Abstract
Regulation of chromatin is a key process in the developmental control of gene expression. Many multi-subunit protein complexes have been found to regulate chromatin through the modification of histone residues. One such complex is the MOF histone acetyltransferase-containing NSL complex. While the composition of the human and Drosophila NSL complexes has been determined and the functions of these complexes investigated, the existence of an equivalent complex in nematodes such as Caenorhabditis elegans has not yet been explored. Here we summarise evidence, from our own work and that of others, that homologues of NSL complex components are found in C. elegans. We review data suggesting that nematode proteins SUMV-1 and SUMV-2 are homologous to NSL2 and NSL3, respectively, and that SUMV-1 and SUMV-2 may form a complex with MYS-2, the worm homolog of MOF. We propose that these interactions suggest the existence of a nematode NSL-like complex and discuss the roles of this putative NSL complex in worms as well as exploring the possibility of crosstalk between NSL and COMPASS complexes via components that are common to both. We present the groundwork from which a full characterization of a nematode NSL complex may begin.
Keywords: chromatin, histone acetyltransferase, MOF, NSL, synMuv, vulval development
Abbreviations
- MOF
males absent on the first
- NSL
non-specific lethal
- synMuv
synthetic multivulva
- KAT8
lysine (K) acetyltransferase
- HDAC
histone deacetylase
- HAT
histone acetyltransferase
- HMT
histone methyltransferase
- COMPASS
complex proteins associated with Set1
- ChIP-seq
chromatin immunoprecipitation sequencing
Introduction
The regulation of chromatin is an important process in the developmental control of gene expression. Chromatin, composed of DNA and histone proteins, regulates the accessibility, and consequently the activity, of genes. Specific residues on the histone proteins can be modified, for instance through acetylation, methylation or phosphorylation. Modifications are made by chromatin modifying complexes, which are composed of multiple protein subunits including a histone modifying enzyme, such as a histone acetyltransferase (HAT), histone deacetylase (HDAC) or histone methyltransferase (HMT). One HAT of recent interest is KAT8/MOF (lysine (K) acetyltransferase/males absent on the first). KAT8/MOF is best known for its role in Drosophila dosage compensation as a component of the MSL (male-specific lethal) complex.1 More recently, however, it was found that KAT8/MOF is also a constituent of a separate complex called NSL (non-specific lethal).
In Drosophila, the NSL complex was found to regulate constitutively expressed genes. ChIP-seq analysis revealed that it binds preferentially to promoters of housekeeping genes and that the NSL complex is required for efficient recruitment of RNA polymerase II to these promoters.2 The NSL complex has similarly been found to regulate housekeeping genes in mouse embryonic stem cells (mESCs)3,4 and to also be required for the expression of key pluripotency factors in these cells.4
Affinity purification and mass spectrometry analyses have revealed that the Drosophila and human NSL complexes are composed of 9 subunits: MOF/hMOF, dHCF/HCFC1, OGT/OGT1, WDS/WDR5, MCRS2/MCRS1, MBD-R2/PHF20, NSL1/KANSL1, NSL2/KANSL2 and NSL3/KANSL3 (Fig. 1).5–7 While the majority of these proteins have homologues in C. elegans, the existence of a functional nematode NSL complex has yet to be determined. Here we describe each of the components of the NSL complex and summarise current knowledge of their homologues in C. elegans before discussing the possible functions of a putative nematode NSL complex.
Figure 1.
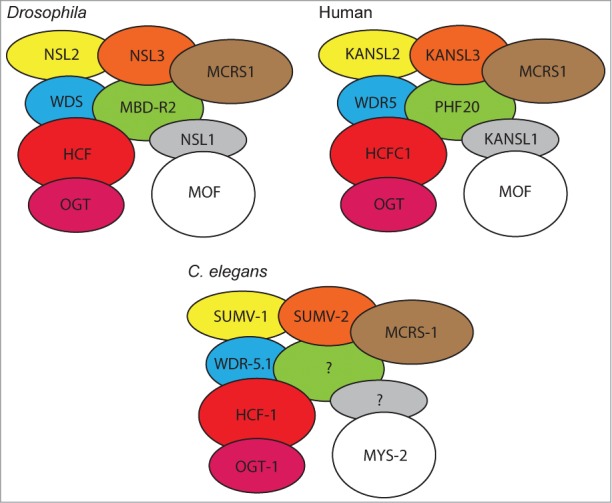
Schematic of Drosophila and human NSL complex compared with putative C. elegans NSL complex. Both Drosophila and human complexes consist of at least 9 members. Seven of these members are known to have C. elegans homologues. Components have been arranged to be adjacent to all other components with predicted interactions. Interactions predicted in STRING database.
MOF
MOF is a member of the MYST family of HATs and is the enzymatic component of the NSL complex. Although in the context of the MSL complex MOF specifically acetylates histone 4 lysine 16 (H4K16ac),1,8 in the NSL complex MOF shows broader substrate specificity, additionally acetylating lysine 5 and 8 of H4.7 In Drosophila, the involvement of MOF in dosage compensation leads to a male-lethal phenotype in mof mutant flies,1 while murine MOF is required for both male and female embryos to develop beyond the expanded blastocyst stage.9 The C. elegans homologue of MOF, called MYS-2, also plays roles in development. For instance, reduction of mys-2 function (in combination with reduction of function of another MYST HAT, mys-1) causes defects in cell fate maintenance.10
HCFC1
Host cell factor C1 (HCFC1) is a transcriptional cofactor formed via the non-covalent association of multiple polypeptides, which are derived from a 2035 amino acid precursor protein through proteolytic cleavage (for a review see ref. 11). Initially identified through its interaction with transcription factor Oct-1 and the viral transactivating protein VP16 in the context of herpes simplex virus immediate early gene expression,12 HCFC1 has since been shown to interact with numerous cellular transcription factors. HCFC1 also associates with histone modification complexes in addition to NSL, including the Sin3 HDAC complex and the Set1/Ash2 HMT complex,13 suggesting that it may serve as a bridge between transcription factors and chromatin modulators.
The C. elegans homologue of HCFC1, called HCF-1, has been primarily studied in the context of longevity and stress responses. Interaction with HCF-1 is proposed to regulate the activity of the forkhead box O (FOXO) transcription factor DAF-16, a key longevity determinant, by sequestering a fraction of DAF-16 away from the promoters of its target genes.14 Additionally, at low temperatures, loss of hcf-1 leads to smaller brood sizes and embryonic lethality due to defects in cell division, phenotypes which have been attributed to a reduction in histone 3 serine 10 phosphorylation (H3S10P).15
OGT
O-linked N-acetylglucosamine (O-GlcNAc) transferase (OGT) catalyzes the O-GlcNAcylation of serine and threonine residues in a variety of nuclear and cytoplasmic proteins and it has been demonstrated that histone proteins are among the nuclear proteins that can be modified by OGT.16,17 The importance of OGT in mammalian development is evidenced by the finding that deletion of OGT compromises the viability of embryonic stem cells.18 While there are no obvious developmental defects associated with reduction of function of the nematode OGT, C. elegans OGT-1 has been implicated in the regulation of macronutrient storage since ogt-1 mutants have increased glycogen levels and decreased lipid stores.19
Interestingly, in human and fly systems, OGT is responsible for the cleavage of the HCFC1 precursor into the subunits which associate to form the active protein.11,20 While the nematode HCF-1 protein, in contrast, does not appear to be subject to proteolytic processing,21 whether it is a substrate for O-GlcNAcylation has yet to be investigated.
WDR5
WDR5 is a WD40 repeat-containing protein. WD40 repeats are short motifs of approximately 40 amino acids that often end with a tryptophan and aspartic acid (WD) and are involved in protein-protein interactions (for a review see ref. 22). Three C. elegans homologues of WDR5 have been identified: WDR-5.1, WDR-5.2, and WDR-5.3.23,24 The best-characterized of these is WDR-5.1, which is required for germline stem cell maintenance and additionally plays a role in the transgenerational regulation of longevity.23,25
Like HCFC1, which interacts with many transcription factors and several histone modification complexes, WDR5 is not uniquely associated with the NSL complex. For instance, WDR5 is also a component of the COMPASS (Complex Proteins Associated with Set1) HMT complex. In this context, C. elegans WDR-5.1 has been found to be required for global di- and trimethylation of H3K4 in early worm embryos.23 Interestingly, coordinated activity of the NSL and COMPASS complexes has been proposed, with WDR5 acting as a bridge between the two.26 Recently, however, it has been reported that the interaction of WDR5 with NSL complex components occurs via the same regions with which WDR5 interfaces with COMPASS complex components, suggesting that the two interactions are mutually exclusive.27
MCRS1
Microspherule protein-1 (MCRS1) contains a forkhead-associated domain and a coiled coil domain, both of which are protein-protein interaction motifs. Like HCFC1 and WDR5, MCRS1 has roles outside the NSL complex, for instance as a component of the INO80 chromatin remodeling complex.28 Human MCRS1 has been associated with a variety of cellular processes, including RNA polymerase I transcription and cell cycle progression.29,30 In Drosophila, MCRS2 is required for optimal recruitment of RNA polymerase II to the promoter regions of cyclin genes and consequently regulates cyclin gene expression.31 Although an MCRS1 homolog has been identified in C. elegans called MCRS-1, the functions of this nematode protein have not yet been examined.
PHF20
Plant Homeodomain (PHD) finger protein 20 (PHF20) is a multidomain protein incorporating two tudor domains, an AT hook motif and a PHD finger, for which it is named. The developmental importance of PHF20 is demonstrated by the perinatal mortality associated with loss of function of murine PHF20.32 Depletion of the Drosophila PHF20 homolog, MBD-R2, severely affects the mRNA levels of NSL target genes in a genome-wide manner.5 A C. elegans PHF20 homolog has not been identified.
The NSL Proteins
The NSL proteins NSL1, NSL2 and NSL3 appear to be unique to the NSL complex. In association with MOF, these proteins regulate housekeeping genes in Drosophila.2,5 NSL1, NSL2 and NSL3 are essential for Drosophila viability with lethality resulting from P-element insertions in the corresponding genes.6
The Drosophila NSL1 protein has been proposed to serve as a scaffold for the NSL complex, interacting directly with MOF, WDS, MCRS2 and MBD-R2.27 NSL1 is predicted to be intrinsically disordered, interacting with its partners through short linear motifs including the PEHE domain for interaction with MOF,33 and a WIN motif for interaction with WDS.27 We have not been able to identify an NSL1 homolog in the nematode proteome. However, since detection of homology for unstructured proteins such as NSL1 is challenging, the existence of a nematode equivalent of NSL1 cannot be excluded at this stage.
Although there is no clear nematode homologue of NSL1, in a recent publication we reported the identification of homologues of NSL2 and NSL3.34 We named these proteins SUMV-1 (C34E10.8) and SUMV-2 (F54D11.2) (suppressor of multivulva), respectively, because mutation of the genes encoding either of these proteins suppresses the multiple vulva phenotype in a synthetic multivulva (synMuv) mutant background. The multiple vulva phenotype of synMuv mutants arises through the simultaneous mutation of two synMuv genes in different classes (e.g. synMuv A and synMuv B) (for a review of synthetic multivulva and synthetic multivulva suppressor genes, see ref. 35).
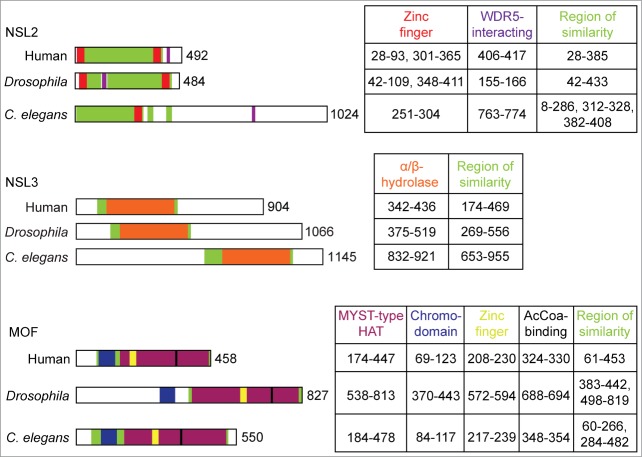
Similarity between C. elegans SUMV-1 and Drosophila NSL2 is apparent primarily in the N-terminal portion of the nematode protein (Fig. 2).34 Within this region, the nematode protein contains a domain called pfam13891, a putative C3HC3H double zinc finger motif annotated as a potential DNA binding domain of chromatin remodeling proteins and helicases,36 although experimental evidence supporting this classification is lacking. It is possible that domains belonging to pfam13891 are, like other small zinc-coordinating domains, instead involved in protein-protein interactions, potentially with modified histones.37,38 While a single pfam13891 domain is found in SUMV-1, NSL2 contains two of these domains, as does Ino80d, a subunit of the INO80 chromatin remodeling complex.28 Recently a WDR5-interacting motif has been identified in NSL227 and we have found a similar motif in SUMV-1 (core sequence DEIDLL, Fig. 2), suggesting that this interaction may be conserved in nematodes.
Figure 2.
Schematic of regions of homology between human, Drosophila and putative C. elegans NSL2, NSL3 and MOF proteins. Black-bordered bars represent entire protein sequence, with the total number of amino acids indicated on the right end. Divisions within each bar correspond to key domains, motifs and regions which are listed in the table to the right of each figure, colored accordingly. Tables list the amino acids that each region has been found, or is predicted, to span. Regions have been positioned based on annotations in UniProt and the conserved domain database where available. Where a region has yet to be specified in either database, putative regions have been identified using alignment prediction tool T-coffee. Region of similarity indicates regions which were found to have substantial conservation, including regions not covered by any other annotation.
Although similarly between C. elegans SUMV-2 and Drosophila NSL3 is only evident in the C-terminal portion of the nematode protein (Fig. 2),34 these proteins are classified as orthologous in OrthoList, as judged by orthology prediction programs InParanoid and Ensemble Compara.39 Within the region of similarity, both proteins are predicted to have an α/β-hydrolase fold.40
A putative nematode NSL-like complex
Prior to our work identifying similarity between SUMV-1 and SUMV-2 and NSL2 and NSL3, respectively, the two nematode genes sumv-1 and sumv-2 had been identified in a genome-wide RNA interference (RNAi) screen for synMuv suppressor genes.41 Interestingly, this screen also identified C. elegans homologues of NSL complex components WDR5 (wdr-5.1) and HCFC1 (hcf-1) as synMuv suppressors. In our study, we additionally showed synMuv suppression with knockdown of mys-2, the worm homologue of MOF.34 That 5 genes which encode proteins that are homologous to components of the Drosophila and mammalian NSL complexes show this same reduction-of-function phenotype, suggests that the proteins that they encode are functionally related. We suggest therefore, that these proteins may act together within a nematode NSL-like HAT complex.
As an initial assessment of this hypothesis, we performed yeast two-hybrid assays and found that SUMV-1 and SUMV-2 interact in this system. We also examined whether these two proteins could interact with MYS-2 and, although we detected no interaction between SUMV-1 and MYS-2, we did find that SUMV-2 was a binding partner of MYS-2.34 Further assays will be required to determine whether there is a physical association between the other putative complex components including HCF-1, WDR-5.1, MCRS-1 and OGT-1, and to test whether reduction of mcrs-1 or ogt-1 function gives rise to a synMuv suppression phenotype, as would be expected if these proteins are functionally linked in an NSL-like complex.
In addition to identifying the genes encoding putative NSL complex components as synMuv suppressor genes, both our study and the earlier RNAi screen also found that RNAi against dpy-30 suppressed the synMuv phenotype.41 The DPY-30 protein is homologous to human protein hDPY-30, which is a member of the COMPASS complex.42 In light of the reported co-ordination of NSL and COMPASS complex activity in humans noted above,26 it is of interest that we also demonstrated a direct interaction between DPY-30 and both SUMV-1 and SUMV-2 using yeast two-hybrid assays.34 The common phenotype of synMuv suppression, combined with evidence of physical association, suggests that DPY-30 might facilitate a functional interaction between the nematode COMPASS complex and putative NSL complex.
Possible roles of a putative NSL complex in C. elegans
Given that the NSL proteins appear to be uniquely involved in the NSL complex, and since SUMV-1 and SUMV-2 are the only identified nematode homologues of the NSL proteins, the functions of these two proteins may shed light on the physiological roles of the putative C. elegans NSL complex. Beyond vulval specification, a further developmental role for this complex is suggested by two observations pertaining to SUMV-2. During normal C. elegans development, germline differentiation is suppressed in somatic cells. However, if the function of a protein called MEP-1 is compromised, the germline-soma distinction is lost, with the result that germline genes such as P-granule components are ectopically expressed in the soma.43 This loss of somatic suppression of germline patterns of gene expression is accompanied by larval lethality in mep-1 mutants grown under standard culture conditions. RNAi of sumv-2 partially rescues the lethality of mep-1 mutants,41 suggesting that SUMV-2 antagonises the activity of MEP-1 in maintaining the germline-soma distinction. mep-1 is a synMuv B gene and, interestingly, loss of function of several other synMuv B genes similarly results in ectopic somatic expression of germline genes but is not accompanied by larval arrest under standard conditions.44 However, when the function of these other synMuv B genes is compromised at high temperature, the ectopic somatic expression of germline genes is enhanced and this is accompanied by larval lethality, with the latter referred to as High Temperature Arrest (HTA).45 Consistent with a role for SUMV-2 in antagonising the activity of the synMuv B genes in the maintenance of the germline-soma distinction, sumv-2 RNAi suppresses the HTA phenotype of several synMuv B mutants.45
There is also evidence implicating SUMV-1 and SUMV-2 in the regulation of transgene expression in C. elegans. The expression of multiple-copy transgenes is normally silenced in the nematode germline and in somatic cells lower levels of expression are detected per gene copy from such transgenes than from the corresponding endogenous gene.46,47 A let-858::GFP transgene is usually expressed in somatic tissues but silenced in the germline, however, knockdown of sumv-2 resulted in germline expression of this transgene,41 suggesting that SUMV-2 is required for germline transgene silencing. In addition, we observed that reduction of sumv-1 function increased hypodermal expression from a multiple-copy ctbp-1::gfp transgene, which may indicate a role for SUMV-1 in the somatic regulation of transgene expression.34
SUMV-1 and SUMV-2 may also regulate expression of the Notch ligand LAG-2. While a number of synMuv genes repress expression of a lag-2::gfp reporter, such that ectopic expression of this reporter is observed in the intestinal cells (among others) of synMuv B mutant animals,48,49 knockdown of either sumv-1 and sumv-2 in a synMuv B mutant background reduced the proportion of animals with intestinal lag-2::GFP expression.41 Thus SUMV-1 and SUMV-2 are required for the ectopic expression of lag-2 and may serve as transcriptional activators of this gene.
Finally, SUMV-1 and SUMV-2 play a role in RNAi. This role was determined in the context of depletion of a gene called mom-2 by RNAi, which usually gives rise to embryonic lethality. When mom-2 RNAi was performed concurrently with either sumv-1 or sumv-2 RNAi, a high proportion of surviving progeny was observed,41 suggesting that sumv-1 and sumv-2 are required for robust RNAi. Interestingly, although a role for the MYS-2 HAT in RNAi has not been examined using this assay, the importance of this protein in transgenerational inheritance of RNAi has been demonstrated. The effect of certain RNAi treatments in C. elegans have been shown to be heritable for as many as 80 generations and this inheritance is dependent on MYS-2, among other factors.50
Conclusion
The characterization of a putative NSL-like complex in C. elegans has begun with our presentation of the first evidence of physical interactions between worm homologues of NSL2, NSL3 and MOF and with our work and that of others showing that putative complex components SUMV-1, SUMV-2, MYS-2, HCF-1 and WDR-5.1 play a common role in vulval development. However, further investigations are required to assess whether these proteins, together with OGT-1 and MCRS-1, genuinely function together in a HAT complex analogous to the Drosophila and mammalian NSL complexes.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by a grant from the Australian Research Council awarded to H.R.N and by an Australian Postgraduate Award awarded to M.H.
References
- 1. Hilfiker A, Hilfiker-Kleiner D, Pannuti A, Lucchesi JC. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J 1997; 16:2054-60; PMID:9155031; http://dx.doi.org/ 10.1093/emboj/16.8.2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lam KC, Muhlpfordt F, Vaquerizas JM, Raja SJ, Holz H, Luscombe NM, Manke T, Akhtar A. The NSL complex regulates housekeeping genes in Drosophila. PLoS Genet 2012; 8:e1002736; PMID:22723752; http://dx.doi.org/ 10.1371/journal.pgen.1002736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ravens S, Fournier M, Ye T, Stierle M, Dembele D, Chavant V, Tora L. Mof-associated complexes have overlapping and unique roles in regulating pluripotency in embryonic stem cells and during differentiation. eLife 2014; 3; PMID:24898753; http://dx.doi.org/ 10.7554/eLife.02104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chelmicki T, Dundar F, Turley MJ, Khanam T, Aktas T, Ramirez F, Gendrel AV, Wright PR, Videm P, Backofen R, et al. . MOF-associated complexes ensure stem cell identity and Xist repression. eLife 2014; 3:e02024; PMID:24842875; http://dx.doi.org/ 10.7554/eLife.02024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Raja SJ, Charapitsa I, Conrad T, Vaquerizas JM, Gebhardt P, Holz H, Kadlec J, Fraterman S, Luscombe NM, Akhtar A. The nonspecific lethal complex is a transcriptional regulator in Drosophila. Mol Cell 2010; 38:827-41; PMID:20620954; http://dx.doi.org/ 10.1016/j.molcel.2010.05.021 [DOI] [PubMed] [Google Scholar]
- 6. Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, Vermeulen M, Buscaino A, Duncan K, Mueller J, et al. . Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell 2006; 21:811-23; PMID:16543150; http://dx.doi.org/ 10.1016/j.molcel.2006.02.007 [DOI] [PubMed] [Google Scholar]
- 7. Cai Y, Jin J, Swanson SK, Cole MD, Choi SH, Florens L, Washburn MP, Conaway JW, Conaway RC. Subunit composition and substrate specificity of a MOF-containing histone acetyltransferase distinct from the male-specific lethal (MSL) complex. J Biol Chem 2010; 285:4268-72; PMID:20018852; http://dx.doi.org/ 10.1074/jbc.C109.087981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Taipale M, Rea S, Richter K, Vilar A, Lichter P, Imhof A, Akhtar A. hMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Mol Cell Biol 2005; 25:6798-810; PMID:16024812; http://dx.doi.org/ 10.1128/MCB.25.15.6798-6810.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thomas T, Dixon MP, Kueh AJ, Voss AK. Mof (MYST1 or KAT8) is essential for progression of embryonic development past the blastocyst stage and required for normal chromatin architecture. Mol Cell Biol 2008; 28:5093-105; PMID:18541669; http://dx.doi.org/ 10.1128/MCB.02202-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shibata Y, Takeshita H, Sasakawa N, Sawa H. Double bromodomain protein BET-1 and MYST HATs establish and maintain stable cell fates in C. elegans. Development 2010; 137:1045-53; PMID:20181741; http://dx.doi.org/ 10.1242/dev.042812 [DOI] [PubMed] [Google Scholar]
- 11. Wysocka J, Herr W. The herpes simplex virus VP16-induced complex: the makings of a regulatory switch. Trends Biochem Sci 2003; 28:294-304; PMID:12826401; http://dx.doi.org/ 10.1016/S0968-0004(03)00088-4 [DOI] [PubMed] [Google Scholar]
- 12. Kristie TM, Sharp PA. Purification of the cellular C1 factor required for the stable recognition of the Oct-1 homeodomain by the herpes simplex virus alpha-trans-induction factor (VP16). J Biol Chem 1993; 268:6525-34; PMID:8454622 [PubMed] [Google Scholar]
- 13. Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev 2003; 17:896-911; PMID:12670868; http://dx.doi.org/ 10.1101/gad.252103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li J, Ebata A, Dong Y, Rizki G, Iwata T, Lee SS. Caenorhabditis elegans HCF-1 Functions in Longevity Maintenance as a DAF-16 Regulator. PLoS biology 2008; 6:e233; PMID:18828672; http://dx.doi.org/ 10.1371/journal.pbio.0060233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee S, Horn V, Julien E, Liu Y, Wysocka J, Bowerman B, Hengartner MO, Herr W. Epigenetic regulation of histone H3 serine 10 phosphorylation status by HCF-1 proteins in C. elegans and mammalian cells. PloS one 2007; 2:e1213; PMID:18043729; http://dx.doi.org/ 10.1371/journal.pone.0001213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang J, Torii M, Liu H, Hart GW, Hu ZZ. dbOGAP - an integrated bioinformatics resource for protein O-GlcNAcylation. BMC Bioinformatics 2011; 12:91; PMID:21466708; http://dx.doi.org/ 10.1186/1471-2105-12-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sakabe K, Wang Z, Hart GW. Beta-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc Natl Acad Sci U S A 2010; 107:19915-20; PMID:21045127; http://dx.doi.org/ 10.1073/pnas.1009023107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shafi R, Iyer SP, Ellies LG, O'Donnell N, Marek KW, Chui D, Hart GW, Marth JD. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci U S A 2000; 97:5735-9; PMID:10801981; http://dx.doi.org/ 10.1073/pnas.100471497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hanover JA, Forsythe ME, Hennessey PT, Brodigan TM, Love DC, Ashwell G, Krause M. A Caenorhabditis elegans model of insulin resistance: altered macronutrient storage and dauer formation in an OGT-1 knockout. Proc Natl Acad Sci U S A 2005; 102:11266-71; PMID:16051707; http://dx.doi.org/ 10.1073/pnas.0408771102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lazarus MB, Jiang J, Kapuria V, Bhuiyan T, Janetzko J, Zandberg WF, Vocadlo DJ, Herr W, Walker S. HCF-1 Is Cleaved in the Active Site of O-GlcNAc Transferase. Science 2013; 342:1235-9; PMID:24311690; http://dx.doi.org/ 10.1126/science.1243990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wysocka J, Liu Y, Kobayashi R, Herr W. Developmental and cell-cycle regulation of Caenorhabditis elegans HCF phosphorylation. Biochemistry 2001; 40:5786-94; PMID:11341844; http://dx.doi.org/ 10.1021/bi010086o [DOI] [PubMed] [Google Scholar]
- 22. Migliori V, Mapelli M, Guccione E. On WD40 proteins: propelling our knowledge of transcriptional control? Epigenetics 2012; 7:815-22; PMID:22810296; http://dx.doi.org/ 10.4161/epi.21140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li T, Kelly WG. A role for Set1/MLL-related components in epigenetic regulation of the Caenorhabditis elegans germ line. PLoS Genet 2011; 7:e1001349; PMID:21455483; http://dx.doi.org/ 10.1371/journal.pgen.1001349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fisher K, Southall SM, Wilson JR, Poulin GB. Methylation and demethylation activities of a C. elegans MLL-like complex attenuate RAS signalling. Dev Biol 2010; 341:142-53; PMID:20188723; http://dx.doi.org/ 10.1016/j.ydbio.2010.02.023 [DOI] [PubMed] [Google Scholar]
- 25. Greer EL, Maures TJ, Ucar D, Hauswirth AG, Mancini E, Lim JP, Benayoun BA, Shi Y, Brunet A. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature 2011; 479:365-71; PMID:22012258; http://dx.doi.org/ 10.1038/nature10572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhao X, Su J, Wang F, Liu D, Ding J, Yang Y, Conaway JW, Conaway RC, Cao L, Wu D, et al. . Crosstalk between NSL histone acetyltransferase and MLL/SET complexes: NSL complex functions in promoting histone H3K4 Di-Methylation activity by MLL/SET complexes. PLoS Genet 2013; 9:e1003940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dias J, Van Nguyen N, Georgiev P, Gaub A, Brettschneider J, Cusack S, Kadlec J, Akhtar A. Structural analysis of the KANSL1/WDR5/KANSL2 complex reveals that WDR5 is required for efficient assembly and chromatin targeting of the NSL complex. Genes Dev 2014; 28:929-42; PMID:24788516; http://dx.doi.org/ 10.1101/gad.240200.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jin J, Cai Y, Yao T, Gottschalk AJ, Florens L, Swanson SK, Gutierrez JL, Coleman MK, Workman JL, Mushegian A, et al. . A mammalian chromatin remodeling complex with similarities to the yeast INO80 complex. J Biol Chem 2005; 280:41207-12; PMID:16230350; http://dx.doi.org/ 10.1074/jbc.M509128200 [DOI] [PubMed] [Google Scholar]
- 29. Shimono K, Shimono Y, Shimokata K, Ishiguro N, Takahashi M. Microspherule protein 1, Mi-2beta, and RET finger protein associate in the nucleolus and up-regulate ribosomal gene transcription. J Biol Chem 2005; 280:39436-47; PMID:16186106; http://dx.doi.org/ 10.1074/jbc.M507356200 [DOI] [PubMed] [Google Scholar]
- 30. Hirohashi Y, Wang Q, Liu Q, Du X, Zhang H, Sato N, Greene MI. p78/MCRS1 forms a complex with centrosomal protein Nde1 and is essential for cell viability. Oncogene 2006; 25:4937-46; PMID:16547491; http://dx.doi.org/ 10.1038/sj.onc.1209500 [DOI] [PubMed] [Google Scholar]
- 31. Andersen DS, Raja SJ, Colombani J, Shaw RL, Langton PF, Akhtar A, Tapon N. Drosophila MCRS2 associates with RNA polymerase II complexes to regulate transcription. Mol Cell Biol 2010; 30:4744-55; PMID:20679484; http://dx.doi.org/ 10.1128/MCB.01586-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Badeaux AI, Yang Y, Cardenas K, Vemulapalli V, Chen K, Kusewitt D, Richie E, Li W, Bedford MT. Loss of the methyl lysine effector protein PHF20 impacts the expression of genes regulated by the lysine acetyltransferase MOF. J Biol Chem 2012; 287:429-37; PMID:22072714; http://dx.doi.org/ 10.1074/jbc.M111.271163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kadlec J, Hallacli E, Lipp M, Holz H, Sanchez-Weatherby J, Cusack S, Akhtar A. Structural basis for MOF and MSL3 recruitment into the dosage compensation complex by MSL1. Nat Struct Mol Biol 2011; 18:142-9; PMID:21217699; http://dx.doi.org/ 10.1038/nsmb.1960 [DOI] [PubMed] [Google Scholar]
- 34. Yücel D, Hoe M, Llamosas E, Kant S, Jamieson C, Young PA, Crossley M, Nicholas HR. SUMV-1 antagonizes the activity of synthetic multivulva genes in Caenorhabditis elegans. Dev Biol 2014; 392:266-82; PMID:24882710; http://dx.doi.org/ 10.1016/j.ydbio.2014.05.018 [DOI] [PubMed] [Google Scholar]
- 35. Cui M, Han M. Roles of chromatin factors in C. elegans development. WormBook 2007:1-16; PMID:18050494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lanczycki CJ, et al. . CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res 2013; 41:D348-52; PMID:23197659; http://dx.doi.org/ 10.1093/nar/gks1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. He F, Umehara T, Saito K, Harada T, Watanabe S, Yabuki T, Kigawa T, Takahashi M, Kuwasako K, Tsuda K, et al. . Structural Insight into the Zinc Finger CW Domain as a Histone Modification Reader. Structure 2010; 18:1127-39; PMID:20826339; http://dx.doi.org/ 10.1016/j.str.2010.06.012 [DOI] [PubMed] [Google Scholar]
- 38. Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 2006; 442:91-5; PMID:16728978; http://dx.doi.org/ 10.1038/nature05020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shaye DD, Greenwald I. OrthoList: a compendium of C. elegans genes with human orthologs. PloS one 2011; 6:e20085; PMID:21647448; http://dx.doi.org/ 10.1371/journal.pone.0020085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kelley LA, Sternberg MJE. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protocols 2009; 4:363-71; http://dx.doi.org/ 10.1038/nprot.2009.2 [DOI] [PubMed] [Google Scholar]
- 41. Cui M, Kim EB, Han M. Diverse chromatin remodeling genes antagonize the Rb-involved SynMuv pathways in C. elegans. PLoS Genet 2006; 2:e74; PMID:16710447; http://dx.doi.org/ 10.1371/journal.pgen.0020074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pferdehirt RR, Kruesi WS, Meyer BJ. An MLL/COMPASS subunit functions in the C. elegans dosage compensation complex to target X chromosomes for transcriptional regulation of gene expression. Genes Dev 2011; 25:499-515; PMID:21363964; http://dx.doi.org/ 10.1101/gad.2016011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Unhavaithaya Y, Shin TH, Miliaras N, Lee J, Oyama T, Mello CC. MEP-1 and a Homolog of the NURD Complex Component Mi-2 Act Together to Maintain Germline-Soma Distinctions in C. elegans. Cell 2002; 111:991-1002; PMID:12507426; http://dx.doi.org/ 10.1016/S0092-8674(02)01202-3 [DOI] [PubMed] [Google Scholar]
- 44. Wang D, Kennedy S, Conte D, Jr., Kim JK, Gabel HW, Kamath RS, Mello CC, Ruvkun G. Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature 2005; 436:593-7; PMID:16049496; http://dx.doi.org/ 10.1038/nature04010 [DOI] [PubMed] [Google Scholar]
- 45. Petrella LN, Wang W, Spike CA, Rechtsteiner A, Reinke V, Strome S. synMuv B proteins antagonize germline fate in the intestine and ensure C. elegans survival. Development 2011; 138:1069-79; PMID:21343362; http://dx.doi.org/ 10.1242/dev.059501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kelly WG, Xu S, Montgomery MK, Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics 1997; 146:227-38; PMID:9136012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mello C, Fire A. DNA transformation. Methods Cell Biol 1995; 48:451-82; PMID:8531738; http://dx.doi.org/ 10.1016/S0091-679X(08)61399-0 [DOI] [PubMed] [Google Scholar]
- 48. Poulin G, Dong Y, Fraser AG, Hopper NA, Ahringer J. Chromatin regulation and sumoylation in the inhibition of Ras-induced vulval development in Caenorhabditis elegans. EMBO J 2005; 24:2613-23; PMID:15990876; http://dx.doi.org/ 10.1038/sj.emboj.7600726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dufourcq P, Victor M, Gay F, Calvo D, Hodgkin J, Shi Y. Functional requirement for histone deacetylase 1 in Caenorhabditis elegans gonadogenesis. Mol Cell Biol 2002; 22:3024-34; PMID:11940660; http://dx.doi.org/ 10.1128/MCB.22.9.3024-3034.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vastenhouw NL, Brunschwig K, Okihara KL, Muller F, Tijsterman M, Plasterk RH. Gene expression: long-term gene silencing by RNAi. Nature 2006; 442:882; PMID:16929289; http://dx.doi.org/ 10.1038/442882a [DOI] [PubMed] [Google Scholar]