Abstract
Caenorhabditis elegans (C. elegans) nematodes transmit small RNAs across generations, a process that enables transgenerational regulation of genes. In contrast to changes to the DNA sequence, transgenerational transmission of small RNA-mediated responses is reversible, and thus enables “soft” or “flexible” inheritance of acquired characteristics. Until very recently only introduction of foreign genetic material (viruses, transposons, transgenes) was shown to directly lead to inheritance of small RNAs. New discoveries however, demonstrate that starvation also triggers inheritance of endogenous small RNAs in C.elegans. Multiple generations of worms inherit starvation-responsive endogenous small RNAs, and starvation also results in heritable extension of the progeny's lifespan. In this Commentary paper we explore the intriguing possibility that large parts of the genome and many additional traits are similarly subjected to heritable small RNA-mediated regulation, and focus on the potential influence of transgenerational RNAi on the worm's physiology. While the universal relevance of this mechanism remains to be discovered, we will examine how the discoveries made in worms already challenge long held dogmas in genetics and evolution.
The Permeability of the Weismann Barrier
The classical view held following the formulation of the Modern Synthesis, which combined population genetics with the ideas of Mendel and Darwin, was that the environment couldn't directly shape the genetic makeup of an organism. The surrounding's only contribution to evolution is in determining the forces of selection.1 An immediate implication of this conviction, which became somewhat outdated due to many developments in the study of epigenetics, is that the door is shut on the possibility of inheriting acquired traits; evolution is “blind,” “clueless,” and “directionless.”
One of the major reasons for the assumption that the interactions with the world cannot directly affect the following generations, hypothesized by Friedrich Leopold August Weismann in the late 19th century,2 is that the germline was thought to be segregated by a theoretically impermeable barrier.3 Acceptance of Weismann's rule (the “Weismann Barrier”) dictates that: “genetic information cannot transfer from the soma to the germline.”4
A polemic discussion over the existence and “permeability” of this barrier stirred the scientific community when the modern theory of evolution was framed. However, due to both scientific and even political reasons, eventually Weismann's principle was recognized as a fundamental truth, and became commonly known as “the second law of biology.”4
Nevertheless, recent research has uncovered epigenetic phenomena (heritable changes that arise independently from changes in the DNA sequence), and in particular mechanisms of RNA interference (RNAi) in Caenorhabditis elegans nematodes, which offer an explanation to how the barrier may be breached, allowing interactions between the soma and the environment to alter heredity, in both transient and stable ways.5 As extensive reviews have very recently elaborated on RNAi mechanisms and RNAi inherita-nce ,6-11 we will only highlight some milestone findings, while our focus will be to examine the unknown: the potential of transgenerational silencing in challenging the current conceptual limits of heredity.
There are still many “unknowns” in the field of epigenetic inheritance; we wish to draw the reader's attention to the 4 fundamental questions that need to be answered, and that will be highlighted throughout the text:
Does the inheritance of a specific non-DNA agent (RNA, chromatin, other) precedes, or establishes, the inheritance of the other epigenetic marks? (“Which comes first?”)
How, to what extent, and depending on which factors, can epigenetic information transfer between the soma and germline?
Which environmental conditions can induce the biogenesis and inheritance of small RNA, and what is typical to these environments?
How is transgenerational information maintained, or reset?
From Soma to Germline in C. elegans
In 1998 Fire and Mello showed that double strand RNA (dsRNA) catalyzes gene silencing in C. elegans, and that the silencing effect of dsRNAs which are injected to different worm somatic tissues spreads systemically, and affects non-treated progeny.12 Soon after, it was shown that RNAi could be induced simply by feeding worms on bacteria which express dsRNA,13 and moreover that the silencing is maintained in the next generation as well. Thus, silencing can pass from digested bacteria in the gut, to other non-gut tissues, including the germline, in clear defiance of Weismann's role.
In worms, dsRNA transporters (most notably the Systemic Interference Deficient gene, SID-1) and proteins that are involved in trafficking of vesicles are required for systemic transfer of small RNAs and acquisition of dsRNA from the environment (e.g. SID-2).14 However, the details regarding the process that leads RNAi from the soma to the germline are not clear, and the factors that mediate such transfer have not been identified. Specifically, it is still not known whether SID-1, which is the best-studied gene involved in systemic RNAi, is required for soma to germline small RNA transfer or transgenerational RNAi. Until now, in worms, only silencing effects that are triggered via exogenous small RNAs where directly shown to transfer between cells. A direct demonstration of cell-to-cell transfer of endogenous small RNAs is still missing in C. elegnas, and it is not known whether SID-1 is involved in the process, assuming that it takes place. In different organisms, including humans, additional methods for spreading RNAi among cells have been found, and certain endogenously-produced small RNA species, for example microRNAs, transfer across tight cell-cell connections (e.g. immunological synapses), and also systemically via vesicles such as exosomes.15,16
Transgenerational Inheritance of RNAi
In worms, in addition to silencing genes in the non-treated offspring, RNAi against a minority of the genes tested (13/171, 7.6%),7 and mostly against germline-experssed genes, can produce a long lasting effect. Here we define “long lasting effects” as RNAi effects which last longer than 2 generations. Our definition is based on the fact that only effects that last longer than 2 generations can be considered as truly “transgenerational” and not “intergenerational.” The exposure of the third generation's germ cells, while in the mother, to the original RNA trigger can be ruled out, since the heritable agents will be diluted by a factor of millions. The Fire lab constructed a sensitive test to assess the durability of heritable RNAi, using injected RNAi that targets a temperature sensitive dominant allele of oma-1. In these experiments it was discovered that RNAi usually peters out after 2-3 generations, but in some cases can last for at least 7 generations.17 Another experiment from the Plasterk group showed that RNAi against some genes was essentially stable, lasting for more than 80 generations.7 Both eggs and sperm can transmit heritable RNAi, and importantly, RNAi inheritance occurs even in the absence of the DNA template of the target gene.18
How can dsRNA-induced silencing persist in worms for multiple generations in spite of a theoretically huge dilution effect? (each worm lays more than 200 eggs on average). Multigenerational RNA-based silencing responses are enabled granted to the action of complementary mechanisms. First, an RNA-dependent RNA polymerase (RdRP)-mediated amplification mechanism generates new (“secondary”) small RNAs in every inheriting generation.19,20 Second, inherited RNA molecules affect the DNA by directing chromatin modifications, which can shut down transcription, and augment the persistency of the silencing effect7 (Fig. 1). Since this paper is focused on small RNA inheritance, we point the reader to comprehensive reviews that discuss Chromatin-mediated inheritance, and other transgenerational epigenetic mechanisms in other systems, most notably in fungi and plants8,21,22 (see also Text Box 1).
Figure 1.
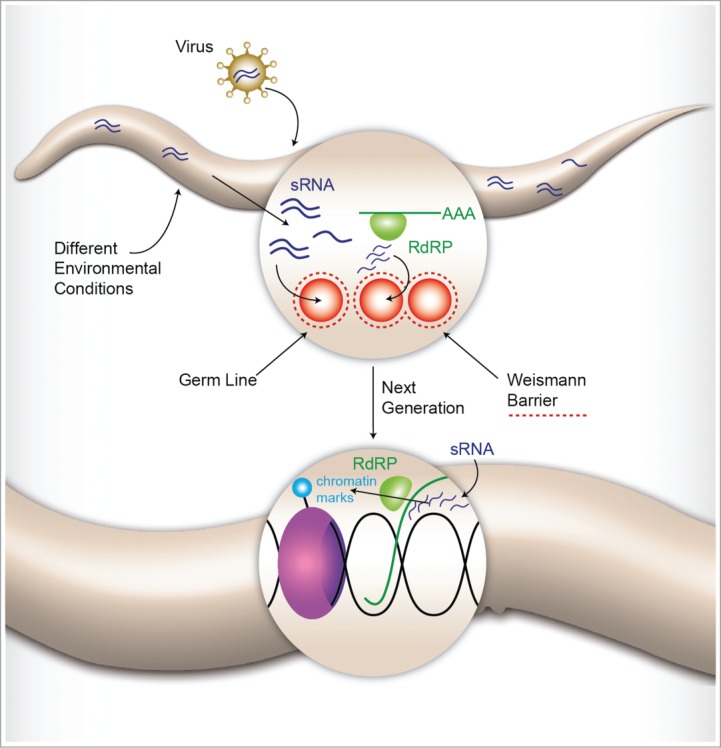
RNAi acts systemically in C. elegans nematodes. Moreover, the RNAi response is amplified via the action of RdRPs, and involves also chromatin remodeling. Transfer of RNAi to the germline produces a heritable response that is re-initiated in the next generations.
RNA and chromatin inheritance are not mutually exclusive processes; on the contrary, small RNA, chromatin, and DNA methylation-based epigenetic phenomena have been shown to be at least partly interdependent in a variety of organisms.22 In worms, RNAi and chromatin-based gene regulation is coordinated by nuclear argonaute proteins that carry 22G small RNAs in the germline, HRDE-1 (Heritable RNAi Deficient-1), and CSR-1 (Chromosome-Segregation and RNAi deficient-1), and NRDE (Nuclear RNAi Deficient) proteins, as detailed elsewhere.6
Both small RNAs and chromatin marks are detectible in the progeny of worms that are treated with RNAi. One study found that the heritable small RNAs are detectable before the chromatin marks, and thus suggested that small RNAs are the primary heritable material, while the chromatin marks are reconstructed de novo in every generation.23 The question of whether heritable small RNAs precede heritable chromatin marks or vice versa is a hotly debated issue, which required more research before it can be resolved. The current models for epigenetic inheritance which take into account feed forward interactions between RNAi factors and chromatin marks and an association between RNAi factors and nascent RNA transcripts (an interface which enables the recruitment of RNA-binding proteins that carry “guiding” small RNAs to the DNA, where interaction with chromatin-modifiers and modification of the chromatin can take place) rely heavily on insights from other organisms, especially fission yeast.24 However, regardless of the exact mechanism, and “who came first,” it is clear that both of these processes act together, and that both are important for epigenetic inheritance in worms as well.
Until very recently it was not clear whether transgenerational transmission of RNAi enables inheritance of physiologically relevant acquired traits, or whether it is triggered only by administration of exogenous dsRNA. However, the discovery of dedicated argonaute proteins that specifically affect inherited RNAi, and not RNAi per se (HRDE-1,25), and traits that are affected by heritable small RNAs (see below), suggests that C.elegans nematodes breach the Weismann barrier in order to complement their genome in an adaptive fashion; Indeed, as described below, heritable small RNAs enable progeny to remember their ancestors' reactions to environmental challenges.
Inheritance of Adaptive Immunity via Transgenerational RNAi
Inherited RNAi was recently linked to several immunological functions.26 Worms are remarkably resistant to viruses because anti-viral small RNAs (viRNAs) destroy viruses with great efficiency.27 Acquired viRNAs, which are amplified by RdRPs, are heritable for multiple generations, and serve as “Inherited vaccines,” to provide the progeny with innate anti-viral protection.20,26,28,29 In addition, heritable small RNAs protect the genome from other intruding elements, such as transposons, by permitting only the expression of endogenous genes. The logic for distinguishing “self” from “foreign” is based on transgenerational memory: genes that should be expressed in the germline are “marked” with heritable endogenous small RNAs that are carried over by the nuclear argonaute CSR-1, and correspond to the previous generations' germline-expressed genes.30,31 If a gene is transcribed in the germline without such heritable “RNA licensing”32,33 the RNAi system recognizes it as “foreign” (as a parasitic mobile element) and silences it by producing heritable PIWI-interacting small RNAs (piRNAs) which shut off the gene's expression at the chromatin level.25,32,34,35 Experimental elimination of these heritable endogenous small RNAs resulted in a “mortal germline” and infertility phenotypes.25,31 Thus, different small RNA species protect the genome by acting as heritable “Guest” and “Black” lists, which restrict expression in the germline.26
Starvation-Induced Small RNA Inheritance
Can C. elegans use heritable small RNAs to regulate different endogenous signaling cascades, and thus to propagate long lasting adaptive physiological responses to challenges, or is the use of RNA inheritance restricted for immunological functions?
As mentioned above, until very recently RNA inheritance was shown to ensue only as a response to the introduction of foreign genetic elements (viruses, transposons, transgenes), which supply the substrate from which small RNAs could be produced. Natural environmental conditions that could trigger the production of endogenous small RNAs are poorly characterized. We were inspired to examine the potential of starvation to induce the production of small RNAs, and to lead to epigenetic effects, since transgenerational effects in response to different diets were shown to occur in diverse animal models, and importantly also in humans, see.36-38 We therefore tested whether heritable small RNAs arise in C. elegans following L1 starvation, and whether a heritable physiological effect can be detected.39 We hypothesized that L1 arrest would dramatically change the worm's endogenous small RNA pools, and thus the levels of small RNAs that could potentially be inherited, because more genes change their expression during L1 starvation than throughout the entire course of larval development.40 In line with our hypothesis, we observed significant changes in the levels of primary endogenous small RNAs (26Gs) and secondary small RNAs (22G) in adults that experienced severe starvation as L1s (6 d without food). In addition, we found 31 genes that were putative targets of both 26Gs and 22Gs, which were differentially expressed following starvation.39
Crucially, we were able to detect a large and highly statistically significant degree of similarity between the small RNA pools of adult animals that experienced starvation as L1s, and the small RNA pools of their F3 progeny that grew Ad libitum for 3 consecutive generations. We focused on the inheritance of 22G Small RNAs as this small RNA species is amplified by RDRPs, and is known to be the cargo of the argonatues that carry the heritable RNAi signal, HRDE-1 and CSR-1.39 The similarity between the small RNA pools of the fed F3 worms and their ancestors that experienced starvation as L1s stemmed primarily from the similarity between the pools of their 22G small RNAs. We examined clusters of 22G small RNAs that align in the antisense orientation to particular genes (Small RNAs that Target specific Genes, or "STGs"). Similar changes in 22G expression was observed for 26.3% of the STGs that were upregulated in the P0 generation following starvation (152/578, p-value < 1.399e-71), 52% of the downregulated STGs (311/597, p-value < 1.256e-292), and 91% of the 22Gs that overlap with 26G STGs (p-value < 5.629e-29, fold-enrichment=26.1).39
When we examined the putative targets of the heritable 22G STGs, we detected a strong enrichment for genes that are involved in nutrition-related functions. Transgenerational regulation of these genes could in theory prepare the progeny for additional hungers. Examination of mRNA levels of genes that were suspected to be targets of heritable 22Gs suggested that in the F3 generation, germline-expressed genes that were previously shown to bind CSR-1 (in immunoprecipitation experiments), increase their expression, while HRDE-1-targets appear to lower their expression.39
It is not yet clear whether starvation initiates small RNA production in the soma or in the germline. However, production of the starvation-responsive small RNAs and regulation of their putative mRNA targets was abrogated in rde-4 mutants (a dsRNA-binding protein, which acts upstream in endo-siRNA production). The heritable changes in the levels of both the starvation-induced small RNAs and their mRNA targets was abolished in hrde-1 mutants.39
Interestingly, another group recently showed that exposing worms to brief periods of starvation is enough to reset the accumulative transgenerational sterility that results when prg-1 mutants are cultivated in 25 degrees (PRG-1 is required for biosynthesis of piRNAs, another small RNA species).41
In addition to the small RNA inheritance that ensues following starvation, we observed that the lifespan of animals that derive from starved great-grandparents is longer than the lifespan of animals whose great-grandparents were continuously fed.39 Previous experiments have already linked longevity with epigenetic inheritance. Greer et al. have shown that inheritance following incomplete reprogramming of chromatin states (in wild type descendants of mutants for ASH-2, WDR-5 or SET-2, which compose the histone H3 lysine 4 trimethylation complex) affects the lifespan of the progeny.42 More research is required in order to understand whether the regulation of genes by inherited small RNAs is causing the heritable increased longevity that is observed upon starvation, and following perturbation of transgenerational chromatin reprogramming.
It is important to understand which other environmental conditions are capable of initiating transgenerational responses, and L1 starvation could be an important case study to learn from. How does starvation trigger the production of specific endo-siRNAs? One intuitive possibility is that during L1 arrest cognate mRNAs, which are transcribed in response to starvation, serve as templates for the synthesis of specific endo-siRNAs.39 For example, bidirectional transcription could produce dsRNA that would then be processed into small RNAs.39 In theory, every environmental condition that would elicit the production of small RNAs that are capable of reaching the germline, or that would affect the pool of already germline-expressed small RNAs, could result in transgenerational effects.
Combining the “Soft” and “Hard” Genomes
Here we suggest that heritable RNA molecules give rise to a rapidly-evolving epigenome, a “Soft RNA Genome” which together with other epigenetic marks, communicates with the stable, and slowly-evolving “Hard” DNA genome, regulates it, and gets regulated by it. The RdRPs that replicate the “Soft” RNA genome have low replication fidelity (error every ∼104 bases), and since RNA pools fluctuate in response to changing environmental signals, the “Soft genome” should increase the interspecies variability on which selection can act. In addition, the “Soft” RNA genome, as the name suggests, is flexible. Since the “Soft” RNA genome represents the reactions of the previous generations to the surroundings, it can either linger transiently, if the environment changes, or establish a stable regulation, when the response is selected due to its relevancy and adaptive nature. Maintenance of the epigenetic effects that are initiated by inherited RNA responses could occur in 2 phases. First, transient epigenetic inheritance could be achieved via RdRP-mediated amplification and feed-forward interactions with chromatin marks.24 Later, if the environmental pressure remains relevant for many generations, the response could be assimilated via mutations or deletions, which accumulate if a gene stays off, by natural selection.43,44 If indeed certain heritable epigenetic responses yield long lasting effects that are “fixed” in the genome, then this type of mechanism could have influenced the rate of the evolutionary process. This is a wild hypothesis that demands serious experimental examination.
Transgenerational RNAi Inheritance Across the Animal Kingdom
Multiple transgenerational epigenetic effects have been convincingly demonstrated in unicellular organisms and plants (see Box.1, and 45), however, unicellular organisms have no germline, and in plants the germline is not segregated. Although the question was not fully addressed in many phyla, most species in the animal kingdom appear to specify their germline by epigenesis without or with very limited deposition of germline-specifying cytoplasmic determinants. In C. elegans, the parental soma communicates with the germline, and the germline is segregated by preformation, which makes it essentially continuous and immortal.46 Thus, if somatic small RNAs get to the nematode's germline, their signaling could propagate for multiple generations. Relevantly, and in line with the hypothesis that epigenetic information accelerates evolution, a recent study showed that protein-coding genes evolve faster in animals that segregate their germline using preformation, in comparison to animals that specify their germline by epigenesis.47
It is possible that in the animal kingdom multigenerational RNAi inheritance is limited to animals with a short generation time, or animals that cannot migrate efficiently, as in these organisms the ancestors' environment is more likely to resemble that of the progeny. Alternatively, the RNA-mediated epigenetic effects found in worms might only be the tip of the iceberg, and may have been primarily discovered due to the fact that C. elegans is genetically tractable, and has a very short life span that allows fast analysis of traits across many generations. While these are still early days, recent studies have shown that inherited RNA may enable transgenerational memorization of stress also in mammals.48
While mammalian genomes do not encode for the canonical RdRPs that are found in nematodes, plants and fungi, it has been recently shown that other polymerases which have RdRP capabilities amplify DICER-substrates.49 There is even functional evidence for the existence of a (yet to be identified) human RdRP: hepatitis delta virus, a human-infecting RNA virus that lacks an RdRP, relies exclusively on host RdRP activity for replication of its RNA genome.50 In addition to perpetuating heritable RNA signaling by RNA amplification, it is also possible that in other organisms an RNA response results in the recruitment of the DNA methylation machinery in the germline, which consolidates the RNA-induced silencing and establishes heritable and stable DNA methylation patterns.22 While it is well established that certain DNA methylation patterns are heritable,51 the epigenetic effects that were recorded in C. elegans nematodes are not mediated by this mechanism, since worms do not seem to have any cytosine methylation.52
Box 1. Epigenetic effects in different organisms.RNAi mediated inheritance across the tree of life
Apart from in C. elegans, non-coding RNAs enable non-Mendelian genetics and inheritance of acquired traits across the tree of life.
In many species of bacteria and arcea incorporation of sliced phages into loci that are termed Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR), enables RNA-mediated transgenerational immunity. CRISPR-derived RNAs guide defense proteins back to the phage's sequence, and the phage is degraded.54
In different ciliates heritable RNA determines which DNA sequences will be maintained or discarded in the next generations, instructs the cell on how to perform massive DNA-rearrangements, and determines gene copy number.55
In plants, RNAi is systemic, and small RNAs that are transferred from somatic cells to the germline, together with DNA methylation, establish transgenerational regulation.51 In plants “Epimutations” can be stable, and require RdRPs for their propagation.56 In flies and fish PIWI-associated RNAs (piRNAs) can establish multigenerational RNAi.57,58
While some evidences exist for RNA inheritance in mammals, the underlying mechanisms are still poorly understood59
Concluding Remarks
It is generally accepted that the first molecule of life, RNA, gave up the throne when DNA came on stage and took over. However, recent findings suggest that discussions regarding “The RNA World”53 should not be carried out in a nostalgic tone since it is very much alive: Even in our current “DNA World,” viruses are not the sole organisms to benefit from the advantages of fast-evolving and flexible RNA genomes. “Soft” RNA-based genomes might be integral to the genetic landscape of higher animals as well, co-existing side-by-side in cooperation with their “hard” and “reliable” DNA-encoded younger brothers.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Eva Jablonka for reading the manuscript, and all members of the Rechavi lab for their helpful comments.
Funding
Writing of this manuscript was made possible by funds and support from the Alon, Bikura, and Yad Hanadiv fellowships and by CBRC, Teva NNE, ISF, JTF, and ERC grants.
References
- 1. Bard JB. The next evolutionary synthesis: from Lamarck and Darwin to genomic variation and systems biology. Cell Commun Signal 2011; 9:30. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3215633&tool=pmcentrez&rendertype=abstract. Accessed 25 May 2013; PMID:22053760; http://dx.doi.org/ 10.1186/1478-811X-9-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Weismann C. Germline Selection: A Weismannian Solution to Lamarckian Problematics. In: Gissis BS, Jablonka E, editors. Transformations of Lamarckism. 2011; pp.57-66. [Google Scholar]
- 3. Poulton EB, Schönland S, Shipley AE, Weismann A. 1889. Essays upon heredity and kindred biological problems. Authorised translation, edited by Poulton Edward B., Schönland Selmar. and Shipley Arthur E. Oxford: Clarendon Press; Available: http://www.biodiversitylibrary.org/bibliography/17713. Accessed 1 June 2013 [Google Scholar]
- 4. Mattick JS. Rocking the foundations of molecular genetics. Proc Natl Acad Sci U S A 2012; 109:16400-01. Available: http://www.pnas.org/content/109/41/16400.long. Accessed 24 September 2013; PMID:23019584; http://dx.doi.org/ 10.1073/pnas.1214129109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jablonka E. Epigenetic inheritance and plasticity: The responsive germline. Prog Biophys Mol Biol 2013; 111:99-107. Available: http://www.ncbi.nlm.nih.gov/pubmed/22975443. Accessed 31 May 2013; PMID:22975443; http://dx.doi.org/ 10.1016/j.pbiomolbio.2012.08.014 [DOI] [PubMed] [Google Scholar]
- 6. Wedeles CJ, Wu MZ, Claycomb JM. Silent no more: Endogenous small RNA pathways promote gene expression. Worm 2014; 3:e28641. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4165530&tool=pmcentrez&rendertype=abstract. Accessed 30 October 2014; PMID:25254148; http://dx.doi.org/ 10.4161/worm.28641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vastenhouw NL, Brunschwig K, Okihara KL, Müller F, Tijsterman M, Plasterk RH. Gene expression: long-term gene silencing by RNAi. Nature 2006; 442:882. Available. Accessed 27 May 2013; PMID:16929289; http://dx.doi.org/ 10.1038/442882a [DOI] [PubMed] [Google Scholar]
- 8. Lim JP, Brunet A Bridging the transgenerational gap with epigenetic memory. Trends Genet 2013; 29:176-86. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3595609&tool=pmcentrez&rendertype=abstract. Accessed 16 October 2014; PMID:23410786; http://dx.doi.org/ 10.1016/j.tig.2012.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hoogstrate SW, Volkers RJ, Sterken MG, Kammenga JE, Snoek LB Nematode endogenous small RNA pathways. Worm 2014; 3:e28234. Available: http://www.ncbi.nlm.nih.gov/pubmed/25340013. Accessed 30 October 2014; PMID:25340013; http://dx.doi.org/ 10.4161/worm.28234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sarkies P, Miska EA. Small RNAs break out: the molecular cell biology of mobile small RNAs. Nat Rev Mol Cell Biol 2014; 15:525-35. Available: http://www.ncbi.nlm.nih.gov/pubmed/25053358. Accessed 23 July 2014; PMID:25053358; http://dx.doi.org/ 10.1038/nrm3840 [DOI] [PubMed] [Google Scholar]
- 11. Grishok A. Biology and Mechanisms of Short RNAs in Caenorhabditis elegans. Adv Genet 2013; 83:1-69. Available: http://www.ncbi.nlm.nih.gov/pubmed/23890211. Accessed 25 March 2014; PMID:23890211; http://dx.doi.org/ 10.1016/B978-0-12-407675-4.00001-8 [DOI] [PubMed] [Google Scholar]
- 12. Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998; 391:806-11. Available. Accessed 27 May 2013; PMID:9486653; http://dx.doi.org/ 10.1038/35888 [DOI] [PubMed] [Google Scholar]
- 13. Timmons L, Court DL, Fire A Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 2001; 263:103-12. Available: http://www.ncbi.nlm.nih.gov/pubmed/11223248. Accessed 1 June 2013; PMID:11223248; http://dx.doi.org/ 10.1016/S0378-1119(00)00579-5 [DOI] [PubMed] [Google Scholar]
- 14. Hunter CP, Winston WM, Molodowitch C, Feinberg EH, Shih J, Sutherlin M, Wright AJ, Fitzgerald MC. Systemic RNAi in Caenorhabditis elegans. Cold Spring Harb Symp Quant Biol 2006; 71:95-100. Available: http://www.ncbi.nlm.nih.gov/pubmed/17381285 Accessed 7 July 2013; PMID:17381285; http://dx.doi.org/ 10.1101/sqb.2006.71.060 [DOI] [PubMed] [Google Scholar]
- 15. Chitwood DH, Timmermans MCP Small RNAs are on the move. Nature 2010; 467:415-19. Available. Accessed 24 May 2013; PMID:20864994; http://dx.doi.org/ 10.1038/nature09351 [DOI] [PubMed] [Google Scholar]
- 16. Rechavi O, Erlich Y, Amram H, Flomenblit L, Karginov F V, Goldstein I, Hannon GJ, Kloog Y. Cell contact-dependent acquisition of cellular and viral nonautonomously encoded small RNAs. Genes Dev 2009; 23:1971-9. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2725935&tool=pmcentrez&rendertype=abstract; PMID:19684116; http://dx.doi.org/ 10.1101/gad.1789609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alcazar RM, Lin R, Fire AZ. Transmission dynamics of heritable silencing induced by double-stranded RNA in Caenorhabditis elegans. Genetics 2008; 180:1275-88. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2581934&tool=pmcentrez&rendertype=abstract. Accessed 1 June 2013; PMID:18757930; http://dx.doi.org/ 10.1534/genetics.108.089433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grishok A, Tabara H, Mello CC. Genetic requirements for inheritance of RNAi in C. elegans. Science 2000; 287:2494-7. Available: http://www.ncbi.nlm.nih.gov/pubmed/10741970. Accessed 1 June 2013; PMID:10741970; http://dx.doi.org/ 10.1126/science.287.5462.2494 [DOI] [PubMed] [Google Scholar]
- 19. Gu SG, Pak J, Guang S, Maniar JM, Kennedy S, Fire A. Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nat Genet 2012; 44:157-64. Available: http://www.ncbi.nlm.nih.gov/pubmed/22231482. Accessed 27 May 2013; PMID:22231482; http://dx.doi.org/ 10.1038/ng.1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rechavi O, Minevich G, Hobert O. Transgenerational Inheritance of an Acquired Small RNA-Based Antiviral Response in C. elegans. Cell 2011; 147:1248-56. Available: http://linkinghub.elsevier.com/retrieve/pii/S0092867411013419; PMID:22119442; http://dx.doi.org/ 10.1016/j.cell.2011.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Claycomb JM. Caenorhabditis elegans small RNA pathways make their mark on chromatin. DNA Cell Biol 2012; 31 Suppl 1:S17-33. Available: http://www.ncbi.nlm.nih.gov/pubmed/23046453. Accessed 30 October 2014; PMID:23046453 [DOI] [PubMed] [Google Scholar]
- 22. Castel SE, Martienssen R a RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet 2013; 14:100-12. Available: http://www.ncbi.nlm.nih.gov/pubmed/23329111. Accessed 25 May 2013; PMID:23329111; http://dx.doi.org/ 10.1038/nrg3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Burton NO, Burkhart KB, Kennedy S Nuclear RNAi maintains heritable gene silencing in Caenorhabditis elegans. Proc Natl Acad Sci U S A 2011; 108:19683-8. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3241819&tool=pmcentrez&rendertype=abstract. Accessed 21 May 2013; PMID:22106253; http://dx.doi.org/ 10.1073/pnas.1113310108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bühler M, Verdel A, Moazed D Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell 2006; 125:873-86. Available: http://www.cell.com/fulltext/S0092-8674(06)00514-9. Accessed 22 May 2013; PMID:16751098; http://dx.doi.org/ 10.1016/j.cell.2006.04.025 [DOI] [PubMed] [Google Scholar]
- 25. Buckley BA, Burkhart KB, Gu SG, Spracklin G, Kershner A, Fritz H, Kimble J, Fire A, Kennedy S. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 2012; 489:447-51. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3509936&tool=pmcentrez&rendertype=abstract. Accessed 1 June 2013; PMID:22810588; http://dx.doi.org/ 10.1038/nature11352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rechavi O. Guest list or black list: heritable small RNAs as immunogenic memories. Trends Cell Biol. 2013; Available: http://www.ncbi.nlm.nih.gov/pubmed/24231398. Accessed 15 November 2013; PMID:24231398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shaham S. Worming into the cell: viral reproduction in Caenorhabditis elegans. Proc Natl Acad Sci U S A 2006; 103:3955-6. Available: http://www.pnas.org/content/103/11/3955.full. Accessed 1 June 2013; PMID:16537467; http://dx.doi.org/ 10.1073/pnas.0600779103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sterken MG, Snoek LB, Bosman KJ, Daamen J, Riksen JAG, Bakker J, Pijlman GP, Kammenga JE. A Heritable Antiviral RNAi Response Limits Orsay Virus Infection in Caenorhabditis elegans N2. PLoS One 2014; 9:e89760. Available: http://dx.plos.org/10.1371/journal.pone.0089760. Accessed 20 March 2014; PMID:24587016; http://dx.doi.org/ 10.1371/journal.pone.0089760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guo X, Li W-X, Lu R Silencing of host genes directed by virus-derived short interfering RNAs in Caenorhabditis elegans. J Virol 2012; 86:11645-53. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3486301&tool=pmcentrez&rendertype=abstract. Accessed 1 June 2013; PMID:22896621; http://dx.doi.org/ 10.1128/JVI.01501-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wedeles CJ, Wu MZ, Claycomb JM. Protection of germline gene expression by the C. elegans Argonaute CSR-1. Dev Cell 2013; 27:664-71. Available: http://www.ncbi.nlm.nih.gov/pubmed/24360783. Accessed 19 March 2014; PMID:24360783; http://dx.doi.org/ 10.1016/j.devcel.2013.11.016 [DOI] [PubMed] [Google Scholar]
- 31. Conine CC, Moresco JJ, Gu W, Shirayama M, Conte D, Jr, Yates JR, 3rd, Mello CC. Argonautes Promote Male Fertility and Provide a Paternal Memory of Germline Gene Expression in C. elegans. Cell 2013; 155:1532-44. Available: http://www.ncbi.nlm.nih.gov/pubmed/24360276. Accessed 21 January 2014; PMID:24360276; http://dx.doi.org/ 10.1016/j.cell.2013.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shirayama M, Seth M, Lee H-C, Gu W, Ishidate T, Conte D, Jr, Mello CC. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 2012; 150:65-77. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3597741&tool=pmcentrez&rendertype=abstract. Accessed 23 May 2013; PMID:22738726; http://dx.doi.org/ 10.1016/j.cell.2012.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Johnson CL, Spence AM. Epigenetic licensing of germline gene expression by maternal RNA in C. elegans. Science 2011; 333:1311-4. Available: http://www.sciencemag.org/content/333/6047/1311.abstract. Accessed 26 May 2013; PMID:21885785; http://dx.doi.org/ 10.1126/science.1208178 [DOI] [PubMed] [Google Scholar]
- 34. Ashe A, Sapetschnig A, Weick E-M, Mitchell J, Bagijn MP, Cording AC, Doebley AL, Goldstein LD, Lehrbach NJ, Le Pen J, et al. . piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 2012; 150:88-99. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3464430&tool=pmcentrez&rendertype=abstract. Accessed 1 June 2013; PMID:22738725; http://dx.doi.org/ 10.1016/j.cell.2012.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Luteijn MJ, van Bergeijk P, Kaaij LJT, Almeida MV, Roovers EF, Berezikov E, Ketting RF. Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. EMBO J 2012; 31:3422-30. Available: http://www.ncbi.nlm.nih.gov/pubmed/22850670. Accessed 1 June 2013; PMID:22850670; http://dx.doi.org/ 10.1038/emboj.2012.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Painter RC, Osmond C, Gluckman P, Hanson M, Phillips DIW, Roseboom TJ. Transgenerational effects of prenatal exposure to the Dutch famine on neonatal adiposity and health in later life. BJOG 2008; 115:1243-49. Available: http://www.ncbi.nlm.nih.gov/pubmed/18715409. Accessed 27 March 2014; PMID:18715409; http://dx.doi.org/ 10.1111/j.1471-0528.2008.01822.x [DOI] [PubMed] [Google Scholar]
- 37. Stanner SA, Bulmer K, Andrès C, Lantseva OE, Borodina V, Poteen VV, Yudkin JS. Does malnutrition in utero determine diabetes and coronary heart disease in adulthood? Results from the Leningrad siege study, a cross sectional study. BMJ 1997; 315:1342-48. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2127836&tool=pmcentrez&rendertype=abstract. Accessed 13 April 2014; PMID:9402775; http://dx.doi.org/ 10.1136/bmj.315.7119.1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Song S, Wang W, Hu P Famine, death, and madness: schizophrenia in early adulthood after prenatal exposure to the Chinese Great Leap Forward Famine. Soc Sci Med 2009; 68:1315-21. Available: http://www.sciencedirect.com/science/article/pii/S0277953609000367. Accessed 13 April 2014; PMID:19232455; http://dx.doi.org/ 10.1016/j.socscimed.2009.01.027 [DOI] [PubMed] [Google Scholar]
- 39. Rechavi O, Houri-Ze'evi L, Anava S, Goh WSS, Kerk SY, Hannon GJ, Hobert O. Starvation-Induced Transgenerational Inheritance of Small RNAs in C. elegans. Cell 2014; 158:277-87. Available: http://www.ncbi.nlm.nih.gov/pubmed/25018105. Accessed 11 July 2014; PMID:25018105; http://dx.doi.org/ 10.1016/j.cell.2014.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Maxwell CS, Antoshechkin I, Kurhanewicz N, Belsky JA, Baugh LR. Nutritional control of mRNA isoform expression during developmental arrest and recovery in C. elegans. Genome Res 2012; 22:1920-9. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3460187&tool=pmcentrez&rendertype=abstract. Accessed 13 April 2014; PMID:22539650; http://dx.doi.org/ 10.1101/gr.133587.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Simon M, Sarkies P, Ikegami K, Doebley A-L, Goldstein LD, Mitchell J, Sakaguchi A, Miska EA, Ahmed S6. Reduced insulin/IGF-1 signaling restores germ cell immortality to caenorhabditis elegans Piwi mutants. Cell Rep 2014; 7:762-73. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4049074&tool=pmcentrez&rendertype=abstract. Accessed 2 November 2014; PMID:24767993; http://dx.doi.org/ 10.1016/j.celrep.2014.03.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Greer EL, Maures TJ, Ucar D, Hauswirth AG, Mancini E, et al. . Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature 2011; 479:365-71. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3368121&tool=pmcentrez&rendertype=abstract. Accessed 9 August 2013; PMID:22012258; http://dx.doi.org/ 10.1038/nature10572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lawrence MS, Stojanov P, Polak P, Kryukov G V, Cibulskis K, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature advance on. 2013; Available: http://dx.doi.org/ 10.1038/nature12213 Accessed 17 June 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Koonin EV. Calorie Restriction à Lamarck. Cell 2014; 158:237-8. Available: http://www.ncbi.nlm.nih.gov/pubmed/25036622. Accessed 18 July 2014; PMID:25036622; http://dx.doi.org/ 10.1016/j.cell.2014.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Grossniklaus U, Kelly B, Ferguson-Smith AC, Pembrey M, Lindquist S Transgenerational epigenetic inheritance: how important is it? Nat Rev Genet 2013; 14:228-35. Available. Accessed 31 May 2013; PMID:23416892; http://dx.doi.org/ 10.1038/nrg3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Extavour CG, Akam M Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development 2003; 130:5869-84. Available: http://dev.biologists.org/content/130/24/5869.short. Accessed 29 May 2013; PMID:14597570; http://dx.doi.org/ 10.1242/dev.00804 [DOI] [PubMed] [Google Scholar]
- 47. Evans T, Wade CM, Chapman FA, Johnson AD, Loose M Acquisition of germ plasm accelerates vertebrate evolution. Science 2014; 344:200-3. Available: http://www.ncbi.nlm.nih.gov/pubmed/24723612. Accessed 19 October 2014; PMID:24723612; http://dx.doi.org/ 10.1126/science.1249325 [DOI] [PubMed] [Google Scholar]
- 48. Gapp K, Jawaid A, Sarkies P, Bohacek J, Pelczar P, et al. . Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat Neurosci advance on. 2014; Available: http://dx.doi.org/ 10.1038/nn.3695 Accessed 14 April 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Maida Y, Yasukawa M, Furuuchi M, Lassmann T, Possemato R, et al. . An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature 2009; 461:230-5. Available; PMID:19701182; http://dx.doi.org/ 10.1038/nature08283. Accessed 1 June 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Greco-Stewart V, Pelchat M Interaction of host cellular proteins with components of the hepatitis delta virus. Viruses 2010; 2:189-212. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3185554&tool=pmcentrez&rendertype=abstract. Accessed 31 May 2013; PMID:21994607; http://dx.doi.org/ 10.3390/v2010189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jones L, Ratcliff F, Baulcombe DC RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance. Curr Biol 2001; 11:747-57ma. Available: http://www.ncbi.nlm.nih.gov/pubmed/11378384. Accessed 1 June 2013; PMID:11378384; http://dx.doi.org/ 10.1016/S0960-9822(01)00226-3 [DOI] [PubMed] [Google Scholar]
- 52. Simpson VJ, Johnson TE, Hammen RF Caenorhabditis elegans DNA does not contain 5-methylcytosine at any time during development or aging. Nucleic Acids Res 1986; 14:6711-9. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=311675&tool=pmcentrez&rendertype=abstract. Accessed 31 May 2013; PMID:3748820; http://dx.doi.org/ 10.1093/nar/14.16.6711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gilbert W. Origin of life: The RNA world. Nature 1986; 319:618 Available: http://www.nature.com/nature/journal/v319/n6055/pdf/319618a0.pdf. Acce-ssed 13 June 2013; http://dx.doi.org/ 10.1038/319618a0 [DOI] [Google Scholar]
- 54. Brouns SJJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJH, et al. . Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 2008; 321:960-4. Available: http://www.ncbi.nlm.nih.gov/pubmed/18703739. Acce-ssed 21 January 2014; PMID:18703739; http://dx.doi.org/ 10.1126/science.1159689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nowacki M, Shetty K, Landweber LF RNA-Mediated Epigenetic Programming of Genome Rearrangements. Annu Rev Genomics Hum Genet 2011; 12:367-89. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3518427&tool=pmcentrez&rendertype=abstract. Accessed 12 December 2013; PMID:21801022; http://dx.doi.org/ 10.1146/annurev-genom-082410-101420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chandler V, Alleman M Paramutation: Epigenetic Instructions Passed Across Generations. Genetics 2008; 178:1839-44. Available: http://www.genetics.org/content/178/4/1839.full#ref-12. Accessed 12 December 2013; PMID:18430919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kaaij LJT, Hoogstrate SW, Berezikov E, Ketting RF piRNA dynamics in divergent zebrafish strains reveal long-lasting maternal influence on zygotic piRNA profiles. RNA 2013; 19:345-56. Available: http://rnajournal.cshlp.org/content/early/2013/01/16/rna.036400.112.abstract. Accessed 31 May 2013; PMID:23335638; http://dx.doi.org/ 10.1261/rna.036400.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. De Vanssay A, Bougé A-L, Boivin A, Hermant C, Teysset L, et al. . Paramutation in Drosophila linked to emergence of a piRNA-producing locus. Nature 2012; 490:112-5. Available: . Accessed 3 June 2013; PMID:22922650; http://dx.doi.org/ 10.1038/nature11416 [DOI] [PubMed] [Google Scholar]
- 59. Rassoulzadegan M, Grandjean V, Gounon P, Cuzin F [Epigenetic heredity in mice: involvement of RNA and miRNas.]. J Soc Bioll 2007; 201:397-9. Available: http://www.ncbi.nlm.nih.gov/pubmed/18533100. Acc-essed 12 December 2013; PMID:18533100; http://dx.doi.org/ 10.1051/jbio:2007911 [DOI] [PubMed] [Google Scholar]


