Abstract
Current antibiotic testing does not include the potential influence of host cell environment on microbial susceptibility and antibiotic resistance, hindering appropriate therapeutic intervention. We devised a strategy to identify the presence of host–pathogen interactions that alter antibiotic efficacy in vivo. Our findings revealed a bacterial mechanism that promotes antibiotic resistance in vivo at concentrations of drug that far exceed dosages determined by standardized antimicrobial testing. This mechanism has escaped prior detection because it is reversible and operates within a subset of host tissues and cells. Bacterial pathogens are thereby protected while their survival promotes the emergence of permanent drug resistance. This host-dependent mechanism of transient antibiotic resistance is applicable to multiple pathogens and has implications for the development of more effective antimicrobial therapies.
Keywords: Antibiotic resistance, MIC testing, Antibiotic-resistant mutants, Multidrug-resistant pathogens, Antimicrobial therapy, Antibiotic susceptibility testing, Salmonella
Graphical abstract
Highlights
-
•
Standard MIC testing does not consider the influence of the host milieu, potentially hindering therapeutic intervention.
-
•
Salmonella induce polymyxin resistance during infection at levels of drug that far exceed dosages determined by MIC testing.
-
•
Polymyxin treatment failed to control Salmonella infection and promotes the emergence of drug-resistant mutants.
Physicians rely on laboratory antimicrobial susceptibility testing of clinical isolates to identify a suitable antibiotic for therapy. Although the recommended antibiotics clear most bacterial infections, some patients fail to respond and require prolonged therapy, higher dosing or different antibiotics. Why does this occur and what are the possible implications? By studying antibiotic resistance in the context of infection, we identified a host-dependent mechanism that promotes antibiotic resistance at concentrations of drug that far exceed dosages determined by standardized antimicrobial testing. These findings question current antibiotic testing methods that have guided physician treatment practices and drug development for the last several decades.
1. Introduction
Recent CDC estimates indicate that one in five pathogens from hospital-acquired infections in the U.S. are multidrug-resistant (Kallen et al., 2010; Sievert et al., 2013), dramatically limiting therapeutic options to antibiotics that may be more toxic, less effective, or more expensive (Centers for Disease Control and Prevention, 2013). In such cases, patients often have longer hospital stays, delayed recuperation, long-term disability, and increased mortality. Deciphering the mechanisms that govern the emergence of multidrug-resistant pathogens is critical to the development of new approaches to control bacterial infections. Many mechanisms of antibiotic resistance have been established, including horizontal gene transfer; genomic mutation; and intrinsic bacterial mechanisms that pre-date antibiotics (Allen et al., 2010; Andersson and Hughes, 2010; Cox and Wright, 2013; D'Costa et al., 2011; Davies and Davies, 2010). Significant advances have been made regarding the generation of antibiotic resistant variants (phenotypic and genotypic) that emerge during infection; e.g., Staphylococcus aureus small colony variants that promote persistent infections (Proctor et al., 2006); antibiotic resistance of Pseudomonas aeruginosa biofilms (Høiby et al., 2010); the evolution and spread of multidrug-resistant pneumococcal variants (Croucher et al., 2011), and heteroresistant subpopulations of vancomycin-susceptible S. aureus (El-Halfawy and Valvano, 2015). Despite this knowledge, the role of host–pathogen interactions in antibiotic resistance is poorly understood, and the use of host models as a primary approach to understanding resistance is not often considered or explored.
For the past several decades, drug development has followed a standard sequential procedure wherein: (i) efficacy is determined in vitro; (ii) pharmacokinetic/pharmacodynamic (PK/PD) parameters are measured in vivo; and (iii) dosing efficacy/toxicity in vivo is established for a limited number of model pathogens (Ambrose et al., 2007; Clinical and Laboratory Standards Institute, 2012; Food and Drug Administration, 2009). However, along with a limited amount of patient-dosing data, physicians rely on in vitro antimicrobial susceptibility testing (AST) of clinical isolates grown on the universal media standard Mueller–Hinton Broth (MHB) for therapeutic intervention (Clinical and Laboratory Standards Institute, 2012; European Committee on Antibiotic Suscepibility Testing, 2014). This standard procedure does not replicate mammalian biochemistry and may not correlate with patient outcome. To overcome these limitations, we investigated antibiotic resistance in the context of animal models of disease, and have identified a mechanism that stimulates bacterial resistance to multiple antibiotics during infection, while promoting the emergence of drug-resistant bacteria.
2. Materials and Methods
2.1. Bacterial Strains and Media
TIVAR + S. Typhimurium ATCC 14028 (CDC 6516–60) and MT2057 (an isogenic KAN resistant derivative of 14028), or TIVAR − S. Typhimurium var. 5 (04)-9639 (a multidrug-resistant isolate) derived from chicken and cow, respectively, were used in these studies (Conner et al., 1998; Heithoff et al., 2008). These strains have identical oral and i.p. lethal dose 50s (LD50) in BALB/c mice, 105 and < 10 colony forming units (CFU), respectively (Heithoff et al., 2008). Salmonella human clinical isolates were obtained from fecal and blood samples derived from patients with gastroenteritis or bacteremia, respectively; animal isolates were derived from different disease outbreaks, individual cases, or surveillance submissions to diagnostic laboratories (Heithoff et al., 2008). Unless otherwise specified, Salmonella were derived from stationary phase cultures aerated at 37 °C containing the Mueller–Hinton broth (MHB) (Clinical and Laboratory Standards Institute, 2012); low phosphate, low magnesium medium (LPM) (Coombes et al., 2004) or N-minimal medium (Nelson and Kennedy, 1971) supplemented with 0.3% glycerol and 0.1% casamino acids; or the Luria–Bertani (LB) medium (Davis et al., 1980). Yersinia pseudotuberculosis IP32953 is a virulent human isolate (Chain et al., 2004), and was assayed from stationary phase cultures aerated at 28 °C containing MHB or LPM media.
2.2. MIC Assays
The minimum inhibitory concentration (MIC) was determined in MHB and LPM media (pH 7 and pH 5.5) according to Clinical and Laboratory Standards Institute (CLSI) guidelines (Clinical and Laboratory Standards Institute, 2012; Wiegand et al., 2008). MIC assays were performed on bacteria obtained from overnight culture, and from bacteria derived from macrophage lysates or host tissues. Bacteria were diluted in 0.15 M NaCl, and a 5 μl volume containing 104 CFU was spotted on agar plates of the media condition indicated, containing two-fold dilutions of each antibiotic. MIC values were derived following 20 h incubation at 37 °C (Salmonella) or 48 h incubation at 28 °C (Y. pseudotuberculosis), and were the result of at least three independent determinations.
2.3. Bacterial Cell Survival Assays
Bacterial cell survival was evaluated after cells derived from a given growth condition were exposed to antibiotics under the same or different growth condition (Groisman et al., 1997). S. Typhimurium 14028 was grown overnight in non-inducing medium for the TIVAR phenotype (N-minimal medium with 10 mM Mg2 + pH 7.7), diluted 1:100 in either inducing medium (N-minimal medium with 10 μM Mg2 + pH 5.8) or non-inducing medium (N-minimal medium with 10 mM Mg2 + pH 7.7), and incubated 4 h at 37 °C. Bacteria were diluted 1:200 and exposed to polymyxin B for 1 h in either inducing medium (N-minimal medium with 10 μM Mg2 + pH 5.8) or non-inducing medium (LB), and plated for CFU on LB medium. Percent survival was calculated as CFU[polymyxin B] / CFU[no drug] × 100 at 1 h post drug exposure; values given are the REML model means ± SEM derived from at least 5 independent determinations.
2.4. Virulence Assays
Intraperitoneal (i.p.) infection. Salmonella grown overnight in LB medium were resuspended in 0.15 M NaCl and administered to mice via the i.p. route of infection (dose 102 or 103 CFU) (Heithoff et al., 1999). Five days post-infection, the bacterial cells were recovered from the spleen and other tissues/fluids of acutely infected animals. Antibiotic treatment. Mice infected i.p. with Salmonella (i.p. dose 102 CFU) were treated (or mock-treated) with a twice-daily polymyxin B or ciprofloxacin dosing regimen (30 mg/kg/day). Bacterial cells were recovered from the spleen of acutely infected animals and enumerated by direct colony count. Mouse survival was assessed for 10 days post-infection. Uninfected mice were also treated with polymyxin B to control for dosing toxicity. Six- to twelve-week old female BALB/c mice were used in all virulence studies. Institutional Animal Care and Use Committee of the University of California, Santa Barbara approved all mouse research protocols undertaken herein.
2.5. Cell Culture
The murine macrophage cell line RAW 264.7 was obtained from the American Type Culture Collection, Rockville, MD., and maintained in minimum essential medium (MEM) supplemented with l-glutamine and 10% heat-inactivated bovine growth-supplemented calf serum (HyClone Laboratories, Logan, UT). Cells were grown in a humidified atmosphere of 5% carbon dioxide and 95% air at 37 °C in 75-cm2 plastic flasks (Corning Glass Works, Corning, NY). Cultured murine macrophages (RAW 264.7) were harvested by scraping with a rubber policeman and plated at a density of 2.5 to 5 × 105 cells/ml in 4 ml of culture medium in 35 mm-diameter, six-well dishes (Corning) and grown for 24 h to approximately 80 to 90% confluence (1 to 5 × 106 cells/well) (adapted from previous methods (Fleckenstein et al., 1996)).
2.6. Bacterial Infection of Cultured Murine Macrophages
Bacterial cells were used to infect cultured murine macrophage (RAW 264.7) monolayers grown in cell culture plates (Corning) at a multiplicity of infection (MOI) of 10:1. The bacteria were centrifuged onto cultured monolayers at 1000 × g for 10 min at room temperature, after which they were incubated for 30 min at 37 °C in a 5% CO2 incubator. The co-culture was washed once with cell culture medium and incubated for 45 min in the presence of gentamicin (100 μg/ml) to kill extracellular bacteria, washed once with pre-warmed cell culture medium, and incubated in 4 ml of culture medium containing polymyxin B at the concentration indicated or 10 μg/ml gentamicin (t = 0 time point) for 24 h (adapted from previous methods (Finlay and Falkow, 1988)).
2.7. Statistical Analyses
Log transformed intracellular CFU and PMBr mutation frequency data were analyzed using ANOVA (ANOVA, GenStat, 15th Edition, VSN International, UK). Intracellular CFU and PMBr mutation frequency data are presented as the means ± standard error of the mean (SEM). Cell survival was analyzed using residual (or restricted) maximum likelihood (REML) analysis (GenStat, 15th Edition, VSN International, UK). A single variate model was used to analyze percentage survival on a log scale. The fixed effects of the model were the factors group, drug concentration, and their interaction. The Wald chi-square test was used to determine significant main effects and/or significant interactions between factors. Any non-significant terms were dropped from the model and analysis repeated. Following analysis, data are presented as predicted model-based means, i.e., predicted means are those obtained from the fitted model rather than the raw sample means. Differences between the individual means calculated using ANOVA and REML were determined by calculating an approximate least significant difference (LSD). A difference of means that exceeded the calculated LSD was considered significant. Statistical significance for difference in proportions of animal survival was calculated using Chi-square (Epi Info 7, CDC). For all statistical analyses, a significance level (P) of less than 0.05 was considered to be statistically significant. Degrees of statistical significance are presented as ***P < 0.001, **P < 0.01, or *P < 0.05.
3. Results
3.1. Bacteria Exhibit High-level Antibiotic Resistance under Conditions That Simulate Host Microenvironments
The biochemical environment within certain host sites may induce transient changes in the bacterium that confer resistance to antibiotics. Thus, we screened a collection of pathogenic strains for antibiotic resistance under experimental conditions that simulated host microenvironments, including those present in the macrophage phagosome, a sub-cellular organelle in which Salmonella resides and replicates (Steele-Mortimer, 2008). Conditions within the phagosome can be mimicked by mildly acidic culture medium (pH 5.5) that is low in phosphate and magnesium (LPM) (Coombes et al., 2004). We compared the efficacy of 16 clinically-relevant antibiotics against Salmonella grown in LPM pH 5.5 medium versus that grown in MHB (unbuffered ~ pH 7.2) via assessment of the minimum inhibitory concentration (MIC) of each antibiotic (Clinical and Laboratory Standards Institute, 2012; European Committee on Antibiotic Suscepibility Testing, 2014). To control for the potential effects of pH and media composition, antibiotic resistance was determined by comparing the MICs in LPM medium versus those obtained in MHB at pH 5.5 and pH 7.
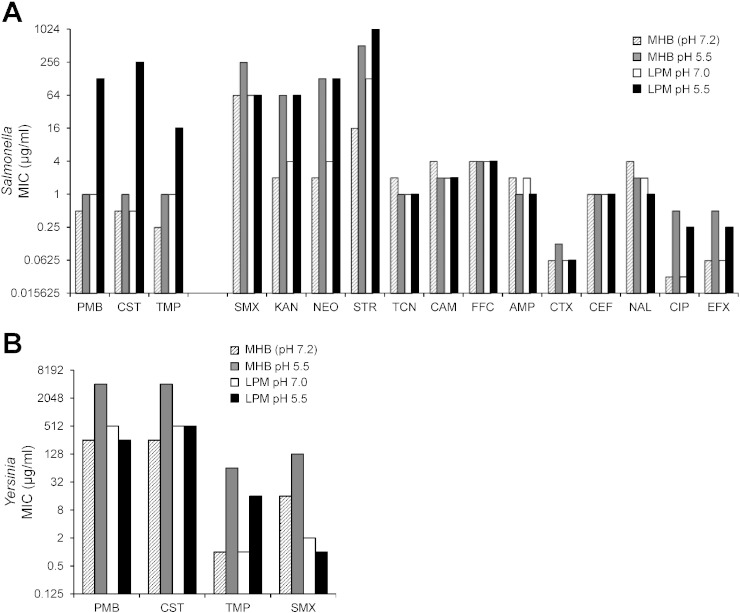
Growth of S. enterica Typhimurium in LPM pH 5.5 medium was linked to high-level resistance to polymyxin B (PMB; 64-fold) and colistin (CST; 256-fold), cationic peptides that disrupt Gram-negative membranes (Bergen et al., 2012; Landman et al., 2008) (Fig. 1A). We also observed a mild resistance to trimethoprim (TMP; 4-fold), an inhibitor of folate metabolism (Burchall, 1973). Under these conditions, Salmonella remains viable, and continues to grow at antibiotic concentrations that far exceed those achieved in treating human infections (0.5 to 2.5 mg/l) (Michalopoulos and Falagas, 2011; National Institutes of Health, 2014b; Sandri et al., 2013; Zavascki et al., 2008). Some other antibiotics examined were subject to pH and/or media composition effects on drug efficacy including kanamycin, streptomycin, and ciprofloxacin. In those cases, we could not determine the induction of antibiotic resistance. In other cases the efficacy of antibiotics was altogether unaffected, including tetracycline, chloramphenicol, and ceftiofur. Many pathogenic Salmonella serovars (serotypic variants) derived from human and livestock infections (Heithoff et al., 2008) exhibited high-level resistance to antibiotics PMB, CST and TMP when grown in LPM pH 5.5 medium: S. Typhimurium (5/6), S. Enteritidis (1/1), S. Dublin (2/2), S. Newport (1/2), S. Bovismorbificans (1/2), and Salmonella C1 K00-670 (1/1) (Supplementary Table 1). These data indicate that induction of antibiotic resistance under conditions simulating the phagosome is not a strain-specific phenomenon.
Fig. 1.
Salmonella and Yersinia exhibit high-level antibiotic resistance under conditions that simulate host microenvironments. (A) S. Typhimurium 14028 was grown in either the Mueller–Hinton Broth (MHB) (Clinical and Laboratory Standards Institute, 2012) or low phosphate, low magnesium medium (LPM) (Coombes et al., 2004) at the pH indicated, and the minimum inhibitory concentration (MIC) of a panel of antibiotics was determined in the same medium (Clinical and Laboratory Standards Institute, 2012; Wiegand et al., 2008). The effect of growth conditions on antibiotic resistance was calculated by comparing the MIC in LPM medium divided by the MIC in MHB medium at both pH 5.5 and pH 7 (unbuffered) (ratio of LPM pH 5.5/pH 7.0 to MHB pH 5.5/pH 7.2). Drugs: polymyxin B, PMB; colistin sulfate, CST; trimethoprim, TMP; sulfamethoxazole, SMX; kanamycin, KAN; neomycin, NEO; streptomycin, STR; tetracycline, TCN; chloramphenicol, CAM; florfenicol, FFC; ampicillin, AMP; ceftriaxone, CTX; ceftiofur, CEF; nalidixic acid, NAL; ciprofloxacin, CIP; enrofloxacin, EFX. (B) The degree of Y. pseudotuberculosis IP32953 susceptibility to antibiotics as a function of growth conditions. MIC values were obtained from at least 3 independent determinations.
To ascertain whether the induction of antibiotic resistance exists in other microbial species, we screened for this phenotype in Y. pseudotuberculosis, a Gram-negative extracellular pathogen that can cause severe disease in humans and livestock (Galindo et al., 2011; Tauxe, 2013). After normalizing for pH and media composition effects on drug efficacy, Y. pseudotuberculosis acquired increased resistance not only to antibiotics PMB, CST, and TMP (32-fold, 16-fold, and 4-fold, respectively), but also to sulfamethoxazole (SMX) (16-fold), another inhibitor of folate metabolism (National Institutes of Health, 2014a) (Fig. 1B; Supplementary Table 2). Further, the induction of Yersinia antibiotic resistance occurred under markedly different environmental conditions compared to Salmonella (in MHB pH 5.5 but not in LPM pH 5.5), perhaps reflecting the dissimilar host intracellular and extracellular trafficking of these bacterial species (Salmonella exposure to defensins within the phagosome (Steele-Mortimer, 2008); Yersinia exposure to cryptdins in the small intestine (Bevins and Salzman, 2011)). Together, these findings suggest that the scope of resistance induction may include a variety of different species; and current methods used to assess the degree and spectrum of antibiotic resistance do not account for environmental influences on microbial susceptibility in vivo.
3.2. Phenotypic Switching to High-level Antibiotic Resistance Is Rapid and Rapidly Reversible
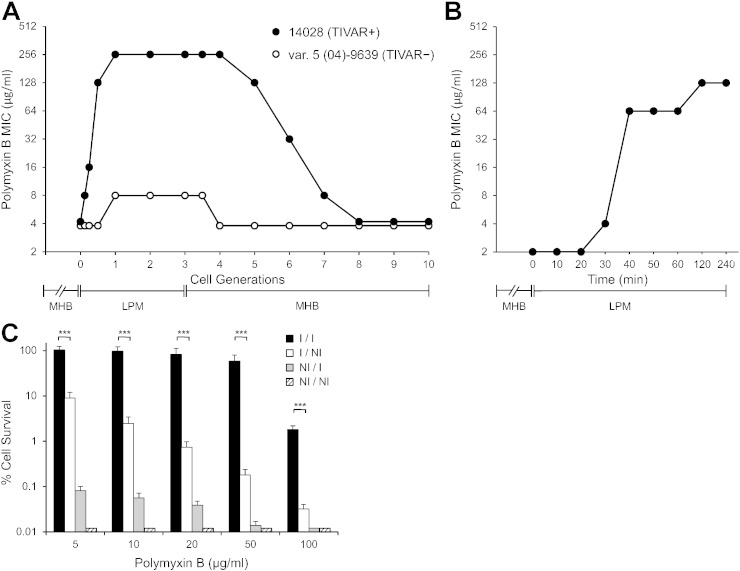
To understand the mechanistic nature of phenotypic switching between antibiotic susceptible to resistant states, the kinetics and degree of antibiotic resistance induction was evaluated upon transfer of bacterial cells from MHB (non-inducing) to LPM pH 5.5 (inducing) medium. For this analysis, comparisons were made between a natural Salmonella isolate that was capable of inducing high-level resistance (S. Typhimurium 14028), with one that was not (S. Typhimurium var. 5 (04)-9639) (Supplementary Table 1). Transfer of S. Typhimurium 14028 from MHB to LPM pH 5.5 medium resulted in a rapid MIC change from a PMB susceptible to resistant phenotype within 1 cell doubling (Fig. 2A). Upon subsequent transfer from LPM pH 5.5 back to MHB medium, bacteria reverted to the susceptible phenotype within 4 to 5 generations. In contrast, transfer of S. Typhimurium var. 5 (04)-9639 from MHB to LPM pH 5.5 medium did not result in high-level PMB resistance. These data indicate that, for isolates capable of resistance induction, phenotypic switching to high-level antibiotic resistance is rapid and rapidly reversible. We then examined whether resistance induction can occur in the absence of rapid cell division and mutational selection via transferring bacterial cells from an overnight MHB culture into LPM pH 5.5 medium. It is anticipated that such a media shift markedly slows bacterial cell division since the final cell density of a saturated culture in rich MHB medium is considerably greater (~ 5-fold) than that in LPM pH 5.5 medium (Heithoff et al., 2012). Transfer of S. Typhimurium 14028 from MHB to LPM pH 5.5 medium, without dilution, resulted in a rapid change in MIC from a PMB susceptible to resistant phenotype after a short incubation period (Fig. 2B). These findings indicate that phenotypic switching between antibiotic susceptibility to resistance does not require rapid cell division and mutational selection. Since this resistance mechanism is rapidly reversible and induced under experimental conditions that simulate host microenvironments, we have termed this mechanism TIVAR, transient in vivo antibiotic resistance.
Fig. 2.
Induction of high-level antibiotic resistance in Salmonella is rapid and rapidly reversible. (A) TIVAR + S. Typhimurium 14028 and TIVAR − S. Typhimurium var. 5 (04)-9639 were grown for 4 cell generations (cell doublings) in MHB (log phase) and transferred to LPM pH 5.5 medium for 3 cell generations; subsequently these cells were transferred back into MHB medium for 7 cell generations (Heithoff et al., 2012). (B) S. Typhimurium 14028 was grown overnight in MHB medium, and bacterial cells from the saturated culture were transferred without dilution to LPM pH 5.5 medium. MIC values were determined in LPM pH 5.5 medium containing polymyxin B from at least three independent determinations. (C) The effect of growth conditions prior to and during drug exposure on Salmonella susceptibility to polymyxin B in bacterial cell survival assays (Groisman et al., 1997). S. Typhimurium 14028 grown (4 h) under inducing (I) or non-inducing (NI) conditions for TIVAR were subsequently exposed to PMB (1 h) under I or NI conditions, with CFU determined on nonselective LB medium. Black bars (I/I), growth and drug exposure (N-minimal medium with 10 μM Mg2 + pH 5.8). White bars (I/NI), growth (N-minimal medium with 10 μM Mg2 + pH 5.8); drug exposure (LB). Gray bars (NI/I), growth (N-minimal medium with 10 mM Mg2 + pH 7.7); drug exposure (N-minimal medium with 10 μM Mg2 + pH 5.8). Hatched bars (NI/NI), growth (N-minimal medium with 10 mM Mg2 + pH 7.7); drug exposure (LB). Percent survival = CFU[polymyxin B] / CFU[no drug] × 100 at 1 h post drug exposure. Statistical significance for cell survival was analyzed using REML analysis. Values given are the REML model means ± SEM derived from at least 5 independent determinations. ***P < 0.001; limit of detection < 0.02%.
Next, the number of resistant cells in the bacterial population was evaluated as a function of environmental conditions in bacterial cell survival assays, wherein cell viability is monitored after drug treatment (Groisman et al., 1997; Gunn and Miller, 1996). Our findings indicate that more than 80% of the bacterial population survived a high-dose PMB treatment (20 μg/ml) only if both prior growth and drug exposure occurred in inducing (I/I) conditions (Fig. 2C); cell survival was markedly decreased if either transpired in non-inducing conditions (I/NI; NI/I; NI/NI). Together, these data indicate that TIVAR induction involves a substantial portion of the bacterial population and requires both prior cell and drug exposure under inducing conditions.
3.3. The role of the environment in microbial susceptibility to antibiotics
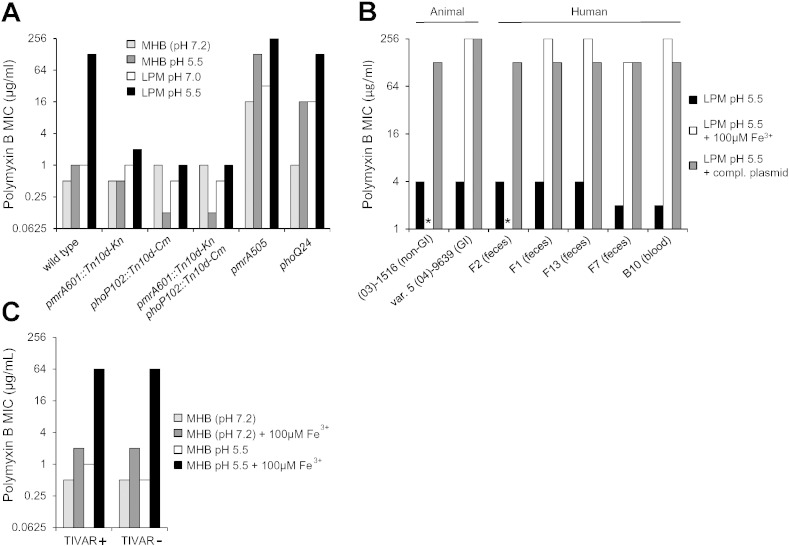
Since the PhoPQ/PmrAB regulons contribute to resistance to cationic peptides via LPS remodeling of Gram-negative membranes (Chen and Groisman, 2013; Gunn, 2008), we tested whether these regulatory functions were required for the TIVAR phenotype. Introduction of pmrA and/or phoP null mutations into TIVAR + S. Typhimurium abrogated PMB resistance under inducing and non-inducing conditions; whereas introduction of constitutively active mutations in pmrA or phoQ (Groisman et al., 1997; Gunn and Miller, 1996; Roland et al., 1993; Tamayo et al., 2005) resulted in high-level PMB resistance under non-inducing conditions (Fig. 3A). None of these mutations affected the induction of trimethoprim or ciprofloxacin resistance, suggesting that the host microenvironment may influence microbial susceptibility through multiple regulatory pathways, each operating in antibiotic-specific fashion.
Fig. 3.
The role of the environment (Fe3 +) in microbial susceptibility to antibiotics. (A) pmrAB and phoPQ null (pmrA601::Tn10d-Kn, phoP102::Tn10d-Cm) and constitutively active mutations (pmrA505, phoQ24) were introduced into S. Typhimurium 14028. Bacteria were grown in either LPM or MHB at pH 5.5 or pH 7 and MIC values were determined in the same medium from at least three independent determinations. (B) Bacteria were grown in LPM pH 5.5 medium in the presence and absence of 100 μM FeSO4, or in the presence of a complementing plasmid containing wild-type sequences of the indicated mutation. * denotes no growth in the presence of 100 μM FeSO4 due to iron toxicity. GI; gastrointestinal. (03)-1516 (horse non-GI) pmrABP1 -72C>T, pmrB H152Y; F2 (human feces) pmrABP1 -72C>T, pmrB H152Y; F1 (human feces) pmrABP2 1599G>A; F13 (human feces) pmrABP2 1408T>C, pmrC E415Q; F7 (human feces) phoQ P83L. Both B10 (human blood) and var. 5 (04)-9639 (cow GI) strains were complemented to TIVAR + with recombinant PmrD + sequences (Roland et al., 1994); corresponding mutations were not found in pmrD, phoPQ, or pmrCAB, suggesting parental mutation(s) are within other genes in the PhoPQ/PmrAB pathway. (C) Bacteria were grown in MHB pH 7.2 (unbuffered) or MHB pH 5.5 media in the presence and absence of 100 μM FeSO4, and the polymyxin B MIC was determined in the same medium from at least three independent determinations.
The PmrAB regulatory system promotes resistance to toxic metals and cationic peptides in response to ex vivo signals (e.g., high Fe3 +), which activate PmrB and downstream genes involved in Gram-negative membrane remodeling (Wösten et al., 2000). Thus, we reasoned that some naturally occurring TIVAR − isolates could potentially be converted to TIVAR + under high Fe3 + conditions via stimulation of the PmrAB regulon and resultant bypassing of inherent mutational deficiencies in membrane stabilization. To test this hypothesis, we screened a collection of Salmonella clinical isolates for susceptibility to PMB under LPM pH 5.5 conditions. These studies revealed that TIVAR − isolates represent a significant subset of natural Salmonella populations derived from human and livestock infections (Heithoff et al., 2008): S. Typhimurium (10/61), S. Newport (2/10), S. Enteritidis (2/8), S. Dublin (0/8), S. Bovismorbificans (2/5), and S. Choleraesuis (3/3). Subsequently, we assessed whether TIVAR − isolates could convert to TIVAR + under high Fe3 + conditions. Growth of TIVAR − S. Typhimurium strains in LPM pH 5.5 medium in the presence of 100 μM FeSO4 resulted in high-level PMB resistance in 5 of 7 strains tested; 2 of 7 strains exhibited no growth in the presence of 100 μM FeSO4 due to iron toxicity (Fig. 3B). Complementation, genetic linkage, and DNA sequence analyses revealed that all (7 of 7) TIVAR − strains contained mutations within the PhoPQ/PmrAB regulatory pathway; and TIVAR − strains contain similar or, in some cases, identical mutations that have circulated among diseased animals and humans. These findings suggest that conversion of TIVAR − strains to the TIVAR + phenotype may occur in high Fe3 + soil/water environments.
Next, we evaluated the role of iron in the susceptibility of microbes to antibiotics in MHB, the standard medium used clinically for antimicrobial susceptibility testing. While growth of TIVAR + and TIVAR − S. Typhimurium strains in MHB in the presence of 100 μM FeSO4 resulted in a mild increase in PMB resistance relative to MHB (2 vs. 0.5 μg/ml), growth in MHB pH 5.5 medium in the presence of 100 μM FeSO4 resulted in a marked increase in PMB resistance relative to MHB pH 5.5 (64 vs. 0.5–1 μg/ml) (Fig. 3C). These levels are similar to those exhibited upon growth in LPM pH 5.5 medium. These findings suggest that drug resistance may also be induced in iron-abundant conditions that may exist within in vivo and ex vivo environs.
3.4. Bacteria Exhibit High-level Antibiotic Resistance Only in a Subset of Host Tissues
The degree of antibiotic resistance as a function of growth within cultured macrophages was evaluated using TIVAR + and TIVAR − bacteria that have comparable virulence in a murine model of typhoid fever (Heithoff et al., 2012). TIVAR + Salmonella derived from infected cultured macrophages exhibited high-level PMB resistance via MIC determination in vitro, whereas TIVAR − Salmonella did not (Fig. 4A). Further, PMB treatment of infected cultured macrophages showed that internalized TIVAR + bacteria were much more resistant to PMB than TIVAR − bacteria (P < 0.001; Fig. 4B) (the inability of PMB to clear TIVAR − bacteria is presumably due to relatively low intracellular activity of cationic antimicrobial peptides (Buyck et al., 2013; Carryn et al., 2003)). These findings indicate that internalized TIVAR + bacteria are refractory to killing by PMB relative to bacteria that do not exhibit the TIVAR phenotype.
Fig. 4.
Salmonella exhibit high-level antibiotic resistance within cultured macrophages and are refractory to killing by antibiotic treatment. (A) Antimicrobial susceptibility of TIVAR + S. Typhimurium 14028 and TIVAR − S. Typhimurium var. 5 (04)-9639 derived from cultured RAW264.7 murine macrophages was evaluated via MIC determination on LPM pH 5.5 medium containing polymyxin B (PMB) or nalidixic acid (NAL). MIC values were the result of 2 to 3 independent determinations performed in triplicate. (B) Efficacy of antibiotic administration on the proliferation of TIVAR + and TIVAR − Salmonella within cultured RAW264.7 murine macrophages (PMB, gentamicin [GNT]). Values given are the average intracellular CFU ± SEM derived from 2 to 4 independent determinations performed in triplicate. Log transformed intracellular CFU data were analyzed for statistical significance using ANOVA; *P < 0.05, ***P < 0.001.
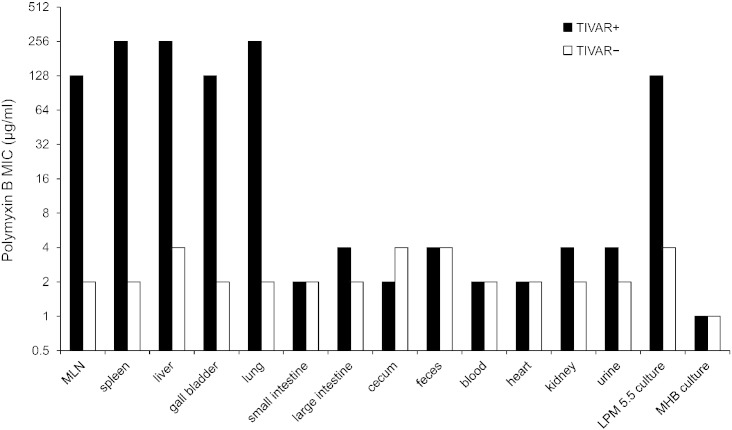
Next, the degree of antibiotic resistance exhibited by bacteria derived from infected mice was evaluated in a murine model of typhoid fever. Mice were intraperitoneally infected with TIVAR + or TIVAR − Salmonella, and bacteria derived from the tissues of septic mice were assessed for antibiotic susceptibility via MIC determination in vitro. TIVAR + Salmonella exhibited high-level PMB resistance (MIC = 128–256 μg/ml) only when derived from a subset of host tissues, including mesenteric lymph nodes, lung, liver, spleen and gall bladder (Fig. 5). In contrast, the TIVAR phenotype was not manifested in TIVAR + bacteria derived from other host tissues and from biological samples routinely used to detect bacterial infection (blood, feces, urine), or in TIVAR − bacteria derived from all tissues and biological samples tested. Since circulating PMB levels in treated patients and experimental rat infections range from 0.5 to 2.5 mg/l (Omri et al., 2002; Sandri et al., 2013; Zavascki et al., 2008), our findings indicate that TIVAR promotes drug resistance at certain host sites even when drug levels in circulation of mice are potentially 100 times higher than in treated patients.
Fig. 5.
Salmonella exhibit high-level antibiotic resistance when derived from a subset of host tissues and cells. BALB/c mice were infected with TIVAR + S. Typhimurium 14028 or TIVAR − S. Typhimurium var. 5 (04)-9639 (i.p. dose 103 CFU), and bacteria were derived from host tissues of septic mice. Antimicrobial susceptibility was assessed by MIC determination under inducing conditions (LPM pH 5.5 medium). TIVAR + S. Typhimurium MT2057 (kanamycin [KAN] resistant derivative of S. Typhimurium 14028) and TIVAR − S. Typhimurium var. 5 (04)-9639 (naturally resistant to chloramphenicol [CAM]) were used in these studies. MIC determinations for these strains contained KAN (40 μg/ml) and CAM (10 μg/ml), respectively, and were obtained from 3 to 13 mice per tissue/fluid. MLN, mesenteric lymph nodes.
3.5. Antibiotic Treatment Fails to Control Bacterial Infection and Promotes the Emergence of Drug-resistant Mutants
Polymyxin B is a last-line therapy to treat infections caused by multidrug-resistant Gram-negative bacteria in critically ill patients (Sandri et al., 2013; Zavascki et al., 2008). Thus, we evaluated whether PMB treatment was able to control TIVAR + Salmonella infection in a murine model of typhoid fever. All untreated mice infected with TIVAR + or TIVAR − bacteria died within 5 days of infection (Fig. 6A). PMB treated mice infected with TIVAR − bacteria survived at least 10 days post-infection, whereas all PMB treated mice infected with TIVAR + bacteria died within 7 days of infection (P < 0.001). Since the PMB dose given (30 mg/kg/day) was 6 to 20-fold higher than that used to treat human or experimental rat infections (1.5 to 5 mg/kg/day (Abdelraouf et al., 2012; Bergen et al., 2012; Landman et al., 2008)), human infection with TIVAR + bacteria would likely not be controlled by prescribed dosages. In contrast, ciprofloxacin (CIP) treated mice infected with TIVAR + Salmonella survived at least 10 days post-infection at dosages used to treat human or experimental mouse infections (30 mg/kg/day (Fantin et al., 2009; Guillard et al., 2013)). These data are consistent with the bacterium's susceptibility to CIP in MIC testing under LPM pH 5.5 conditions (Fig. 1A), and the drug's established intracellular activity (Carryn et al., 2003).
Fig. 6.
Antibiotic treatment is ineffective at controlling Salmonella infection and promotes the emergence of drug-resistant mutants. (A) BALB/c mice infected with TIVAR + S. Typhimurium 14028 or TIVAR − S. Typhimurium var. 5 (04)-9639 (i.p. dose 102 CFU) were treated (or mock-treated) with a twice-daily PMB or ciprofloxacin dosing regimen (30 mg/kg/day; i.p. route; 10 mice/cohort). Mouse survival was assessed for 10 days post-infection. Statistical significance for difference in proportions of animal survival was calculated using Chi-square; ***P < 0.001. (B) TIVAR + or TIVAR − Salmonella derived from overnight LB culture (in vitro) or from the spleens of septic mice (in vivo), obtained from untreated (day 5) and treated animals (TIVAR +, day 7; TIVAR −, day 10), were plated on LB medium containing PMB (16 μg/ml). TIVAR + S. Typhimurium MT2057 (kanamycin resistant derivative of S. Typhimurium 14028) and TIVAR − S. Typhimurium var. 5 (04)-9639 were used in these studies. Values given are the average mutation frequency (no. of PMBr colonies/total no. of colonies plated) ± SEM derived from 10 independent determinations. NA (not applicable): successful antibiotic treatment of mice infected with TIVAR − bacteria precluded recovery of PMBr mutants from these animals. Log transformed intracellular PMBr mutation frequency data were analyzed for statistical significance using ANOVA; ***P < 0.001.
Bacterial genetic mutagenesis occurs with increased frequency during infection of the host (Giraud et al., 2001; Martinez and Baquero, 2000; Nilsson et al., 2004). Here, we compared the frequency at which permanent PMB resistant mutants (PMBr) arise in Salmonella derived from cell culture versus that from antibiotic-treated or untreated animals. TIVAR + or TIVAR − Salmonella derived from overnight LB culture (in vitro) or from the spleens of septic mice (in vivo), obtained from untreated (day 5) and treated animals (TIVAR +, day 7; TIVAR −, day 10), were plated on LB medium containing PMB. Both TIVAR + and TIVAR − bacteria derived from the spleens of untreated mice exhibited a 60-fold increase in frequency of PMBr mutants relative to in vitro grown bacteria (Fig. 6B). Furthermore, TIVAR + bacteria exhibited an additional ~ 5-fold increase in mutation frequency when derived from treated (day 7) versus untreated (day 5) mice (P < 0.001). This implies that the increased lifespan of treated mice allows for additional rounds of bacterial replication, selection, and mutagenesis, ultimately resulting in an increased frequency of permanent drug-resistant mutants (note that successful antibiotic treatment of mice infected with TIVAR − bacteria precluded recovery of PMBr mutants from these animals (day 10)). DNA sequence analysis revealed that all PMBr mutants derived from TIVAR + bacteria in vitro (10/10) and in vivo (4/4) harbor mutations within pmrAB genes, which are known to confer PMB resistance via lipopolysaccharide (LPS) modifications that lead to membrane stabilization (Gunn, 2008; Kawasaki, 2012). Together, these findings indicate that antibiotic treatment fails to control bacterial infection caused by TIVAR + bacteria, and promotes the emergence of permanent drug-resistant mutants due to bacterial survival and growth during treatment.
4. Discussion
Since the discovery of penicillin by Alexander Fleming in 1928, antibiotic resistance has become widespread and plagues our standard of care today. We have identified a mechanism of antibiotic resistance (TIVAR) that promotes resistance in vivo at levels of drug that far exceed clinical doses routinely used for treatment. This mechanism has been overlooked in part because it is reversible and only operates within a subset of host tissues and cells, whereby the host milieu induces changes in the bacterium such that pathogens may become transiently resistant to high doses of certain antibiotics. TIVAR leads to treatment failure in animals, and may lead to patient treatment failure with the emergence of drug-resistant mutants due to bacterial survival and growth during antibiotic exposure. These findings call into question standardized MIC testing that has guided physician antibiotic treatment practices and drug development for the last several decades as it may not correlate with patient outcome.
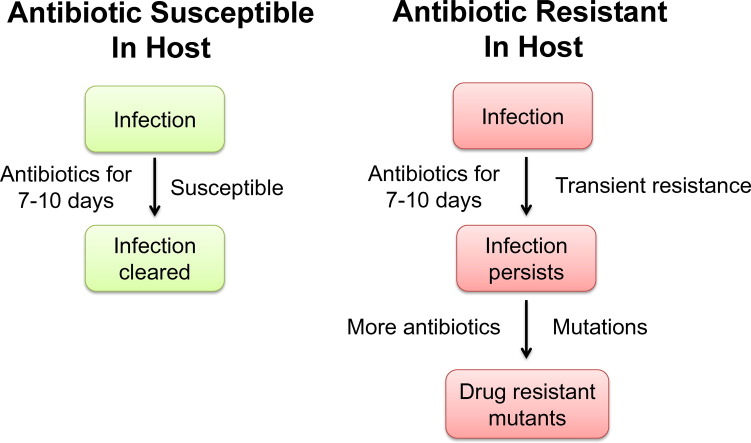
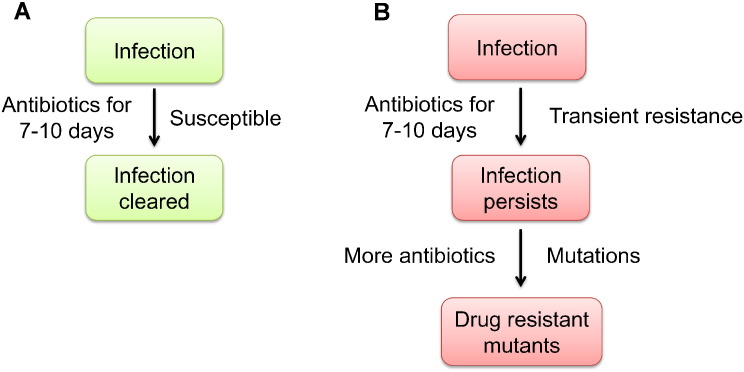
Although the standard 7–10 day antibiotic treatment regimen is usually sufficient to clear microbial infections, some patients fail to respond and require prolonged therapy, higher dosing or alternative antibiotics. This is particularly confounding in cases involving immune competent individuals where bacterial ID and AST results predict drug sensitivity. Why does this occur and what are the possible implications? Although bacteria may be susceptible to antibiotics in the laboratory, certain host sites may present a unique biochemical environment that induces changes in the bacterium so they become transiently resistant to high doses of certain antibiotics (Fig. 7). This results in a large, transiently-resistant bacterial population from which permanent drug-resistant mutants may arise via standard mutational mechanisms. Supporting this hypothesis, Salmonella exhibited PMB resistance only when derived from a subset of host tissues (lung, liver, spleen), but not from other host tissues or from biological samples routinely used to detect bacterial infection (blood, feces, urine). PMB treatment failed to control bacterial infection in a murine model of typhoid fever and was associated with an increased frequency of permanent drug-resistant bacteria. This was presumably due to the increased lifespan of treated mice, allowing for additional rounds of bacterial replication, selection, and mutagenesis during antibiotic exposure. Additionally, due to cross resistance between polymyxins and host antimicrobials (Band and Weiss, 2014), exposure to host cationic antimicrobial peptides may provide selective pressure for drug-resistant mutants to arise as soon as a bacterial infection is established and long before antibiotic therapy is even started.
Fig. 7.
TIVAR overview. (A) The standard 7–10 day antibiotic treatment regimen is sufficient to clear most bacterial infections. (B) Antibiotic treatment of persistent infections may be ineffective at controlling bacterial proliferation as certain host microenvironments may stimulate changes in the bacterium that result in the induction of transient resistance to high doses of antibiotics. This creates a major bacterial population that is transiently resistant to certain antibiotics, from which drug-resistant mutants may arise via established mutational mechanisms.
The TIVAR mechanism of resistance to cationic peptides is mediated by the PhoPQ/PmrAB regulatory system. Phenotypic switching from susceptibility to resistance was found to be more rapid than that of resistance to susceptibility, providing a means for retention of resistance during bacterial dissemination from permissive host sites (e.g., macrophages) in treated animals. Thus, macrophages may serve as potential reservoirs for persistent infections caused by intracellular bacteria. Additionally, the presence of Fe3 + triggered resistance in otherwise non-permissive conditions, indicating that resistance induction is not limited to the macrophage phagosome, but may also occur in iron-abundant environments that may exist ex vivo (soil/water) or in vivo (stomach) after ingestion of iron-rich foods (Wösten et al., 2000). This raises the possibility that resistance may be inadvertently triggered by diet; underlying conditions in the patient (e.g., iron overload during hemochromatosis (National Institutes of Health, 2015)); or by clinical interventions that may counteract drug efficacy (e.g., treatment of uncomplicated urinary tract infections to lower the pH of urine via ascorbic acid administration (Carlsson et al., 2001)).
A large proportion of antimicrobial use is directed to therapeutic and prophylactic use in livestock, which has been associated with the emergence of multidrug-resistant bacteria that have disseminated worldwide (Cloeckaert and Schwarz, 2001). As such, TIVAR has potential negative impacts on livestock production, with an increased risk of zoonotic transmission of multidrug-resistant pathogens to humans whereby therapeutic options would be even further constrained (Singer et al., 2003; Wegener, 2012). Moreover, since the induction of resistance was responsive to subtle changes in environmental conditions, management practices and environmental conditions inherent to livestock production have the potential to inadvertently trigger antibiotic resistance and the emergence of multidrug-resistant mutants; e.g., diet; the use of antibiotics in feeds as growth promotants; and/or exposure to environmental variables that may induce TIVAR resistance (livestock waste, subtherapeutic concentration of antimicrobials, and/or passage through different classes of stock).
TIVAR was evident in pathogenic serotypes of Salmonella and Yersinia, and similar mechanisms of resistance may be prevalent across the microbial realm. While it is likely that evolution of TIVAR predates antibiotic use, it is likely to play into the alarming rates of emerging antibiotic resistant bacteria. Even in the most advanced hospitals, high drug doses are given to infected patients without the knowledge that the host milieu may render bacteria inherently resistant to the antibiotics prescribed to control them. Nonetheless, for physicians managing cases where an antibiotic fails to clear an infection, rather than extending the current treatment or increasing the dose, a potentially more effective therapeutic option is prescription of another antibiotic indicated for the suspected pathogen. Unfortunately, standard in vitro testing methods may also inadvertently exclude antibiotics with potent efficacy, thereby unnecessarily limiting therapeutic options for multidrug-resistant pathogens as has been recently shown with azithromycin (Lin et al., 2015). Additionally, in vitro assessments of antibiotic resistance mutation frequency are not accurate predictors of clinical resistance (Thulin et al., 2015). Our findings support the use of host models as an important adjunct approach for antibiotic drug development and susceptibility testing, and may further help to modify current treatment guidelines to improve therapeutic intervention and diminish the emergence of multidrug-resistant pathogens.
Declaration of Interests
The authors have no competing interests to declare.
Author Contributions
Author contributions: J.Z.K., D.M.H., W.R.S, and M.J.M designed research; J.Z.K., D.M.H., W.R.S., and S.C.E. performed research; J.Z.K., D.M.H., W.R.S., S.C.E., J.K.H., J.D.M., J.W.S., and M.J.M. analyzed data; and J.W.S. and M.J.M. wrote the paper.
Funding
This work was supported by G. Harold & Leila Y. Mathers Foundation and Santa Barbara Cottage Hospital Research Program (2012–265; 2014–290) (to M.J.M); NIH grants HL135251 and GM100192 (to J.D.M); and National Science Foundation Graduate Research Fellowship DGE-1144085 (to J.Z.K.).
Acknowledgments
We thank Barbara A. Byrne for Salmonella clinical isolates used in this study, Lucien Barnes V for DNA sequence analysis, and Robert L. Sinsheimer for critically reading the manuscript.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2015.08.012.
Appendix A. Supplementary data
Supplementary tables.
References
- Abdelraouf K., He J., Ledesma K., Hu M., Tam V. Pharmacokinetics and renal disposition of polymyxin B in an animal model. Antimicrob. Agents Chemother. 2012;56:5724–5727. doi: 10.1128/AAC.01333-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen H., Donato J., Wang H., Cloud-Hansen K., Davies J., Handelsman J. Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010;8:251–259. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- Ambrose P., Bhavnani S., Rubino C., Louie A., Gumbo T., Forrest A., Drusano G. Pharmacokinetics-pharmacodynamics of antimicrobial therapy: it's not just for mice anymore. Clin. Infect. Dis. 2007;44:79–86. doi: 10.1086/510079. [DOI] [PubMed] [Google Scholar]
- Andersson D., Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat. Rev. Microbiol. 2010;8:260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- Band V., Weiss D. Mechanisms of antimicrobial peptide resistance in Gram-negative bacteria. Antibiotics. 2014;4:18–41. doi: 10.3390/antibiotics4010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergen P.J., Landersdorfer C.B., Zhang J., Zhao M., Lee H.J., Nation R.L., Li J. Pharmacokinetics and pharmacodynamics of ‘old’ polymyxins: what is new? Diagn. Microbiol. Infect. Dis. 2012;74:213–223. doi: 10.1016/j.diagmicrobio.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevins C., Salzman N. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol. 2011;9:356–368. doi: 10.1038/nrmicro2546. [DOI] [PubMed] [Google Scholar]
- Burchall J.J. Mechanism of action of trimethoprim-sulfamethoxazole. II. Int. J. Infect. Dis. 1973;128(Suppl.):437–441. doi: 10.1093/infdis/128.supplement_3.s437. [DOI] [PubMed] [Google Scholar]
- Buyck J., Tulkens P., Van Bambeke F. Pharmacodynamic evaluation of the intracellular activity of antibiotics towards Pseudomonas aeruginosa PAO1 in a model of THP-1 human monocytes. Antimicrob. Agents Chemother. 2013;57:2310–2318. doi: 10.1128/AAC.02609-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson S., Wiklund N., Engstrand L., Weitzberg E., Lundberg J. Effects of pH, nitrite, and ascorbic acid on nonenzymatic nitric oxide generation and bacterial growth in urine. Nitric Oxide. 2001;5:580–586. doi: 10.1006/niox.2001.0371. [DOI] [PubMed] [Google Scholar]
- Carryn S., Chanteux H., Seral C., Mingeot-Leclercq M.-P., Van Bambeke F., Tulkens P. Intracellular pharmacodynamics of antibiotics. Infect. Dis. Clin. N. Am. 2003;17:615–634. doi: 10.1016/s0891-5520(03)00066-7. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention . 2013. Antibiotic Resistance Threats in the United States, 2013. [Google Scholar]
- Chain P., Carniel E., Larimer F., Lamerdin J., Stoutland P., Regala W., Georgescu A., Vergez L., Land M., Motin V. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. U. S. A. 2004;101:13826–13831. doi: 10.1073/pnas.0404012101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Groisman E. The biology of the PmrA/PmrB two-component system: the major regulator of lipopolysaccharide modifications. Annu. Rev. Microbiol. 2013;67:83–112. doi: 10.1146/annurev-micro-092412-155751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute . Approved Standard-Ninth Edition. Clinical and Laboratory Standards Institute; 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. [Google Scholar]
- Cloeckaert A., Schwarz S. Molecular characterization, spread and evolution of multidrug resistance in Salmonella enterica Typhimurium DT104. Vet. Res. 2001;32:301–310. doi: 10.1051/vetres:2001126. [DOI] [PubMed] [Google Scholar]
- Conner C.P., Heithoff D.M., Julio S.M., Sinsheimer R.L., Mahan M.J. Differential patterns of acquired virulence genes distinguish Salmonella strains. Proc. Natl. Acad. Sci. U. S. A. 1998;95:4641–4645. doi: 10.1073/pnas.95.8.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes B., Brown N., Valdez Y., Brumell J., Finlay B. Expression and secretion of Salmonella pathogenicity island-2 virulence genes in response to acidification exhibit differential requirements of a functional type III secretion apparatus and SsaL. J. Biol. Chem. 2004;279:49804. doi: 10.1074/jbc.M404299200. [DOI] [PubMed] [Google Scholar]
- Cox G., Wright G. Intrinsic antibiotic resistance: mechanisms, origins, challenges and solutions. Int. J. Med. Microbiol. 2013;303:287–292. doi: 10.1016/j.ijmm.2013.02.009. [DOI] [PubMed] [Google Scholar]
- Croucher N., Harris S., Fraser C., Quail M., Burton J., van der Linden M., McGee L., von Gottberg A., Song J., Ko K. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Costa V., King C., Kalan L., Morar M., Sung W., Schwarz C., Froese D., Zazula G., Calmels F., Debruyne R. Antibiotic resistance is ancient. Nature. 2011;477:457–461. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- Davies J., Davies D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R.W., Botstein D., Roth J.R. Cold Spring Harbor Laboratory Press; Plainview, N. Y.: 1980. Advanced Bacterial Genetics. [Google Scholar]
- El-Halfawy O., Valvano M. Antimicrobial heteroresistance: an emerging field in need of clarity. Clin. Microbiol. Rev. 2015;28:191–207. doi: 10.1128/CMR.00058-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Committee on Antibiotic Suscepibility Testing EUCAST definitions of clinical breakpoints and epidemiological cut-off values. 2014. http://www.srga.org/Eucastwt/eucastdefinitions.htm
- Fantin B., Duval X., Massias L., Alavoine L., Chau F., Retout S., Andremont A., Mentré F. Ciprofloxacin dosage and emergence of resistance in human commensal bacteria. J. Infect. Dis. 2009;200:390–398. doi: 10.1086/600122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay B., Falkow S. Comparison of the invasion strategies used by Salmonella cholerae-suis, Shigella flexneri and Yersinia enterocolitica to enter cultured animal cells: endosome acidification is not required for bacterial invasion or intracellular replication. Biochimie. 1988;70:1089–1099. doi: 10.1016/0300-9084(88)90271-4. [DOI] [PubMed] [Google Scholar]
- Fleckenstein J., Kopecko D., Warren R., Elsinghorst E. Molecular characterization of the tia invasion locus from enterotoxigenic Escherichia coli. Infect. Immun. 1996;64:2256–2265. doi: 10.1128/iai.64.6.2256-2265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Food and Drug Administration . Center for Drug Evaluation and Research (CDER), U.S. Department of Health and Human Services; 2009. Guidance for Industry Microbiological Data for Systemic Antibacterial Drug Products — Development, Analysis, and Presentation. [Google Scholar]
- Galindo C., Rosenzweig J., Kirtley M., Chopra A. Pathogenesis of Y. enterocolitica and Y. pseudotuberculosis in human yersiniosis. J. Pathog. 2011;2011:1–16. doi: 10.4061/2011/182051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud A., Matic I., Tenaillon O., Clara A., Radman M., Fons M., Taddei F. Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science. 2001;291:2606–2608. doi: 10.1126/science.1056421. [DOI] [PubMed] [Google Scholar]
- Groisman E., Kayser J., Soncini F. Regulation of polymyxin resistance and adaptation to low-Mg2 + environments. J. Bacteriol. 1997;179:7040–7045. doi: 10.1128/jb.179.22.7040-7045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillard T., Cambau E., Chau F., Massias L., De Champs C., Fantin B. Ciprofloxacin treatment failure in a murine model of pyelonephritis due to an AAC (6′)-Ib-cr-producing Escherichia coli strain susceptible to ciprofloxacin in vitro. Antimicrob. Agents Chemother. 2013;57:5830–5835. doi: 10.1128/AAC.01489-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn J. The Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol. 2008;16:284–290. doi: 10.1016/j.tim.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Gunn J., Miller S. PhoP–PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J. Bacteriol. 1996;178:6857–6864. doi: 10.1128/jb.178.23.6857-6864.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heithoff D.M., Sinsheimer R.L., Low D.A., Mahan M.J. An essential role for DNA adenine methylation in bacterial virulence. Science. 1999;284:967–970. doi: 10.1126/science.284.5416.967. [DOI] [PubMed] [Google Scholar]
- Heithoff D.M., Shimp W.R., Lau P.W., Badie G., Enioutina E.Y., Daynes R.A., Byrne B.A., House J.K., Mahan M.J. Human Salmonella clinical isolates distinct from those of animal origin. Appl. Environ. Microbiol. 2008;74:1757–1766. doi: 10.1128/AEM.02740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heithoff D.M., Shimp W.R., House J.K., Xie Y., Weimer B.C., Sinsheimer R.L., Mahan M.J. Intraspecies variation in the emergence of hyperinfectious bacterial strains in nature. PLoS Pathog. 2012;8:e1002647. doi: 10.1371/journal.ppat.1002647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høiby N., Bjarnsholt T., Givskov M., Molin S., Ciofu O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents. 2010;35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- Kallen A., Hidron A., Patel J., Srinivasan A. Multidrug resistance among Gram‐negative pathogens that caused healthcare‐associated infections reported to the National Healthcare Safety Network, 2006–2008. Infect. Control Hosp. Epidemiol. 2010;31:528–531. doi: 10.1086/652152. [DOI] [PubMed] [Google Scholar]
- Kawasaki K. Complexity of lipopolysaccharide modifications in Salmonella enterica: its effects on endotoxin activity, membrane permeability, and resistance to antimicrobial peptides. Food Res. Int. 2012;45:493–501. [Google Scholar]
- Landman D., Georgescu C., Martin D.A., Quale J. Polymyxins revisited. Clin. Microbiol. Rev. 2008;21:449–465. doi: 10.1128/CMR.00006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Nonejuie P., Munguia J., Hollands A., Olson J., Dam Q., Kumaraswamy M., Rivera H., Corriden R., Rohde M. Azithromycin synergizes with cationic antimicrobial peptides to exert bactericidal and therapeutic activity against highly multidrug-resistant Gram-negative bacterial pathogens. EBioMedicine. 2015;2:690–698. doi: 10.1016/j.ebiom.2015.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J.L., Baquero F. Mutation frequencies and antibiotic resistance. Antimicrob. Agents Chemother. 2000;44:1771–1777. doi: 10.1128/aac.44.7.1771-1777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalopoulos A., Falagas M. Colistin: recent data on pharmacodynamics properties and clinical efficacy in critically ill patients. Ann. Intensive Care. 2011;1:1–6. doi: 10.1186/2110-5820-1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institutes of Health, 2014a. Co-trimoxazole Oral, in: http://www.nlm.nih.gov/medlineplus/druginfo/meds/a684026.html (Ed.), Medline Plus Drug Information. NIH.
- National Institutes of Health, 2014b. Trimethoprim (trimethoprim) Tablet, in: http://dailymed.nlm.nih.gov/dailymed/lookup.cfm?setid = a4e9183f-d0eb-4ba7-9204-760b1fd62010 (Ed.), Daily Med, Current Medical Information. NIH.
- National Institutes of Health, 2015. Hemachromatosis, in: http://www.nlm.nih.gov/medlineplus/hemochromatosis.html (Ed.), Medline Plus. NIH, U.S. National Library of Medicine
- Nelson D., Kennedy E. Magnesium transport in Escherichia coli inhibition by cobaltous ion. J. Biol. Chem. 1971;246:3042–3049. [PubMed] [Google Scholar]
- Nilsson A.I., Kugelberg E., Berg O.G., Andersson D.I. Experimental adaptation of Salmonella typhimurium to mice. Genetics. 2004;168:1119–1130. doi: 10.1534/genetics.104.030304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omri A., Suntres Z., Shek P. Enhanced activity of liposomal polymyxin B against Pseudomonas aeruginosa in a rat model of lung infection. Biochem. Pharmacol. 2002;64:1407–1413. doi: 10.1016/s0006-2952(02)01346-1. [DOI] [PubMed] [Google Scholar]
- Proctor R., Von Eiff C., Kahl B., Becker K., McNamara P., Herrmann M., Peters G. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 2006;4:295–305. doi: 10.1038/nrmicro1384. [DOI] [PubMed] [Google Scholar]
- Roland K., Martin L., Esther C., Spitznagel J. Spontaneous pmrA mutants of Salmonella typhimurium LT2 define a new two-component regulatory system with a possible role in virulence. J. Bacteriol. 1993;175:4154–4164. doi: 10.1128/jb.175.13.4154-4164.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roland K., Esther C., Spitznagel J. Isolation and characterization of a gene, pmrD, from Salmonella typhimurium that confers resistance to polymyxin when expressed in multiple copies. J. Bacteriol. 1994;176:3589–3597. doi: 10.1128/jb.176.12.3589-3597.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri A., Landersdorfer C., Jacob J., Boniatti M., Dalarosa M., Falci D., Behle T., Bordinhão R., Wang J., Forrest A. Population pharmacokinetics of intravenous polymyxin B in critically ill patients: implications for selection of dosage regimens. Clin. Infect. Dis. 2013;57:524–531. doi: 10.1093/cid/cit334. [DOI] [PubMed] [Google Scholar]
- Sievert D., Ricks P., Edwards J., Schneider A., Patel J., Srinivasan A., Kallen A., Limbago B., Fridkin S. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect. Control Hosp. Epidemiol. 2013;34:1–14. doi: 10.1086/668770. [DOI] [PubMed] [Google Scholar]
- Singer R., Finch R., Wegener H., Bywater R., Walters J., Lipsitch M. Antibiotic resistance—the interplay between antibiotic use in animals and human beings. Lancet Infect. Dis. 2003;3:47–51. doi: 10.1016/s1473-3099(03)00490-0. [DOI] [PubMed] [Google Scholar]
- Steele-Mortimer O. The Salmonella-containing vacuole—moving with the times. Curr. Opin. Microbiol. 2008;11:38–45. doi: 10.1016/j.mib.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo R., Prouty A., Gunn J. Identification and functional analysis of Salmonella enterica serovar Typhimurium PmrA‐regulated genes. FEMS Immunol. Med. Microbiol. 2005;43:249–258. doi: 10.1016/j.femsim.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Tauxe, R., 2013. Epidemiology of yersiniosis, in: Calderwood, S.B., section editor, Bloom, A., deputy editor (Eds.), http://www.uptodate.com/contents/epidemiology-of-yersiniosis, Accessed: May 23, 2013.
- Thulin E., Sundqvist M., Andersson D. Amdinocillin (mecillinam) resistance mutations in clinical isolates and laboratory-selected mutants of Escherichia. Antimicrob. Agents Chemother. 2015;59:1718–1727. doi: 10.1128/AAC.04819-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener H. Improving Food Safety Through A One Health Approach: Workshop Summary. 2012. Antibitiotic resistance — linking humans and animal health. [Google Scholar]
- Wiegand I., Hilpert K., Hancock R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008;3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- Wösten M.M., Kox L.F., Chamnongpol S., Soncini F., Groisman E. A signal transduction system that responds to extracellular iron. Cell. 2000;103:113–125. doi: 10.1016/s0092-8674(00)00092-1. [DOI] [PubMed] [Google Scholar]
- Zavascki A., Goldani L., Cao G., Superti S., Lutz L., Barth A., Ramos F., Boniatti M., Nation R., Li J. Pharmacokinetics of intravenous polymyxin B in critically ill patients. Clin. Infect. Dis. 2008;47:1298–1304. doi: 10.1086/592577. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables.