Abstract
Conjugative plasmids of the A/C incompatibility group (IncA/C) have become substantial players in the dissemination of multidrug resistance. These large conjugative plasmids are characterized by their broad host-range, extended spectrum of antimicrobials resistance, and prevalence in enteric bacteria recovered from both environmental and clinical settings. Until recently, relatively little was known about the basic biology of IncA/C plasmids, mostly because of the hindrance of multidrug resistance for molecular biology experiments. To circumvent this issue, we previously developed pVCR94ΔX, a convenient prototype that codes for a reduced set of antibiotic resistances. Using pVCR94ΔX, we then characterized the regulatory pathway governing IncA/C plasmid dissemination. We found that the expression of roughly 2 thirds of the genes encoded by this plasmid, including large operons involved in the conjugation process, depends on an FlhCD-like master activator called AcaCD. Beyond the mobility of IncA/C plasmids, AcaCD was also shown to play a key role in the mobilization of different classes of genomic islands (GIs) identified in various pathogenic bacteria. By doing so, IncA/C plasmids can have a considerable impact on bacterial genomes plasticity and evolution.
Keywords: AcaCD, antibiotic resistance, IncA/C plasmids, mobilizable genomic islands, pVCR94, regulation, Salmonella, SGI1, Vibrio
Abbreviations
- ChIP-exo
chromatin immunoprecipitation coupled to exonuclease digestion
- GI
genomic island
- ICE
integrative and conjugative element
- IncA/C
incompatibility group A/C
- MGE
mobile genetic element
- RDF
recombination directionality factor
- SGI1
Salmonella genomic island 1
- T4SS
type IV secretion system
- VR
variable region.
IncA/C conjugative plasmids are large (ca. 140 to 200 kb) plasmids that disseminate among several species of Enterobacteriaceae and occasionally, in Vibrio cholerae.1-3 IncA/C plasmids can efficiently mobilize genes conferring resistance to a large spectrum of antibiotics, including β-lactams, aminoglycosides, chloramphenicol, folate pathway inhibitors (sulfonamides and trimethoprim), quinolones and tetracycline.4,5 Research conducted to date on IncA/C conjugative plasmids has been mostly focusing on epidemiological aspects and was restricted to typing, antibiotic resistance profiling, sequencing and comparative genomics.2,6-9 Consequently, the basic biology and regulatory mechanisms of the dissemination of these mobile genetic elements (MGEs) in bacterial populations remain poorly understood.
pVCR94 is an IncA/C plasmid originating from a V. cholerae O1 strain isolated during the cholera outbreak in Democratic Republic of Congo in 1994.10 We recently created and utilized pVCR94ΔX, a derivative of pVCR94 lacking most of the multidrug resistance genes, to investigate the transcriptional regulation of IncA/C conjugative transfer functions.11,12 Because of the high conservation of the IncA/C core sequence (>98 % of nucleotide identity over 110-kb), our results should apply to virtually any plasmid from this group.4,7,11,13
Transcriptional Regulation by AcaCD
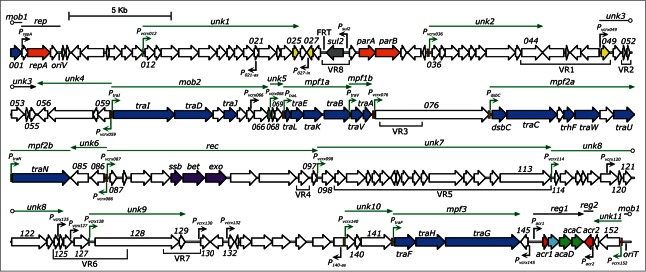
The transcriptional regulatory network that governs IncA/C plasmid mobility is composed of 4 major players: Acr1 and Acr2 both repress the expression of a heteromeric master activator encoded by the acaC and acaD genes (Fig. 1).12 We have characterized the complete plasmid-borne regulon of the master regulator AcaCD, and identified DNA-binding motifs recognized by this activator complex using chromatin immunoprecipitation coupled to exonuclease digestion (ChIP-exo) and RNA sequencing.12 Our experiments uncovered 18 AcaCD-regulated promoters that together drive the expression of about 2 thirds of the genes in pVCR94ΔX (Fig. 2). As expected from the conjugation-deficient phenotype resulting from the deletion of acaCD, the expression of genes predicted to code for components of the conjugation apparatus is dependent of AcaCD (Figs. 1 and 2).12 However, this represents only a fourth of AcaCD-regulated promoters in the plasmid, and approximately 75 additional genes – the vast majority of yet unknown function – also appear to be part of the AcaCD regulon. The possibility that a significant fraction of these genes plays an essential role in conjugation seems unlikely given that the basic conjugation machinery is already well characterized in the distantly related integrative and conjugative elements (ICEs) of the SXT/R391 family.11,14,15 Auxiliary factors increasing conjugation efficiency under specific conditions could be present, especially in the case of unknown function genes found within the mob2 and mpf2a operons (Fig. 2), but this represents a relatively small number of genes. What could be the role of other AcaCD-regulated genes and why are there so many? Part of the answer could be that AcaCD regulates more than the strict conjugation process. We do not yet know what is the biological signal triggering the activity of AcaCD but this could potentially help understand its role in a broader context. Another possibility is that the genomic organization of IncA/C plasmids constitutes a mosaic that could have led to the accumulation of AcaCD promoters separated by regions of cargo DNA. This hypothesis is supported by the fact that 20 genes found in putative operons transcribed from AcaCD-regulated promoters are part of variable regions (VR) that are not conserved between IncA/C plasmids (Fig. 2). For example, AcaCD-activated operon unk7 is composed of 16 genes, of which 15 are not conserved between IncA/C plasmids.11 Furthermore, it is likely that the number and/or nature of VRs are currently underestimated by the relatively low number of complete IncA/C sequences. Comparative genomics using a greater number of IncA/C representatives will help to better define the genuine core genes in these plasmids.
Figure 1.
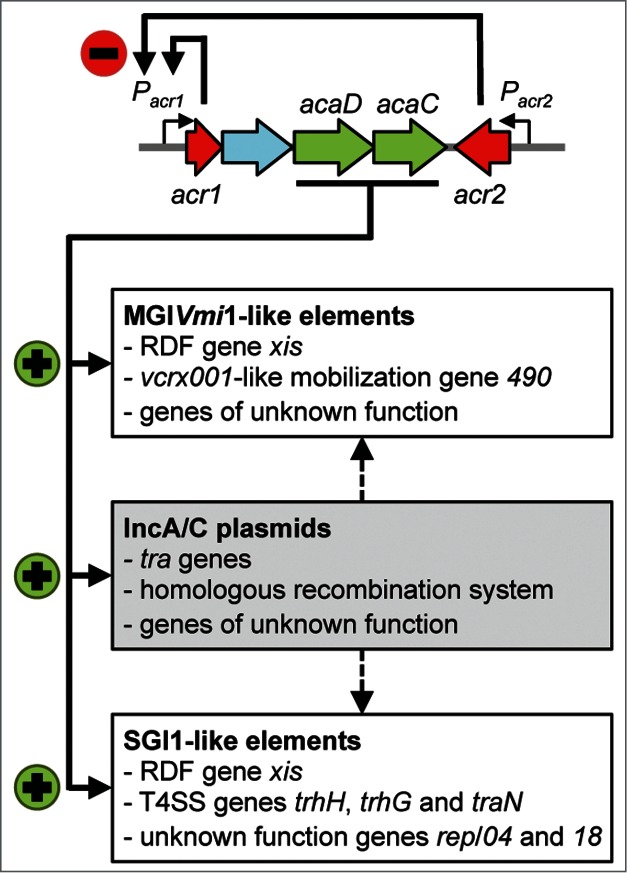
Overview of IncA/C plasmid regulation and impact on GIs. Acr1 and Acr2 directly repress (red minus sign) the expression of the master activator complex AcaCD from the promoter of Pacr1. AcaCD directly activates (green plus signs) the expression of the transfer genes of pVCR94, as well as the expression of the bet/exo homologous recombination system and numerous genes of unknown function. AcaCD triggered the excision of SGI1-like and MGIVmi1-like elements by directly activating the expression of the RDF gene xis. AcaCD activates the expression of a mobilization gene and other genes of unknown function in MGIVmi1-like elements. AcaCD also activates the expression of 3 genes coding for putative component of T4SS, as well as genes of unknown function such as S004, rep and S018 in conserved backbone of SGI1-like elements. IncA/C plasmids likely provide additional functions for GIs dissemination such as oriT recognition and processing, and formation of the mating apparatus (black hatched arrows). (Adapted from Ref. 12).
Figure 2.
pVCR94ΔX functional map. Schematic representation of the genetic organization and transcriptional units of pVCR94ΔX adapted from Carraro et al.12 The circular map of the plasmid was linearized at the start position of gene vcrx001. Genes are represented by arrows and color coded according to their function: white, unknown; blue, conjugative transfer; light blue, lytic transglycosylase; orange, replication and segregation; gray, antibiotic resistance; yellow, putative regulatory function; purple, recombination; green, activator; red, repressor. vcrx genes are indicated by their corresponding number. Variable DNA regions inserted in the conserved core of IncA/C plasmids are indicated below genes (VR1 to VR8).11 The origin of replication (oriV) and the origin of transfer (oriT) are indicated. The position of the FRT site resulting from the deletion of the antibiotic resistance gene cluster is also shown. AcaCD-binding motifs located on the positive and negative DNA strands are represented by light green and red boxes, respectively. Notable DNA clusters and operons are indicated by straight lines and arrows positioned above represented genes, respectively. Open circles mark operons and DNA clusters interruptions generated by the map format. AcaCD-regulated promoters, clusters, and operons are colored in green. mob1–2, DNA processing; rep, replication; unk1–11, unknown; mpf1–3, mating pore formation; rec, recombination; reg1–2, regulation. P021-as and P140-as: vcrx021 and vcrx140 antisens promoters, respectively. P027-in: vcrx027 internal promoter.
We have deliberately restricted our analyses to the IncA/C sequence to understand the role of AcaCD but one can suspect that genes encoded by the host chromosome could also be regulated to increase survival or dissemination of the plasmid. A simple search using the AcaCD-binding motif effectively reveals putative target genes in natural IncA/C host strains such as Escherichia coli, Vibrio sp. and Salmonella sp. It will be interesting to determine if these motifs are genuine AcaCD targets and what is the impact of the regulation of the downstream genes. However, we have already provided experimental evidence that the regulon of AcaCD extends beyond plasmid-borne genes, thereby promoting the mobility of GIs that encode multidrug resistance and other adaptive traits in various pathogenic bacteria.12 Although we initially reported the analysis of only 2 unrelated classes of GIs, it is likely that reliance on IncA/C plasmids for dissemination constitutes a more general strategy for many MGEs (Fig. 1).
Occurrence of AcaCD Motifs in GIs from Diverse Microorganisms
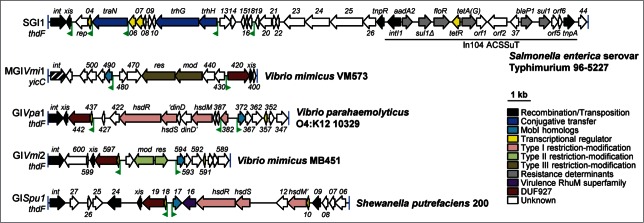
It has been known for years that IncA/C conjugative plasmids are capable of high efficiency in trans mobilization of the Salmonella genomic island 1 (SGI1), which carries the multidrug resistance determinant In104 (Fig. 3).16-18 Heretofore, the mechanism and regulation of this phenomenon remained unknown. We have now established that trans-mobilization of SGI1 by pVCR94ΔX is conditional to the presence of acaCD (Fig. 1).12 Although this dependency partially relies on the AcaCD-mediated expression of pVCR94ΔX conjugative functions, excision of SGI1 from the chromosome of its host, a key step prior to SGI1 dissemination, does not occur in the absence of AcaCD. In fact, expression of xis, which codes for the recombination directionality factor (RDF) of SGI1, is directly under control of an AcaCD-dependent promoter (Figs. 1 and 3). Analysis of SGI1 sequence allowed us to predict AcaCD binding sites upstream of 4 other genes, including tra genes coding for putative components of a type IV secretion system (T4SS) (TraN, TrhH and TrhG). Although the role of these genes in SGI1 mobilization by IncA/C plasmids is not clear, they may provide an advantage to SGI1, which transfers better than its helper plasmid. Whether SGI1 replaces the TraN, TraH and TraG proteins of the IncA/C plasmids-encoded conjugative apparatus by its own alternative homologs, and if such substitutions boost SGI1 transfer remain to be determined. Intriguingly, co-acquisition of both SGI1 and pVCR94ΔX is a very rare event (104 to 105 lower than acquisition of either element), suggesting that strong negative interactions take place between the 2 MGEs and highlighting the parasitic nature of SGI1 toward IncA/C plasmids.12
Figure 3.
AcaCD-activated GIs. Schematic representation of the genetic map of GIs bearing predicted AcaCD binding sites. GIs are drawn to scale. The position and orientation of the predicted AcaCD boxes are shown by green flags. Gene numbers correspond to the last digits of respective locus tags in Genbank accession numbers AF261825 (SGI1), NZ_ACYV01000005 (MGIVmi1), AFBW01000022 (GIVpa1), ADAF01000001 (GIVmi2), CP002457 (GISpu1). The insertion site of the GIs is indicated below GI names. A, ampicillin; C, chloramphenicol; S, streptomycin; Su, sulfamethoxazole; T, tetracycline. The integrase (int) gene of MGIVmi1, which belong to a different family of int genes compared to other GIs represented in the figure, is shown by a black hatched arrow.
Seeking for AcaCD-binding sites in the genomes of several species of Gammaproteobacteria, we have identified MGIVmi1, a mobilizable GI integrated into the 3′ end of yicC in Vibrio mimicus (Figs. 1 and 3).12 Unlike SGI1, only 2 AcaCD boxes are present in this GI. The first one is located upstream of 2 genes, 420 and xis, coding for a DUF927-containing protein and a predicted RDF, respectively. The second one is found upstream of gene 490 coding for a predicted protein of unknown function. Expression of acaCD is sufficient to allow expression of xis and 490, and to trigger the excision of MGIVmi1 from the chromosome.12 In the presence of pVCR94ΔX, the GI transfers to new recipient cells, although not as efficiently as SGI1 or the helper plasmid. The absence of tra gene homologs in MGIVmi1 suggests that MGIVmi1 and SGI1 use different mechanisms of mobilization by IncA/C plasmids. This is further supported by similarities found between the translation product of 490 with MobI of SXT and Vcrx001 of pVCR94. Since MobI and Vcrx001 are crucial for the initiation of conjugative transfer of SXT and pVCR94,11,19 we surmise that their homolog encoded by 490 could similarly be essential for MGIVmi1 mobilization. In addition, because the origin of transfer (oriT) is respectively located upstream of mobI and vcrx001 in SXT and pVCR94,11,19 we predict that the region located upstream of 490 acts as an oriT for MGIVmi1.
Based on these observations, we have searched in microbial genomes for homologs of protein 490 and found 3 additional putative GIs integrated, like SGI1, into the 3′ end of thdF in the genome of 3 strains of V. mimicus, V. parahaemolyticus, and Shewanella putrefaciens (Fig. 3). All three GIs exhibit AcaCD boxes in contexts similar to those observed for MGIVmi1, that is upstream of a gene coding for a 490 homolog and 2 genes coding for a DUF927-containing protein and a predicted RDF. Together with MGIVmi1, all these GIs also code for type I, type II or type III restriction-modification systems (Fig. 3), likely conferring a strong selective advantage against viral attacks to the host of these GIs, which thrive in aquatic environments.
Concluding Remarks
Further experiments are needed to fully understand the molecular mechanisms regulating the dissemination of IncA/C plasmids. For example, we currently do not know what signal triggers the activation of AcaCD, and if IncA/C plasmids have developed mechanisms to manipulate their host to their advantage. Another significant limitation comes from the lack of information regarding the function of most IncA/C encoded genes and their relative importance. Gene inactivation by transposon mutagenesis would constitute a powerful tool to help uncover novel genes essential for the replication, stability, and conjugation of IncA/C elements. These approaches could pave the way for their size reduction, and help create minimal IncA/C elements to study their biology in greater details.
The study of IncA/C plasmids regulation revealed the diversity of GIs that they are susceptible to mobilize. Further investigation will unravel the interactions between the different classes of mobilizable GIs and their helper IncA/C plasmids. Given the diversity of gene content and organization of these GIs, one is left wondering what are the mechanisms mediating their mobilization? Beyond transcriptional activation of GIs by the master activator of IncA/C plasmids, how do such mobile elements interact with the DNA processing functions and mating pore generated by IncA/C plasmids? Moreover, characterization of the positive and negative interactions between SGI1-like elements and IncA/C plasmids during their dissemination is an interesting aspect of cooperative and parasitic relationships between MGEs.
Our study on IncA/C plasmids represents a proof of concept for detecting the action of transcription activators through cognate target sequences in multidrug resistance disseminating MGEs.12 It also provides a glimpse about their potential impact on genomes dynamics and bacterial evolution. Further studies on other MGEs will provide a detailed network of the interconnections linking numerous classes of autonomous and non-autonomous MGEs. Ultimately, it will help to take measures and discover targets for the development of new drugs aiming to overcome the problem of the emergence of highly pathogenic and multiresistant bacteria.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Alain Lavigueur for critical reading of the manuscript.
Funding
This work was supported by the Fonds de recherche du Québec – Nature et technologies (DM, SR and VB), and a Discovery Grant and Discovery Acceleration Supplement from the Natural Sciences and Engineering Council of Canada (VB). VB holds a Canada Research Chair in molecular bacterial genetics. SR is a Chercheur-boursier Junior 1 from the Fonds de recherche du Québec – Santé.
References
- 1.Johnson TJ, Lang KS.. IncA/C plasmids: An emerging threat to human and animal health? Mob Genet Elements 2012; 2:55-8; PMID:22754754; http://dx.doi.org/ 10.4161/mge.19626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang R, Yu D, Zhu L, Li J, Yue J, Kan B.. IncA/C plasmids harboured in serious multidrug-resistant Vibrio cholerae serogroup O139 strains in China. Int J Antimicrob Agents 2015; 45:249-54; PMID:25532743; http://dx.doi.org/ 10.1016/j.ijantimicag.2014.10.021 [DOI] [PubMed] [Google Scholar]
- 3.Folster JP, Katz L, McCullough A, Parsons MB, Knipe K, Sammons SA, Boncy J, Tarr CL, Whichard JM.. Multidrug-resistant IncA/C plasmid in vibrio cholerae from Haiti. Emerg Infect Dis 2014; 20:1951-3; PMID:25340576; http://dx.doi.org/ 10.3201/eid2011.140889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fricke WF, Welch TJ, McDermott PF, Mammel MK, LeClerc JE, White DG, Cebula TA, Ravel J.. Comparative genomics of the IncA/C multidrug resistance plasmid family. J Bacteriol 2009; 191:4750-7; PMID:19482926; http://dx.doi.org/ 10.1128/JB.00189-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Welch TJ, Fricke WF, McDermott PF, White DG, Rosso M-L, Rasko DA, Mammel MK, Eppinger M, Rosovitz MJ, Wagner D, et al.. Multiple antimicrobial resistance in plague: an emerging public health risk. PLoS One 2007; 2:e309; PMID:17375195; http://dx.doi.org/ 10.1371/journal.pone.0000309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao G, Allard MW, Hoffmann M, Monday SR, Muruvanda T, Luo Y, Payne J, Rump L, Meng K, Zhao S, et al.. Complete sequences of six IncA/C plasmids of multidrug-resistant salmonella enterica subsp. enterica serotype newport. Genome Announc 2015; 3:e00027-15; PMID:25720681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernández-Alarcón C, Singer RS, Johnson TJ.. Comparative genomics of multidrug resistance-encoding IncA/C plasmids from commensal and pathogenic Escherichia coli from multiple animal sources. PLoS One 2011; 6:e23415; http://dx.doi.org/ 10.1371/journal.pone.0023415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hazen TH, Zhao L, Boutin MA, Stancil A, Robinson G, Harris AD, Rasko DA, Johnson JK.. Comparative genomics of an IncA/C multidrug resistance plasmid from escherichia coli and klebsiella isolates from intensive care unit patients and the utility of whole-genome sequencing in health care settings. Antimicrob Agents Chemother 2014; 58:4814-25; PMID:24914121; http://dx.doi.org/ 10.1128/AAC.02573-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo Y-F, Zhang W-H, Ren S-Q, Yang L, Lü D-H, Zeng Z-L, Liu Y-H, Jiang H-X.. IncA/C plasmid-mediated spread of CMY-2 in multidrug-resistant Escherichia coli from food animals in China. PLoS One 2014; 9:e96738; PMID:24816748; http://dx.doi.org/ 10.1371/journal.pone.0096738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siddique AK, Salam A, Islam MS, Akram K, Majumdar RN, Zaman K, Fronczak N, Laston S.. Why treatment centres failed to prevent cholera deaths among Rwandan refugees in Goma, Zaire. Lancet 1995; 345:359-61; PMID:7646639; http://dx.doi.org/ 10.1016/S0140-6736(95)90344-5 [DOI] [PubMed] [Google Scholar]
- 11.Carraro N, Sauvé M, Matteau D, Lauzon G, Rodrigue S, Burrus V.. Development of pVCR94ΔX from Vibrio cholerae, a prototype for studying multidrug resistant IncA/C conjugative plasmids. Front Microbiol 2014; 5:44; PMID:24567731; http://dx.doi.org/ 10.3389/fmicb.2014.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carraro N, Matteau D, Luo P, Rodrigue S, Vincent B.. The master activator of IncA/C conjugative plasmids stimulates genomic islands and multidrug resistance dissemination. PLoS Genet 2014; 10: e1004714; PMID:25340549; http://dx.doi.org/ 10.1371/journal.pgen.1004714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Del Castillo CS, Hikima J-I, Jang H-B, Nho S-W, Jung T-S, Wongtavatchai J, Kondo H, Hirono I, Takeyama H, Aoki T.. Comparative sequence analysis of a multidrug-resistant plasmid from Aeromonas hydrophila. Antimicrob Agents Chemother 2013; 57:120-9; PMID:23070174; http://dx.doi.org/ 10.1128/AAC.01239-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wozniak RAF, Fouts DE, Spagnoletti M, Colombo MM, Ceccarelli D, Garriss G, Déry C, Burrus V, Waldor MK.. Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet 2009; 5:e1000786; PMID:20041216; http://dx.doi.org/ 10.1371/journal.pgen.1000786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carraro N, Burrus V.. Biology of Three ICE Families: SXT/R391, ICEBs1, and ICESt1/ICESt3. Microbiol Spectr 2014; 2:MDNA3-0008-2014. [DOI] [PubMed] [Google Scholar]
- 16.Doublet B, Boyd D, Mulvey MR, Cloeckaert A.. The Salmonella genomic island 1 is an integrative mobilizable element. Mol Microbiol 2005; 55:1911-24; PMID:15752209; http://dx.doi.org/ 10.1111/j.1365-2958.2005.04520.x [DOI] [PubMed] [Google Scholar]
- 17.Douard G, Praud K, Cloeckaert A, Doublet B.. The Salmonella genomic island 1 is specifically mobilized in trans by the IncA/C multidrug resistance plasmid family. PLoS One 2010; 5:e15302; PMID:21187963; http://dx.doi.org/ 10.1371/journal.pone.0015302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levings RS, Lightfoot D, Partridge SR, Hall RM, Djordjevic SP.. The genomic island SGI1, containing the multiple antibiotic resistance region of Salmonella enterica serovar Typhimurium DT104 or variants of it, is widely distributed in other S. enterica serovars. J Bacteriol 2005; 187:4401-9; PMID:15968049; http://dx.doi.org/ 10.1128/JB.187.13.4401-4409.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ceccarelli D, Daccord A, René M, Burrus V.. Identification of the origin of transfer (oriT) and a new gene required for mobilization of the SXT/R391 family of integrating conjugative elements. J Bacteriol 2008; 190:5328-38; PMID:18539733; http://dx.doi.org/ 10.1128/JB.00150-08 [DOI] [PMC free article] [PubMed] [Google Scholar]