Abstract
Endogenous retroelements (EREs) are essential motors of evolution yet require careful control to prevent genomic catastrophes, notably during the vulnerable phases of epigenetic reprogramming that occur immediately after fertilization and in germ cells. Accordingly, a variety of mechanisms restrict these mobile genetic units. Previous studies have revealed the importance of KRAB-containing zinc finger proteins (KRAB-ZFPs) and their cofactor, KAP1, in the early embryonic silencing of endogenous retroviruses and so-called SVAs, but the implication of this transcriptional repression system in the control of LINE-1, the only known active autonomous retrotransposon in the human genome, was thought to be marginal. Two recent studies straighten the record by revealing that the KRAB/KAP system is key to the control of L1 in embryonic stem (ES) cells, and go further in demonstrating that DNA methylation and KRAB/KAP1-induced repression contribute to this process in an evolutionally dynamic fashion. These results shed light on the delicate equilibrium between higher vertebrates and endogenous retroelements, which are not just genetic invaders calling for strict control but rather a constantly renewed and nicely exploitable source of evolutionary potential.
Keywords: ERE, embryonic stem cells, endogenous retroelements, genome evolution, KRAB-ZFPs or KRAB-ZNFs, KAP1, LINE-1 or L1, retrotransposition
Barbara McClintock's discovery of transposable elements in maize in the early 1950s 1 challenged the prevailing dogma, which considered genomes as static entities passed essentially unchanged from one generation to the next. As such, it was met with scepticism, as was even more her suggestion that mobile genetic elements played important roles in regulating gene expression. Yet this was ultimately recognized as a formidable intuition, and served as one of the bases for Roy Britten and Eric Davidson's visionary model on gene regulation in eukaryotic cells.2 Today, it has become obvious that transposable elements are not only essential motors of evolution that remodel genome architecture, but also key components of transcriptional networks that govern processes as crucial as embryonic stem cell pluripotency.3-8
Transposons in the LINE light
Transposons account for a readily identifiable 50% of the human and mouse genomic DNA,9,10 far more than the 1.5% encoding for protein.11,12 And taking into account the decay of older elements beyond recognition, genomic rearrangements occurring over the course of evolution, and the challenge of sequencing and assembling repetitive sequences, the true proportion of the genomes of these and other higher species contributed by mobile genetic elements is most likely much greater.13
DNA transposons, which replicate by a cut-and-paste mechanism, are rare and inactive in the genomes of higher vertebrates, representing less than 2% of the human DNA. The very vast majority of mobile elements in both humans and mice are indeed retroelements, which spread by reverse transcription of an RNA intermediate and integration of its DNA copy, a copy-and-paste process that progressively amplifies their representation in the host genome. Whether in mouse or human, about a quarter of the retroelement-derived DNA stems from LTR-containing retroviruses, which became part of the genome of these species or their ancestors after infecting the germ line.14–16 The rest originates in non-LTR retrotransposons, mobile elements without known extracellular equivalent that can be either the autonomous Long Interspersed Element or LINE (e.g. LINE-1 or L1), or the non-autonomous Short Interspersed Element SINE (e.g., Alu) and SVA, dependent for their retrotransposition on LINE-provided trans-acting functions. Non-LTR retroelements are the only class of transposons still active in humans, accounting for an estimated one new germ line integrant every 50 human births.17
LINE-1 makes up approximately 17% of the human DNA (with around 500’000 copies), and is the only currently active autonomous transposon in humans, with 80 to 100 copies still retrotransposition-competent.11,18–20 LINE can also mobilize non-autonomous retrotransposons and cellular RNAs, including non-coding and mRNAs, which at times results in the formation of processed pseudogenes.21–24 A full-length LINE-1 is typically 6.0 Kb-long with an approximately 900bp 5’ untranslated region (UTR), 2 open reading frames (ORF1 and ORF2) and a 200bp 3’UTR ending in a poly(A) tail. The 5’UTR drives L1 transcription as it contains binding sites for many transcription factors,25–29 and further contains both sense and antisense promoters. ORF1 and ORF2 encode for proteins (ORF1p and ORF2p) that are essential for retrotransposition. ORF1p is a 40-kDa, RNA binding, nucleo-cytoplasmic protein with nucleic acid chaperone activity.30,31 Importantly, L1 ORF1p preferentially associates with the mRNA it originates from, a phenomenon called cis-preference that leads to the formation of ribonucleoprotein particles (RNPs).32 ORF2p (150 kDa) has 2 domains carrying out enzymatic functions essential for L1 retrotransposition: endonuclease (EN) and reverse transcriptase (RT).33,34 L1 retrotransposition occurs by a mechanism known as Target Primed Reverse Transcription (TPRT), whereby reverse transcription and integration are coupled.35 It is a qualitatively inefficient process, as the vast majority of L1 integrants present in mammalian genomes are 5’ truncated, that is, devoid of promoter.36
L1 originated at least 170 million years ago (mya), before the marsupial/eutherian divergence. Therefore the human genome contains L1 insertions predating the origin of primates, as well as more recent, primate- and even human-specific integrants.37 L1 can be sorted into subfamilies based on their age, which can be estimated by sequence divergence analysis and by comparing the genomes of different species.38–40 Since the emergence of apes (Hominoidea), some 25 mya, 5 major L1 subfamilies have amplified, named L1PA5 to L1PA1, the latter human-specific hence also called L1Hs. In primates and other mammals, L1s have usually evolved with a single subfamily active at any given time, amplifying to thousands of copies before its replacement by another subfamily likely under selective pressure from host defense mechanisms.37,41,42 This pattern of evolution is remarkably different from that of non-LTR retrotransposons in other organisms such as Drosophila and fish, where multiple L1 lineages have usually evolved in parallel.43,44
L1 can affect genome structure and function in multiple ways, sometimes leading to pathology. There is to date at least 96 retrotransposition events (from L1s and non-autonomous non-LTR retroelements) known to have resulted in human monogenic disorders.45 While this illustrates the deleterious potential of retrotransposons, it has become increasingly evident that these are also essential to the evolution of higher species. Indeed, as postulated by Barbara McClintock, mobile genetic elements shape the structural and transcriptional landscape of the genome through their ability to generate new genes, to influence the expression of existing ones via enhancer, insulator or repressor effects,46 and to serve as platforms for recombination events that lead to chromosomal rearrangements.47,48
Host Maneuvers to Cut the LINE
Given the diverse effects of L1 and other EREs on the genome, their hosts have unsurprisingly evolved molecular barriers to prevent the uncontrolled spread of these elements. These defense mechanisms can target various steps of the ERE life cycle, including transcription, post-transcriptional processing, reverse transcription and integration. Transcriptional silencing by DNA methylation, post-transcriptional repression by RNA interference, and poisoning of reverse transcripts by cytidine deamination are the main known mechanisms of control of endogenous retroelements.49 As expected from the high error rate of all reverse transcriptases, EREs overcome these restrictions through the emergence of escape mutants, which in turn impose new selective pressures for the host to adapt its defense mechanisms.
It has been proposed that gene silencing by DNA methylation initially evolved as a defense mechanism against endogenous retrotransposons.50 It is often preceded by or coupled with the induction of heterochromatin through histone modifications, particularly during the vulnerable phases of genome-wide demethylation that occurs in early development and in germ cells. The corepressor KAP1/TRIM28, which serves as a scaffold for heterochromatin- and DNA methylation-inducing factors, plays a central role in the control of many endogenous retroelements (EREs) in human and mouse embryonic stem cells.51,52 The KAP1-nucleated repressor complex is commonly tethered to DNA by members of the KRAB zinc finger (KRAB-ZNF or KRAB-ZFP) protein family, which can bind DNA in a highly sequence-specific manner through a C-terminal array of zinc fingers.53 Supporting a role for the KRAB-ZNF gene family in the control of transposable elements, its rapid expansion went parallel to an increase in the abundance of EREs in tetrapod genomes, and a large fraction of both KRAB-ZNFs and EREs are species-specific.54
Previous studies had revealed the role of KAP1 in the repression of exogenous and endogenous retroviruses during early embryonic development.51,52,55 We recently demonstrated that it also controls L1 retrotransposons in both mouse and human ES cells.56 Moreover, we interestingly found that KAP1 is recruited only to a discrete set of L1 subfamilies, namely L1PA6 to L1PA3. A previous study on the evolution of L1 retrotransposons, which examined the timing of emergence of the human L1 subfamilies during primate evolution, estimated that these families amplified in our ancestral genome between 25 and 7.6 mya.41 Older L1 elements, likely due to their complete inactivation by mutations accumulated over time, seem not to be presently targeted by any silencing mechanism. Younger L1 elements, mostly human-specific, are not recognized by KAP1 but instead repressed by DNA methylation. For those subjected to KAP1-induced silencing, recruitment of the corepressor must occur via KRAB-ZNFs, as indicated by our identification of Gm6871 as a mouse-specific KRAB-ZFP responsible for tethering KAP1 to a temporally discrete subset of murine L1 elements. These data, coupled with the recent demonstration that the PIWI2 protein partakes in the regulation of L1HS in pluripotent cells,57 strongly support an evolutionary model in which the transcription of newly emerged L1 lineages is first repressed by small RNA-induced DNA methylation, before KAP1-mediated silencing takes over through the selection of KRAB-ZFPs capable of tethering the master corepressor to their sequence. The observed dynamics of KAP1 binding to L1 subfamilies and the identification of Gm6871 as an L1-specific KRAB-ZFP support a model whereby species-specific KRAB-ZFPs have evolved as host defense factors restricting specific groups of L1 and other EREs.
Fulfilling this prediction, another recent study identified ZNF91 and ZNF93 as 2 primate-specific KRAB-ZFPs repressing the activity of primate-specific L1 and SVA subfamilies in hES cells.58 By using trans-chromosomal mouse ES cells that contained a copy of human chromosome 11 (TC11-mESC), Jacobs and collaborators elegantly determined the epigenetic and transcriptional fate of primate-specific retrotransposons in a non-primate background. They first observed that in TC11-mESC, a subset of SVA and L1 carried by the human genomic fragment, which were normally repressed by KAP1 in human ES cells, were derepressed and enriched for the active mark H3K4me3. They then set up a luciferase-based repression assay to screen a set of 14 primate-specific KRAB-ZFPs highly expressed in hES for their ability to target these cis-acting SVA and L1 sequences. Finally, they validated the most repressive candidates by complementation in the TC11-mESC. This lead to the identification of ZNF91 and ZNF93 as responsible for silencing these SVA and L1, respectively. Further in silico analyses aimed at reconstructing the evolutionary history of these KRAB-ZNFs, complemented with in vitro repression assays, indicated that mutations and structural alterations had accumulated in the zinc fingers of these proteins, which led to their ability to recognize their current ERE targets. In turn, the authors observed that L1 elements descending from ZNF93-blocked L1 had a deletion within the ZNF93 binding site in the 5’UTR, which allowed them to escape repression by the KRAB-ZNF. In summary, these 2 studies shed a ray of light on the genetic and epigenetic bases of the evolutionally dynamic events that govern interactions between L1 and higher vertebrates.
Repression of the Already Suppressed
An intriguing observation is that EREs keep being repressed by KAP1 long after losing their retrotransposition potential through mutations. What are the bases of this phenomenon? Is it just the vestige of a now obsolete control mechanism, or rather due to the need to maintain repression for reasons unrelated to transposition, and the manifestation of the progressive co-option of targeted retroelements?
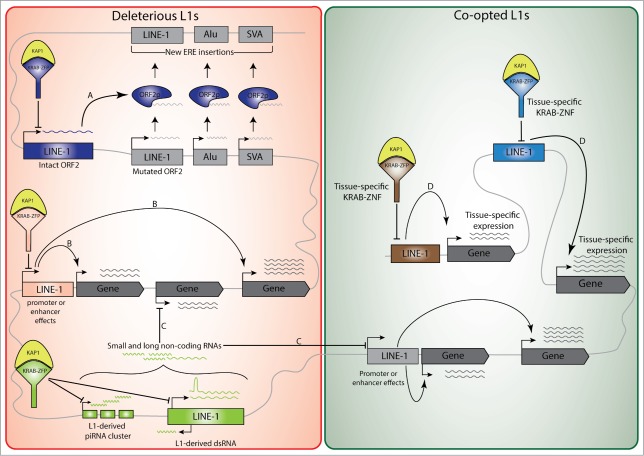
The production of an intact ORF2 protein by an otherwise defective L1 could keep promoting the mobilization of non-autonomous elements or other L1s still endowed with functional cis-acting sequences and ORF1 (Fig. 1A). However, we determined that only 1.5% of KAP1-bound L1 elements still have an intact ORF2. Thus, for the vast majority of retrotransposition-defective L1, the risk of trans-stimulation of other retroelements does not justify persistent repression, even though it is conceivable that rare truncated ORF2s retaining a functional endonuclease domain might cause DNA damage through the generation of DNA double-strand breaks.59–62
Figure 1.
KRAB-ZFP-mediated transcriptional regulation of L1 elements and its long-term impact on the host transcriptional landscape and genome architecture. Detrimental influences exerted by retrotransposition-defective L1 elements on the host could explain the maintenance of their KAP1-mediated repression (red area): For example, the production of an intact ORF2 protein by an otherwise defective L1 could promote the mobilization of non-autonomous Alu and SVA elements, as well as other L1s with a mutated ORF2 but endowed with functional 5’UTR and ORF1 (A). In addition, KAP1-mediated control of L1 could be required to avoid transcriptional perturbations induced either in cis via promoter or enhancer effects (B), or in trans by L1-derived small and long non-coding RNAs (C). Some L1 sequences can undergo positive selection and be co-opted by the host as promoters or enhancers (green area), which coupled to the tissue-specific expression of KRAB-ZNFs, may serve as a platform for context-specific gene regulation (D).
Less anecdotally, there is a growing recognition that many cis-acting regulatory sequences reside in EREs, and influence cellular genes expression by acting as promoters, enhancers or insulators (Fig. 1B).46 Furthermore, KAP1-mediated epigenetic repression can spread over tens of kilobases,63 and tethering of the KAP1 complex to genomic loci during the early embryonic period can trigger the methylation of adjacent CpG islands.64 Consistent with these findings, we could observe the KAP1-dependent silencing in human ES cells of a reporter construct containing a KAP1-targeted L1 sequence upstream of its promoter.56 Examining the transcriptome of murine and human ES cells, in which depletion of KAP1 or a specific KRAB-ZFP was used to incapacitate the KRAB/KAP1 pathway, more globally revealed the cis-acting impact of ERE-nucleated KAP1-mediated repression. This demonstrated that not only a wide range of EREs were upregulated, but also a high number of ERE-close genes, the transcription of which was stimulated via either promoter or enhancer effects.52,58,65However, most of these local influences stemmed from the de-repression of ERVs or SVAs, whereas no significant association was recorded for L1.56 How to explain this difference? First, it could be that KAP1-bound L1s, but not ERVs and SVAs, lack DNA binding sites for transcription factors highly active in ES cells. This hypothesis is consistent with the stronger upregulation of ERVs and SVAs, compared with L1, in KAP1-depleted human and murine ES cells.51,52,56 Second, L1-residing cis-acting regulatory sequences that are inert in ES cells could exert their influence later in differentiation. By performing KAP1 ChiP-seq analyses in primary CD4+ T lymphocytes and CD34+ haematopoietic stem cells, we indeed noted that between 4 and 7% of full-length L1 copies bearing KAP1 in ES cells remained KAP1-bound in these other cellular environments (unpublished). It is tempting to speculate that these L1 integrants contain tissue-specific enhancers or promoters, which become active when KAP1 is released or post-translationally modified at these loci by differentiation-induced signals (Fig. 1D). Comparing the genetic and epigenetic features of KAP1-bearing L1 integrants and of their genomic neighborhood in ES cells and various somatic cells will constitute a first step toward addressing this hypothesis.
Moving from a local to a more global scale, it could be that the KAP1-mediated control of L1 in ES cells avoids transcriptional perturbations induced in trans by L1-driven RNA interference (Fig. 1C). RNA interference (RNAi) designates a group of pathways, in which usually short noncoding RNAs serve as guides to protein complexes that down-regulate target genes either by blocking translation, triggering RNA degradation or inhibiting transcription. Various classes of small RNAs can be distinguished, including endogenous small interfering RNAs (endo-siRNAs), microRNAs (miRNAs) and PIWI-interacting RNAs (piRNAs).66 The link between RNA interference and endogenous retroelements is manifold and bidirectional. First, a significant fraction of small RNAs, in particular endo-siRNAs and piRNAs, are produced by transposable elements, suggesting their relevance in the sequence-based production, recognition and regulation of these transposons. Notably, the simultaneous activity of L1 sense and antisense promoters leads to “self-inflicted” L1 control via the generation of endo-siRNAs.67 Second, in silico analyses reveal that a subset of mammalian miRNAs is derived from ancient LINE-2,68 and the production of L1 5’UTR-derived miRNAs has also been demonstrated.69,70 Third, it was recently described that Microprocessor, a nuclear protein complex involved in miRNA biogenesis, can recognize secondary structures formed within the 5’UTR of L1 mRNAs transcribed from evolutionary older L1 subfamilies.71 Fourth, PIWI-interacting small RNAs, which are generated from clusters of transposable elements, have been implicated in the control of L1 retrotransposition in human pluripotent stem cells.57 Finally, EREs have been major contributors to the generation and diversification of long noncoding (lnc) RNAs, another group of RNAs involved in the regulation of cellular gene expression.72,73
It is predicted that the recruitment of KAP1 to L1 elements will repress the expression of small and long noncoding RNAs by these transposons, although this needs to be experimentally verified. The relevance of this effect, for integrants that are themselves retrotransposition-incompetent, may be in the possible targeting of other genomic regions by these interfering RNAs. This appears to be a distinct possibility, considering that a remarkably important fraction (17%) of the human genome is L1-derived, including 1 to 4% of coding sequences,10 and that, for instance, a majority of expressed full-length L1 elements in hES cells are located within genes.74 Additionally, due to mutations accumulated in these elements over time, they could produce degenerate interfering RNAs with off-target effects on cellular genes. In support of a role for ERE-derived interfering RNAs in the control of physiological processes, recent data indicate that ERE-produced lncRNAs are key to the maintenance of pluripotency.3,5,8,75,76 This warrants experiments aimed at determining whether KAP1 and specific KRAB-ZNFs regulate the production of L1-derived small and long noncoding RNAs, and at identifying the possibly non-ERE targets of these effectors.
Concluding remarks
Some fifty years after Barbara McClintock's original description of mobile genetic elements,1 their prominence in the genetic make up of higher species came to light with the first draft of the human genome.10,11 Since then, the genomics revolution has started unveiling the formidable intricacy of the relationship between transposable elements and their hosts. This relationship is not just an escalating arms race, but rather a subtle game of give-and-take, where hosts are protected from genomic disasters by restriction mechanisms, yet these controls are imposed with enough subtlety to preserve the production of genetic diversity, and what is more exploited at times to help domesticate a selected subset of transposons for adaptive purposes, by using them as platforms for modulating transcription networks. Evolutionary biology at its best…
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
Work in the authors’ laboratory is supported by the Swiss National Science Foundation, the European Research Council, and a Marie Curie network training grant (INGENIUM) from the seventh framework program of the European Union.
References
- 1. McClintock B. The origin and behavior of mutable loci in maize. Proc Nl Acad Sci U S A (1950); 36:344-355; PMID:15430309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Britten RJ, Davidson EH. Gene regulation for higher cells: a theory. Science (1969); 165:349-357; PMID:5789433 [DOI] [PubMed] [Google Scholar]
- 3. Fort A, Hashimoto K, Yamada D, Salimullah M, Keya CA, Saxena A, Bonetti A, Voineagu I, Bertin N, Kratz A, et al. Deep transcriptome profiling of mammalian stem cells supports a regulatory role for retrotransposons in pluripotency maintenance. Nat Gen (2014); 46:558-566; PMID:24777452; doi: 10.1038/ng.2965 [DOI] [PubMed] [Google Scholar]
- 4. Koyanagi-Aoi M, Ohnuki M, Takahashi K, Okita K, Noma H, Sawamura Y, Teramoto I, Narita M, Sato Y, Ichisaka T, et al. Differentiation-defective phenotypes revealed by large-scale analyses of human pluripotent stem cells. Proc Natl Acad Sci U S A (2013); 110:20569-20574; PMID:24259714; doi: 10.1073/pnas.1319061110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lu X, Sachs F, Ramsay L, Jacques PÉ, Göke J, Bourque G, Ng HH. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat Struct Mol Biol (2014); 21:423-425; PMID:24681886; doi: 10.1038/nsmb.2799 [DOI] [PubMed] [Google Scholar]
- 6. Macfarlan TS. et al. in Nature (2012); Vol. 487 57-63; PMID:22722858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ohnuki M, Tanabe K, Sutou K, Teramoto I, Sawamura Y, Narita M, Nakamura M, Tokunaga Y, Nakamura M, Watanabe A, et al. Dynamic regulation of human endogenous retroviruses mediates factor-induced reprogramming and differentiation potential. Proc Natl Acad Sci U S A (2014); 111:12426-12431; PMID:25097266; doi: 10.1073/pnas.1413299111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Santoni FA, Guerra J, Luban J. HERV-H RNA is abundant in human embryonic stem cells and a precise marker for pluripotency. Retrovirology (2012); 9:111; PMID:23253934; doi: 10.1186/1742-4690-9-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bannert N, Kurth R. Retroelements and the human genome: new perspectives on an old relation. Proc Natl Acad Sci U S A (2004); 101 Suppl 2:14572-14579; PMID:15310846; doi: 10.1073/pnas.0404838101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature (2001); 409:860-921; PMID:11237011; doi: 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- 11. Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science (2001); 291:1304-1351; PMID:11181995; doi: 10.1126/science.1058040 [DOI] [PubMed] [Google Scholar]
- 12. Waterston RH, Lander ES, Sulston JE. On the sequencing of the human genome. Proc Natl Acad Sci U S A (2002); 99:3712-3716; PMID:11880605; doi: 10.1073/pnas.042692499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS Gen (2011); 7:e1002384; PMID:22144907; doi: 10.1371/journal.pgen.1002384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cordaux R, Batzer MA. in Nat Rev. Gen (2009); Vol. 10:691-703; PMID:19763152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jern, P. & Coffin, J. M. Effects of retroviruses on host genome function. Annual review of genetics 42, 709-732, doi: 10.1146/annurev.genet.42.110807.091501 (2008). [DOI] [PubMed] [Google Scholar]
- 16. Mandal PK, Kazazian HH, Jr. SnapShot: Vertebrate transposons. Cell (2008); 135:192-192 e191; PMID:18854165; doi: 10.1016/j.cell.2008.09.028 [DOI] [PubMed] [Google Scholar]
- 17. Beck CR, Garcia-Perez JL, Badge RM, Moran JV. in Annu Rev Genomics Hum Genet (2011); Vol. 12:187-215; PMID:21801021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, Badge RM, Moran JV, in Cell (2010); Vol. 141:1159-1170; PMID:20602998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH, Jr. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci U S A (2003); 100:5280-5285; PMID:12682288; doi: 10.1073/pnas.0831042100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Furano AV. The biological properties and evolutionary dynamics of mammalian LINE-1 retrotransposons. Prog Nucl Acid Res Mol Biol (2000); 64:255-294; PMID:10697412 [DOI] [PubMed] [Google Scholar]
- 21. Buzdin A, Khodosevich K, Mamedov I, Vinogradova T, Lebedev Y, Hunsmann G, Sverdlov E, A technique for genome-wide identification of differences in the interspersed repeats integrations between closely related genomes and its application to detection of human-specific integrations of HERV-K LTRs. Genomics (2002); 79:413-422; PMID:11863371; doi: 10.1006/geno.2002.6705 [DOI] [PubMed] [Google Scholar]
- 22. Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet (2003); 35:41-48; PMID:12897783; doi: 10.1038/ng1223 [DOI] [PubMed] [Google Scholar]
- 23. Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet (2000); 24:363-367; PMID:10742098; doi: 10.1038/74184 [DOI] [PubMed] [Google Scholar]
- 24. Garcia-Perez JL, Doucet AJ, Bucheton A, Moran JV, Gilbert N. Distinct mechanisms for trans-mediated mobilization of cellular RNAs by the LINE-1 reverse transcriptase. Genome Res (2007); 17:602-611; PMID:17416749; doi: 10.1101/gr.5870107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Athanikar JN, Badge RM, Moran JV. A YY1-binding site is required for accurate human LINE-1 transcription initiation. Nucleic Acids Res (2004); 32:3846-3855; PMID:15272086; doi: 10.1093/nar/gkh698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Becker KG, Swergold GD, Ozato K, Thayer RE. Binding of the ubiquitous nuclear transcription factor YY1 to a cis regulatory sequence in the human LINE-1 transposable element. Hum Mol Genet (1993); 2:1697-1702; PMID:8268924 [DOI] [PubMed] [Google Scholar]
- 27. Kuwabara T, Hsieh J, Muotri A, Yeo G, Warashina M, Lie DC, Moore L, Nakashima K, Asashima M, Gage FH, Wnt-mediated activation of NeuroD1 and retro-elements during adult neurogenesis. Nat Neurosci (2009); 12:1097-1105; PMID:19701198; doi: 10.1038/nn.2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tchenio T, Casella JF, Heidmann T. Members of the SRY family regulate the human LINE retrotransposons. Nucleic Acids Res (2000); 28:411-415; PMID:10606637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang N, Zhang L, Zhang Y, Kazazian HH, Jr. An important role for RUNX3 in human L1 transcription and retrotransposition. Nucleic Acids Res (2003); 31:4929-4940; PMID:12907736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hohjoh H, Singer MF. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J (1996); 15:630-639; PMID:8599946 [PMC free article] [PubMed] [Google Scholar]
- 31. Martin SL. Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Mol Cell Biol (1991); 11:4804-4807; PMID:1715025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV, Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol (2001); 21:1429-1439; PMID:11158327; doi: 10.1128/MCB.21.4.1429-1439.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell (1996); 87:905-916; PMID:8945517 [DOI] [PubMed] [Google Scholar]
- 34. Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science (1991); 254:1808-1810; PMID:1722352 [DOI] [PubMed] [Google Scholar]
- 35. Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. EMBO J (2002); 21:5899-5910; PMID:12411507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kazazian HH, Jr, Moran JV. The impact of L1 retrotransposons on the human genome. Nat Genet (1998); 19:19-24; PMID:9590283; http://dx.doi.org/ 10.1038/ng0598-19 [DOI] [PubMed] [Google Scholar]
- 37. Smit AF, Tóth G, Riggs AD, Jurka J. in J Mol Biol (1995); Vol. 246 401-417; PMID:7877164 [DOI] [PubMed] [Google Scholar]
- 38. Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics (2000); 156:297-304; PMID:10978293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xing J, Hedges DJ, Han K, Wang H, Cordaux R, Batzer MA, Alu element mutation spectra: molecular clocks and the effect of DNA methylation. J Mol Biol (2004); 344:675-682; PMID:15533437; http://dx.doi.org/ 10.1016/j.jmb.2004.09.058 [DOI] [PubMed] [Google Scholar]
- 40. Boissinot S, Furano AV. The recent evolution of human L1 retrotransposons. Cytogenet Genome Res (2005); 110:402-406; PMID:16093692; http://dx.doi.org/ 10.1159/000084972 [DOI] [PubMed] [Google Scholar]
- 41. Khan H, Smit A, Boissinot S. in Genome Res (2006); Vol. 16:78-87; PMID:16344559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sookdeo A, Hepp CM, McClure MA, Boissinot S. in Mob DNA (2013); Vol. 4:3; PMID:23286374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Eickbush TH, Furano AV. Fruit flies and humans respond differently to retrotransposons. Curr Opin Genet Dev (2002); 12:669-674; PMID:12433580 [DOI] [PubMed] [Google Scholar]
- 44. Furano AV, Duvernell DD, Boissinot S. L1 (LINE-1) retrotransposon diversity differs dramatically between mammals and fish. Trends Genet (2004); 20:9-14; PMID:14698614; http://dx.doi.org/ 10.1016/j.tig.2003.11.006 [DOI] [PubMed] [Google Scholar]
- 45. Hancks DC, Kazazian J, Haig H. in Curr Opin Genet; Dev (2012); Vol. 22:191-203; PMID:22406018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gifford WD, Pfaff SL, Macfarlan TS. Transposable elements as genetic regulatory substrates in early development. Trends Cell Biol (2013); 23:218-226; PMID:23411159; http://dx.doi.org/ 10.1016/j.tcb.2013.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fitch DH, Bailey WJ, Tagle DA, Goodman M, Sieu L, Slightom JL, Duplication of the gamma-globin gene mediated by L1 long interspersed repetitive elements in an early ancestor of simian primates. Proc Natl Acad Sci U S A (1991); 88:7396-7400; PMID:1908094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schwartz A, Chan DC, Brown LG, Alagappan R, Pettay D, Disteche C, McGillivray B, de la Chapelle A, Page DC. Reconstructing hominid Y evolution: X-homologous block, created by X-Y transposition, was disrupted by Yp inversion through LINE-LINE recombination. Hum Mol Genet (1998); 7:1-11; PMID:9384598 [DOI] [PubMed] [Google Scholar]
- 49. Goodier JL, Kazazian HH, Jr. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell (2008); 135:23-35; PMID:18854152; http://dx.doi.org/ 10.1016/j.cell.2008.09.022 [DOI] [PubMed] [Google Scholar]
- 50. Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genetics (1997); 13:335-340; PMID:9260521 [DOI] [PubMed] [Google Scholar]
- 51. Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. in Nature (2010); Vol. 463 237-240; PMID:20075919 [DOI] [PubMed] [Google Scholar]
- 52. Turelli P, Castro-Diaz N, Marzetta F, Kapopoulou A, Raclot C, Duc J, Tieng V, Quenneville S, Trono D. Interplay of TRIM28 and DNA methylation in controlling human endogenous retroelements. Genome Res (2014); 24:1260-1270; PMID:24879559; http://dx.doi.org/ 10.1101/gr.172833.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Friedman JR, Fredericks WJ, Jensen DE, Speicher DW, Huang XP, Neilson EG, Rauscher FJ, 3rd. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Gen Dev (1996); 10:2067-2078; PMID:8769649 [DOI] [PubMed] [Google Scholar]
- 54. Thomas JH, Schneider S. in Genome Res (2011); Vol. 21 1800-1812; PMID:21784874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wolf D, Goff SP. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature (2009); 458:1201-1204; PMID:19270682; http://dx.doi.org/ 10.1038/nature07844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Castro-Diaz N, Ecco G, Coluccio A, Kapopoulou A, Yazdanpanah B, Friedli M, Duc J, Jang SM, Turelli P, Trono D. Evolutionally dynamic L1 regulation in embryonic stem cells. Gen Dev (2014); 28:1397-1409; PMID:24939876; http://dx.doi.org/ 10.1101/gad.241661.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Marchetto MCN, Narvaiza I, Denli AM, Benner C, Lazzarini TA, Nathanson JL, Paquola AC, Desai KN, Herai RH, Weitzman MD, et al. in Nature (2013); Vol. 503:525-529; PMID:24153179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jacobs FMJ. et al. in Nature (2014). [Google Scholar]
- 59. Belancio VP, Roy-Engel AM, Pochampally RR, Deininger P. Somatic expression of LINE-1 elements in human tissues. Nucleic Acids Res (2010); 38:3909-3922; PMID:20215437; http://dx.doi.org/ 10.1093/nar/gkq132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Belgnaoui SM, Gosden RG, Semmes OJ, Haoudi A. Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells. Cancer Cell Int (2006); 6:13; PMID:16670018; http://dx.doi.org/ 10.1186/1475-2867-6-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gasior SL, Wakeman TP, Xu B, Deininger PL. The human LINE-1 retrotransposon creates DNA double-strand breaks. J Mol Biol (2006); 357:1383-1393; PMID:16490214; http://dx.doi.org/ 10.1016/j.jmb.2006.01.089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kines KJ, Sokolowski M, deHaro DL, Christian CM, Belancio VP. Potential for genomic instability associated with retrotranspositionally-incompetent L1 loci. Nucleic Acids Res (2014); 42:10488-10502; PMID:25143528; http://dx.doi.org/ 10.1093/nar/gku687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Groner AC, Meylan S, Ciuffi A, Zangger N, Ambrosini G, Dénervaud N, Bucher P, Trono D. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genet (2010); 6:e1000869; PMID:20221260; http://dx.doi.org/ 10.1371/journal.pgen.1000869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Quenneville S, Turelli P, Bojkowska K, Raclot C, Offner S, Kapopoulou A, Trono D. The KRAB-ZFP/KAP1 system contributes to the early embryonic establishment of site-specific DNA methylation patterns maintained during development. Cell Rep (2012); 2:766-773; PMID:23041315; http://dx.doi.org/ 10.1016/j.celrep.2012.08.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rowe HM, Friedli M, Offner S, Verp S, Mesnard D, Marquis J, Aktas T, Trono D. De novo DNA methylation of endogenous retroviruses is shaped by KRAB-ZFPs/KAP1 and ESET. Development (2013); 140:519-529; PMID:23293284; http://dx.doi.org/ 10.1242/dev.087585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat Rev. Genet (2007); 8:884-896; PMID:17943195; http://dx.doi.org/ 10.1038/nrg2179 [DOI] [PubMed] [Google Scholar]
- 67. Yang N, Kazazian HH, Jr. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol (2006); 13:763-771; PMID:16936727; http://dx.doi.org/ 10.1038/nsmb1141 [DOI] [PubMed] [Google Scholar]
- 68. Smalheiser NR, Torvik VI. Mammalian microRNAs derived from genomic repeats. Trends Genet (2005); 21:322-326; PMID:15922829; http://dx.doi.org/ 10.1016/j.tig.2005.04.008 [DOI] [PubMed] [Google Scholar]
- 69. Ciaudo C, Jay F, Okamoto I, Chen CJ, Sarazin A, Servant N, Barillot E, Heard E, Voinnet O. in PLoS Genet (2013); Vol. 9:e1003791; PMID:24244175 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 70. Faulkner GJ. Retrotransposon silencing during embryogenesis: dicer cuts in LINE. PLoS Genet (2013); 9:e1003944; PMID:24244199; http://dx.doi.org/ 10.1371/journal.pgen.1003944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Heras S.R. et al. The Microprocessor controls the activity of mammalian retrotransposons. Nature structural & molecular biology 20, 1173-1181, doi: 10.1038/nsmb.2658 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet (2013); 9:e1003470; PMID:23637635; http://dx.doi.org/ 10.1371/journal.pgen.1003470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev. Genet (2009); 10:155-159; PMID:19188922; http://dx.doi.org/ 10.1038/nrg2521 [DOI] [PubMed] [Google Scholar]
- 74. Macía A, Muñoz-Lopez M, Cortes JL, Hastings RK, Morell S, Lucena-Aguilar G, Marchal JA, Badge RM, Garcia-Perez JL. in Mol Cel Biol (2011); Vol. 31:300-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature (2011); 477:295-300; PMID:21874018; http://dx.doi.org/ 10.1038/nature10398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kelley D, Rinn J. Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol (2012); 13:R107; PMID:23181609; http://dx.doi.org/ 10.1186/gb-2012-13-11-r107 [DOI] [PMC free article] [PubMed] [Google Scholar]