predicted to form tumor-specific mutant antigens for CD8+ T cells (37), but it would take four years before experimental support could be obtained to corroborate their computational predictions.
In 2012, two independent reports provided the first unequivocal evidence that genomics and bioinformatics approaches could be used to identify tumor-specific mutant proteins that function as TSAs. Using a combination of next generation sequencing, in silico epitope prediction, and immunological approaches, Sahin and colleagues (38) and our group (39) independently identified and validated distinct TSAs in murine B16-F10 melanoma tumor cells and in highly immunogenic methylcholanthrene-induced (MCA-induced) sarcoma cells, respectively. Importantly, these studies showed that the time frame needed to identify TSAs could be shortened to only a few weeks as opposed to the months required using conventional antigen-cloning approaches. Moreover, our study (39) and an independent study by Dupage and Jacks (40) showed that TSAs were also key targets of cancer immunoediting — the immunological process that not only protects against cancer development, but also sculpts the immunogenicity of cancers that form in an immunocompetent individual (41–43).
The following year, this work was extended to human cancers. Robbins and Rosenberg showed that exome sequencing could be used to identify mutated human TSAs recognized by adoptively transferred tumor-reactive T cells (44). Concomitantly, Schumacher used a combination of exome sequencing and high-throughput MHC tetramer screening to show that checkpoint blockade immunotherapy facilitated expansion of preexisting T cells specific for tumor neoantigens in a human melanoma patient (45). Checkpoint blockade therapy, originally developed by Allison and colleagues, is based on the capacity of antibodies to block inhibitory receptors (such as cytoxic T lymphocyte–associated protein 4 [CTLA-4]) on T cells or deplete inhibitory receptor–bearing T cells, thereby unleashing suppressed T cell–dependent antitumor effector functions in tumor-bearing hosts (46–48). The capacity of epitope prediction algorithms to identify human TSAs was further demonstrated in both prospective and retrospective analyses performed by Fritsch, Hacohen, and Wu (49, 50), Schumacher and colleagues (51), and Nelson et al. (52). Together, the mouse and human studies suggested that it was indeed possible to use genomics and bioinformatics approaches to rapidly identify mutant proteins expressed exclusively in cancer cells that function as tumor neoantigens.
Identifying tumor-specific mutations and predicting their capacity to function as TSAs
Whereas these initial studies revealed the power of combining genomics, bioinformatics, and immunological approaches to identify mutant TSAs, additional refinements have been made over the last few years that further improved the accuracy of the process.
Next generation sequencing.
Advances in sequencing technology have transformed our ability to decode cancer-specific mutations by coupling the sequencing reaction with detection of nucleotide incorporation events for hundreds of millions of genomic fragments in the same instrument run (53). In particular, tumor-specific or “somatic” mutations can be identified using massively parallel sequencing (MPS) (17) approaches to compare DNA isolated from tumor versus normal sources. Similar to DNA-based assays using MPS, RNA from tumors can be analyzed by conversion to cDNA and construction of a library suitable for sequencing. Since the genome is large (3 billion base pairs) and its analysis complex, the advent of hybrid capture technology has permitted investigators to focus only on the 1% of the genome that comprises the coding exons of known genes, (i.e., the “exome”) (54–56). Here, probes designed to bind the exon sequences of annotated genes are synthesized, biotinylated, and hybridized in solution with a fragmented whole genome library. The probe-bound library fragments are subsequently captured and isolated using streptavidin-coated magnetic beads. After release from the beads by denaturation, the library fragments are amplified, quantitated, and sequenced.
Exome-capture can be used in a clinical setting, but challenges include (a) obtaining information in a clinically relevant time frame, (b) the small amounts of DNA/RNA available from a core biopsy procedure, (c) tissue preservation in formalin and paraffin (formalin-fixed paraffin embedded [FFPE]), which promotes the degradation of nucleic acids via backbone crosslinking, and (d) data interpretation. Recent technical innovations have reduced the time for this approach from approximately one week to around two hours for hybrid capture. It is now feasible to generate exome-capture data and produce a list of somatic mutations in about three days. Hybrid capture also enhances the sequencing data quality obtained from tumor RNA (cDNA) sequencing, especially for low yield and/or FFPE-derived samples (57).
Detecting somatic mutations.
Mutation calling from exome-capture sequencing data is achieved by aligning sequence reads to reference genomes, which serve as the keystone for analyzing the short read lengths (~100 bp) produced by MPS platforms. Once reads are aligned to the genome, variants are identified using several algorithms to interpret different types of mutations, including point mutations (or single nucleotide variants [SNVs]) and focused insertion or deletion variants (indels). Tumor variant calls are then compared with data from a matched normal tissue DNA obtained using a similar capture reagent in order to identify tumor-unique (“somatic”) mutations. Subsequent annotation steps convert variations in nucleic acid sequence to changes in amino acid sequence, thereby providing the initial data required to identify and rank order tumor neoantigens.
One aspect of this process that needs further work is the ability to predict neoantigens arising from more “extreme” mutations. In principle, variants that add or delete an amino acid or truncate or extend open reading frames or fusion genes arising from translocations or inversions could be a source of highly antigenic novel epitopes; however, indel variants have, in the past, been difficult to detect with high certainty, even when algorithms are employed that are specifically tuned to detect this type of mutation. However, recent advances now permit the identification of some indels with a fairly high degree of certainty, although indels containing highly repetitive regions remain a difficulty. Structural variants also are difficult to identify, especially from exome-capture data, and hence are not likely to be easily detected unless RNA sequencing (RNA-Seq) data can be evaluated for fusion transcripts (which has a correlative high false-positive rate). In all cases, the use of RNA data from cDNA capture sequencing (cDNA Cap-Seq) or RNA-Seq to identify and/or confirm such variants is critically important.
Predicting tumor neoepitopes.
Currently, the most useful epitope prediction algorithms are those that focus on binding of peptides to MHC class I (MHCI) molecules. In both humans and mice, the MHCI antigen presentation pathway is responsible for presenting peptides derived from endogenous cell-intrinsic proteins to CD8+ CTL (58). Endogenous proteins are processed by the proteasome (59) and the resulting 8–11 amino acid peptides transported into the ER by the transporter associated with antigen processing (TAP) (60), where they are loaded onto newly synthesized class I molecules and the stabilized peptide–MHCI (p–MHCI) complexes are transported to the cell surface. MHCI alleles are remarkably polymorphic, and the number of potential self and foreign peptides processed by normal, infected, or transformed cells is very large (61). There are nearly 2,500 human MHCI allelic sequences and, because human cells can express as many as six distinct MHCI alleles, the capacity to accurately predict which tumor-derived mutant peptide will bind a particular MHCI is challenging.
Multiple tools exist to predict peptide binding to MHCI. A comprehensive list of prediction tools is available (http://cancerimmunity.org/resources/webtools/), and the bioinformatics and biochemical aspects of these programs have been extensively reviewed elsewhere (62–64). Whereas SYFPEITHI (65), Rankpep (66), and BIMAS (67) were the first such tools to be developed, more accurate prediction algorithms have now come on line, and some have been incorporated into the Immune Epitope Database and Analysis Resource (IEDB) (68). A subset of these algorithms predicts peptide binding to different MHCI variants based on artificial neural networks (ANN), providing predicted IC50 as an output (69). In this category, NetMHC (70) is one of the most commonly used and best validated prediction programs (71, 72). Neural network–based approaches depend on the quality and size of the training set and therefore are likely to be more accurate for the more common alleles. A modified form of NetMHC, NetMHCpan (73, 74), expands the training set by including data from other species, leading to improved accuracy for rare MHC alleles. SMM (75) and SMMPMBEC (76) are examples of a second subset of prediction algorithms that use position-weight matrices to describe statistical preferences from p–MHCI binding data. This approach suppresses noise caused by both experimental error and a limited number of data points present in the training set.
Neoepitope prioritization.
For identifying tumor-derived mutant epitopes, most studies use predicted p–MHCI binding affinity as the primary criterion for generating an initial prioritized list of candidate epitopes. Most of the reported studies indicate that natural immune responses to tumor neoantigens are selectively directed to epitopes within a group predicted to have the strongest MHCI binding affinities (44, 77, 78). Peptide/MHCI binding is influenced by two additional parameters — epitope abundance and antigen processing (i.e., protein degradation and peptide transport). Although mass spectrometry could potentially provide information on epitope abundance and is rapidly becoming more sensitive, the current sensitivity and requirement for large numbers of tumor cells remain as important impediments in employing this approach to identifying the entire peptidome expressed on MHCI (79). Therefore, epitope abundance is currently estimated indirectly by quantitating RNA expression levels. In one approach, mutations defined by tumor-to-normal DNA comparisons are subjected to bioinformatic analysis to predict their immunogenicity and the levels of candidate immune stimulatory peptides are estimated by RNA-Seq. RNA evaluation provides information regarding (a) whether the variant is expressed in the RNA and (b) the mutant allele’s expression level relative to other genes. In a second approach, cDNA capture is performed from tumor RNA and compared with normal DNA to provide a list of mutated peptides. Due to the error rate of reverse transcriptase and sources of false positivity in variant calling in RNA versus DNA, the predicted mutations are refined further by applying a series of filters to remove known sources of false-positive variants (e.g., low coverage in tumor or normal, or low numbers of variant-containing reads) and to eliminate nonexpressed genes and/or alleles. A final filter eliminates genes with low expression that have a fragment per kilobase of million mapped reads (FPKM) of less than one. Downstream steps of immunogenicity prediction can then be made for this filtered set of mutant peptides. Additional algorithms exist to refine epitope predictions including those focused on defining proteasomal cleavage (i.e., NetChop) (80) and/or TAP transport (i.e., NetCTL and NetCTLpan) (81, 82). However, because the latter are currently not as reliable as the MHC-affinity prediction algorithms, they should be used with caution.
In contrast to predicting MHCI epitopes, the accuracy of predicting MHCII epitopes has been problematic. Whereas the MHCI binding groove is closed at both ends, MHCII has a peptide-binding groove that is open, leading to considerable variation in both the length of peptides that can bind to MHCII and the location of the binding core. Multiple MHCII binding prediction algorithms are available (such as TEPITOPE, ref. 83; netMHCII, ref. 84; and SMM-align, ref. 85); however, they have not been extensively used in the past to identify tumor-specific mutant MHCII epitopes.
Recent preclinical studies illustrating the use of personalized TSAs in immunotherapy
MHCI neoepitopes.
Two recent preclinical studies demonstrated the feasibility of developing personalized neoantigen-based cancer immunotherapies. Using a combination of mass spectrometry, next generation sequencing, and bioinformatics approaches, Yadav, Lill, Delamarre, and colleagues identified seven mutant neoepitopes bound to MHCI expressed on MC38 mouse colon adenocarcinoma cells (78). Two of the stronger H-2Db–binding epitopes, derived from point mutations of Reps1 and Adpgk, induced antigen-specific CD8+ T cell responses in naive WT syngeneic mice when administered together with polyinosinic:polycytidylic acid (poly I:C) and agonistic anti-CD40. One H-2Kb epitope, derived from a point mutation of Dpagt1, also induced a weak T cell response. Therapeutic vaccines comprising synthetic long peptides (SLP) containing the three positive epitopes provided mice bearing established MC38 tumors with a significant degree of therapeutic protection.
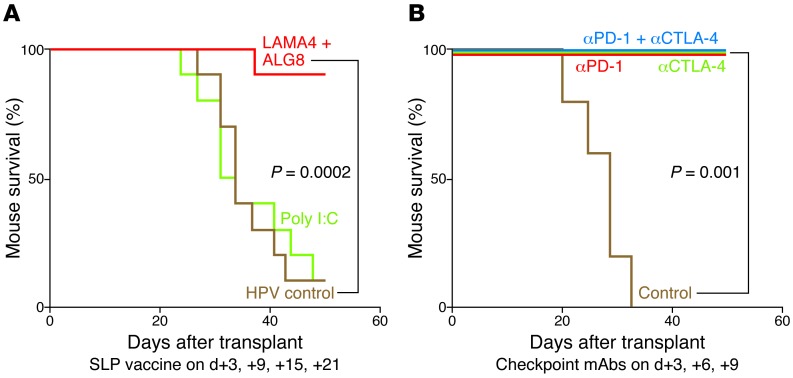
An independent study published concomitantly by our group took the approach of determining the tumor antigen specificities of CD8+ T cells responsible for rejecting tumors in mice undergoing checkpoint blockade immunotherapy (77). In this study, we used an edited mouse MCA sarcoma line (T3) that formed progressively growing tumors in naive syngeneic WT mice, which were rejected upon treatment with mAbs specific for two different immune checkpoints — CTLA-4 and programmed death-1 (PD-1) (Figure 1). As assessed by cDNA–Cap-Seq and a recent refinement to calling mutations, T3 expressed approximately 2,200 nonsynonymous point mutations. Putative epitopes were predicted using three MHCI binding algorithms (SMM, NetMHC, and NetMHCpan) and calculating the median binding affinities for each epitope. Additionally, we applied filters for proteasomal processing (NetChop), neoepitopes (i.e., where the mutant epitope bound to MHCI in a manner equal or greater to WT sequence), and deprioritization of hypothetical proteins. Using this approach, two MHCI H-2Kb–restricted tumor neoantigens were identified: point mutations in α-1,3-glucosyltransferase (Alg8) and laminin α subunit 4 (Lama4). These predictions were validated by the following observations. First, primary TILs from T3 tumors were specifically stained by H-2Kb tetramers carrying either ALG8 or LAMA4 epitopes (but not other predicted epitopes), accumulated in tumors in a time-dependent manner, and produced IFN-γ and/or TNF-α when stimulated with irradiated splenocytes pulsed with either ALG8 or LAMA4 (but not other predicted epitopes). Second, the same two mutant epitopes were recognized by cloned T cells from mice that had rejected T3 sarcomas following anti–PD-1 treatment. Third, ALG8 and LAMA4 were detected by mass spectrometry bound to H-2Kb on T3 sarcoma cells. Fourth, ALG8 or LAMA4 peptide vaccines induced T cell responses in naive syngeneic WT mice. Fifth, vaccines comprising ALG8/LAMA4 SLPs plus poly I:C prophylactically protected mice against subsequent challenge with T3 tumor cells and therapeutically induced rejection in mice bearing established T3 tumors. Importantly, the degree of therapeutic protection by the combined neoantigen SLP vaccine was comparable to that afforded by checkpoint blockade therapy (Figure 2). Together, these studies revealed the therapeutic potential of TSA-based personalized cancer immunotherapy.
Figure 1. Genomics-based identification of neoepitopes.
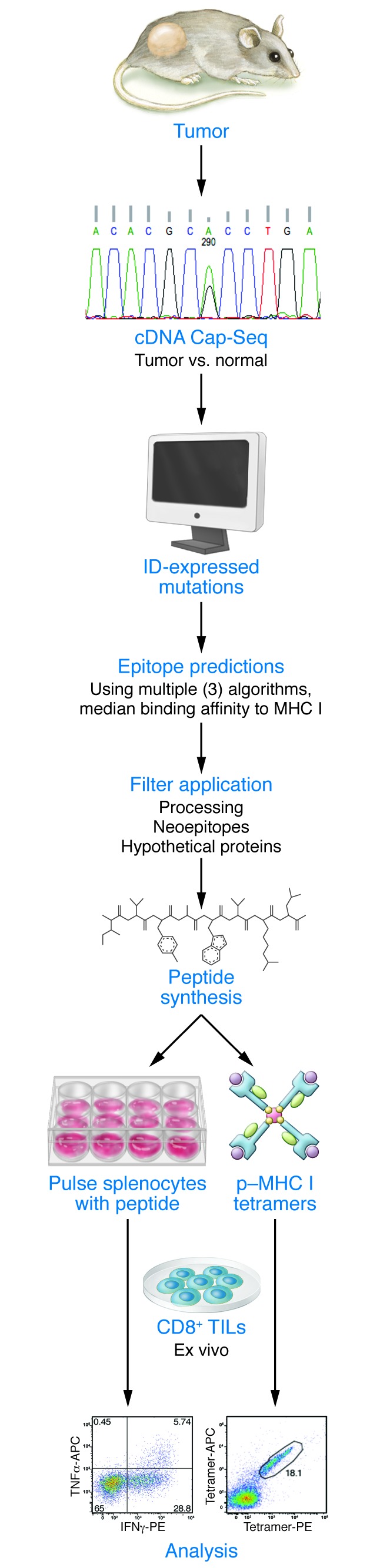
Tumor cells and normal tissue were subjected to cDNA Cap-Seq to identify expressed, nonsynonymous somatic mutations. Corresponding mutant epitopes were then analyzed in silico for MHCI binding. Filters were applied for antigen processing, neoepitopes, and deprioritization of hypothetical proteins. Peptides corresponding to predicted epitopes were then synthesized and used to identify mutant neoantigen–specific T cells in freshly explanted TIL using MHC multimer–based screens or cytokine induction by peptide stimulation.
Figure 2. Mutant neoantigen–specific peptide vaccines induce therapeutic effects comparable to those of checkpoint blockade therapy.
Kaplan-Meier survival curves of tumor-bearing mice therapeutically vaccinated with a vaccine comprising poly I:C plus either ALG8 plus LAMA4 SLP, control SLP (HPV peptide), or buffer (A) or therapeutically treated with mAbs to CTLA-4 and/or PD-1 immune checkpoints (B). Adapted with permission from Nature (ref. 77; Figure 1A and Figure 2, D and E)
Srivastava and colleagues concomitantly proposed an alternative prediction method based exclusively on the enhancement of neoepitope binding to MHCI alleles while discounting mutations that affect TCR binding, which identified unique tumor neoepitopes that provided protection in a prophylactic vaccine setting (86). Intriguingly, many of these displayed MHCI binding affinities that were orders of magnitude weaker than other validated MHCI–restricted epitopes. It remains unclear how such weak epitopes could function in a therapeutic setting, and additional work is needed to explore this issue.
MHCII neoepitopes.
In many mouse tumor models, spontaneous or therapeutically induced tumor rejection is inhibited following depletion of either CD4+ or CD8+ T cells (39, 40, 77). Moreover, the presence of tumor-specific IgG antibodies in cancer patients and tumor-bearing mice implies that CD4+ T cells play important roles in the antitumor response (87). Using humanized MHC transgenic mice, Platten and colleagues demonstrated that SLP vaccines containing a mutant MHCII epitope from isocitrate dehydrogenase 1 (IDH1; a previously identified mutation in many human glioblastoma patients) controlled outgrowth of mouse tumors engineered to express mutant human IDH1 (88). Furthermore, using exome sequencing along with a high-throughput MHCII epitope screening method that did not require prediction of MHCII epitopes, Schumacher and colleagues identified and validated mutant MHCII–restricted neoepitopes in melanomas from several patients (89). They further showed that, whereas MHCII epitopes were identified in 4 of 5 different human melanomas, they represented only 0.5% of the melanoma mutanome, thus translating into only one to two MHCII epitopes per tumor.
In a more recent study, the Sahin group vaccinated naive WT mice with synthetic 27 mer peptides containing putative MHCI epitopes they had identified in a previous study from B16-F10 melanoma cells and found that nearly 30% of these peptides induced both strong T cell responses and strong antitumor activity when injected into tumor-bearing mice (90). Surprisingly, 95% of the epitope-specific lymphocytes induced by vaccination were CD4+ rather than CD8+ T cells. A predominance of MHCII neoepitopes was also observed for CT26 (a colorectal adenocarcinoma) and 4T1 (a breast adenocarcinoma) (89). In the case of CT26, the neoepitopes were selected using bioinformatics approaches based exclusively on the use of expression level predictions and MHCII–binding capacity. RNA-based vaccines encoding mutant MHCII epitopes induced complete rejection of established, aggressively growing tumors. By applying the same predictive algorithm to human cancers, the investigators observed a high abundance of MHCII neoepitopes. The number of MHCII epitopes per tumor cell appears to be vastly different between the Schumacher and Sahin studies (89, 90). At least in part, this difference could reflect the methods used to reveal the epitopes (checkpoint blockade versus active vaccination, respectively), but could also reflect the use of paired tumor and normal samples in the former study versus use of a half-century old tumor where no corresponding normal tissue of similar age was available. Nevertheless, the two studies support the conclusion that T cell–dependent immune control of cancer is dependent on the presence of both mutant MHCII and mutant MHCI tumor neoepitopes.
Therapeutic use of tumor-specific mutant antigens in human cancer
In the past year, the first examples of TSA-based personalized cancer immunotherapies have begun to emerge. In 2014, the Rosenberg group used a novel TSA-based personalized adoptive cell therapy (ACT) method to treat a patient with metastatic cholangiocarcinoma (91). Specifically, 26 nonsynonymous mutations were identified in a lung metastasis from the patient by whole-exome sequencing, and candidate minigene constructs for each mutation were transfected into patient-derived APCs. TILs from lung metastases were screened for reactivity with transfected APCs, leading to identification of a point mutant ERBB2-interacting protein as a neoantigen recognized by CD4+ T cells within the TIL population. TILs enriched to 25% for reactivity to the neoantigen were then infused back into the patient, who showed a partial response to the first ACT treatment and showed improved responses to subsequent treatments with TIL preparations enriched to 95% of neoantigen-specific CD4+ T cells. These results not only demonstrated a therapeutic advantage to personalizing ACT therapy, but also specifically documented the therapeutic efficacy of neoantigen-specific CD4+ T cells.
Using more classical vaccine approaches, Carreno et al. recently showed that tumor neoantigens, when administered as a vaccine to three melanoma patients, can enhance both preexisting anti-tumor T cell responses and induce responses to neoepitopes that were undetectable prior to vaccination (92). Thus, these studies support the preclinical work that preceded them and show that vaccines comprising IFN-γ/CD40 ligand–activated dendritic cells pulsed with tumor neoepitopes represent yet another method to deliver personalized cancer vaccines to tumor-bearing individuals (Figure 3).
Figure 3. Neoantigen-specific T cell therapy.
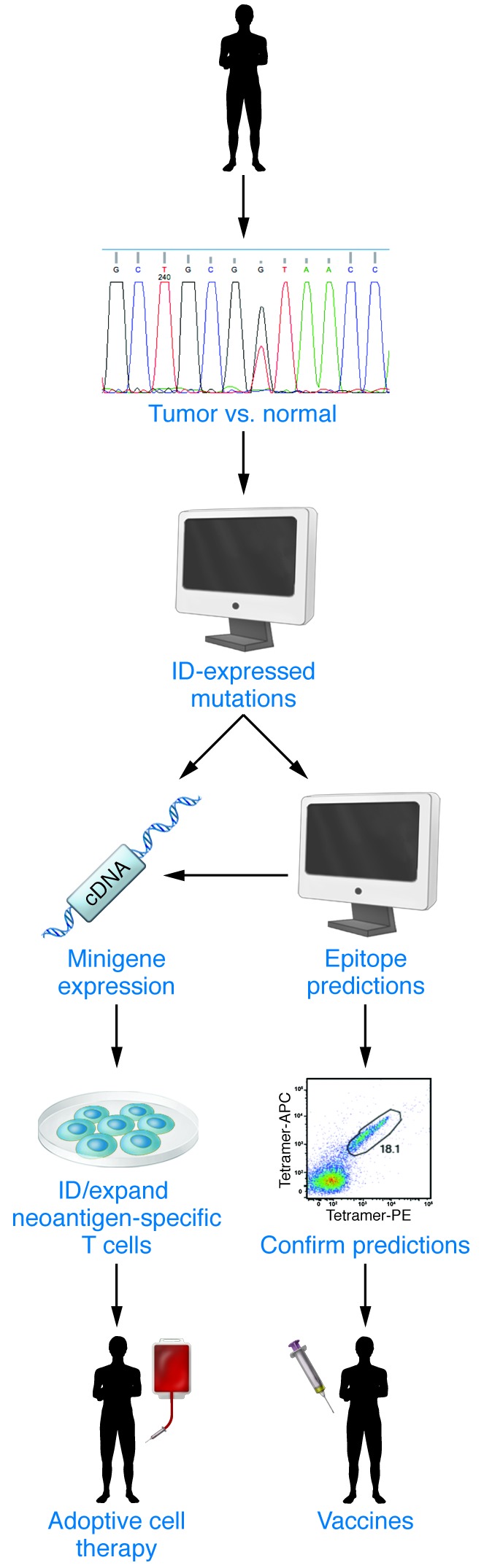
Patient tumor cells and normal tissue are subjected to whole-exome sequencing and RNA-Seq to identify expressed nonsynonymous somatic mutations. These mutations are pipelined into MHCI epitope prediction algorithms to prioritize a list of candidate antigens and/or may be expressed as minigenes used for the identification and expansion of mutant neoantigen–specific autologous T cells isolated from blood or tumor of the same patient. Ex vivo–expanded T cells are then infused back into the cancer patient. Alternatively, expressed mutations predicted to form neoantigens by MHCI epitope–binding algorithms are confirmed and then used to generate neoantigen vaccines.
The idea that the neoantigen landscape might predict clinical responses to checkpoint blockade therapy was proposed in recent work from Snyder, Wolchok, and Chan, where a mutational gene signature was inferred that correlated with long-term clinical benefit of anti–CTLA-4 (ipilimumab) therapy (93). While the mutational load alone was found to partially predict long-term clinical benefit, specific four–amino acid motifs within the NetMHC-predicted neoepitopes were claimed to be even better predictors of clinical response. However, this claim has been recently challenged because other investigators have not been able to find such motifs in validated neoepitopes. This same group together with the Schumacher group subsequently demonstrated that higher mutational burden was also associated with improved responses and progression-free survival in patients with non–small cell lung cancer treated with anti–PD-1 (pembrolizumab) (94). Response was also correlated with mutations in genes encoding components in the DNA-repair pathway that resulted in a higher number of predicted neoantigens. For the one patient in the cohort whose tumor neoantigen was defined, neoantigen-specific T cell responses paralleled tumor regression, implicating a link between the T cell responses and the antitumor effects of anti–PD-1 therapy. A recent report by Chowell et al. used bioinformatics approaches to support an argument that the hydrophobicity of TCR contact residues form a hallmark of immunogenic epitopes (95). Thus it is clear that more work is needed to clarify whether there are amino acid motifs that play important roles in determining neoantigenicity.
Concluding remarks
The last decade has seen an explosive growth in our appreciation of the role of immunity in cancer. We now know that the immune system recognizes, shapes, and/or promotes cancer development or outgrowth (22, 41–43, 96, 97). Based on our enhanced insight into immune system/cancer interactions, new immunologic approaches to treating cancer have been developed, often leading to durable clinical responses in a subset of cancer patients (47, 98–100).
Two particular characteristics have brought cancer immunotherapy to the forefront of modern molecular medicine (101). First, this therapy can be staged such that it induces complete responses or stable disease with less collateral damage than conventional chemotherapy or radiation therapy. Second, because the immune response can continuously evolve new specificities, cancer immunotherapy has the capacity to respond reactively to genetically unstable, continuously changing cancer cell target populations. However, current cancer immunotherapies still display inherent toxicities because they function within the narrow therapeutic space between tumor immunity and autoimmunity and induce responses only in a subset of patients. The use of TSAs as the basis for personalized cancer immunotherapy offers the potential to make the therapy more specific, more effective, and safer compared with the cancer immunotherapies that we have available today. However, key questions remain to be answered. Although we have achieved a level of competency in identifying point mutations and indels that can form MHCI epitopes, there remain several other potential tumor neoantigens (such as products of gene fusions, proteins arising from mistakes in translation, or mutant proteins that form MHCII antigens) that are currently not easy to predict with bioinformatics tools. We need to resolve the important questions of whether tumor-specific mutant antigens (or the closely related group of shared driver neoantigens that could reduce the chances of escape variant formation) really offer unique opportunities to target tumor cells compared with shared, nonmutated TAAs. Furthermore, we need to determine what minimum number of neoepitopes must be employed to reduce the likelihood of formation of antigen-loss variants. Finally, we need to experimentally establish that therapeutically targeting tumor neoantigens does not lead to unanticipated autoimmune consequences. Even after we resolve these issues, we still need to determine how best to use this information therapeutically. Is it through defining the best types of personalized cancer vaccines, individualized adoptive cell therapies, some combination of these together with checkpoint blockade immunotherapy, or even combinations of immunologic and standard cancer immunotherapies? Strategies targeting different aspects of the “cancer-immunity cycle” (102) (i.e., priming and/or effector immunity) with checkpoint blockade may be helpful in contextualizing the role of targeting TSAs within the multistep process of effective antitumor immunity. Will the methods that are ultimately used need to be employed in a manner that is based on the type of cancer being treated? We envisage that these types of questions will be answered in the next decade of cancer immunology, genomics, and immunotherapy research. It is clear that we now stand at an important crossroads of opportunities to enhance our ability to use the immune system to make cancer a controllable and, in some cases, curable disease. We predict that the capacity to personalize cancer immunotherapy will contribute significantly to this important endeavor.
Acknowledgments
We are grateful to K. Sheehan, G. Dunn, A. Miceli, T. Noguchi, J. Ward, and E. Alspach for constructive criticism and comments. R.D. Schreiber receives research support from the National Cancer Institute (RO1 CA043059, RO1 CA190700, U01 CA141541), the Cancer Research Institute, the WWWW Foundation, the Siteman Cancer Center/Barnes-Jewish Hospital (Cancer Frontier Fund), Bristol-Myers Squibb Inc., and Stand Up to Cancer. E.R. Mardis is supported by an NIH large-scale centers grant (U54 HG003079). M.M. Gubin is supported by a postdoctoral training grant (Irvington Postdoctoral Fellowship) from the Cancer Research Institute.
Footnotes
Conflict of interest: Robert D. Schreiber is a co-founder of Igenica Biotherapeutics and Jounce Therapeutics. He has a paid advisory relationship with Third Rock Ventures and received research support from Bristol-Myers Squibb.
Reference information:J Clin Invest. 2015;125(9):3413–3421. doi:10.1172/JCI80008.
References
- 1.Gross L. Intradermal immunization of C3H mice against a sarcoma that originated in an animal of the same line. Cancer Res. 1943;3(5):326–333. [Google Scholar]
- 2.Foley EJ. Antigenic properties of methylcholanthrene-induced tumors in mice of the strain of origin. Cancer Res. 1953;13(12):835–837. [PubMed] [Google Scholar]
- 3.Prehn RT, Main JM. Immunity to methylcholanthrene-induced sarcomas. J Natl Cancer Inst. 1957;18(6):769–778. [PubMed] [Google Scholar]
- 4.Old LJ. Cancer immunology: the search for specificity. Natl Cancer Inst Monogr. 1982;60:193–209. [PubMed] [Google Scholar]
- 5.Babbitt BP, Allen PM, Matsueda G, Haber E, Unanue ER. Binding of immunogenic peptides to Ia histocompatibility molecules. Nature. 1985;317(6035):359–361. doi: 10.1038/317359a0. [DOI] [PubMed] [Google Scholar]
- 6.Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987;329(6139):506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- 7.Cerottini JC, Engers HD, Macdonald HR, Brunner T. Generation of cytotoxic T lymphocytes in vitro. I. Response of normal and immune mouse spleen cells in mixed leukocyte cultures. J Exp Med. 1974;140(3):703–717. doi: 10.1084/jem.140.3.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gillis S, Smith KA. Long term culture of tumour-specific cytotoxic T cells. Nature. 1977;268(5616):154–156. doi: 10.1038/268154a0. [DOI] [PubMed] [Google Scholar]
- 9.De Plaen E, et al. Immunogenic (tum-) variants of mouse tumor P815: cloning of the gene of tum- antigen P91A and identification of the tum- mutation. Proc Natl Acad Sci U S A. 1988;85(7):2274–2278. doi: 10.1073/pnas.85.7.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monach PA, Meredith SC, Siegel CT, Schreiber H. A unique tumor antigen produced by a single amino acid substitution. Immunity. 1995;2(1):45–59. doi: 10.1016/1074-7613(95)90078-0. [DOI] [PubMed] [Google Scholar]
- 11.Dubey P, et al. The immunodominant antigen of an ultraviolet-induced regressor tumor is generated by a somatic point mutation in the DEAD box helicase p68. J Exp Med. 1997;185(4):695–705. doi: 10.1084/jem.185.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knuth A, Danowski B, Oettgen HF, Old LJ. T-cell-mediated cytotoxicity against autologous malignant melanoma: analysis with interleukin 2-dependent T-cell cultures. Proc Natl Acad Sci U S A. 1984;81(11):3511–3515. doi: 10.1073/pnas.81.11.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robbins PF, et al. A mutated β-catenin gene encodes a melanoma-specific antigen recognized by tumor infiltrating lymphocytes. J Exp Med. 1996;183(3):1185–1192. doi: 10.1084/jem.183.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Bruggen P, et al. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254(5038):1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- 15.Coulie PG, Van den Eynde BJ, van der Bruggen P, Boon T. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat Rev Cancer. 2014;14(2):135–146. doi: 10.1038/nrc3670. [DOI] [PubMed] [Google Scholar]
- 16.Sahin U, et al. Human neoplasms elicit multiple specific immune responses in the autologous host. Proc Natl Acad Sci U S A. 1995;92(25):11810–11813. doi: 10.1073/pnas.92.25.11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5(8):615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 18.Vigneron N, Stroobant V, Van den Eynde BJ, van der Bruggen P. Database of T cell-defined human tumor antigens: the 2013 update. Cancer Immun. 2013;13:15. [PMC free article] [PubMed] [Google Scholar]
- 19.Heemskerk B, Kvistborg P, Schumacher TN. The cancer antigenome. EMBO J. 2013;32(2):194–203. doi: 10.1038/emboj.2012.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 21.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446(7132):153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 23.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348(6230):69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 25.Ohminami H, Yasukawa M, Fujita S. HLA class I-restricted lysis of leukemia cells by a CD8(+) cytotoxic T-lymphocyte clone specific for WT1 peptide. Blood. 2000;95(1):286–293. [PubMed] [Google Scholar]
- 26.Kawashima I, Tsai V, Southwood S, Takesako K, Sette A, Celis E. Identification of HLA-A3-restricted cytotoxic T lymphocyte epitopes from carcinoembryonic antigen and HER-2/neu by primary in vitro immunization with peptide-pulsed dendritic cells. Cancer Res. 1999;59(2):431–435. [PubMed] [Google Scholar]
- 27.Doyle HA, et al. Isoaspartyl post-translational modification triggers anti-tumor T and B lymphocyte immunity. J Biol Chem. 2006;281(43):32676–32683. doi: 10.1074/jbc.M604847200. [DOI] [PubMed] [Google Scholar]
- 28.Cobbold M, et al. MHC class I-associated phosphopeptides are the targets of memory-like immunity in leukemia. Sci Transl Med. 2013;5(203):203ra125. doi: 10.1126/scitranslmed.3006061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pardoll D. Does the immune system see tumors as foreign or self? Annu Rev Immunol. 2003;21:807–839. doi: 10.1146/annurev.immunol.21.120601.141135. [DOI] [PubMed] [Google Scholar]
- 30.Hogquist KA, Baldwin TA, Jameson SC. Central tolerance: learning self-control in the thymus. Nat Rev Immunol. 2005;5(10):772–782. doi: 10.1038/nri1707. [DOI] [PubMed] [Google Scholar]
- 31.Stone JD, Harris DT, Kranz DM. TCR affinity for p/MHC formed by tumor antigens that are self-proteins: impact on efficacy and toxicity. Curr Opin Immunol. 2015;33:16–22. doi: 10.1016/j.coi.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scanlan MJ, Gure AO, Jungbluth AA, Old LJ, Chen YT. Cancer/testis antigens: an expanding family of targets for cancer immunotherapy. Immunol Rev. 2002;188:22–32. doi: 10.1034/j.1600-065X.2002.18803.x. [DOI] [PubMed] [Google Scholar]
- 33.Lennerz V, et al. The response of autologous T cells to a human melanoma is dominated by mutated neoantigens. Proc Natl Acad Sci U S A. 2005;102(44):16013–16018. doi: 10.1073/pnas.0500090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou J, Dudley ME, Rosenberg SA, Robbins PF. Persistence of multiple tumor-specific T-cell clones is associated with complete tumor regression in a melanoma patient receiving adoptive cell transfer therapy. J Immunother. 2005;28(1):53–62. doi: 10.1097/00002371-200501000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 36.Ley TJ, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456(7218):66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Segal NH, et al. Epitope landscape in breast and colorectal cancer. Cancer Res. 2008;68(3):889–892. doi: 10.1158/0008-5472.CAN-07-3095. [DOI] [PubMed] [Google Scholar]
- 38.Castle JC, et al. Exploiting the mutanome for tumor vaccination. Cancer Res. 2012;72(5):1081–1091. doi: 10.1158/0008-5472.CAN-11-3722. [DOI] [PubMed] [Google Scholar]
- 39.Matsushita H, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482(7385):400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DuPage M, Mazumdar C, Schmidt LM, Cheung AF, Jacks T. Expression of tumour-specific antigens underlies cancer immunoediting. Nature. 2012;482(7385):405–409. doi: 10.1038/nature10803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shankaran V, et al. IFNγ and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410(6832):1107–1111. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 42.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3(11):991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 43.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331(6024):1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 44.Robbins PF, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19(6):747–752. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Rooij N, et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol. 2013;31(32):e439–e442. doi: 10.1200/JCO.2012.47.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leach DR, Krummel MF, Allison JP. Enhancement of antitumor immunity by CTLA-4 blockade. Science. 1996;271(5256):1734–1736. doi: 10.1126/science.271.5256.1734. [DOI] [PubMed] [Google Scholar]
- 47.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12(4):252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharma P, Allison JP. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell. 2015;161(2):205–214. doi: 10.1016/j.cell.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fritsch EF, Rajasagi M, Ott PA, Brusic V, Hacohen N, Wu CJ. HLA-binding properties of tumor neoepitopes in humans. Cancer Immunol Res. 2014;2(6):522–529. doi: 10.1158/2326-6066.CIR-13-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rajasagi M, et al. Systematic identification of personal tumor-specific neoantigens in chronic lymphocytic leukemia. Blood. 2014;124(3):453–462. doi: 10.1182/blood-2014-04-567933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Buuren MM, Calis JJ, Schumacher TN. High sensitivity of cancer exome-based CD8 T cell neo-antigen identification. Oncoimmunology. 2014;3:e28836. doi: 10.4161/onci.28836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wick DA, et al. Surveillance of the tumor mutanome by T cells during progression from primary to recurrent ovarian cancer. Clin Cancer Res. 2014;20(5):1125–1134. doi: 10.1158/1078-0432.CCR-13-2147. [DOI] [PubMed] [Google Scholar]
- 53.Mardis ER. Next-generation sequencing platforms. Annu Rev Anal Chem (Palo Alto Calif) 2013;6:287–303. doi: 10.1146/annurev-anchem-062012-092628. [DOI] [PubMed] [Google Scholar]
- 54.Albert TJ, et al. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4(11):903–905. doi: 10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- 55.Hodges E, et al. Hybrid selection of discrete genomic intervals on custom-designed microarrays for massively parallel sequencing. Nat Protoc. 2009;4(6):960–974. doi: 10.1038/nprot.2009.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blumenstiel B, et al. Targeted exon sequencing by in-solution hybrid selection. Curr Protoc Hum Genet. 2010;Chapter 18:Unit 18.4. doi: 10.1002/0471142905.hg1804s66. [DOI] [PubMed] [Google Scholar]
- 57.Cabanski CR, et al. cDNA hybrid capture improves transcriptome analysis on low-input and archived samples. J Mol Diagn. 2014;16(4):440–451. doi: 10.1016/j.jmoldx.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blum JS, Wearsch PA, Cresswell P. Pathways of antigen processing. Annu Rev Immunol. 2013;31:443–473. doi: 10.1146/annurev-immunol-032712-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rock KL, et al. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78(5):761–771. doi: 10.1016/S0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 60.Van Kaer L, Ashton-Rickardt PG, Ploegh HL, Tonegawa S. TAP1 mutant mice are deficient in antigen presentation, surface class I molecules, and CD4-8+ T cells. Cell. 1992;71(7):1205–1214. doi: 10.1016/S0092-8674(05)80068-6. [DOI] [PubMed] [Google Scholar]
- 61.Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SG. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 2015;43(Database issue):D423–D431. doi: 10.1093/nar/gku1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lundegaard C, Lund O, Kesmir C, Brunak S, Nielsen M. Modeling the adaptive immune system: predictions and simulations. Bioinformatics. 2007;23(24):3265–3275. doi: 10.1093/bioinformatics/btm471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin HH, Ray S, Tongchusak S, Reinherz EL, Brusic V. Evaluation of MHC class I peptide binding prediction servers: applications for vaccine research. BMC Immunol. 2008;9:8. doi: 10.1186/1471-2172-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang L, Udaka K, Mamitsuka H, Zhu S. Toward more accurate pan-specific MHC-peptide binding prediction: a review of current methods and tools. Brief Bioinform. 2012;13(3):350–364. doi: 10.1093/bib/bbr060. [DOI] [PubMed] [Google Scholar]
- 65.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50(3–4):213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 66.Reche PA, Glutting JP, Reinherz EL. Prediction of MHC class I binding peptides using profile motifs. Hum Immunol. 2002;63(9):701–709. doi: 10.1016/S0198-8859(02)00432-9. [DOI] [PubMed] [Google Scholar]
- 67.Parker KC, Bednarek MA, Coligan JE. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J Immunol. 1994;152(1):163–175. [PubMed] [Google Scholar]
- 68.Vita R, et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015;43(Database issue):D405–D412. doi: 10.1093/nar/gku938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lundegaard C, Lund O, Nielsen M. Prediction of epitopes using neural network based methods. J Immunol Methods. 2011;374(1–2):26–34. doi: 10.1016/j.jim.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lundegaard C, Lamberth K, Harndahl M, Buus S, Lund O, Nielsen M. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8-11. Nucleic Acids Res. 2008;36(Web Server issue):W509–W512. doi: 10.1093/nar/gkn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Honeyman MC, Brusic V, Stone NL, Harrison LC. Neural network-based prediction of candidate T-cell epitopes. Nat Biotechnol. 1998;16(10):966–969. doi: 10.1038/nbt1098-966. [DOI] [PubMed] [Google Scholar]
- 72.Lundegaard C, Nielsen M, Lund O. The validity of predicted T-cell epitopes. Trends Biotechnol. 2006;24(12):537–538. doi: 10.1016/j.tibtech.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 73.Nielsen M, et al. NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS One. 2007;2(8):e796. doi: 10.1371/journal.pone.0000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hoof I, et al. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics. 2009;61(1):1–13. doi: 10.1007/s00251-008-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Peters B, Sette A. Generating quantitative models describing the sequence specificity of biological processes with the stabilized matrix method. BMC Bioinformatics. 2005;6:132. doi: 10.1186/1471-2105-6-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim Y, Sidney J, Pinilla C, Sette A, Peters B. Derivation of an amino acid similarity matrix for peptide: MHC binding and its application as a Bayesian prior. BMC Bioinformatics. 2009;10:394. doi: 10.1186/1471-2105-10-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gubin MM, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515(7528):577–581. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yadav M, et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature. 2014;515(7528):572–576. doi: 10.1038/nature14001. [DOI] [PubMed] [Google Scholar]
- 79.Rammensee HG, Singh-Jasuja H. HLA ligandome tumor antigen discovery for personalized vaccine approach. Expert Rev Vaccines. 2013;12(10):1211–1217. doi: 10.1586/14760584.2013.836911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nielsen M, Lundegaard C, Lund O, Kesmir C. The role of the proteasome in generating cytotoxic T-cell epitopes: insights obtained from improved predictions of proteasomal cleavage. Immunogenetics. 2005;57(1–2):33–41. doi: 10.1007/s00251-005-0781-7. [DOI] [PubMed] [Google Scholar]
- 81.Peters B, Bulik S, Tampe R, Van Endert PM, Holzhutter HG. Identifying MHC class I epitopes by predicting the TAP transport efficiency of epitope precursors. J Immunol. 2003;171(4):1741–1749. doi: 10.4049/jimmunol.171.4.1741. [DOI] [PubMed] [Google Scholar]
- 82.Larsen MV, Lundegaard C, Lamberth K, Buus S, Lund O, Nielsen M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinformatics. 2007;8:424. doi: 10.1186/1471-2105-8-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hammer J, Bono E, Gallazzi F, Belunis C, Nagy Z, Sinigaglia F. Precise prediction of major histocompatibility complex class II-peptide interaction based on peptide side chain scanning. J Exp Med. 1994;180(6):2353–2358. doi: 10.1084/jem.180.6.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nielsen M, Lund O. NN-align. An artificial neural network-based alignment algorithm for MHC class II peptide binding prediction. BMC Bioinformatics. 2009;10:296. doi: 10.1186/1471-2105-10-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nielsen M, Lundegaard C, Lund O. Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method. BMC Bioinformatics. 2007;8:238. doi: 10.1186/1471-2105-8-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Duan F, et al. Genomic and bioinformatic profiling of mutational neoepitopes reveals new rules to predict anticancer immunogenicity. J Exp Med. 2014;211(11):2231–2248. doi: 10.1084/jem.20141308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gnjatic S, et al. Survey of naturally occurring CD4+ T cell responses against NY-ESO-1 in cancer patients: correlation with antibody responses. Proc Natl Acad Sci U S A. 2003;100(15):8862–8867. doi: 10.1073/pnas.1133324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schumacher T, et al. A vaccine targeting mutant IDH1 induces antitumour immunity. Nature. 2014;512(7514):324–327. doi: 10.1038/nature13387. [DOI] [PubMed] [Google Scholar]
- 89.Linnemann C, et al. High-throughput epitope discovery reveals frequent recognition of neo-antigens by CD4+ T cells in human melanoma. Nat Med. 2015;21(1):81–85. doi: 10.1038/nm.3773. [DOI] [PubMed] [Google Scholar]
- 90.Kreiter S, et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature. 2015;520(7549):692–696. doi: 10.1038/nature14426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tran E, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344(6184):641–645. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Carreno BM, et al. A dendritic cell vaccine increases the breadth diversity of melanoma neoantigen-specific T cells. Science. 2015;348(6236):803–808. doi: 10.1126/science.aaa3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Snyder A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rizvi NA, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chowell D, et al. TCR contact residue hydrophobicity is a hallmark of immunogenic CD8+ T cell epitopes. Proc Natl Acad Sci U S A. 2015;112(14):E1754–E1762. doi: 10.1073/pnas.1500973112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Trinchieri G. Cancer and inflammation: an old intuition with rapidly evolving new concepts. Annu Rev Immunol. 2012;30:677–706. doi: 10.1146/annurev-immunol-020711-075008. [DOI] [PubMed] [Google Scholar]
- 98.Hodi FS, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363(8):711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Topalian SL, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348(6230):56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 101.Mellman I, Coukos G, Dranoff G. Cancer immunotherapy comes of age. Nature. 2011;480(7378):480–489. doi: 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39(1):1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]