Abstract
Background
The quartan malaria parasite Plasmodium malariae is the widest spread and best adapted human malaria parasite. The simian Plasmodium brasilianum causes quartan fever in New World monkeys and resembles P. malariae morphologically. Since the genetics of the two parasites are nearly identical, differing only in a range of mutations expected within a species, it has long been speculated that the two are the same. However, no naturally acquired infection with parasites termed as P. brasilianum has been found in humans until now.
Methods
We investigated malaria cases from remote Yanomami indigenous communities of the Venezuelan Amazon and analyzed the genes coding for the circumsporozoite protein (CSP) and the small subunit of ribosomes (18S) by species-specific PCR and capillary based-DNA sequencing.
Findings
Based on 18S rRNA gene sequencing, we identified 12 patients harboring malaria parasites which were 100% identical with P. brasilianum isolated from the monkey, Alouatta seniculus. Translated amino acid sequences of the CS protein gene showed identical immunodominant repeat units between quartan malaria parasites isolated from both humans and monkeys.
Interpretation
This study reports, for the first time, naturally acquired infections in humans with parasites termed as P. brasilianum. We conclude that quartan malaria parasites are easily exchanged between humans and monkeys in Latin America. We hypothesize a lack of host specificity in mammalian hosts and consider quartan malaria to be a true anthropozoonosis. Since the name P. brasilianum suggests a malaria species distinct from P. malariae, we propose that P. brasilianum should have a nomenclatorial revision in case further research confirms our findings. The expansive reservoir of mammalian hosts discriminates quartan malaria from other Plasmodium spp. and requires particular research efforts.
Keywords: Quartan malaria, Yanomami, Venezuela, Plasmodium malariae, Plasmodium brasilianum, New World monkey, Anthropozoonosis, Anthroponosis, Zoonosis, Circumsporozoite protein, CSP, Small subunit ribosomal RNA, 18S rRNA, Polymerase change reaction, Sequencing, PCR
Highlights
-
•
We found human infections with ‘Plasmodium brasilianum’, a quartan malaria parasite of New World monkeys in South America
-
•
We show that in areas of close contact humans and non-human primates are concurrently infected with quartan malaria parasites
-
•
We conclude that quartan malaria parasites can transcend host species boundaries with impunity
We found naturally acquired infections in humans with Plasmodium brasilianum parasites, a quartan malaria parasite which usually infects more than 35 monkey species in South America. This confirms that malaria parasites, which cause the quartan type of fever (two days without fever between fever peaks), are easily exchanged between humans and monkeys in Latin America. The wide host reservoir of quartan malaria parasites requires particular malaria research efforts.
1. Introduction
Since malaria eradication is on the global health agenda again, non-human primates as source for Plasmodium infections in humans have received increased attention (Ramasamy, 2014). In this context, the simian Plasmodium brasilianum is particularly interesting. In 1908, a quartan malaria parasite was identified by Gonder and von Berenberg-Gossler in an imported ‘bald-headed uakari’ (Cacajao calvus) and named P. brasilianum, the quartan malaria parasite of New World monkeys in Latin America (Gonder and Von Berenberg-Gossler, 1908). P. brasilianum resembles the human quartan parasite Plasmodium malariae under the microscope, but early cross-species experimental infections by subcutaneous transfer of parasitized blood from black spider monkeys in the 1930s were unsuccessful. Hence, the names of two distinct parasites were maintained (Coatney et al., 2003).
Later investigations in the 1960s demonstrated that humans could very well be experimentally infected with P. brasilianum from monkeys, and, vice versa, New World monkeys could be experimentally infected with P. malariae from humans (Coatney et al., 2003; Geiman and Siddiqui, 1969). Moreover, studies in the 1980s showed that monoclonal antibodies against the circumsporozoite protein (CSP) of P. malariae cross-reacted and even neutralized the infectivity of P. brasilianum sporozoites to monkeys and vice versa (Cochrane et al., 1985). Sequencing of the gene coding for CSP confirmed the identity of this otherwise species-specific epitope in the two parasites (Lal et al., 1988).
Another common tool for the molecular species identification of malaria parasites is the gene for the small subunit (18S) of ribosomes (Snounou et al., 1993). In 1999, Fandeur et al. analyzed the 18S gene sequences from quartan malaria parasites (P. brasilianum) found in four monkey species from French Guiana with the highest prevalence in Alouatta monkeys (Fandeur et al., 2000). The similarity between 18S sequences from P. brasilianum and P. malariae is more than 99% differing only in single nucleotide polymorphisms (SNPs). SNPs are distributed at random like in the genetic pool of one single species, and no distinctive marker has been identified so far.
Unlike other human Plasmodium species, no whole genome sequence is available for quartan malaria parasites. We reviewed other published gene targets (msp-1, dhfr, cytochrome b, microsatellite DNA markers) and found striking homologies in all the markers without any specific identifying SNPs between the two parasite types (Fandeur et al., 2000; Guimarães et al., 2012; Tanomsing et al., 2007). The two parasites are nowadays perceived as variants of the same species, which had specialized on different hosts. Or, in practical terms, when quartan malaria parasites were identified in monkeys, they were designated as P. brasilianum. Conversely, when quartan malaria parasites were detected in humans, they were classified as P. malariae. Thus, the infected host determined the Plasmodium designation.
At the time of writing, altogether thirteen 18S sequences of P. brasilianum and thirty-four 18S sequences of P. malariae were registered in the NCBI GenBank nucleotide database. Sero-epidemiological studies in Brazil and French Guyana already suggested that non-human primates might constitute a natural reservoir for human malaria, and may contribute to the maintenance of foci for P. malariae (Volney et al., 2002). However, as naturally-acquired infections with parasites termed as P. brasilianum were never described in humans (Baird, 2009), the idea of host specificity was upheld and the classification of P. brasilianum as an independent Plasmodium species was retained.
Amazonas, the most southern federal state of Venezuela, is bordering Colombia to the west and Brazil to the east. Half of the population belongs to one of eighteen indigenous ethnic groups with the Yanomami representing one of the largest Amerindian communities. These seminomadic Indians live on both sides of the frontier between Venezuela and Brazil. On the Venezuelan side, about 12,000 Yanomami inhabit the vast forest area where the Orinoco originates and the Casiquiare river bifurcates towards the southern Amazon (Metzger et al., 2008; Humboldt, 1812).
In a traditional Yanomami village, all persons live under one common roof, shabono, consisting of a circular open wooden construction that accommodates up to 400 people. Daily life takes place “open air” and the night is spent in hammocks. Many shabonos are difficult to reach and are several days walking distance to the nearest health post. It is estimated that around 5000 Yanomami have retreated into the deep jungle with little or no contact to Western culture (Metzger et al., 2008). As forest-dwelling people, Yanomami hunt monkeys as a food source and incorporate them as household's pets.
Overall, P. vivax is predominant in Amazonas with roughly 85% of all detected parasites, but the distribution pattern of the malaria species is variable and contingent upon the geographic settlement of the ethnic groups. For example, a pilot study conducted in Yanomami communities from the Upper Orinoco revealed that nearly half of the malaria positive samples were P. malariae (Metzger et al., 2008). In contrast, no infections caused by P. malariae were detected among indigenous Piaroa from the Middle Orinoco basin (Rodulfo et al., 2007). Interestingly, Yanomami communities have also the highest P. falciparum rates (40.3%) compared to other ethnic groups in the region (8.7–22.4%) (Metzger et al., 2009).
The current study was carried out to identify and characterize Plasmodium species in the Venezuelan Amazon. Specifically, we investigated quartan malaria cases in Yanomami communities living in remote areas of the Alto Orinoco Casiquiare Biosphere Reserve where humans and non-humans live in such close vicinity that they could be concurrent reservoirs of transmission.
2. Materials and methods
2.1. Samples
Samples for this study originate from surveys in the Yanomami communities of Ocamo, Mavaca, Koyowe, and Platanal situated in the Upper Orinoco area near the Brazilian border, which were carried out as part of governmental malaria and onchocerciasis control activities in the region between 2005 and 2007. Ethical approval was obtained by the Ethical Committees of the Servicio Autonomo Centro Amazónico de Investigación y Control de Enfermedades Tropicales ‘Simon Bolivar’ (SA-CAICET), Puerto Ayacucho, Venezuela, and the London School of Hygiene & Tropical Medicine, London (LSHTM), UK.
When the team—consisting of medical doctors, scientists and health workers—arrived in a shabono, people were invited for a gathering. Malaria control and research activities were explained with the help of translators. Special importance was given to the presence of elders and leaders of the community. Informed consent was obtained orally. All individuals who felt sick were examined, diagnosed and treated for malaria, or the respective disease, according to the guidelines of the Venezuelan Health Ministry (Metzger et al., 2008). The ages of the patients were estimated as the Yanomami have no counting system.
Thick and thin blood smears were taken from individuals who presented with a history of fever and/or headache and/or malaise. Blood samples were collected by finger prick and stored on filter papers. 633 samples were used for the retrospective screening to investigate the molecular genetics of P. malariae parasites.
2.2. DNA extraction and PCR diagnosis
Parasite DNA from field samples was extracted from dried blood spots on filter paper using a commercial extraction kit (QIAamp DNA Blood Mini Kit, Qiagen). Screening for Plasmodium spp. infection was carried out by conventional nested-PCR assay with genus and species-specific primers based on the small subunit ribosomal RNA genes (18S) described previously by Snounou et al. (1993).
Genomic DNA of the P. brasilianum Peruvian III strain (MR4-349) was obtained from the Malaria Research and Reference Resource Center (MR4) to generate reference sequences for analyses at the University of New Mexico School of Medicine, Albuquerque, NM, USA.
2.3. 18S: development of new primers and sequencing
Though a P. malariae-specific PCR assay by Snounou et al. (1993) is sensitive and typically employed in the differential diagnosis of species, the 145 bp product of this primer set is too short for extensive sequence analysis. Therefore, we designed new sequencing primers targeting an amplicon spanning the entire variable region 5 (V5), one of the eight highly variable regions in the 18S gene which has considerable sequence variations among Plasmodium species. In addition we amplified three variable domains (V4, V5, V7) from the genomic DNA of P. brasilianum Peruvian III strain obtained from MR4.
Primer sequences were selected from unique and common regions for P. malariae and P. brasilianum species. The first primer pair (Pm18S Outer-F and Pm18S Outer-R) amplifies an 808-bp fragment. The second set of nested primers (Pm18S Inner-F and Pm18S Outer-R) amplifies a 763-bp fragment. All amplified samples were purified using Exo-SAP It Kit (USB) and sequenced bi-directionally using forward and reverse primers. Details of primer sequences, PCR amplification, and sequencing methods are shown in the appendix.
2.4. CSP: primers and sequencing
For CSP gene amplification, primers were used from the conserved regions flanking the central repeat region that contains immunodominant epitopes. The amplified CSP gene fragment from each isolate was purified using Exo-Sap It kit (USB), cloned using a TOPO TA cloning kit (Invitrogen), and transformed into TOP10 competent Escherichia coli cells (Invitrogen). We sequenced at least 3 clones from each isolate using M13 primers and gene specific internal primers. Details of the methods are given in the appendix.
2.5. Sequence analysis of 18S and CSP gene
For the 18S gene, sequences were aligned and edited using the Vector NTI ContigExpress program version 10 (Invitrogen). Nucleotide sequences generated from each isolate were queried against the NCBI GenBank nucleotide database using BLASTN for similarity search. Out of the total 47 published sequences of P. malariae and P. brasilianum, 23 sequences overlapped with the amplified target. The remaining 24 sequences were amplified from regions different from Snounou et al. primer site. Detailed information about source, origin, and accession number of all published 18S gene sequences is listed in the supplementary (appendix). A Plasmodium phylogenetic tree based on the 18S gene was constructed using the Kimura 3-parameter implemented in MEGA software ver. 6.0 (Tamura et al., 2013). Theileria sp. (GenBank Ac. AF162432) was used as outgroup. The reliability of the tree was assessed by the bootstrap significance test with 1000 replications. The final tree was refined using the program FigTree version 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/). The nucleotide diversities were calculated using DnaSP (Librado and Rozas, 2009). The maximum likelihood genetic distances based on Kimura 3-parameter, modeled with gamma distribution (shape parameter = 0.21) were calculated with MEGA ver. 6.0. For distance calculation, the best suited maximum likelihood model was obtained by using Modeltest implemented in MEGA ver.6.0. The comparable 18S gene (A-type) sequences of other Plasmodium spp. for the analyses were obtained from NCBI GenBank and listed in the appendix. The sequence alignments that were used to infer phylogenetic relationships are available from the authors on request.
For the CSP gene, each plasmid sequence containing the CSP gene fragment was aligned using the Vector NTI ContigExpress program version 10 (Invitrogen). The edited sequences of the CSP gene from each isolate were translated to corresponding amino acid sequences using the ExPASy Translate tool (http://web.expasy.org/translate/). Deduced amino acid sequences were compared to CS protein sequences of P. malariae and P. brasilianum isolates available in the database.
3. Results
3.1. Diagnosis of P. malariae infection
The determination of malaria by conventional nested-PCR detected the presence of P. malariae DNA in 75 of 633 samples collected from different individuals in Yanomami villages, constituting an 11.8% carrier rate in this survey. 25 of 75 samples (33%) were co-infected with P. vivax (n = 7), P. falciparum (n = 12), or triple infections (n = 6) while the remaining 50 had mono-infections with quartan malaria parasites.
3.2. Differentiating P. brasilianum from P. malariae
Out of the 75 samples PCR-positive for P. malariae, the 18S gene from 33 samples (27 mono-infections and 6 mixed infections) was successfully amplified and the resulting 763 bp product was analyzed by sequencing. Upon sequence analysis, 12 of the 33 samples had 18S gene sequences that were 100% identical with a P. brasilianum strain (GenBank AF130735) isolated from an infected monkey (Alouatta seniculus) in French Guiana (Fandeur et al., 2000). The twelve P. brasilianum infected individuals were from five different shabonos. The estimated ages of the patients were from 6 to 60 years.
In addition, isolates from four patients (n = 4) were 100% identical with the P. malariae Myanmar strain 1 (GenBank AF487999); six isolates (n = 6) were 100% identical with the P. malariae Myanmar strain 2 (GenBank AF488000); and one isolate (n = 1) was 100% identical with the P. malariae PNG strain (GenBank AF145336). The remaining ten isolates (n = 10) were 99% identical to either P. malariae Myanmar strain 2 or P. malariae PNG strain. Variation in the ten samples was represented by nucleotide polymorphisms (i.e., substitutions, insertions and/or deletions). Among these ten isolates, four new variants were identified (appendix).
Alignment of the twenty-three published 18S sequences and newly generated sequences of quartan malaria parasites showed 27 polymorphic sites in the 496-nucleotide sequences spanning V5 region. The overall nucleotide diversity (Pi) was 6.5 × 10− 3 (SD 0.001), with an average number of nucleotide differences (k) of 2.935.
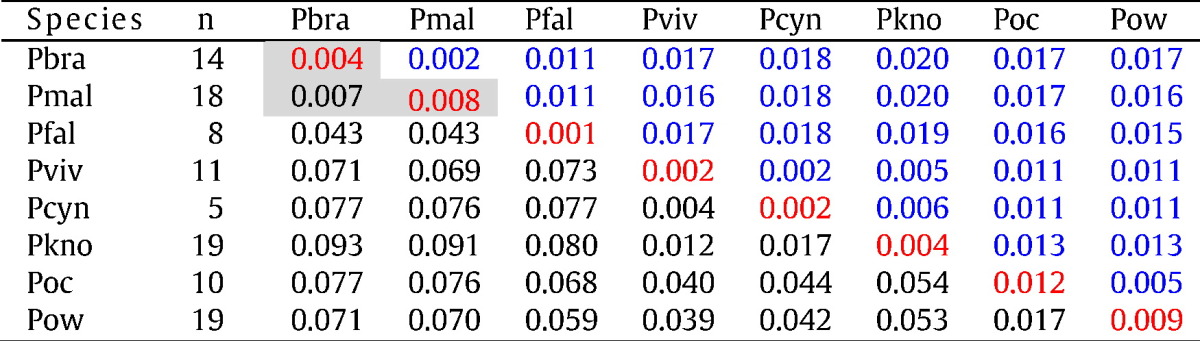
Table 1 shows the extent of divergence in the genetic distances between Plasmodium species, as calculated from the 18S gene spanning variable domain 5. Although the quartan parasites were genetically distinct from the other Plasmodium species, there was no genetic differentiation between the P. malariae and P. brasilianum isolates (distance, d = 0.005). The average genetic distance between all quartan isolates from human and monkey is comparable to intra-species genetic distance in other Plasmodium spp. (Table 1).
Table 1.
Estimates of the average genetic distance between and within Plasmodium spp.
The table shows average genetic distances based on variable domain 5 (V5) of the 18S gene sequences between (black) and within (red) Plasmodium spp. Standard errors (SE) for interspecies values (black) are shown above the diagonal (blue). The number of sequences (n) in each group of Plasmodium spp. is indicated in the second column. All V5 sequences generated in this study (including the MR4 sequence) and all comparable sequences from GenBank were included into the analysis. Sequences for P. malariae and P. brasilianum are listed in the supplementary appendix. Data show that genetic divergence between P. brasilianum and P. malariae is not more than within a species (gray boxes). Pmal, P. malariae; Pbra, P. brasilianum; Pfal, P. falciparum; Pviv, P. vivax; Pkno, P. knowlesi; Pcyn, P. cynomolgi; Poc, P. ovale curtisi; Pow, P. ovale wallikeri.
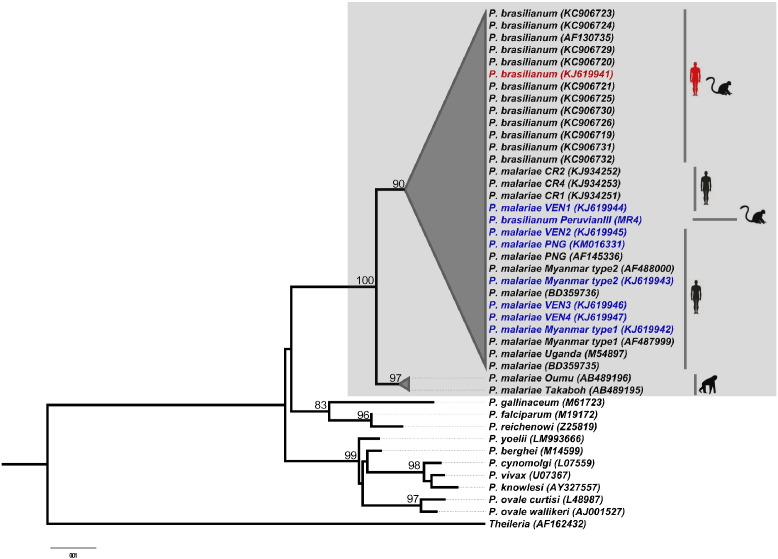
The phylogenetic analysis by the Neighbor joining method confirmed the similarity of all quartan malaria parasites (irrespective of the source of isolation, P. brasilianum or P. malariae) by clustering into a single monophyletic clade with a high bootstrap support of 100% (Fig. 1).
Fig. 1.
Neighbor Joining Tree based on 18S gene sequences of Plasmodium species. The tree shows that all quartan malaria parasites from humans and monkeys cluster into a monophyletic clade supported by a high bootstrap value of 99%. P. brasilianum and P. malariae sequences from this study are shown in color (red and blue). Hosts (non-human primate, human) are indicated by graphic symbols beside the taxa names. The sequence highlighted in red was found by Fandeur et al. in Alouatta monkeys, and in this study in humans. The two additional P. malariae isolates from human infections from Bangladesh (GenBank Ac. KF906514 & GenBank Ac. KF906514); Fuehrer et al. showed that these two isolates were 100% identical with a P. malariae-like isolate from the Chimpanzee Takaboh (GenBank Ac. AB489195).
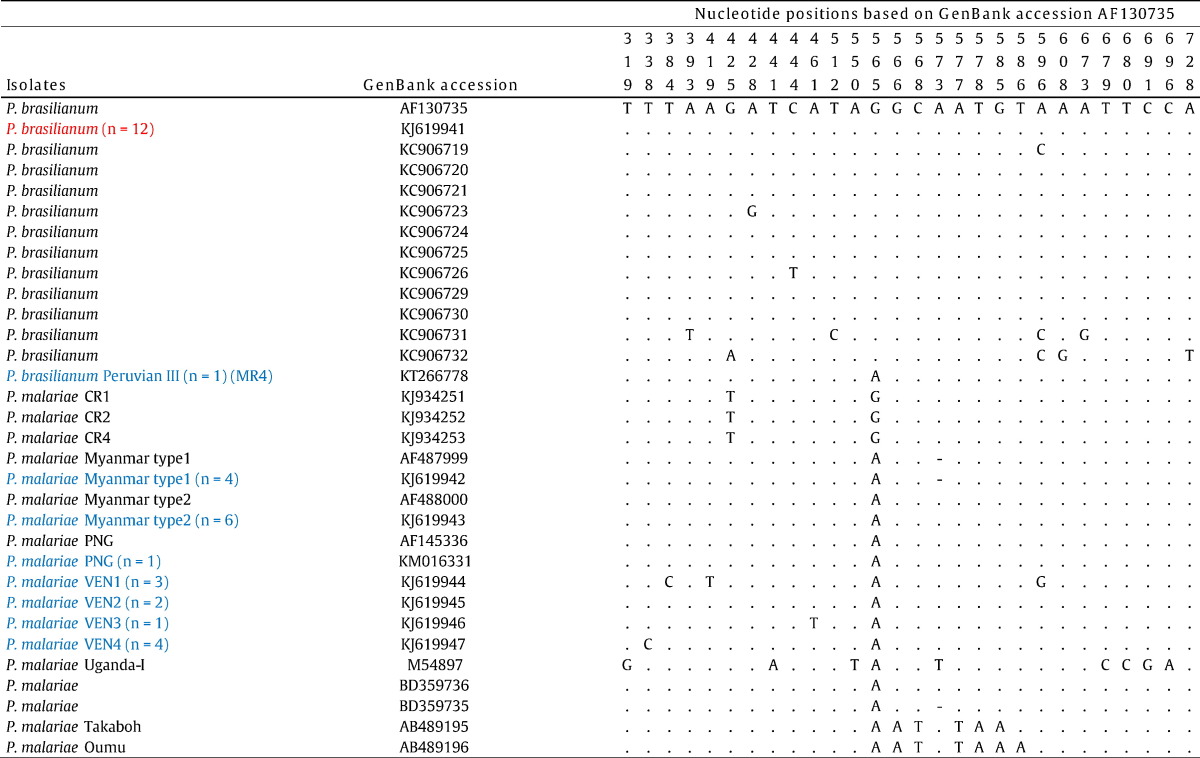
No identifying SNPs specific to either P. malariae or P. brasilianum was identified in the gene locus conventionally employed for differential diagnosis of malaria species. For example, all published P. brasilianum sequences isolated from monkeys and the twelve P. brasilianum sequences isolated from humans and sequenced in this study displayed G at position 565. However, the P. brasilianum isolate from Peru (MR4) sequenced in this study displayed an A, and three recently sequenced P. malariae isolates from humans in Costa Rica displayed a G at position 565 (Table 2).
Table 2.
Nucleotide polymorphisms in 18S gene sequences of P. brasilianum and P. malariae isolates.
The table shows that no distinctive marker for P. brasilianum could be identified. The SNP positions given vertically above are numbered according to the nucleotide sequences of P. brasilianum (GenBank AF130735). Dots represent identical residues; dashes represent deletions. P. brasilianum and P. malariae sequences from this study are shown in red and blue, respectively. The numbers in brackets denote the number of identical isolates found in this study. Apart of the four P. malariae strains VEN1, VEN2, VEN3, and VEN4, all isolates identified in this study are identical to pre-existing P. malariae/P. brasilianum sequences.
The new sequences identified in this study were submitted to the NCBI GenBank with accession numbers KJ619941–KJ619947KJ619941KJ619942KJ619943KJ619944KJ619945KJ619946KJ619947 and KM016331–KM016338KM016331KM016332KM016333KM016334KM016335KM016336KM016337KM016338.
3.3. Analysis of the CSP gene
The central immunodominant repeat region of the CSP gene was amplified and sequenced from three isolates, each representing P. brasilianum, P. malariae Myanmar strain 1, and P. malariae Myanmar strain 2, identified by 18S gene sequencing. Translated amino acid sequences showed that all three isolates constituted the minor tandem tetrapeptide repeat unit NDAG (N, asparagine; D, aspartic acid; A, alanine; G, glycine) and the major unit NAAG (N, asparagine; A, alanine; G, glycine) varying only by the number of repeat units.
All published isolates from South America, including P. brasilianum and P. malariae, from this and other studies, started the repeat region with the tetrapeptide NDEG, which is similar to P. malariae isolates from Asia. In contrast, African isolates from Uganda, Cameroon, and Cote d'Ivoire started the repeat region with NDAG (Table 3).
Table 3.
Comparison and characterization of the circumsporozoite protein (CSP) central repeat region among quartan malaria species.
The table shows that the otherwise species-specific immunodominant repeat region of the CS protein is the same for all quartan malaria parasites. It consists of the major repeat unit NAAG and the minor repeat unit NDAG. Numbers of the repeat unit can change between isolates; this has been shown for all Plasmodium spp. Clones of PCR amplified CSP alleles from three isolates are displayed each representing P. brasilianum, P. malariae Myanmar strain 1, and P. malariae Myanmar strain 2, identified by 18S gene sequencing in this study. African isolates start with a NDAG unit, whereas isolates from Asia and South America (P. malariae and P. brasilianum) start with NDEG.
| Central immunodominant repeat units | ||||||||
|---|---|---|---|---|---|---|---|---|
| Protein ID | Species (strain) | Origins (authors) | Repeat start | Minor unit | No. | Major unit | No. | Repeat size |
| KM016332 | P. brasilianum | Venezuela (this study) | NDEG | NDAG | 4 | NAAG | 50 | 54 |
| KM016333 | P. brasilianum | Venezuela (this study) | NDEG | NDAG | 4 | NAAG | 50 | 54 |
| KM016334 | P. brasilianum | Venezuela (this study) | NDEG | NDAG | 4 | NAAG | 50 | 54 |
| KM016337 | P. malariae (Myanmar type 1) | Venezuela (this study) | NDEG | NDAG | 4 | NAAG | 50 | 54 |
| KM016338 | P. malariae (Myanmar type 1) | Venezuela (this study) | NDEG | NDAG | 4 | NAAG | 49 | 53 |
| KM016335 | P. malariae (Myanmar type 2) | Venezuela (this study) | NDEG | NDAG | 5 | NAAG | 50 | 55 |
| KM016336 | P. malariae (Myanmar type 2) | Venezuela (this study) | NDEG | NDAG | 5 | NAAG | 51 | 56 |
| AAA29553 | P. brasilianum | Unknown (Lal et al., 1988) | NDEG | NDAG | 5 | NAAG | 58 | 63 |
| AGO33295 | P. brasilianum | Brazil (Araújo et.al. 2013) | NDEG | NDAG | 2 | NAAG | 21 | 23 |
| AAA18618 | P. malariae (China-1 CDC) | China (Qari et.al. 1994) | NDEG | NDAG | 7 | NAAG | 46 | 53 |
| AAA29557 | P. malariae (Uganda-1 CDC) | Uganda (Lal et al., 1988) | NDAG | NDAG | 6 | NAAG | 45 | 51 |
| CAA04809 | P. malariae | Cameroon (Tahar et al., 1998) | NDAG | NDAG | 7 | NAAG | 44 | 51 |
| CAA04812 | P. malariae | Cote d'Ivoire (Tahar et al., 1998) | NDAG | NDAG | 6 | NAAG | 46 | 52 |
4. Discussion
All quartan malaria parasites analyzed in this study would be P. malariae, if they had been found fifteen years earlier. However, as Fandeur et al. (2000) detected some 100% identical strains in monkeys in 1999, twelve of the 33 parasites had to be named P. brasilianum.
Based on phylogenetic analysis standards, P. malariae and P. brasilianum are one species (Fig. 1), and the punctual differences are not more than differences between strains of other Plasmodium species (Tables 1 and 2). So far, the host made the difference, as the infected host has been the classification determinant for P. malariae and P. brasilianum. Our findings show that this distinction criterion no longer applies.
Malaria history surmises that 500 years ago, Old World Humans introduced P. malariae to the New World; some of the parasites crossed the species barrier, adapted to New World monkeys and became P. brasilianum, the simian quartan malaria parasite of Latin America (Coatney et al., 2003). This hypothesis was challenged later; based on genetic diversity assumptions, it was reasoned that quartan parasites jumped from monkey to man and became human P. malariae (Tazi and Ayala, 2011). Though directions of the cross-species transfer are opposing, the two hypotheses share the common element of “host switching” which implies host specificity of P. brasilianum and P. malariae to monkey and man, respectively.
Our results allow an alternative view. For the first time, quartan malaria parasites, which are identical to those found in naturally infected primates of Latin America, were detected in naturally infected humans. Thus, our results provide evidence that quartan parasites are able to cross host species boundaries with impunity and that humans and non-human primates—in conditions of close contact—share quartan parasites without host specificity. Moreover, it can be speculated that P. brasilianum and P. malariae neither are distinct species, as the name suggests, nor are they distinct variants of one species, which became specialized on different hosts after switching, but are rather the same quartan malaria parasite species—an anthropozoonosis—circulating freely between monkeys and humans.
This confirmation of host sharing characteristics has long been pending Coatney, 1971; Escalante et al., 1995 and might give a reason to re-appraise quartan malaria, a largely neglected disease thus far, with the qualities to become an emerging infection.
In 1890, P. malariae was the first malaria species to get a scientific name (Collins and Jeffery, 2007). To date, the causative agent of quartan malaria has been largely understudied, mainly because—due to low levels of parasitaemia—in most epidemiological surveys only a few infections were detected (Mueller et al., 2007). Nonetheless, in studies with improved detection limits, P. malariae has been demonstrated in all malaria-endemic regions of the world Autino et al., 2012 and therefore—strictly speaking—might be the malaria parasite with the widest geographical distribution.
Furthermore, quartan malaria parasites might represent the best adapted malaria parasites. The adaption of these protozoans to its hosts results in a mainly chronic clinical outcome with many carriers suffering no symptoms. It is well-known that quartan parasites can persist in dormancy for decades in the host without causing symptoms (Collins and Jeffery, 2007). For example, two chimpanzees, Takaboh and Oumu, acquired quartan P. malariae-like parasites in the African rain forest when they were babies, and the parasites remained undiscovered until detection 30 years after their first day of confinement in a Japanese zoo (Hayakawa et al., 2009). This is perplexing because “hypnozoites”—as in P. vivax and P. ovale—have never been discovered or have not been well investigated.
What discriminates quartan malaria parasites from other Plasmodium spp. is their expansive reservoir of mammalian hosts. Besides the human host, there have been studies showing quartan malaria parasites in several dozens of monkey species. Merely in South America, quartan parasites were described in 35 monkey species (as P. brasilianum) (Fandeur et al., 2000). In Asia, monkeys are infected by the quartan Plasmodium inui (Coatney et al., 2003), and in the African rain forest, great apes are infected by quartan parasites (as Plasmodium rodhaini or P. malariae, and P. malariae-like) (Ramasamy, 2014). To our knowledge, this is by far the widest host reservoir compared with malaria of other periodicity.
Clinically, quartan malaria is considered as relatively harmless, but sufficient data are lacking to substantiate this assessment. Many investigators assume that P. malariae is the causal factor—perhaps with co-factors—for renal pathologies (Ehrich and Eke, 2007). Generally, it can be assumed that the clinical outcome of P. malariae infections is misinterpreted (McKenzie et al., 2001). Due to the chronic nature of the infection, capturing the true burden of the disease would require large longitudinal studies to assess the impact of the infection on occupational and social life of an individual.
Globally, the disease burden of quartan malaria is difficult to assess, because data on the incidence of P. malariae are faulty. The main reason is that P. malariae is principally underdiagnosed, because it thrives with a few hard-to-detect parasites, which are indistinguishable from P. vivax on the thick blood smear and which can also be misinterpreted as P. falciparum. Therefore, time-consuming reading of a thin smear or molecular methods would be necessary to identify this species. This explains why in the 1980s the Brazilian Health Ministry had “eradicated” P. malariae from Brazil by simply switching the official method of diagnosis from thin to thick blood smear (Oliveira-Ferreira et al., 2010).
In areas with marked variation in seasonal climate, P. malariae may account for 50% of the malaria episodes during the low-transmission season (Greenwood et al., 1987). As P. malariae is commonly found in sympatry with other Plasmodium species of humans, better understanding of species-interaction is necessary. Especially the interactions with P. falciparum in mixed infections is a controversially discussed topic (Mueller et al., 2007). Recent seroepidemiological and biomolecular surveys indicated that the prevalence of P. malariae are underestimated Mueller et al., 2007; Autino et al., 2012 and high prevalences of P. malariae have been reported from Africa (Doderer-Lang et al., 2014), Asia (Bharti et al., 2013), and Latin America (Volney et al., 2002). These findings correspond well with results of our pilot study of the Upper Orinoco Metzger et al., 2008 which prompted us to undertake the current investigation.
Initially, we hypothesized that P. brasilianum and P. malariae might be discernible by 18S gene sequencing since we noticed that the SNP at position 565 was shared by all hitherto published P. brasilianum isolates. Consequently, we identified this SNP as a possible distinctive marker. However, when we sequenced the P. brasilianum Peruvian III strain (provided by MR4), our hypothesis was rebutted. It was rebutted a second time, when three P. malariae isolates from humans in Costa Rica were published during the revision process of this article displaying a G at position 565. It appeared that SNPs were distributed at random, and none was specific for P. malariae or P. brasilianum (see Table 2).
To further ensure what was found in the 18S sequence, we analyzed the CS protein sequence, which also is used for species distinction, because the amino acid composition of the CSP gene is an adaptation for eliciting species specific antibody response. For example P. vivax, P. falciparum and P. knowlesi differ considerably in the CSP repeat region: P. vivax: DRAGGQPAG, P. falciparum: NANP, and P. knowlesi: GQPQAQGDGANA (Verra and Hughes, 1999). The CS protein sequence was identical between P. brasilianum and P. malariae, consistent with results by other investigators (see Table 3) (Escalante et al., 1995; Tahar et al., 1998).
So, unlike the zoonotic parasite P. knowlesi, which is genetically different from other Plasmodium spp. in several typical marker genes such as CSP and 18S genes (see Table 1) (Escalante et al., 1995; Escalante and Ayala, 1994), P. brasilianum is indistinguishable from P. malariae and infects the same hosts. And other than the two recently distinguished sympatric P. ovale curtisi and P. ovale wallikeri (Sutherland et al., 2010), the quartan parasites P. malariae and P. brasilianum do not segregate into distinct types in all genomic markers used so far (see Fig. 1) (Fandeur et al., 2000; Guimarães et al., 2012; Tanomsing et al., 2007).
It has to be adverted, that logic does not allow demonstration of identity, or non-difference, even if a few whole genome sequences of the parasites were available. Hence, the best approach to show that the parasites are indeed one species would be to do cross-mating experiments. However, if no distinctive markers are known whereby the off-spring can be identified, these experiments are very difficult to design and it is questionable if they are ethically justified.
So far, the evidence of naturally acquired human infections with parasites termed as P. brasilianum was a missing piece in the puzzle of the identity of this quartan parasite (Baird, 2009). Based on the results presented here, we conclude that anthropozoonotic transmission of quartan malaria occurs in areas where the habitat of man and monkey overlap. Similar transmission scenarios wit simian participation have been reported in areas outside the Amazonian region, for example in the Atlantic Forest of Brazil (De Pina-Costa et al., 2014).
We wonder if P. brasilianum will undergo a nomenclatorial revision, if further research confirms our findings. This would not be the first time that the name of a quartan malaria parasite is revised: In the 1940s, when P. rodhaini, the quartan parasite of apes in Africa, was identified to be P. malariae, the former name was soon “sinking into synonymy with P. malariae” (Coatney et al., 2003) and is not anymore in use today. This was justified just recently: Alignment of the 18S sequences of P. malariae from humans in Bangladesh with the parasite sequences of one of the two simian immigrants in the Japanese zoo, Takaboh (GenBank Ac. AB489195), resulted in 100% identity (see also legend of Fig. 1) (Fuehrer et al., 2014).
Malariologists know for some time that simian malaria will play an important role when human malaria eradication is envisaged (Bruce-Chwatt, 1968). As quartan parasites are the only global human malaria parasites successfully infecting multiple mammalian hosts, they could evade control measures that do not account for the animal reservoir. These hidden and possibly emerging parasites could, therefore, represent an important area for future research efforts.
Contributors
WGM, MM, and SVM carried out the primary data collection and supervised field and laboratory work. AL, MK, and PK performed the molecular experiments. BM, ME, SJ, and DJP supervised the laboratory work, facilitated the collaboration and gave overall input. WGM, MM, and AL conceived the study and designed the experiments. WGM and AL did the analysis, drafted and revised the manuscript. All authors interpreted the results, revised and approved the final manuscript.
Declaration of interests
We declare no competing interests.
Funding
The study was funded by WHO (grant No. HQ/05/131401) and the Ministry of Sciences and Technology of Venezuela (grant No. PC125-018) and institutional core funding of the Institute of Tropical Medicine of the University of Tübingen.
Acknowledgement
The study was funded by WHO (grant no. HQ/05/131401), the Ministry of Sciences and Technology of Venezuela (grant no. PC125-018), and institutional core funding of the Institute of Tropical Medicine of the University of Tübingen. The following reagent was obtained through the MR4 as part of the BEI Resources Repository, NIAID, NIH: MRA-349G, P. brasilianum Peruvian III genomic DNA (deposited by William Collins). We acknowledge the support by the Deutsche Forschungsgemeinschaft and Open Access Publishing fund of the University of Tuebingen. We thank the regional health authorities for their logistic support and the Yanomami communities of Ocamo, Mavaca, Koyowe, and Platanal.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2015.07.033.
Appendix A. Supplementary data
Supplementary material.
References
- Araújo M.S. Natural Plasmodium infection in monkeys in the state of Rondônia (Brazilian Western Amazon) Malar. J. 2013;12:180. doi: 10.1186/1475-2875-12-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autino B., Noris A., Russo R., Castelli F. Epidemiology of malaria in endemic areas. Mediterr. J. Hematol. Infect. Dis. 2012;4:e2012060. doi: 10.4084/MJHID.2012.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird J.K. Malaria zoonoses. Travel Med. Infect. Dis. 2009;7:269–277. doi: 10.1016/j.tmaid.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Bharti P.K. Emergence of a new focus of Plasmodium malariae in forest villages of district Balaghat, Central India: implications for the diagnosis of malaria and its control. Tropical Med. Int. Health TM IH. 2013;18:12–17. doi: 10.1111/tmi.12005. [DOI] [PubMed] [Google Scholar]
- Bruce-Chwatt L.J. Malaria zoonosis in relation to malaria eradication. Trop. Geogr. Med. 1968;20:50–87. [PubMed] [Google Scholar]
- Coatney G.R. The simian malarias: zoonoses, anthroponoses, or both? Am. J. Trop. Med. Hyg. 1971;20:795–803. doi: 10.4269/ajtmh.1971.20.795. [DOI] [PubMed] [Google Scholar]
- Coatney G.R., Collins W.E., Warren M., Contacos P.G. Division of Parasitic Disease, Producers. Version 1.0. CDC; Atlanta, GA: 2003. CD-ROM. The primate malarias [Original book published 1971] [Google Scholar]
- Cochrane A.H., Barnwell J.W., Collins W.E., Nussenzweig R.S. Monoclonal antibodies produced against sporozoites of the human parasite Plasmodium malariae abolish infectivity of sporozoites of the simian parasite Plasmodium brasilianum. Infect. Immun. 1985;50:58–61. doi: 10.1128/iai.50.1.58-61.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins W.E., Jeffery G.M. Plasmodium malariae: parasite and disease. Clin. Microbiol. Rev. 2007;20:579–592. doi: 10.1128/CMR.00027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pina-Costa A. Malaria in Brazil: what happens outside the Amazonian endemic region. Mem. Inst. Oswaldo Cruz. 2014;109:618–633. doi: 10.1590/0074-0276140228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doderer-Lang C. The ears of the African elephant: unexpected high seroprevalence of Plasmodium ovale and Plasmodium malariae in healthy populations in Western Africa. Malar. J. 2014;13:240. doi: 10.1186/1475-2875-13-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrich J.H.H., Eke F.U. Malaria-induced renal damage: facts and myths. Pediatr. Nephrol. Berl. Ger. 2007;22:626–637. doi: 10.1007/s00467-006-0332-y. [DOI] [PubMed] [Google Scholar]
- Escalante A.A., Ayala F.J. Phylogeny of the malarial genus Plasmodium, derived from rRNA gene sequences. Proc. Natl. Acad. Sci. U. S. A. 1994;91:11373–11377. doi: 10.1073/pnas.91.24.11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante A.A., Barrio E., Ayala F.J. Evolutionary origin of human and primate malarias: evidence from the circumsporozoite protein gene. Mol. Biol. Evol. 1995;12:616–626. doi: 10.1093/oxfordjournals.molbev.a040241. [DOI] [PubMed] [Google Scholar]
- Fandeur T., Volney B., Peneau C., de Thoisy B. Monkeys of the rainforest in French Guiana are natural reservoirs for P. brasilianum/P. malariae malaria. Parasitology. 2000;120(Pt 1):11–21. doi: 10.1017/s0031182099005168. [DOI] [PubMed] [Google Scholar]
- Fuehrer H.-P. High prevalence and genetic diversity of Plasmodium malariae and no evidence of Plasmodium knowlesi in Bangladesh. Parasitol. Res. 2014;113:1537–1543. doi: 10.1007/s00436-014-3798-8. [DOI] [PubMed] [Google Scholar]
- Geiman Q.M., Siddiqui W.A. Susceptibility of a New World monkey to Plasmodium malariae from aman. Am. J. Trop. Med. Hyg. 1969;18:351–354. doi: 10.4269/ajtmh.1969.18.351. [DOI] [PubMed] [Google Scholar]
- Gonder R., Von Berenberg-Gossler Untersuchungen über malaria-plasmodien der affen. Malar. Int. Arch. Leipzig. 1908;1:47–50. [Google Scholar]
- Greenwood B.M. Mortality and morbidity from malaria among children in a rural area of The Gambia, West Africa. Trans. R. Soc. Trop. Med. Hyg. 1987;81:478–486. doi: 10.1016/0035-9203(87)90170-2. [DOI] [PubMed] [Google Scholar]
- Guimarães L.O. The genetic diversity of Plasmodium malariae and Plasmodium brasilianum from human, simian and mosquito hosts in Brazil. Acta Trop. 2012;124:27–32. doi: 10.1016/j.actatropica.2012.05.016. [DOI] [PubMed] [Google Scholar]
- Hayakawa T. Identification of Plasmodium malariae, a human malaria parasite, in imported chimpanzees. PLoS One. 2009;4:e7412. doi: 10.1371/journal.pone.0007412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humboldt A. v. Über die Verbindung zwischen dem Orinoko und dem Amazonenfluss. Mon. Corresp. Zur Beförd. Erd- Himmels-Kunde. 1812;26:S.230–S.235. [Google Scholar]
- Lal A.A. Circumsporozoite protein gene from Plasmodium brasilianum. Animal reservoirs for human malaria parasites? J. Biol. Chem. 1988;263:5495–5498. [PubMed] [Google Scholar]
- Librado P., Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinforma. Oxf. Engl. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- McKenzie F.E., Jeffery G.M., Collins W.E. Plasmodium malariae blood-stage dynamics. J. Parasitol. 2001;87:626–637. doi: 10.1645/0022-3395(2001)087[0626:PMBSD]2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger W.G. Malaria diagnosis under field conditions in the Venezuelan Amazon. Trans. R. Soc. Trop. Med. Hyg. 2008;102:20–24. doi: 10.1016/j.trstmh.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Metzger W.G. A rapid malaria appraisal in the Venezuelan Amazon. Malar. J. 2009;8:291. doi: 10.1186/1475-2875-8-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller I., Zimmerman P.A., Reeder J.C. Plasmodium malariae and Plasmodium ovale—the ‘bashful’ malaria parasites. Trends Parasitol. 2007;23:278–283. doi: 10.1016/j.pt.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira-Ferreira J. Malaria in Brazil: an overview. Malar. J. 2010;9:115. doi: 10.1186/1475-2875-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qari S.H., Collins W.E., Lobel H.O., Taylor F., Lal A.A. A study of polymorphism in the circumsporozoite protein of human malaria parasites. Am. J. Trop. Med. Hyg. 1994;50(1):45–51.c. doi: 10.4269/ajtmh.1994.50.45. [DOI] [PubMed] [Google Scholar]
- Ramasamy R. Zoonotic malaria — global overview and research and policy needs. Front. Public Health. 2014;2:123. doi: 10.3389/fpubh.2014.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodulfo H., De Donato M., Mora R., González L., Contreras C.E. Comparison of the diagnosis of malaria by microscopy, immunochromatography and PCR in endemic areas of Venezuela. Braz. J. Med. Biol. Res. Rev. Bras. Pesqui. Médicas E Biológicas Soc. Bras. Biofísica Al. 2007;40:535–543. doi: 10.1590/s0100-879x2007000400012. [DOI] [PubMed] [Google Scholar]
- Snounou G. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol. Biochem. Parasitol. 1993;61:315–320. doi: 10.1016/0166-6851(93)90077-b. [DOI] [PubMed] [Google Scholar]
- Sutherland C.J. Two nonrecombining sympatric forms of the human malaria parasite Plasmodium ovale occur globally. J. Infect. Dis. 2010;201:1544–1550. doi: 10.1086/652240. [DOI] [PubMed] [Google Scholar]
- Tahar R., Ringwald P., Basco L.K. Heterogeneity in the circumsporozoite protein gene of Plasmodium malariae isolates from sub-Saharan Africa. Mol. Biochem. Parasitol. 1998;92:71–78. doi: 10.1016/s0166-6851(97)00226-0. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanomsing N. Genetic analysis of the dihydrofolate reductase–thymidylate synthase gene from geographically diverse isolates of Plasmodium malariae. Antimicrob. Agents Chemother. 2007;51:3523–3530. doi: 10.1128/AAC.00234-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazi L., Ayala F.J. Unresolved direction of host transfer of Plasmodium vivax v. P. simium and P. malariae v. P. brasilianum. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2011;11:209–221. doi: 10.1016/j.meegid.2010.08.007. [DOI] [PubMed] [Google Scholar]
- Verra F., Hughes A.L. Biased amino acid composition in repeat regions of Plasmodium antigens. Mol. Biol. Evol. 1999;16:627–633. doi: 10.1093/oxfordjournals.molbev.a026145. [DOI] [PubMed] [Google Scholar]
- Volney B., Pouliquen J.-F., De Thoisy B., Fandeur T. A sero-epidemiological study of malaria in human and monkey populations in French Guiana. Acta Trop. 2002;82:11–23. doi: 10.1016/s0001-706x(02)00036-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.