Abstract
Tenofovir (TFV) is used in combination with other antiretroviral drugs for human immunodeficiency virus (HIV) treatment and prevention. TFV requires two phosphorylation steps to become pharmacologically active; however, the kinases that activate TFV in cells and tissues susceptible to HIV infection have yet to be identified. Peripheral blood mononuclear cells (PBMC), vaginal, and colorectal tissues were transfected with siRNA targeting nucleotide kinases, incubated with TFV, and TFV-monophosphate (TFV-MP) and TFV-diphosphate (TFV-DP) were measured using mass spectrometry–liquid chromatography. Adenylate kinase 2 (AK2) performed the first TFV phosphorylation step in PBMC, vaginal, and colorectal tissues. Interestingly, both pyruvate kinase isozymes, muscle (PKM) or liver and red blood cell (PKLR), were able to phosphorylate TFV-MP to TFV-DP in PBMC and vaginal tissue, while creatine kinase, muscle (CKM) catalyzed this conversion in colorectal tissue. In addition, next-generation sequencing of the Microbicide Trials Network MTN-001 clinical samples detected 71 previously unreported genetic variants in the genes encoding these kinases. In conclusion, our results demonstrate that TFV is activated in a compartment-specific manner. Further, genetic variants have been identified that could negatively impact TFV activation, thereby compromising TFV efficacy in HIV treatment and prevention.
Abbreviations: HIV, human immunodeficiency virus; PrEP, pre-exposure prophylaxis; TFV, tenofovir; PBMC, peripheral blood mononuclear cells; TFV-MP, tenofovir-monophosphate; TFV-DP, tenofovir-diphosphate; MTN-001, Microbicide Trials Network Study MTN-001; AK2, adenylate kinase 2; PKM, pyruvate kinase, muscle; PKLR, pyruvate kinase, liver and red blood cell; CKM, creatine kinase, muscle; NME1, NME/NM23 nucleoside diphosphate kinase 1; GUK1, guanylate kinase 1; SNV, single-nucleotide variant
Keywords: HIV, HIV pre-exposure prophylaxis, Tenofovir activation, Nucleotide kinases, Microbicide Trials Network study MTN-001, Targeted next-generation sequencing
Highlights
-
•
The anti-HIV drug tenofovir is activated in a tissue-specific manner.
-
•
AK2 phosphorylates tenofovir to tenofovir-monophosphate in PBMC, vagina, and colon.
-
•
PKM, PKLR phosphorylate tenofovir-monophosphate to diphosphate in PBMC and vagina.
-
•
CKM phosphorylates tenofovir-monophosphate to diphosphate in colon.
-
•
Because these enzymes are polymorphic and may be dysfunctional in some individuals, these findings suggest that tenofovir-based HIV PrEP may not be protective for all individuals.
1. Introduction
Within the last decade, tenofovir (TFV), prescribed as tenofovir disoproxil fumarate in its prodrug formulation, has emerged as a critical component of antiretroviral combination therapy for the treatment of human immunodeficiency virus (HIV) (Schooley et al., 2002; Robbins et al., 1998). More recently, oral as well as vaginal and rectal microbicide gel preparations of TFV have been investigated for use in pre-exposure prophylaxis (PrEP) as an HIV prevention strategy for individuals at high-risk of viral exposure (Baeten et al., 2012; Mayer et al., 2006; Anton et al., 2012). TFV is a desirable drug candidate for PrEP due to the long half-life of active drug TFV-diphosphate (TFV-DP), reported to be 53 h in vaginal tissue homogenate and up to 139 h in vaginal CD4 + cells following an oral dosing of HIV-uninfected women (Derdelinckx et al., 2006; Louissaint et al., 2013). That being said, there has been discrepancy in the prophylactic effect observed for TFV-based regimens. For example, the Partners in Prevention study demonstrated a 67–75% reduction in HIV acquisition in serodiscordant heterosexual couples, iPrEx demonstrated a 44% reduction in men or transgender women who have sex with men, whereas FEM-PrEP and VOICE trials showed no significant reduction in the rate of infection in heterosexual women (Grant et al., 2010; Van Damme et al., 2012; Marrazzo et al., 2015). This disparity has been primarily due to poor adherence. Adjusting for adherence, the differences among these clinical trials have been attributed to the increased accumulation of active drug in colorectal versus vaginal tissue. As such, these findings suggest that the enzymes responsible for TFV activation may differ between colorectal and vaginal tissue, however, this has not been tested thus far. Further, while yet to be explored, it can be envisioned that genetic variation in the nucleotide kinases that activate TFV could underlie observed inter-individual differences in tissue TFV-DP concentrations that has been noted even when adherence is high (Louissaint et al., 2013; Hendrix et al., 2013; Patterson et al., 2011).
As TFV requires phosphorylation by nucleotide kinases in order to become pharmacologically active, both local mucosal tissue cell phosphorylation and distant peripheral blood CD4 + cell phosphorylation with secondary migration to mucosal tissue may be critical in achieving effective TFV-DP concentrations. If mucosal tissue phosphorylation is an essential contribution to mucosal tissue cell TFV-DP concentrations, then the kinases responsible for TFV transformation are expressed in cells and tissues associated with HIV infection (Robbins et al., 1998; Kearney et al., 2004). To date, the expression profiles of nucleotide kinases in peripheral blood mononuclear cells (PBMC), vaginal tissue, and colorectal tissue have not been characterized and data demonstrating kinase activity towards TFV and TFV-MP are lacking. In vitro studies using human T-lymphoid cells demonstrated the mitochondrial and cytosolic enzyme adenylate kinase isoform 2 (AK2) could catalyze the phosphorylation of TFV to form TFV-monophosphate (TFV-MP) (Nobumoto et al., 1998; Robbins et al., 1995; Topalis et al., 2008). Experimental evidence is deficient, however, with regard to the nucleotide kinase(s) capable of transforming TFV-MP to TFV-DP. TFV-MP has been reported as a substrate of NME/NM23 nucleoside diphosphate kinase 1 (NME1) as this enzyme demonstrates broad activity towards purine nucleoside diphosphates (Robbins et al., 1998; Bourdais et al., 1996). Yet, in separate studies, NME1 exhibited low to non-detectable rates of catalysis, while creatine kinase, muscle (CKM) efficiently phosphorylated TFV-MP and minor catalytic efficiency towards TFV-MP was demonstrated by rabbit pyruvate kinase, muscle (PKM) (Koch et al., 2009; Varga et al., 2013). Nevertheless, although these kinases exhibit activity towards TFV or TFV-MP using in vitro model systems, whether or not they are expressed in cells and tissues susceptible to HIV infection is unknown.
In the present study, we characterized nucleotide kinase expression in PBMC, vaginal tissue, and colorectal tissue in order to identify those that catalyze the phosphorylation of TFV to TFV-MP and TFV-MP to TFV-DP. Towards this end, we knocked down the protein expression of AK2, CKM, PKM, pyruvate kinase, liver and red blood cell (PKLR), and guanylate kinase 1 (GUK1) using siRNA. Neither PKLR nor GUK1 have been previously reported to activate TFV; however, PKLR exhibits an approximate 70% amino acid sequence identity to the isozyme PKM and GUK1 has been demonstrated to phosphorylate the antiviral drug adefovir which is structurally similar to TFV (Gentry et al., 2011). Taking these studies a step further, we wanted to leverage our identification of the nucleotide kinases that activate TFV and test for the existence of genetic variants in AK2, CKM, PKM, and PKLR. To do so, we performed next-generation targeted sequencing of genomic DNA isolated from the plasma of 142 HIV-uninfected female participants of the Microbicide Trials Network study MTN-001 (Hendrix et al., 2013). In this work, we have demonstrated at an enzymatic level that TFV is activated in a tissue-specific manner. We also put forth an innovative concept that variation in the genes that encode the nucleotide kinases that activate TFV may contribute to inter-individual differences observed clinically. Taken together, these findings represent a shift in thinking about the factors that govern variability in TFV efficacy and pharmacokinetics.
2. Materials and Methods
2.1. siRNA Knockdown of Nucleotide Kinases
PBMC were obtained from Bioreclamation (Westbury, NY), and fresh vaginal and colorectal biopsies were obtained from the Johns Hopkins University School of Medicine Tissue Bank (Baltimore, MD). Donor information is as follows: PBMC (n = 3; 38 y.o. male, 24 y.o. female, and 27 y.o. female); vaginal tissue (n = 3; 31 y.o. female, 48 y.o. female, and 37 y.o. female); colorectal tissue (n = 3; 35 y.o. male, 39 y.o. female, 42 y.o. male). All donors were healthy, HIV-uninfected individuals. PBMC, vaginal, and colorectal tissues were transfected with siRNA and incubated with TFV. Homogenized cells and tissues were then immunoblotted for protein expression and intracellular metabolites detected using ultra-high performance liquid chromatography–tandem mass spectrometry (uHPLC–MS/MS). Electroporation, immunoblotting, and uHPLC–MS/MS assay conditions are described in the supplementary methods.
2.2. Clinical Samples
Plasma was obtained from HIV-uninfected females (n = 142) enrolled in the Microbicide Trials Network study MTN-001 across seven study sites: Umkomaas and Botha's Hill, Durban, South Africa; Makerere University–Johns Hopkins University Research Collaboration, Kampala, Uganda; Case Western Reserve University in Cleveland, OH, United States of America (USA); University of Pittsburgh, Pittsburgh, PA, USA; University of Alabama at Birmingham, Birmingham, AL, USA; Bronx-Lebanon Hospital Center, New York City, NY, USA (Hendrix et al., 2013). The current analysis was approved by the Johns Hopkins Medicine IRB (NA_00016287).
2.3. Genomic DNA Isolation
Genomic DNA was isolated from 200 μL of plasma using the GeneJET Whole Blood Genomic DNA Purification Mini Kit (Thermo Fisher Scientific, Waltham, MA). Purified DNA was eluted using 50 μL of elution buffer.
2.4. Next-generation Sequencing Targeted Enrichment Design
Sequencing was performed using the Illumina TruSeq custom amplicon v1.5 kit (San Diego, CA). Custom probes targeting the exonic regions of AK2, CKM, PKM, and PKLR were generated in silico using Illumina DesignStudio software. The chromosomal coordinates used were as follows: AK2 1:33473531–1:33502522; CKM 19:45809661–19:45826243; PKM 15:72492805–15:72523694; and PKLR 1:155259074–1:155271235. The start and stop coordinates for each target region is detailed in Supplementary Table 1. The final design included 102 amplicons listed in Supplementary Table 2. Sample preparation, sequencing, and data analyses are detailed in the supplementary methods. All genetic variants reported in this study have been submitted to the SNP database under the submitter handle “BUMPUSLAB.” The phenotypic consequence of missense variants was assigned using SIFT (sorts intolerant from tolerant substitutions; J. Craig Venter Institute online tool) and PolyPhen (polymorphism phenotyping; Harvard University online tool) in silico prediction tools where amino acid substitutions were scored (Ng and Henikoff, 2001; Ramensky et al., 2002).
2.5. Statistical Analysis
Statistical analyses were performed using GraphPad Prism (San Diego, CA). Two-tailed unpaired t tests were performed and significance was denoted as follows: *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001.
2.6. Funding
This work was supported by the NIH grants UM1 AI106707 (Microbicide Trials Network), UM1AI068613 (HIV Prevention Trials Network), P30 AI094189 (Johns Hopkins University Center for AIDS Research), R01 GM103853 (awarded to N.N.B.) and by a 2015 PhRMA Foundation Pre Doctoral Fellowship in Pharmacology (awarded to J.M.L.). The Microbicide Trials Network is funded by NIAID (UM1AI068633, UM1AI068615, UM1AI106707), with co-funding from the Eunice Kennedy Shriver NICHD and NIMH, all components of the NIH.
The funding sponsors had no role in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; and in the decision to submit the paper for publication.
3. Results
3.1. Nucleotide Kinase Activation of TFV in Cells and Tissues Susceptible to HIV Infection
In order to identify the nucleotide kinases that activate TFV in PBMC, vaginal, and colorectal tissue, we delivered siRNA targeted to AK2, GUK1, PKM, PKLR, and CKM to cells and tissues followed by incubation with TFV to test the impact on drug activation. The experiments described herein were performed in cells and tissues from healthy, HIV-uninfected donors that were not administered TFV. Candidate kinases were screened using immunoblotting in order to test for their expression in PBMC, vaginal, and colorectal tissues shown in Fig. 1. The non-targeting, or no target, siRNA lane in the representative immunoblot is commensurate with basal expression of each kinase in PBMC, colorectal, and vaginal tissues, respectively. We found that AK2, which has been previously reported to catalyze the phosphorylation of TFV to TFV-MP, was expressed ubiquitously in the cells and tissues investigated in this study. A similar protein expression profile was observed for the nucleotide kinase GUK1. Interestingly, of the candidates examined for the transformation of TFV-MP to TFV-DP, PKM and PKLR were detectable in both PBMC and vaginal tissue, while basal expression of either isoenzyme was not detectable in colorectal tissue using immunoblot analyses. In contrast, of the kinases tested, CKM was observed to be exclusively expressed in colorectal tissue and not detectable at the protein level in PBMC or vaginal tissue. It is important to note that NME1, which has been reported to transform TFV-MP to TFV-DP with weak catalytic efficiency, was not detectable at the protein level in PBMC, vaginal tissue, or colorectal tissue (data not shown).
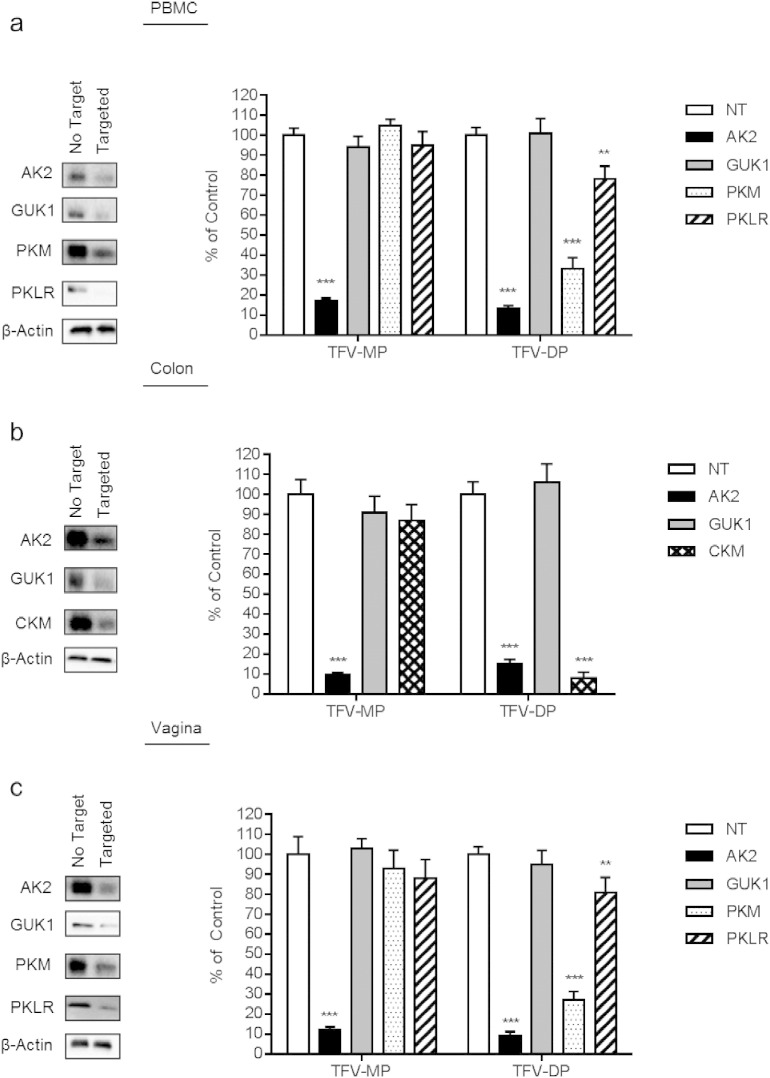
Fig. 1.
Targeted siRNA knockdown of nucleotide kinases in PBMC, colorectal tissue, and vaginal tissue and the resulting impact on TFV-MP and TFV-DP intracellular formation.
PBMC, colorectal tissue, and vaginal tissue were electroporated with 500 nM non-targeting siRNA or siRNA targeting AK2, GUK1, PKM, PKLR, and CKM and incubated for 24 h or 48 h for tissue or PBMC, respectively. Representative immunoblots demonstrate decreased nucleotide kinase expression with each targeted siRNA treatment relative to the non-targeting siRNA control. siRNA treated (a) PBMC, (b) colorectal tissue, and (c) vaginal tissue were incubated with 10 μM TFV for 12 h (n = 3 per treatment). Intracellular anabolites were extracted from which TFV-MP and TFV-DP were detected using uHPLC–MS/MS as depicted in the corresponding bar graphs showing mean ± standard deviation for each treatment. Statistical analyses were performed using a two-tailed unpaired t test to compare relative levels of anabolite production between non-targeted and targeted siRNA conditions; ** = p ≤ 0.01; *** = p ≤ 0.001.
Guided by our preliminary findings, siRNA was used to knockdown the following nucleotide kinases AK2, GUK1, PKM, and PKLR in PBMC (n = 3), AK2, GUK1, and CKM in colorectal tissue (n = 3), and AK2, GUK1, PKM, and PKLR in vaginal tissue (n = 3). Decreased expression of each targeted kinase was evaluated relative to the non-targeting siRNA control using immunoblot analysis (Fig. 1). The intracellular formation of TFV metabolites was detected using uHPLC–MS/MS and the impact of enzyme loss-of-function on TFV-MP and TFV-DP formation following incubation with TFV are shown as bar graphs in Fig. 1. When AK2 was knocked down, TFV-MP decreased to 17 ± 1.4% (17 / 100 ± 1.4 / 100), 9.5 ± 1.1% (9.5 / 100 ± 1.1 / 100), and 12 ± 1.5% (12 / 100 ± 1.5 / 100; p-value = 3.4E − 6, 3.6E − 5, and 7.2E − 5) of the non-targeting siRNA control in PBMC, colorectal tissue, and vaginal tissue, respectively. Knockdown of AK2 protein expression also resulted in a decrease of TFV-DP to 13 ± 1.7% (13 / 100 ± 1.7/100), 15 ± 2.3% (15 / 100 ± 2.3 / 100), and 9 ± 2.2% (9 / 100 ± 2.2 / 100; p-value = 3.9E − 6, 4.5E − 6, and 2.7E − 5) of non-targeting control in PBMC, colorectal tissue, and vaginal tissue, respectively. Knockdown of PKM decreased TFV-DP to 33 ± 5.8% (33 / 100 ± 5.8 / 100) and 27 ± 4.3% (27 / 100 ± 4.3 / 100; p-value = 2.7E − 5 and 8.2E − 5) of control in PBMC and vaginal tissue, respectively. Additionally, knockdown of PKLR decreased TFV-DP to 78 ± 6.6% (78 / 100 ± 6.6 / 100) and 81 ± 7.4% (81 / 100 ± 7.4 / 100; p-value = 0.008 and 0.017) of control in PBMC and vaginal tissue, respectively. When CKM was knocked down in colorectal tissue, TFV-DP was decreased to 8 ± 2.9% (8 / 100 ± 2.9 / 100; p-value = 2.2E − 5) of control. The observed tissue-specific activation of TFV has been summarized in Fig. 2.
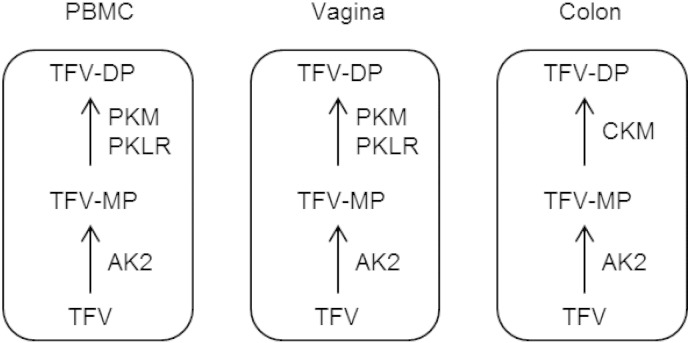
Fig. 2.
Schematic summarizing the intracellular activation of the antiretroviral drug TFV in cells and tissues susceptible to HIV infection.
AK2 can transform TFV to TFV-MP in PBMC, colorectal, and vaginal tissues. PKM and PKLR can transform TFV-MP to the active anabolite TFV-DP in PBMC and vaginal tissue, while CKM can transform TFV-MP to TFV-DP in colorectal tissue.
3.2. MTN-001 Participant Demographics and Ethnicities
For this retrospective study, we obtained plasma from 142 of the total 144 participants of the Microbicide Trials Network study MTN-001, and their demographics and self-identified ethnicities are summarized in Table 1. An approximate half of these participants were enrolled at the United States study sites (USA; n = 72) and the remaining participants were enrolled at either the South Africa (SA; n = 46) or Uganda (UGA; n = 24) study sites.
Table 1.
MTN-001 participant demographics and self-identified ethnicities.
Plasma samples were obtained from 142 HIV-uninfected female participants of the MTN-001 clinical trial for genomic DNA isolation. Participants were enrolled in study sites across the United States (USA; n = 72), South Africa (SA; n = 46), and Uganda (UGA; n = 24).
| MTN-001 participant demographics and ethnicities n = 142 | ||
|---|---|---|
| Study site | Self-identified ethnicity | n (% of n) |
| USA | 72 | |
| African American | 35 (49) | |
| European American | 32 (44) | |
| Asian | 2 (3) | |
| Multi-ethnic | 2 (3) | |
| Hispanic | 1 (1) | |
| SA | 46 | |
| African | 37 (80) | |
| Asian | 5 (11) | |
| Multi-ethnic | 3 (7) | |
| Bhaca | 1 (2) | |
| UGA | 24 | |
| African | 24 (100) | |
3.3. Next-generation Sequencing of MTN-001 Clinical Samples
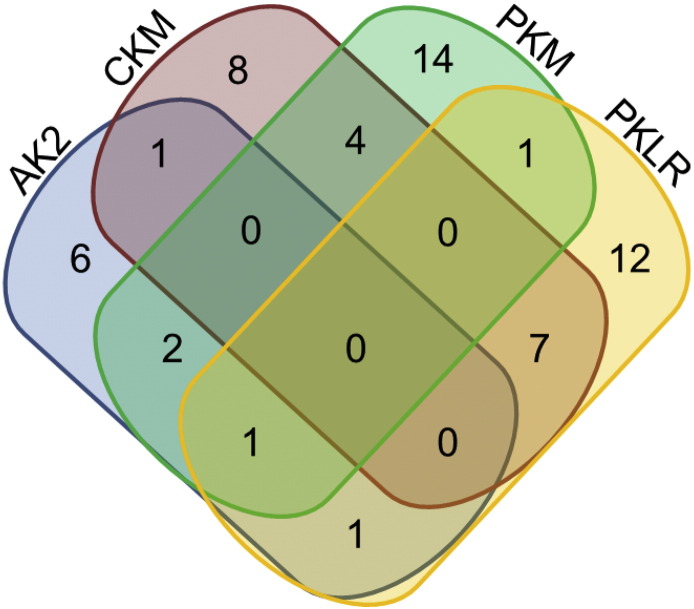
In order to test for the existence of genetic variants within the nucleotide kinases that activate TFV, we designed a targeted assay to sequence the exonic regions of AK2, CKM, PKM, and PKLR. GUK1 was not sequenced in the experiments described herein as we demonstrated siRNA knockdown of this enzyme had no significant impact on the formation of TFV-MP for PBMC, vaginal, or colorectal tissues. Of the 142 MTN-001 participants sequenced in this study, we observed 57 subjects (40%, 57/142) to carry single-base variations or deletions in their DNA that may result in a mutation at the amino acid level. Though this assay was targeted to exonic regions, we detected genetic variation within flanking intronic regions as well. The distribution of the variants and deletions detected for each kinase are depicted using a Venn diagram in Fig. 3. In 40 participants, we observed genetic variants within only one nucleotide kinase, while 17 individuals carried variants within two (n = 16) or three (n = 1) nucleotide kinases.
Fig. 3.
Distribution of nucleotide kinase genetic variants detected in 57 MTN-001 participants.
Each colored oval is representative of a nucleotide kinase AK2 (blue), CKM (red), PKM (green), and PKLR (yellow). Non-overlapping regions demonstrate the number of participants observed to carry single nucleotide variants (SNVs) and deletions within only one gene (n = 40; AK2 n = 6, CKM n = 8, PKM n = 14, PKLR n = 12). Overlapping regions demonstrate the number of participants that were observed to carry SNVs and deletions in more than one gene (n = 17). For example, moving down the left-hand side of the diagram, six participants carried AK2 genetic variants alone, two participants carried variants for AK2 and PKM, one participant carried variants for AK2, PKM, and PKLR, and one participant carried variants for AK2 and PKLR.
3.4. Targeted Sequencing of AK2
A total of 12 previously unreported single nucleotide variants (SNVs) and deletions that may impact the protein-coding potential of the AK2 mRNA transcript were detected in 11 participants, six enrolled in USA study sites, two in SA, and three in UGA. The observed SNVs and deletions for AK2 DNA reference sequence NM_001625.3 are listed in Supplementary Table 3. Of the detected AK2 variants, seven missense variants, which cause a mutation at the amino acid level for the encoded protein, were detected in seven heterozygous individuals at a frequency of one individual per variant (Table 2). Using in silico tools, three of the observed missense variants were predicted to have a deleterious and damaging impact on protein function with a frequency of 2% (3/142 individuals).
Table 2.
AK2 missense variants detected in MTN-001 participants.
A total of seven previously unreported AK2 missense variants were detected in seven heterozygous MTN-001 participants for the coding DNA reference sequence NM_001625.3. The functional consequence of resulting amino acid mutations were predicted using SIFT and PolyPhen in silico tools. A SIFT score < 0.05 was suggestive of a damaging amino acid substitution and > 0.05 a tolerated substitution. A PolyPhen score > 0.908 was suggestive of a probably damaging, 0.447–0.908 a possibly damaging, or < 0.447 a benign amino acid substitution. Deleterious and probably damaging AK2 missense variants was observed at a frequency of 2% (3/142 individuals).
| Study site | Ethnicity | AK2 variant (ref. > alt.) | cDNA position | Coding DNA sequence position | Protein position | Amino acid (ref. > alt.) | Exon | SIFT prediction | PolyPhen prediction |
|---|---|---|---|---|---|---|---|---|---|
| USA | African American | A > G | 249 | 166 | 56 | S > P | 2/6 | Deleterious (0.02) | Probably damaging (0.95) |
| USA | European American | T > C | 451 | 368 | 123 | D > G | 4/6 | Deleterious (0) | Possibly damaging (0.452) |
| SA | African | A > G | 511 | 428 | 143 | L > P | 5/6 | Deleterious (0) | Probably damaging (1) |
| SA | African | G > A | 516 | 433 | 145 | H > Y | 5/6 | Deleterious (0) | Probably damaging (1) |
| USA | African American | A > G | 583 | 500 | 167 | I > T | 6/6 | Deleterious (0.03) | Possibly damaging (0.632) |
| UGA | African | T > A | 622 | 539 | 180 | E > V | 6/6 | Tolerated (0.27) | Benign (0.009) |
| UGA | African | G > T | 743 | 660 | 220 | F > L | 6/6 | Tolerated (0.08) | Benign (0.079) |
3.5. Targeted Sequencing of CKM
A total of 18 previously unreported SNVs and deletions that may impact the protein-coding potential of the CKM mRNA transcript were detected in 17 participants, eight enrolled in USA study sites, four in SA, and five in UGA. The observed variants for CKM DNA reference sequence NM_001824.4 are listed in Supplementary Table 4. Of the detected SNVs, 15 heterozygous participants were found to carry 15 CKM missense variants with a frequency of one individual per variant (Table 3). Using in silico tools, four of the observed missense variants were predicted to have a deleterious and damaging impact on protein function with a frequency of 3% (4/142 individuals). Further, the following reference single nucleotide polymorphism (SNP) missense variants rs11559024 (NM_001824.4:c.248T > C) and rs17357122 (NM_001824.4:c.497G > A) were detected in two heterozygous USA participants (one individual per variant), and rs17875625 (NM_001824.4:c.728C > A) was detected in one heterozygous SA participant. Of the detected reference SNPs, only rs17875625 was predicted to have a deleterious and damaging impact on protein function.
Table 3.
CKM missense variants detected in MTN-001 participants.
A total of 15 previously unreported CKM missense variants were detected in 15 heterozygous MTN-001 participants for the coding DNA reference sequence NM_001824.4. The functional consequence of resulting amino acid mutations were predicted using SIFT and PolyPhen in silico tools. A SIFT score < 0.05 was suggestive of a damaging amino acid substitution and > 0.05 a tolerated substitution. A PolyPhen score > 0.908 was suggestive of a probably damaging, 0.447–0.908 a possibly damaging, or < 0.447 a benign amino acid substitution. Deleterious and probably damaging CKM missense variants was observed at a frequency of 3% (4/142 individuals).
| Study site | Ethnicity | CKM variant (ref. > alt.) | cDNA position | Coding DNA sequence position | Protein position | Amino acid (ref. > alt.) | Exon | SIFT prediction | PolyPhen prediction |
|---|---|---|---|---|---|---|---|---|---|
| USA | European American | A > G | 419 | 244 | 82 | Y > H | 3/8 | Deleterious (0.02) | Possibly damaging (0.474) |
| USA | African American | T > C | 435 | 260 | 87 | E > G | 3/8 | Deleterious (0.04) | Possibly damaging (0.467) |
| SA | African | C > T | 462 | 287 | 96 | R > H | 3/8 | Tolerated (0.07) | Probably damaging (0.976) |
| UGA | African | G > A | 464 | 289 | 97 | H > Y | 3/8 | Deleterious (0) | Probably damaging (0.985) |
| UGA | African | C > T | 564 | 389 | 130 | R > H | 4/8 | Deleterious (0) | Probably damaging (0.966) |
| USA | European American | C > T | 674 | 499 | 167 | G > S | 5/8 | Deleterious (0.01) | Benign (0.307) |
| SA | African | A > G | 680 | 505 | 169 | F > L | 5/8 | Tolerated (1) | Benign (0.003) |
| USA | African American | A > G | 692 | 517 | 173 | Y > H | 5/8 | Deleterious (0) | Possibly Damaging (0.725) |
| USA | Hispanic | C > T | 758 | 583 | 195 | D > N | 5/8 | Tolerated (0.06) | Benign (0.099) |
| UGA | African | G > C | 779 | 604 | 202 | L > V | 5/8 | Deleterious (0.03) | Possibly damaging (0.878) |
| UGA | African | T > A | 969 | 794 | 265 | K > M | 7/8 | Deleterious (0) | Possibly damaging (0.799) |
| USA | African American | T > C | 981 | 806 | 269 | H > R | 7/8 | Tolerated (0.27) | Benign (0.002) |
| SA | African | A > T | 1080 | 905 | 302 | L > Q | 7/8 | Deleterious (0) | Probably damaging (0.919) |
| USA | African American | A > G | 1125 | 950 | 317 | L > P | 7/8 | Deleterious (0) | Probably damaging (0.999) |
| USA | European American | T > C | 1280 | 1105 | 369 | K > E | 8/8 | Tolerated (0.1) | Benign (0.001) |
3.6. Targeted Sequencing of PKM
A total of 19 previously unreported SNVs and deletions that may impact the protein-coding potential of the PKM mRNA transcript were detected in 19 participants, nine enrolled in USA study sites, five in SA, and five in UGA. The detected variants and deletions for PKM DNA reference sequence NM_001206796.1 are listed in Supplementary Table 5. Of the detected SNVs, 14 PKM missense variants were detected in 14 heterozygous participants at a frequency of one individual per variant (Table 4). In silico tools were unable to predict the functional impact of the observed missense variants due to the lack of sequence diversity required for the performed multiple sequence alignments. In addition to the observed PKM SNVs, three heterozygous USA participants (one individual per variant) were found to carry the following reference SNPs: stop gained variants rs180716407 (NM_001206796.1:c.14G > C) and rs151078084 (NM_001206796.1:c.1354G > A) and the missense variant rs147939689 (NM_001206796.1:c.389C > A).
Table 4.
PKM missense variants detected in MTN-001 participants.
A total of 14 previously unreported PKM missense variants were detected in 14 heterozygous MTN-001 participants for the coding DNA reference sequence NM_001206796.1. SIFT and PolyPhen in silico tools could not predict the functional impact of the observed missense variants with a high degree of confidence due to the lack of sequence diversity that is required for the performed multiple sequence alignments.
| Study site | Ethnicity | PKM variant (ref. > alt.) | cDNA position | Coding DNA sequence position | Protein position | Amino acid (ref. > alt.) | Exon |
|---|---|---|---|---|---|---|---|
| USA | African American | C > T | 643 | 244 | 82 | A > T | 3/12 |
| USA | Asian | G > C | 753 | 354 | 118 | N > K | 3/12 |
| SA | African | T > A | 754 | 355 | 119 | T > S | 3/12 |
| USA | European American | A > T | 919 | 520 | 174 | S > T | 5/12 |
| SA | African | A > T | 1052 | 653 | 218 | L > Q | 6/12 |
| UGA | African | T > C | 1103 | 704 | 235 | Y > C | 6/12 |
| UGA | African | C > A | 1483 | 1084 | 362 | D > Y | 8/12 |
| USA | Multiracial | T > C | 1489 | 1090 | 364 | I > V | 8/12 |
| UGA | African | C > T | 1600 | 1201 | 401 | A > T | 8/12 |
| UGA | African | T > C | 1628 | 1229 | 410 | K > R | 9/12 |
| USA | African American | T > G | 1642 | 1243 | 415 | T > P | 9/12 |
| SA | Asian | C > T | 1820 | 1421 | 474 | R > Q | 10/12 |
| UGA | African | C > T | 1897 | 1498 | 500 | A > T | 10/12 |
| USA | European American | T > C | 2114 | 1715 | 572 | K > R | 12/12 |
3.7. Targeted Sequencing of PKLR
A total of 22 previously unreported SNVs and deletions that may impact the protein-coding potential of the PKLR mRNA transcript were detected in 21 participants, 14 enrolled in USA study sites, four in SA, and three in UGA. The variants and deletions for PKLR DNA reference sequence NM_000298.5 are listed in Supplementary Table 6. Of the observed SNVs, 15 PKLR missense variants were detected in 15 heterozygous participants at a frequency of one individual per variant (Table 5). Using in silico tools, four of the detected missense variants were predicted to have both a deleterious and damaging impact on protein function with a frequency of 3% (4/142 individuals). The reference SNP missense variant rs147689373 (NM_000298.5:c.829C > T) was detected in one heterozygous SA participant.
Table 5.
PKLR missense variants detected in MTN-001 participants.
A total of 15 previously unreported PKLR missense variants were observed in 15 heterozygous MTN-001 participants for the coding DNA reference sequence NM_000298.5. The functional consequence of resulting amino acid substitutions were predicted using SIFT and PolyPhen in silico tools. A SIFT score < 0.05 was suggestive of a damaging amino acid substitution and > 0.05 a tolerated substitution. A PolyPhen score > 0.908 was suggestive of a probably damaging, 0.447–0.908 a possibly damaging, or < 0.447 a benign amino acid substitution. Deleterious and probably damaging PKLR missense variants was observed at a frequency of 3% (4/142 individuals).
| Study site | Ethnicity | PKLR variant (ref. > alt.) | cDNA position | Coding DNA sequence position | Protein position | Amino acid (ref. > alt.) | Exon | SIFT prediction | PolyPhen prediction |
|---|---|---|---|---|---|---|---|---|---|
| SA | African | C > T | 128 | 89 | 30 | G > E | 1/11 | Deleterious (0.01) | Probably damaging (0.995) |
| SA | African | A > T | 206 | 167 | 56 | F > Y | 2/11 | Tolerated (0.29) | Benign (0.075) |
| USA | African American | T > A | 251 | 212 | 71 | E > V | 2/11 | Deleterious (0) | Probably damaging (0.919) |
| USA | African American | A > T | 277 | 238 | 80 | S > T | 2/11 | Tolerated (0.19) | Benign (0.087) |
| UGA | African | A > G | 287 | 248 | 83 | V > A | 2/11 | Tolerated (0.97) | Benign (0.001) |
| USA | African American | A > T | 337 | 298 | 100 | S > T | 3/11 | Tolerated (0.06) | Benign (0.32) |
| USA | European American | C > T | 386 | 347 | 116 | R > Q | 3/11 | Deleterious (0) | Probably damaging (0.996) |
| USA | Hispanic | G > A | 674 | 635 | 212 | P > L | 5/11 | Deleterious (0.03) | Benign (0.094) |
| USA | African American | T > C | 730 | 691 | 231 | I > V | 5/11 | Tolerated (0.86) | Benign (0.002) |
| USA | African American | C > T | 814 | 775 | 259 | V > M | 6/11 | Deleterious (0.03) | Possibly damaging (0.892) |
| UGA | African | T > C | 908 | 869 | 290 | K > R | 6/11 | Tolerated (0.24) | Possibly damaging (0.879) |
| USA | European American | G > T | 990 | 951 | 317 | H > Q | 6/11 | Tolerated (0.98) | Possibly damaging (0.577) |
| USA | African American | C > T | 1066 | 1027 | 343 | E > K | 7/11 | Deleterious (0) | Probably damaging (0.992) |
| USA | European American | T > C | 1346 | 1307 | 436 | Q > R | 9/11 | Tolerated (0.27) | Benign (0.423) |
| SA | African | C > T | 1357 | 1318 | 440 | E > K | 9/11 | Deleterious (0.05) | Possibly damaging (0.858) |
4. Discussion
This study identified the nucleotide kinases that activate TFV in cells and tissues susceptible to HIV infection. Our data suggests that AK2 may be a key contributor to systemic and localized TFV activation as siRNA knockdown in PBMC, vaginal tissue, and colorectal tissue resulted in significantly decreased formation of TFV metabolites. Additionally, we demonstrated that PKLR can contribute to TFV-DP formation in PBMC and vaginal tissue. PKLR is highly expressed in the liver and therefore may be critical for the activation of oral TFV preparations that undergo first-pass metabolism. Interestingly, we observed CKM protein expression to be limited to colorectal tissue. TFV-DP concentrations in colorectal tissue exceed those in vaginal tissue with an oral dose of TFV and tissue-specific expression of CKM could contribute to this phenomenon. Additionally in this study, we examined a potential role for the nucleotide diphosphate kinase NME1. We did not detect NME1 expression at the protein level in PBMC, vaginal, or colorectal tissue, suggesting NME1 is unlikely to contribute to the activation of TFV in the cells and tissues investigated.
Through our work, we identified AK2, CKM, PKM, and PKLR as candidates for examination in a retrospective clinical study. In doing so, we detected 71 previously unreported variants in the genes encoding these kinases. The coverage threshold implemented in our analyses was in line with the American College of Medical Genetics and Genomics clinical laboratory standards for next-generation sequencing, increasing the confidence in the variants called (Rehm et al., 2013). These discovered variants, however, were not validated in parallel using Sanger sequencing. We detected seven genetic variants in CKM, PKM, and PKLR that have been reported in the SNP database. The CKM SNPs rs11559024 and rs17357122 detected in MTN-001 European American participants have frequencies of 1–3% in Western European populations as reported by the 1000 Genomes Project (Abecasis et al., 2012). These populations also exhibit a comparable frequency for the PKM SNP rs180716407 detected in one European American participant, and African sub-populations of Nigeria, Kenya, and Sierra Leone have a frequency of 1–2% for the PKLR SNP rs147689373 detected in one South African participant. Population genetics aside, the impact of these discovered and reported genetic variants on enzymatic activity is unknown. For the detected missense variants, however, we were able to predict the phenotypic consequence of resulting amino acid mutations using the in silico bioinformatic tools SIFT and PolyPhen. For example, one MTN-001 participant carried the CKM missense variant NM_001824.4:c.389G > A, causing a deleterious and damaging amino acid mutation of R130H. In line with this, a study characterizing TFV-MP binding within the active site of CKM observed this particular amino acid residue R130 to form strong ionic interactions with its substrates (Varga et al., 2013). Moreover, one can envision that such a mutation may interfere with substrate binding and, therefore, decrease rates of phosphorylation. That being said, the variants predicted in our study to negatively impact kinase function ultimately need to be tested in vitro using site-directed mutagenesis in order to elucidate their effect on activity towards TFV and TFV-MP.
Due to the frequency of the genetic variants detected within the MTN-001 participants, we were unable to make strong correlations with our published TFV pharmacokinetic data (Hendrix et al., 2013). Additionally, adherence is a confounding issue for HIV PrEP studies making it particularly difficult to execute these analyses in a retrospective study. As a proof of concept, however, it is worth noting that an African American participant carrying a predicted dysfunctional missense variant for PKLR exhibited a three-fold greater ratio of TFV to TFV-DP in vaginal tissue compared to homozygous wild-type participants with daily use of a 1% TFV microbicide gel and is commensurate with lower TFV-DP tissue concentrations. In order to perform a powerful analysis, a pharmacokinetic study design is required in which healthy volunteers would be enrolled based on genotype in the same manner used for our recent study of the anti-HIV drug maraviroc (Lu et al., 2014). Further, as this preliminary pharmacogenomics study was limited to females it would also be of interest to perform a similar analysis in men in order to investigate potential sexual dimorphisms in allele frequencies.
In conclusion, our results present a fundamental shift when considering the factors that govern TFV disposition as differing routes of activation could contribute to tissue-specific pharmacokinetic–pharmacodynamic relationships. Further, in identifying the nucleotide kinases responsible for TFV activation in PBMC, vaginal tissue, and colorectal tissue, we were able to perform targeted sequencing of the genes encoding these kinases using clinical samples of the MTN-001 Microbicide Trials Network study. We detected genetic variants in these kinases predicted to have deleterious and damaging phenotypes, demonstrating that there could be a genetic basis for inter-individual variation in TFV drug levels. Taken together, the results from this study are pushing an important concept forward in that HIV PrEP is not a one-size-fits-all preventative approach, and these data can guide future clinical studies to investigate the impact of nucleotide kinase genetic variants on TFV pharmacokinetics.
Author Contributions
CWH and NNB conceived and designed the experiments; JML, EET, CWH, and NNB acquired the data. JML and NNB performed the statistical analyses; JML and NNB drafted the manuscript; JML, EET, CWH, and NNB provided critical revisions and approved the final version of the manuscript for submission.
Declaration of Interests
We declare no competing interests.
Acknowledgments
The authors wish to acknowledge the generous contributions made by the research participants of this study and we wish to acknowledge the contributions of the entire MTN-001 Study Group.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2015.07.008.
Contributor Information
Julie M. Lade, Email: jmaylor1@jhmi.edu.
Elaine E. To, Email: to.elaine.e@gmail.com.
Craig W. Hendrix, Email: chendrix@jhmi.edu.
Namandjé N. Bumpus, Email: nbumpus1@jhmi.edu.
Appendix A. Supplementary data
Supplementary materials.
References
- Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton P.A., Cranston R.D., Kashuba A., Hendrix C.W., Bumpus N.N., Richardson-Harman N. RMP-02/MTN-006: a phase 1 rectal safety, acceptability, pharmacokinetic, and pharmacodynamic study of tenofovir 1% gel compared with oral tenofovir disoproxil fumarate. AIDS Res. Hum. Retrovir. 2012;28(11):1412–1421. doi: 10.1089/aid.2012.0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeten J.M., Donnell D., Ndase P., Mugo N.R., Campbell J.D., Wangisi J. Antiretroviral prophylaxis for HIV prevention in heterosexual men and women. N. Engl. J. Med. 2012;367(5):399–410. doi: 10.1056/NEJMoa1108524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdais J., Biondi R., Sarfati S., Guerreiro C., Lascu I., Janin J. Cellular phosphorylation of anti-HIV nucleosides. Role of nucleoside diphosphate kinase. J. Biol. Chem. 1996;271(14):7887–7890. doi: 10.1074/jbc.271.14.7887. [DOI] [PubMed] [Google Scholar]
- Derdelinckx I., Wainberg M.A., Lange J.M., Hill A., Halima Y., Boucher C.A. Criteria for drugs used in pre-exposure prophylaxis trials against HIV infection. PLoS Med. 2006;3(11):e454. doi: 10.1371/journal.pmed.0030454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry B.G., Gentry S.N., Jackson T.L., Zemlicka J., Drach J.C. Phosphorylation of antiviral and endogenous nucleotides to di- and triphosphates by guanosine monophosphate kinase. Biochem. Pharmacol. 2011;81(1):43–49. doi: 10.1016/j.bcp.2010.09.005. [DOI] [PubMed] [Google Scholar]
- Grant R.M., Lama J.R., Anderson P.L., McMahan V., Liu A.Y., Vargas L. Preexposure chemoprophylaxis for HIV prevention in men who have sex with men. N. Engl. J. Med. 2010;363(27):2587–2599. doi: 10.1056/NEJMoa1011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix C.W., Chen B.A., Guddera V., Hoesley C., Justman J., Nakabiito C. MTN-001: randomized pharmacokinetic cross-over study comparing tenofovir vaginal gel and oral tablets in vaginal tissue and other compartments. PLoS One. 2013;8(1):e55013. doi: 10.1371/journal.pone.0055013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney B.P., Flaherty J.F., Shah J. Tenofovir disoproxil fumarate: clinical pharmacology and pharmacokinetics. Clin. Pharmacokinet. 2004;43(9):595–612. doi: 10.2165/00003088-200443090-00003. [DOI] [PubMed] [Google Scholar]
- Koch K., Chen Y., Feng J.Y., Borroto-Esoda K., Deville-Bonne D., Gallois-Montbrun S. Nucleoside diphosphate kinase and the activation of antiviral phosphonate analogs of nucleotides: binding mode and phosphorylation of tenofovir derivatives. Nucleosides Nucleotides Nucleic Acids. 2009;28(8):776–792. doi: 10.1080/15257770903155899. [DOI] [PubMed] [Google Scholar]
- Louissaint N.A., Cao Y.J., Skipper P.L., Liberman R.G., Tannenbaum S.R., Nimmagadda S. Single dose pharmacokinetics of oral tenofovir in plasma, peripheral blood mononuclear cells, colonic tissue, and vaginal tissue. AIDS Res. Hum. Retrovir. 2013;29(11):1443–1450. doi: 10.1089/aid.2013.0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Fuchs E.J., Hendrix C.W., Bumpus N.N. CYP3A5 genotype impacts maraviroc concentrations in healthy volunteers. Drug Metab. Dispos. 2014;42(11):1796–1802. doi: 10.1124/dmd.114.060194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrazzo J.M., Ramjee G., Richardson B.A., Gomez K., Mgodi N., Nair G. Tenofovir-based preexposure prophylaxis for HIV infection among African women. N. Engl. J. Med. 2015;372(6):509–518. doi: 10.1056/NEJMoa1402269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer K.H., Maslankowski L.A., Gai F., El-Sadr W.M., Justman J., Kwiecien A. Safety and tolerability of tenofovir vaginal gel in abstinent and sexually active HIV-infected and uninfected women. AIDS. 2006;20(4):543–551. doi: 10.1097/01.aids.0000210608.70762.c3. [DOI] [PubMed] [Google Scholar]
- Ng P.C., Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11(5):863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobumoto M., Yamada M., Song S., Inouye S., Nakazawa A. Mechanism of mitochondrial import of adenylate kinase isozymes. J. Biochem. 1998;123(1):128–135. doi: 10.1093/oxfordjournals.jbchem.a021899. [DOI] [PubMed] [Google Scholar]
- Patterson K.B., Prince H.A., Kraft E., Jenkins A.J., Shaheen N.J., Rooney J.F. Penetration of tenofovir and emtricitabine in mucosal tissues: implications for prevention of HIV-1 transmission. Sci. Transl. Med. 2011;3(112):112re4. doi: 10.1126/scitranslmed.3003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramensky V., Bork P., Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30(17):3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm H.L., Bale S.J., Bayrak-Toydemir P., Berg J.S., Brown K.K., Deignan J.L. ACMG clinical laboratory standards for next-generation sequencing. Genet. Med. 2013;15(9):733–747. doi: 10.1038/gim.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins B.L., Greenhaw J., Connelly M.C., Fridland A. Metabolic pathways for activation of the antiviral agent 9-(2-phosphonylmethoxyethyl)adenine in human lymphoid cells. Antimicrob. Agents Chemother. 1995;39(10):2304–2308. doi: 10.1128/aac.39.10.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins B.L., Srinivas R.V., Kim C., Bischofberger N., Fridland A. Anti-human immunodeficiency virus activity and cellular metabolism of a potential prodrug of the acyclic nucleoside phosphonate 9-R-(2-phosphonomethoxypropyl)adenine (PMPA), Bis(isopropyloxymethylcarbonyl)PMPA. Antimicrob. Agents Chemother. 1998;42(3):612–617. doi: 10.1128/aac.42.3.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schooley R.T., Ruane P., Myers R.A., Beall G., Lampiris H., Berger D. Tenofovir DF in antiretroviral-experienced patients: results from a 48-week, randomized, double-blind study. AIDS. 2002;16(9):1257–1263. doi: 10.1097/00002030-200206140-00008. [DOI] [PubMed] [Google Scholar]
- Topalis D., Alvarez K., Barral K., Munier-Lehmann H., Schneider B., Veron M. Acyclic phosphonate nucleotides and human adenylate kinases: impact of a borano group on alpha-P position. Nucleosides Nucleotides Nucleic Acids. 2008;27(4):319–331. doi: 10.1080/15257770801941952. [DOI] [PubMed] [Google Scholar]
- Van Damme L., Corneli A., Ahmed K., Agot K., Lombaard J., Kapiga S. Preexposure prophylaxis for HIV infection among African women. N. Engl. J. Med. 2012;367(5):411–422. doi: 10.1056/NEJMoa1202614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga A., Graczer E., Chaloin L., Liliom K., Zavodszky P., Lionne C. Selectivity of kinases on the activation of tenofovir, an anti-HIV agent. Eur. J. Pharm. Sci. 2013;48(1–2):307–315. doi: 10.1016/j.ejps.2012.11.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials.