Abstract
Of the two cultivated species of allopolyploid cotton, Gossypium barbadense produces extra-long fibers for the production of superior textiles. We sequenced its genome (AD)2 and performed a comparative analysis. We identified three bursts of retrotransposons from 20 million years ago (Mya) and a genome-wide uneven pseudogenization peak at 11–20 Mya, which likely contributed to genomic divergences. Among the 2,483 genes preferentially expressed in fiber, a cell elongation regulator, PRE1, is strikingly At biased and fiber specific, echoing the A-genome origin of spinnable fiber. The expansion of the PRE members implies a genetic factor that underlies fiber elongation. Mature cotton fiber consists of nearly pure cellulose. G. barbadense and G. hirsutum contain 29 and 30 cellulose synthase (CesA) genes, respectively; whereas most of these genes (>25) are expressed in fiber, genes for secondary cell wall biosynthesis exhibited a delayed and higher degree of up-regulation in G. barbadense compared with G. hirsutum, conferring an extended elongation stage and highly active secondary wall deposition during extra-long fiber development. The rapid diversification of sesquiterpene synthase genes in the gossypol pathway exemplifies the chemical diversity of lineage-specific secondary metabolites. The G. barbadense genome advances our understanding of allopolyploidy, which will help improve cotton fiber quality.
Whole-genome duplication (WGD) or polyploidy is a primary driving force in the evolution of many eukaryotic organisms, especially flowering plants1,2,3,4. Many crops are neo-allopolyploids that harbor different sets of genomes5,6, including the cultivated Upland cotton Gossypium hirsutum (AD)1 and the extra-long staple (ELS) cotton Gossypium barbadense (AD)2. However, our understanding of the molecular mechanism that facilitates the success of allopolyploids and the formation of agronomic traits remains limited.
Cotton provides the most important raw material for the textile industry and consequently profoundly affects the world economy and daily human life. The cotton genus Gossypium contains 45 diploid (2n = 26) and six tetraploid (2n = 52) species7,8, among which only four species, including two tetraploids (G. hirsutum and G. barbadense) and two diploids (G. herbaceum and G. arboreum), produce spinnable fiber. Diploid cottons are divided into eight cytogenetic genome groups, A-G and K. The sizes of genomes vary between groups due to the lineage-specific proliferation of retrotransposons7. The D-group species have the smallest genome with G. raimondii (D5) of less than 880 Mb9,10,11, whereas the genome of G. arboreum (A2) in the A-group is approximately 1,700 Mb12. G. hirsutum and G. barbadense are considered classic natural allotetraploids that originated in the New World approximately 2 million years ago (Mya) from trans-oceanic hybridization between an A-genome ancestral African species, G. herbaceum (A1) or G. arboreum (A2), and a native D-genome species, G. raimondii or G. gossypioides (D6)13, followed by divergence from their common ancestor (Fig. 1). These two allotetraploids are likely the oldest major allopolyploid crops10,14,15.
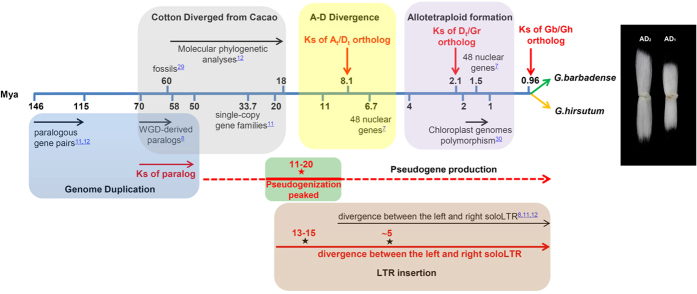
Figure 1. A schematic map of the evolution of allotetraploid cottons.
Allotetraploid cotton evolved from the natural hybridization between A- and D-genome species and has split into six species, including the widely cultivated G. barbadense (AD2) and G. hirsutum (AD1). Evolutionary time (in Mya) is indicated by a numbered axis; major evolutionary events are represented by arrows and concluded in boxes. A black star indicates a retrotransposon burst, and a red star indicates a boom in pseudogene production. Gr, G. raimondii, a diploid species (D5); Gb, G. barbadense; Gh, G. hirsutum. Mature cotton fiber is shown for extra-long stable (ELS) cotton (G. barbadense, AD2) and Upland cotton (G. hirsutum, AD1).
Cotton fiber is derived from single-celled, seed-borne hair (trichrome), and the development of fiber cells is largely synchronized in a cotton ball (fruit) in four overlapping stages: initiation, elongation, secondary cell wall synthesis and maturation16. These processes provide an excellent model to dissect cell differentiation, elongation and cellulose biosynthesis. The rate and duration of the elongation stage determines fiber length, and the secondary cell wall biosynthesis affects fiber strength and fineness17,18. The Upland cotton G. hirsutum constitutes ~90% of the annual cotton output and is characterized by its high yield yet moderate fiber qualities, whereas the ELS cotton G. barbadense produces over 5% of the world’s cotton and is famous for its superior quality fiber, as based on the length, strength and fineness of its fibers (Fig. 1). Therefore, G. barbadense is preferred for the production of high-grade or special cotton textiles.
Although G. barbadense and G. hirsutum may share a common progenitor, the two species substantially differ, which has hindered the transfer of the superior fiber traits of G. barbadense to G. hirsutum via inter-species hybridization. This transfer has been particularly hindered by distorted segregation19. The recently released genome sequences of G. hirsutum20,21 and the two extant diploid progenitor species, G. raimondii10,11 and G. arboreum12, have provided insight into cotton evolution and a wealth of resources for fiber improvement. A genome sequence of G. barbadense will further our understanding of the dynamics of genome structures and the genetic driving force associated with allotetraploids, particularly the molecular basis of the formation of fibers with superior traits.
Results
Genome sequence and assembly
We adopted a progressive strategy to sequence the allotetraploid genome of G. barbadense cv. Xinhai21 (AD)2. First, the genomes of the extant diploid species of G. arboreum (A2) and G. raimondii (D5) were separately sequenced and assembled. These sequences, together with their published genomes10,12, were used as references for early assortments of the primary reads into At and Dt subgenomes. Then the sequences were assembled into At and Dt contigs and scaffolds (Supplementary Table 1). A total of 471 Gb (188× genome equivalent) of data were separately produced using the Roche 454, Illumina Hiseq2000 and PacBio SMRT sequencing platforms (Supplementary Table 2). The particularly long reads (22.67 Gb) obtained from PacBio SMRT and the assembled 53-Gb contigs of the BAC pool further reduced the effects of repeats in the assembly, yielding a gap reduction of 63.4% (Supplementary Fig. 1). Finally, we used the ultra-dense linkage map consisting of 4,999,048 single-nucleotide polymorphism (SNP) loci22 to assign and orient the 26 chromosomes and validate the polyploidy genome of G. barbadense (Supplementary Fig. 2). We detected only 20 Mb sequences in which the subgenome classification of homoeologous sequences conflicted between the sequence assembly and the linkage mapping strategies, which was likely due to sequence conversions between the two subgenomes. A total of 208 Mb sequences with erroneous inter-chromosomal joins in the At or Dt subgenome were detected and then corrected.
The combination of these methods resulted in a draft genome for G. barbadense with an overall contig N50 of 72 kilobases (kb) and scaffold N50 of 503 kb covering 1.395 Gigabases (Gb) of the A subgenome (At) and 0.776 Gb of the D subgenome (Dt) (Table 1 and Fig. 2). In total, ~88% of the 2.470 Gb genome was based on k-mer estimation (Supplementary Fig. 3). The genome contains at least 63.2% repeated sequences (Supplementary Table 3), half of which are transposable elements (TEs) that primarily consist of long-terminal-repeat retrotransposons (LTR retrons) (Supplementary Fig. 4).
Table 1. Statistics of G. barbadense genome features.
| Category | At | Dt |
|---|---|---|
| Genome Size (bp) | 1,394,663,696 | 775,997,401 |
| Gene Number | 40,502 | 37,024 |
| Ave. Gene Size (bp) | 2,601 | 2,553 |
| Total Gene Region | 123,247,562 | 104,783,505 |
| Ave. CDS Size (bp) | 1,099 | 1,111 |
| Max. CDS Length | 19,647 | 16,596 |
| Total Coding Region (bp) | 52,095,402 | 45,586,340 |
| Total Exon Number | 240,755 | 208,290 |
| Exon Number per Gene | 5 | 5 |
| Ave. Exon Size (bp) | 216 | 219 |
| Max. Exon Size (bp) | 5,651 | 6,031 |
| Total Intron Number | 193,370 | 167,253 |
| Ave. Intron Size (bp) | 368 | 354 |
| Max. Intron Size (bp) | 85,091 | 86,599 |
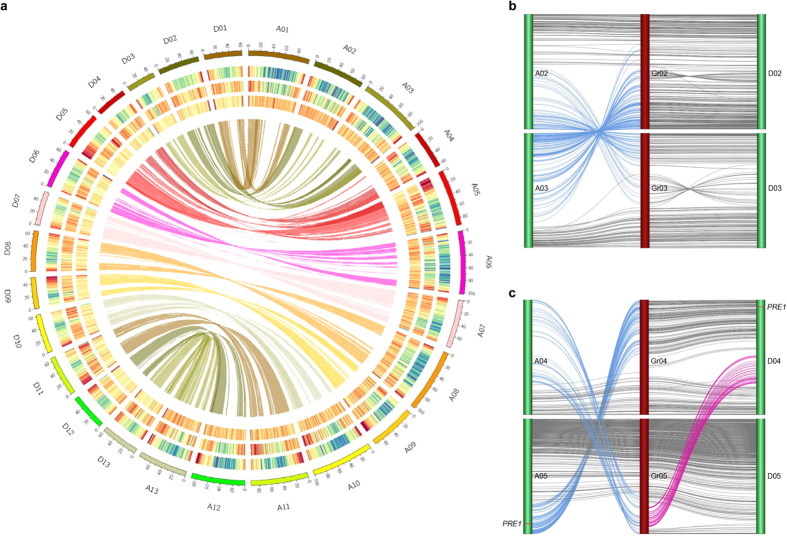
Figure 2. G. barbadense genome atlas and chromosome-level translocations.
(a) Genome atlas. The outermost circle represents the numbered chromosomes of At and Dt, and chromosome sizes are marked by a scale plate. The three tracks moving inside successively represent gene, peudogene and repeat densities (calculated with 1 Mb windows) across the chromosomes. The core ribbon-link shows collinearity between At and Dt. (b,c) chromosomal translocations. The translocations among chromosome 2 and chromosome 3 of either At or Dt are indicated with blue lines (b) and those among chromosome 4 and chromosome 5 with blue and purple lines (c). The vertical colored lines from left to right represent chromosomes. The loci of PRE1 implicated in fiber cell elongation are specifically marked with red in the chromosomes A05 and D04. Digits (01 to 13) after A, D or Gr indicate the chromosome of the At/Dt subgenome of G. barbadense or of G. raimondii, respectively.
Gene annotation
To initiate gene prediction, ~1 million expressed sequence tags (ESTs) that were generated using Roche 454 from a combination of 28 samples of eight tissues/organs collected at different development stages were mapped to the genome as gene models, which resulted in 40,502 and 37,024 protein-coding genes (CDSs) with an average length of 1,077 and 1,123 bp in the G. barbadense At and Dt subgenomes, respectively (Table 1), and falling in the same range as the number and length of CDSs of G. raimondii10,11. Further evaluation using the 70-Gb RNA-Seq data via Illumina supported 96.6% of the predicted CDSs. The 77,526 predicted genes were annotated, which revealed 62,966 functional genes, excluding 8,518 At and 6,042 Dt genes (~20%) that lacked clear biological functions.
To examine the influence of allopolyploidy on gene contents, we classified cotton genes into domain families. The composition and family size of the assigned Pfam domain families are overall identical in G. barbadense At and Dt, G. raimondii and, to a lesser extent, G. arboreum. Protein domains whose function was clearly annotated, such as protein kinase, cytochrome P450, and pentatrico-peptide repeat (PPR), were commonly over-represented as large families (Supplementary Table 4 and Supplementary Fig. 5) as in other angiosperm plants23,24,25. Although most domains (3,039 out of 3,674) were maintained in each subgenome after the two were merged, pronounced changes in family size occurred, as exemplified by more ring finger domain (PF13639) and leucine rich repeat (PF13855) genes in the diploid D genome than in either At or Dt (Supplementary Table 4). This finding suggested that super-large families have evolved faster than others and tended to lose members in polyploids26.
Genome evolution
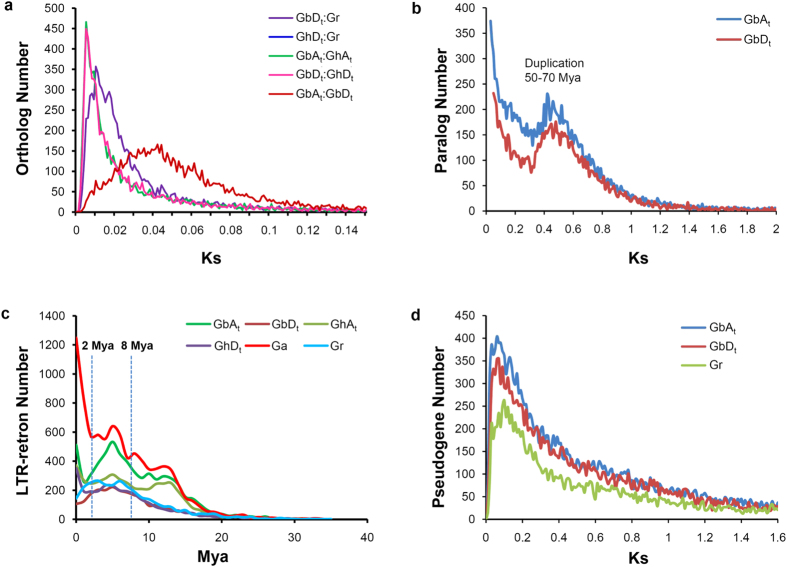
A total of 21,639 pairs of orthologs were identified between At and Dt. We compared the Ks values of orthologous gene pairs among G. barbadense (Gb), G. hirsutum (Gh) and G. raimondii (Gr) at the whole-genome level (Fig. 3a and Supplementary Table 5). A peak of 0.011 in both GbDt:GrD5 and GhDt:GrD5 indicates that the Dt subgenome in of both allotetraploids originated from a G. raimondii-like progenitor27. The peak values for GbAt:GaA2 and GhAt:GaA2 are lower but again similar, presumably due to a shorter time since divergence compared to that between D-genome species. In addition, unlike G. raimondii, which is a wild species, G. arboreum has long been cultivated in African and Asian countries. Another pair of similar Ks peaks (0.005) of GbAt:GhAt and GbDt:GhDt further supports the common origin of the two allotetraploid cottons and suggests their later divergence approximately 1 Mya (Fig. 3a). Based on the larger Ks value (0.04) for At:Dt, we estimated the divergence time between the Gossypium A- and D-genome species to be approximately 8 Mya, consistent with previous estimates that were based on a few single-copy genes13,27. The Ks values of paralogs in the two subgenomes of G. barbadense both peak at 0.4–0.5, which indicate ancient WGD event(s) that occurred 50–70 Mya (Fig. 3b), which were responsible for the repeated genome expansion in Gossypium after divergence from the Theobroma cacao lineage more than 60 Mya10.
Figure 3. Evolutionary analysis of the G. barbadense genome.
(a) Ks distribution of orthologs in cotton genomes. Data are grouped into 0.001 Ks units. (b) Ks distribution of paralogs in the G. barbadense genome. Data are grouped into 0.01 Ks units, and the peak region corresponds to 50–70 million years. (c) The distribution curve of the insertion times in the LTR retrons in the G. barbadense genome. The LTR retrons bursts are separated by dashed lines. (d) Ks distribution of pseudogenes with their closest functional paralogous genes. Data are grouped into 0.001-Ks units. The genomes of allotetraploid cottons are labeled using At/Dt, and the genomes of G. arboreum (A2) and G. raimondii (D5) are labeled using Ga and Gr.
Both the At and Dt subgenomes of G. barbadense demonstrate a high level of co-linearity with the G. raimondii genome10,11 (Supplementary Fig. 6). A total of 21 Megabase (Mb) sequences in the Dt and 7.4 Mb in the At were identified as inter-subgenome translocation regions (Supplementary Fig. 7). Two of three major intra-subgenomic rearrangements between chrA2/chrA3 and chrA4/chrA528,29 were observed in the At of both of the allotetraploid cottons but absent in the Dt or G. raimondii genome (Fig. 2), suggesting that the two translocations likely occurred after the separation of the A and D genomes.
Genomic plasticity and evolution
We identified 6,014/2,422 complete LTR retrons with an average length of 9,256/8,130 bp in At/Dt (Supplementary Tables 6 and 7), similar to the numbers of LTR retrons in G. hirsutum At and Dt, G. arboreum and G. raimondii (Supplementary Table 8). The singleton LTR retrons ratio is 83.5% in At and 82.2% in Dt (compared with 85.4% in G. raimondii and 73.2% in G. arboreum), close to that (86%) in the genome of a gymnosperm tree, Picea abies30 (an indication of high divergence).
The TE proliferations in G. barbadense and G. hirsutum20,21, represented by insertions of LTR retrons based on estimations according to the sequence divergence between the left and right soloLTR31, have increased since 20 Mya, and three distinct bursts were identified. Interestingly, the first two bursts appear to successively pre-date the divergence and the re-unification of the diploid A/D genomes (Fig. 3c). The LTR retrons clearly show type-specific and subgenome-biased proliferations (Fig. 3c). Their insertion rates in the A genome appear consistently higher than those in the D genome. For example, a large number (9.15%) of LTR retrons burst at 5 Mya and decreased thereafter in At, whereas a substantially lower and flat peak appeared 3–5 Mya in Dt (Fig. 3c). This peak at least partly accounts for the 1.7-fold more LTR retrons in the former genome. However, the faster loss of LTR retrons in the D genome may also be responsible for genome size variations and the different rates of genome expansion32. Notably, the third asymmetric activities of transposons differ between G. barbadense and G. hirsutum (Fig. 3c), which suggests a possible cause of subgenome divergence that may have promoted the speciation of allotetraploid cottons beginning approximately 1 Mya (Fig. 1). These observations indicate that the genome-specific differential dynamics of TE proliferations could be a major force that has driven the rapid evolution and diversification of Gossypium species, which may also be inferred in other flowering plants.
Pseudogenization prior to and after polyploidization
Pseudogenes are disabled copies resembling functional genes that have been retained in the genome26,33. They can be grouped into three categories: duplicated (derived from gene duplication), processed (generated by the integration of reversed-transcribed cDNAs into genomes) and fragmented (neither processed nor duplicated)33. To further investigate the influence of TE bursts and polyploidization on the cotton genomic architecture, we predicted pseudogenes in G. barbadense (Supplementary Table 9) and classified them into the three categories (Supplementary Fig. 8), most of which are silenced without any detectable transcripts in all tissues examined.
Each subgenome of G. barbadense contains more predicted pseudogenes than the diploid genome of G. raimondii (Supplementary Table 9 and Supplementary Fig. 8), implying an accelerated pseudogenization after allopolyploid formation. A substantial portion of the pseudogenes in At and Dt showed a high sequence identity (above 90%, for example) with their parental genes (Supplementary Fig. 9), suggesting an insufficient duration for degeneration in recently formed pseudogenes. As expected, the Ka/Ks distributions indicate a substantially weaker natural selection on pseudogenes than on protein-coding genes (Supplementary Fig. 10), which is likely due to a loss of function in pseudogenes. The Ks value peaks at 0.06–0.1 corresponding to 11–20 Mya (Fig. 3d) and this boom of pseudogenization correlates with an LTR retron burst prior to the divergence of the A and D genomes (Fig. 3c). The average expression levels of the genes with LTR retron insertion within a 20-kb region upstream of the start codon are generally lower (RPKM = 7.72) than those of genes lacking this insertion (RPKM = 13) (Supplementary Table 10). Therefore, LTR retrons negatively affect the expression of nearby genes, which may promote pseudogenization. These results suggest that cotton progenitors likely lost genes and experienced LTR retron bursts following the ancient WGD, which promoted diversification in Gossypium genomes; however, the role of TE-associated pseudogenization in the stabilization of subgenomes in polyploids requires a more detailed analysis.
Extra-long staple fiber formation
We identified 2,483 and 1,879 genes that are specifically or preferentially expressed in fibers and the ovule, respectively (Supplementary Tables 11 and 12). The highly active genes in the ovule are abundant in the protein families of nucleic acid binding/transcription factor activity and nutrient reservoir activity, whereas the up-regulated genes of fibers are enriched in several categories, such as those related to cytoskeleton, carbohydrate metabolism, cell wall biosynthesis and cellulose biosynthesis function (Supplementary Tables 13 and 14).
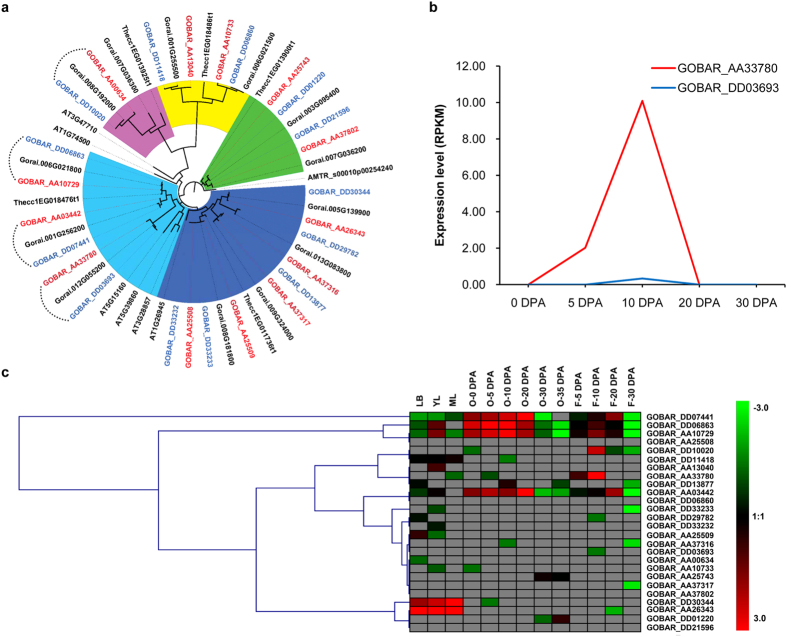
Consistent with a previous report34, equal numbers of genes in the At and Dt subgenomes demonstrated biased expression patterns (Supplementary Tables 15 and 16). Transcription factors play an important role in controlling agronomic novelty, and the MYB and homeodomain-containing factors have been shown to be key regulators of cotton fiber traits development10,35,36,37. We then analyzed transcription factor genes expressed in G. barbadense fiber in detail (Supplementary Table 17 and Supplementary Fig. 11). Paclobutrazol Resistance (PRE) genes encode a group of transcription regulators known in other plants to promote cell elongation38,39,40. We identified 13 PRE family genes in G. raimondii; their 26 orthologous genes were recovered in G. barbadense. Analyzing the PRE-containing synteny blocks in plants revealed that cacao41 has five PRE genes, each of which has at least two orthologs in the Gossypium diploid genomes or the allotetraploid subgenomes (Fig. 4a and Supplementary Fig. 12). This expansion of PRE genes in cotton may have occurred during a complex 5–6-fold polyploidy process10,11, which was followed by differential gene loss but the retention of the ancient orthologs. Interestingly, two PRE genes are located in the two At translocation regions (chrA2/chrA3 and chrA4/chrA5) (Fig. 2c and Supplementary Fig. 12). In cotton, PRE genes are preferentially expressed in young tissues (Fig. 4b,c), which is consistent with their role in controlling cell size. Moreover, the expression of At and Dt PRE homoeologous genes was biased in G. barbadense (Supplementary Tables 11–12). In particular, the expression level of At-subgenome PRE1 was high and fiber specific, whereas the expression the Dt homoeolog was nearly undetectable (Fig. 4b). The At-specific expression of a cell growth regulator provides a clue to support the origin or early evolution of spinnable fiber in the A-genome species10,11. The expansion and subsequent selection11,34 of PRE genes in Gossypium may have increased their regulatory activity and recruited specific member(s) for the rapid and extensive elongation of cotton fiber (Figs 1 and 4c).
Figure 4. Expansion and diversification of PRE genes in Gossypium.
(a) Phylogenetic analysis of PRE family genes in Amborella trichopoda, Arabidopsis thaliana, G. raimondii and G. barbadense. Subfamilies are overlaid with different colors, and the curved dotted lines indicate homoeologous gene pairs expressed in fiber. (b) GbPRE1 (GOBAR_AA33780, GOBAR_DD03693) is a fiber-specific gene with strong At bias expression. The expression levels (RPKM) in ovules (0 DPA) and fiber cells (5, 10, 20, and 30 DPA) are shown. Detailed expression data are provided in Supplementary Table 10. (c) Hierarchical clustering analysis of expression of PRE genes in G. barbadense. LB, leaf bud; YL, young leaf; ML, mature leaf; O, ovule; F, fiber; DPA, days post-anthesis.
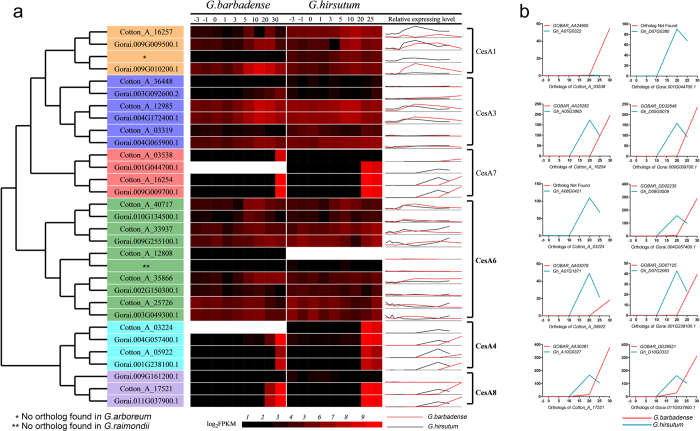
Cellulose, which consists of linear chains of β (1–4)-linked D-glucose, is the major component of higher plant cell walls and the most abundant biopolymer on land. Plants express multiple cellulose synthases (CesAs) that, together with CesA-associated proteins, form the cellulose synthase complex42,43. Cotton fiber is distinct not only in its extensive elongation (ELS cotton fiber is longer than 35 mm) but also in its exceptionally high amount of cellulose, which constitutes more than 95% of the dry weight of mature fiber16,44. Notably, the first higher plant cellulose synthase gene was cloned from cotton45. Ten, 14 and 15 CesA genes are expressed in Arabidopsis thaliana42,43, G. arboreum12 and G. raimondii10, respectively (Fig. 5 and Table 2). We identified 29 CesA genes, including 14 At and 15 Dt, in the G. barbadense genome, whereas 30 (14 At and 16 Dt) CesA genes were identified in G. hirsutum; most CesA genes had been retained after the merger of the A and D genomes (Table 2 and Supplementary Fig. 13). Compared to Arabidopsis, each cotton genome or subgenome contains more genes in the CesA3, CesA4, CesA7 and CesA8 clades. Notably, chromosome 5 of both the At and Dt subgenomes of G. barbadense (GOBAR_AA25282, GOBAR_AA25287/GOBAR_DD32643, GOBAR_DD32648 and GOBAR_DD32650) and G. hirsutum (Gh_A05G3959, Gh_A05G3965, Gh_A05G3967/Gh_D05G0077, Gh_D05G0079 and Gh_D05G0084) as well as G. arboreum and G. raimondii contain a CesA cluster composed of 3 or, rarely, 2 genes, in addition to the CesA-like (CSL) genes (Table 2); thus, the duplication(s) occurred in the ancient cotton genome.
Figure 5. Cotton CesA genes and their expression in developing fiber cells of G. barbadense and G. hirsutum.
(a) CesA genes from four cotton species were clustered (left) via MAGE5 using the maximum likelihood method. G. arboreum (Cotton_A) and G. raimondii (Gorai) contain 14 and 15 CesA genes, respectively, which are shown in the left column. The heat map (middle) shows the transcript level (FPKM, Reads Per Kilobase of exon model per Million mapped reads) of each homeologous gene in G. barbadense and G. hirsutum (Table 2) fibers at different DPA. The relative expression level in the two allotetraploid cottons was compared (right). CesA1, CesA3 and CesA6 are implicated in primary cell wall biosynthesis, and CesA4, CesA7 and CesA8 are implicated in secondary cell wall biosynthesis. (b) Temporal expression patterns of secondary cell wall CesA genes (CesA7, CesA4 and CesA8 clades) in G. barbadense and G. hirsutum fiber. Note that the expression was generally delayed in G. barbadense fiber. X-axis: day post-anthesis. Y-axis: FPKM.
Table 2. Cellulose synthase (CesA) genes in four cotton species.
| Homologs in G. arboreumand G. raimondii | Homologs in G. hirsutum | Homologs in G. barbadense |
|---|---|---|
| Genes related to the synthesis of cellulose in prototypical primarycell walls (CESA1, CESA3, CESA6 clades) | ||
| CESA1 Clade | ||
| no apparent ortholog | Gh_A05G3959 | no apparent ortholog |
| Gorai.009G010200.1 | Gh_D05G0077 | GOBAR_DD32650 |
| Cotton_A_16257 | Gh_A05G3967 | GOBAR_AA25287 |
| Gorai.009G009500.1 | Gh_D05G0084 | GOBAR_DD32643 |
| CESA3 Clade | ||
| Cotton_A_03319 | Gh_A08G0498 | GOBAR_AA12453 |
| Gorai.004G065900.1 | Gh_D08G0584 | GOBAR_DD11497 |
| Cotton_A_12985 | Gh_A08G1305 | GOBAR_AA08823 |
| Gorai.004G172400.1 | Gh_D08G1597 | GOBAR_DD05460 |
| Cotton_A_36448 | Gh_A02G1066 | GOBAR_AA03569 |
| Gorai.003G092600.2 | Gh_D03G0611 | GOBAR_DD02554 |
| CESA6 Clade | ||
| Cotton_A_25726 | Gh_A02G1317 | GOBAR_AA16276 |
| Gorai.003G049300.1 | Gh_D03G0455 | GOBAR_DD10475 |
| Cotton_A_33937 | Gh_A05G3694 | GOBAR_AA32700 |
| Gorai.009G255100.1 | Gh_D05G2313 | GOBAR_DD35549 |
| Cotton_A_40717 | Gh_A06G1017 | GOBAR_AA04815 |
| Gorai.010G134500.1 | Gh_D06G1219 | GOBAR_DD30509 |
| Cotton_A_35866 | Gh_A11G3209 | GOBAR_AA22611 |
| Gorai.002G150300.1 | Gh_D11G2235 | GOBAR_DD13415 |
| Cotton_A_12808 | no apparent ortholog | GOBAR_AA34523 |
| no apparent ortholog | Gh_D12G0885 | GOBAR_DD19420 |
| Genes related to the synthesis of cellulose in prototypicalsecondary cell walls (CESA4, CESA7, CESA8 clades) | ||
| CESA4 Clade | ||
| Cotton_A_05922 | Gh_A07G1871 | GOBAR_AA03078 |
| Gorai.001G238100.1 | Gh_D07G2083 | GOBAR_DD07125 |
| Cotton_A_03224 | Gh_A08G0421 | no apparent ortholog |
| Gorai.004G057400.1 | Gh_D08G0509 | GOBAR_DD02235 |
| CESA7 Clade | ||
| Cotton_A_03538 | Gh_A07G0322 | GOBAR_AA24905 |
| Gorai.001G044700.1 | Gh_D07G0380 | no apparent ortholog |
| Cotton_A_16254 | Gh_A05G3965 | GOBAR_AA25282 |
| Gorai.009G009700.1 | Gh_D05G0079 | GOBAR_DD32648 |
| CESA8 Clade | ||
| Gorai.009G161200.1 | Gh_D05G1460 | GOBAR_AA30803* |
| Cotton_A_17521 | Gh_A10G0327 | GOBAR_AA30381 |
| Gorai.011G037900.1 | Gh_D10G0333 | GOBAR_DD29521 |
*May have translocated.
Although not exclusively, plant CesAs have functionally diverged into two major classes responsible for either primary cell wall or secondary cell wall biosynthesis42,43. Whereas spinnable cotton fiber evolved in the A-genome species and further developed in AD allotetraploids, the CesA gene family has not undergone expansion in any of the three cultivated cotton species sequenced. However, cotton fiber expresses many (at least 25) CesA genes (Fig. 5), demonstrating an enrichment of cellulose synthases in fiber cells. A comparison of the two allotetraploid cottons revealed that the secondary cell wall genes CesA4, CesA7 and CesA8 showed a delayed (>5 days) and more drastic up-regulation in G. barbadense fiber than in G. hirsutum fiber (Fig. 5), which indicates a prolonged duration of fiber elongation and a high activity of cellulose biosynthesis in the secondary cell wall formation stage. Additionally, this temporal expression pattern suggests that the functional allocation of CesA members to primary and secondary wall biosynthesis, which is primarily based on Arabidopsis research42,43,46, are likely conserved in angiosperms. Thus, both the retention of CesA family members and the expression pattern of functionally specialized genes in G. barbadense support the formation of extra-long and high-grade cotton fiber.
Terpene synthases and the evolution of cotton phytoalexins
Terpenoids constitute a large family of natural compounds and play diverse roles in plant-environment interactions. Cotton plants accumulate a specialized group of cadinene-type sesquiterpenoids (including gossypol) that function as phytoalexins against pathogens and pests47,48. However, these sesquiterpenoids also reduce the value of cotton seeds that are rich in oil and proteins. Terpene synthases (TPSs) are a family of enzymes responsible for the synthesis of various terpenes from the 10-, 15-, and 20-carbon precursors assembled from the 5-carbon building blocks of IPP and its isomer DMAPP49. A manual search of the G. barbadense genome with TPS N- and C-terminal domains (PF01397 and PF03936) identified 115 TPS genes, including 44 monoterpene, 59 sesquiterpene and 8 diterpene synthases, as well as 4 triterpene (squalene) synthases. This number is higher than that in T. cacao (43), Arabidopsis thaliana (34) and Vitis vinifera (98) and similar to that in G. hirsutum (110) but slightly less than twice that in G. raimondii (69).
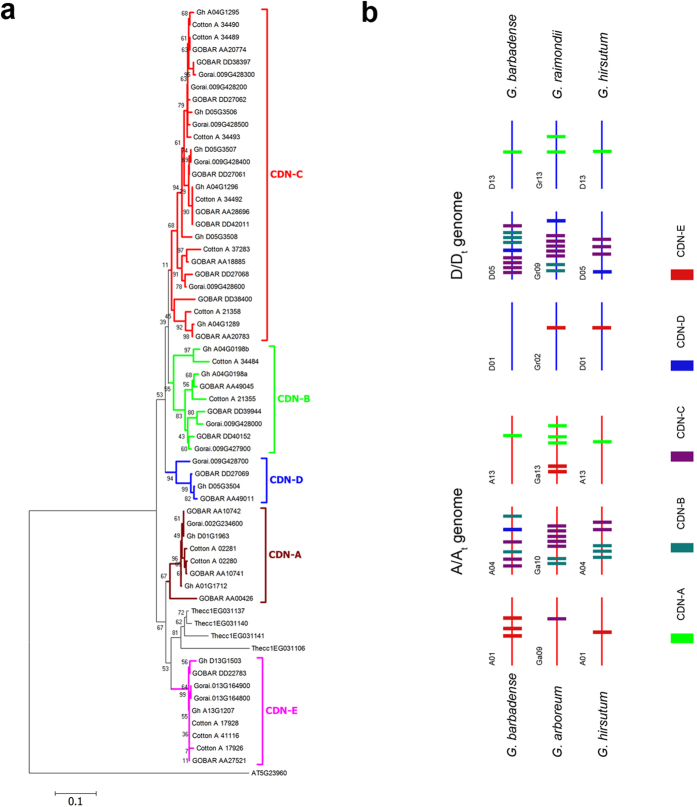
The cotton sesquiterpene synthase (+)-δ-cadinene synthase (CDN) catalyzes the first step of gossypol biosynthesis50. The G. barbadense genome harbors 19 CDN family genes (sharing >80% nucleotide identity), whereas G. raimondii, G. arboreum and G. hirsutum harbor 11, 14 and 13 of these genes, respectively (Fig. 6 and Supplementary Table 18). These genes evolved faster than cotton speciation; thus, the CDN family evolved approximately 60 Mya based on the phylogenetics of cotton plants (Fig. 1). The CDN subfamilies A and E were found closer to the ancient type and duplicated after the divergence of the cotton and cacao lineages (Fig. 6 and Supplementary Fig. 14). The variable CDN gene numbers in cotton species possibly refer to recent small-scale duplication events, e.g., CDN-A member duplication in the D genome ~1 Mya (Supplementary Table 18 and Supplementary Fig. 14). Thus, the CDN subfamilies in Gossypium represent an example of the rapid lineage-specific evolution of critical genes for specialized metabolites.
Figure 6. Phylogenetic analysis of (+)-δ-cadinene synthase (CDN) family genes and their genome distribution.
(a) The amino acid sequences of CDNs of G. arboreum (Cotton_A), G. raimondii (Gorai), G. hirsutum (Gh) and G. barbadense (GOBAR) and T. cacao (Thecc) were used to build the phylogenetic tree using a neighbor-joining algorithm via the MEGA software. The Arabidopsis thaliana sesquiterpene synthase gene At5g23960 was used as a phylogenetic outgroup. (b) Chromosomal locations of the CDN genes in four Gossypium species as indicated.
Discussion
ELS cotton likely produces one of the most resilient fibers in the plant kingdom; they are highly elongated and contain nearly pure cellulose. This draft sequence of the G. barbadense genome provides valuable genomic resources for studying various aspects of cotton. This draft sequence also facilitates breeding practices aimed at improving cotton fiber traits and increasing the production of high-quality biomass (cellulose).
The genomes of two or more parental species have combined to significantly change the genome structure and function of allopolyploid plants38,51,52. Inter-genomic chromosomal rearrangements, differential gene loss (the loss of some duplicates), gene conversion, divergence and the functional diversification of duplicated genes often arise with the onset of polyploidization53. Our comparative analysis of cotton genomes also provides new insight into dynamic allopolyploidy processes, such as the mechanism via TE (LTR retrons) bursts and pseudogenization, which have significantly contributed to plant genome evolution and trait formation.
Methods
Plant materials
Young leaves of Gossypium barbadense cv. Xinhai21, G. arboreum cv. Qingyangxiaozi and G. raimondii were collected from a single plant of each species for genomic DNA extraction and sequencing. For transcriptome sequencing, 28 samples from G. barbadense roots, stems, flowers, leaves, ovules and fibers were collected for total RNA extraction (Supplementary Table 19).
DNA isolation, library construction and sequencing
Genomic DNA was isolated from fresh cotton leaves using a previously described method54. The shotgun library (300–800 bp fragments) was prepared from 5 μg of DNA using a standard protocol, and a total of 55,296,227 reads with an average length of 542 bp were produced via Roche 454 GS FLX to provide a 12-fold coverage of the genome. The paired-end libraries of different insertion sizes were constructed, and 1,325,215,140 pairs of 100-bp reads were produced via Illumina Hiseq2000 (Illumina, San Diego, CA) to provide 105-fold coverage of the genome. The 3-, 5-, 8 and 20-kb mate-pair libraries were constructed by combining the GS FLX and Illumina mate-pair protocol, and a total of 773,715,534 mate-pair reads were produced via Illumina Hiseq2000 to provide 61.9-fold sequencing coverage. The BAC library (insert, 80–120 kb) was constructed using the pCC1BAC vector (Epicentre Inc.) and Hind III enzyme digestion. The BAC clones were both-end sequenced using ABI 3730, and 20 BACs at a time were pooled and sequenced on Illumina Hiseq2000 to generate a 300-bp paired-end library.
For the PacBio library construction and sequencing, genomic DNA was sheared using a Covaris g-TUBE followed by purification via binding to pre-washed AMPure XP beads (Beckman Coulter Inc.). After end-repair, the blunt adapters were ligated, followed by exonuclease incubation to remove all un-ligated adapters and DNA. The final “SMRT bells” were annealed with primers and bound to the proprietary polymerase using the PacBio DNA/Polymerase Binding Kit P4 (Part Number 100–236–500) to form the “Binding Complex”. After dilution, the library was loaded onto the instrument with DNA Sequencing Kit 2.0 (Part Number 100–216–400) and a SMRT Cell 8Pac for sequencing. A primary filtering analysis was performed with the RS instrument, and the secondary analysis was performed using the SMRT analysis pipeline version 2.1.0.
Genome assembly
The genomes of two diploid cotton species, G. arboreum and G. raimondii, were each sequenced at 100-fold coverage using Illumina Hiseq2000. The assembly resulted in 3,767,593 contigs of 1.5 Gb for G. arboreum and 1,111,300 contigs of 788 Mb for G. raimondii. These contigs, together with the published genomic data of G. raimondii10 and G. arboreum12, were used as template for grouping of G. barbadense sequencing reads into subgenomes, which resulted in totally 44.9% of the reads being At-unique, 26.9% being Dt-unique and 9.7% being both sharing. The remaining 18.5% none hit reads were further grouped during subgenome during sequence assembly.
After subgenome grouping, the At and Dt subgenomes of G. barbadense were assembled separately using a combined strategy. The Roche 454 reads were first assembled using Newbler v2.3. In total, 773,548 contigs with an average length of 2.5 kb were produced. Illumina pair-end reads, mate-pair reads, PacBio SMRT reads and BAC ends were then successively mapped to the contigs to improve quality. The 59,868 contigs (BACtigs) with an N50 of 23.8 kb from 515 BAC pools were merged. These approaches resulted in 4,586 At scaffolds and 2,186 Dt scaffolds with a total size of 2.2 Gb and maximum length of 3.4 Mb. Data statistics are given in Supplementary Table 2 and Table 1.
Finally, a high-density genetic map of G. hirsutum cv. TM-1 × G. barbadense cv. Hai7124 containing 4,999,048 SNPs22 was mapped to the G. barbadense assembly using the BWA program, which anchored 1.95 Gb or 88% of the assembled sequences and produced 26 pseudo-molecules (chromosomes).
Gene prediction and annotation
Three gene prediction programs, GeneMark (v2.3a)54, Augustus (v2.5)55 and FgeneSH56, were used to predict protein-coding genes in the G. barbadense genome. A final gene model was produced by combining the three prediction results with an in-house developed program (GLAD), a tool that creates consensus gene lists by integrating evidence from homology, de novo prediction, and RNA-Seq/EST data. Annotation was performed by comparing the predicted proteins with non-redundant proteins (nr) and the UniProt and KEGG databases. Blast2go57 was used to assign preliminary GO terms to the predicted gene models. Transcription factors were predicted using PlantTFDB v3.058. Protein domain predictions were performed using RPS-BLAST with a coverage >90%. The metabolic pathways were constructed using the KEGG database59.
Ortholog identification and Ks calculation
Genes were classified into ortholog groups with OrthoMCL60 against OrthoMCL proteins (default parameters) [PMID: 12952885]. The orthologs between species, or homoeologs between the At and Dt subgenomes of G. barbadense, were defined using BLASTP based on the Bidirectional Best Hit (BBH) method with a sequence coverage >30% and identity >30%, followed by selection of the best match. The Ka and Ks between orthologs were calculated using the KaKs_Calculator61 via model averaging. The unique gene in each subgenome was defined using the following parameters: 1. protein sequence with no match according to BLASTP against proteins of the other subgenome with E-value 1E-3; and 2. the sum of the length of the high-scoring segment pairs (HSP) was less than 1/3 of the CDS length (via BLASTN) against the genome sequence of the other subgenome.
Repeat and LTR retrotransposon analysis
Repetitive sequences were identified using RepeatScout with default parameters. The consensus sequences of each repeat family were used to identify repeat compositions in the genome via Censor. The complete LTR retron structures were predicted using LTR_finder62, and miniature inverted-repeat transposable elements (MITEs) were identified using MITE-Hunter63. Individual LTR retrotransposons were clustered into the same family using the 80–80–80 rule: If two TIR sequences share 80% or higher similarity in at least 80% of their length with an alignment length longer than 80 bp, the two sequences were clustered into the same family64.
The insertion ages of each full-length LTR retron were calculated based on the divergence between the left and right solo-LTR sequences using distmat from EMBOSS with the Kimura-2-parameter distance option, and insertion ages were calculated according to the formula T = K/(2r) (K = Kimura distance value, average substitution rate r = 2.6 × 10–9 in cotton).
Pseudogene identification
Pseudogenes were predicted using Pseudopipe65 with default parameters. The predicted protein-coding gene sequences from both G. barbadense subgenomes were used as queries to search repeat-masked intergenic regions. Putative pseudogenes were filtered by excluding genes that significantly overlapped with functional gene annotations, genes with parental genes annotated as transposon elements or plastid genes, and genes with sequence lengths shorter than 150 bp.
RNA extraction and transcriptome sequencing
The total RNA from each sample was extracted using TRIzol reagent (Invitrogen) following a standard protocol. The mRNAs were purified with the MicroPoly(A) Purist Kit (Ambion), fragmented and converted into an RNA-Seq library using the mRNAseq library construction kit (Illumina Inc.) and sequenced via Illumina Hiseq2000. The mRNAs of 28 samples were also pooled and sequenced on the 454 Genome Sequencer FLX instrument.
Sequence reads from all samples were cleaned using the FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). All reads containing ‘N’ were discarded. Adapter sequences were then removed using the fastx_clipper program, followed by the removal of low-quality (Q < 5) bases from the 3′ end with fastq_quality_trimmer while requiring a minimum sequence length of 50 bp.
The RNA-Seq reads of each sample were mapped to the At and Dt genes using bowtie266 with a mismatch in seed alignment of 0. Differentially expressed genes were identified via the DEGseq package using the MARS method (MA-plot-based method with Random Sampling model)67 based on their RPKM (Reads Per Kilobases per Million reads) or FPKM (reads per kilobase of exon model per million mapped reads) values68 with an FDR≤0.001 and |log2 Ratio |≥ 1 as the threshold. KEGG pathway enrichment was performed with a corrected P-value of < 0.05 as a threshold. GO enrichment was performed using Blast2go57.
Additional Information
Accession numbers: The G. barbadense genome assembly contigs and scaffolds have been deposited in GenBank under PRJNA251673. The sequences and functional annotation of G. barbadense protein encoding genes, including predicted genes and transcriptome data, are available from the website. (http://database.chgc.sh.cn/cotton/index.html).
How to cite this article: Liu, X. et al. Gossypium barbadense genome sequence provides insight into the evolution of extra-long staple fiber and specialized metabolites. Sci. Rep. 5, 14139; doi: 10.1038/srep14139 (2015).
Supplementary Material
Acknowledgments
This project was funded by Esquel Group. The work was supported by grants from The Strategic Priority Research Program of the Chinese Academy of Sciences (XDB11030300), National Science Foundation of China (31330058), Shanghai Municipal Commission for Science and Technology (11DZ2292600 and 13DZ2291800), the Ministry of Agriculture of China (2014ZX08009001), The Science and Technology Support Program of Science and Technology Office of Xinjiang province (201311106), Scientific Cooperation and Guidance Program of Science and Technology Office of Foshan City (2012HY100034). The Fudan University High-End Computing Center kindly provided computation facilities for part of the data analysis.
Footnotes
The authors declare no competing financial interests.
Author Contributions X.-Y.C., T.-Z.Z., G.-P.Z., J.-J.C., X.L., S.-Y.W. and Y.-G.Z., coordinated the project. X.L., Y.H., B.Z., L.-J.W., X.-X.S.G., Y.-L.B, D.M., W.-H.L., S.W. and X.-F.Z. collected the samples and extracted genomic DNA and RNA. S.-Y.W., Y.-Q.Z., L.J., H.K., L.-J.W., J.C., S.-T.Y., S.-T.C, B.-Y.W., J.W. and W.-Y.G. performed the genome, transcriptome and BAC clone sequencing. G.L., J.-D.C., D.M. and S.-Y.W. assembled the genome and EST sequence data. W.-Z.G., L.F., C.-P.C., B.-L.Z., Y.-Z.W., L.Z., J.-P.J., Y.-Q.Z., X.-L.H., R.W., S.-C.S., X.-Y.L. and Z.-C.G. performed the gene prediction, annotation and validation. H.-J.Z., B.Z., C.-Q.Y., D.Y.C., J.-X.R., G.L., C.-M.S., Y.-B.M., Y.Z., L.Z., J.-P.J., X.D., X.L., S.-H.X., S.-Y.W., T.-Z.Z., G.-P.Z. and X.-Y.C. performed the genome analysis. X.-Y.C., G.-P.Z., H.-J.Z., C.-Q.Y., B.Z., G.L., Z.J.C., T.-Z.Z., S.-Y.W., Y.Z. and X.L. wrote the manuscript.
References
- Soltis D. E., Visger C. J. & Soltis P. S. The polyploidy revolution then and now: Stebbins revisited. Am J Bot. 101, 1057–1078 (2014). [DOI] [PubMed] [Google Scholar]
- Hegarty M. J. & Hiscock S. J. Genomic clues to the evolutionary success of polyploid plants. Curr Biol. 18, R435–444 (2008). [DOI] [PubMed] [Google Scholar]
- Otto S. P. The evolutionary consequences of polyploidy. Cell. 131, 452–462 (2007). [DOI] [PubMed] [Google Scholar]
- Jiao Y. et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 473, 97–100 (2011). [DOI] [PubMed] [Google Scholar]
- Renny-Byfield S. & Wendel J. F. Doubling down on genomes: polyploidy and crop plants. Am J Bot. 101, 1711–1725 (2014). [DOI] [PubMed] [Google Scholar]
- Gong L., Kakrana A., Arikit S., Meyers B. C. & Wendel J. F. Composition and expression of conserved microRNA genes in diploid cotton (Gossypium) species. Genome biol Evol. 5, 2449–2459 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins J. S., Kim H., Nason J. D., Wing R. A. & Wendel J. F. Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Res. 16, 1252–1261 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover C. et al. Molecular confirmation of species status for the allopolyploid cotton species, Gossypium ekmanianum Wittmack. Genet Resour Crop Ev. 62, 103–114 (2015). [Google Scholar]
- Hendrix B. & Stewart J. M. Estimation of the nuclear DNA content of gossypium species. Ann Bot. 95, 789–797 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A. H. et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature. 492, 423–427 (2012). [DOI] [PubMed] [Google Scholar]
- Wang K. et al. The draft genome of a diploid cotton Gossypium raimondii. Nat Genet. 44, 1098–1103 (2012). [DOI] [PubMed] [Google Scholar]
- Li F. et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat Genet. 46, 567–572 (2014). [DOI] [PubMed] [Google Scholar]
- Senchina D. S. et al. Rate variation among nuclear genes and the age of polyploidy in Gossypium. Mol Biol Evol. 20, 633–643 (2003). [DOI] [PubMed] [Google Scholar]
- Marcussen T. et al. Ancient hybridizations among the ancestral genomes of bread wheat. Science. 345, 1250092 (2014). [DOI] [PubMed] [Google Scholar]
- Chalhoub B. et al. Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 345, 950–953 (2014). [DOI] [PubMed] [Google Scholar]
- Kim H. J. & Triplett B. A. Cotton fiber growth in planta and in vitro. Models for plant cell elongation and cell wall biogenesis. Plant Physiol. 127, 1361–1366 (2001). [PMC free article] [PubMed] [Google Scholar]
- Smart L. B., Vojdani F., Maeshima M. & Wilkins T. A. Genes involved in osmoregulation during turgor-driven cell expansion of developing cotton fibers are differentially regulated. Plant Physiol. 116, 1539–1549 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y. L., Llewellyn D. J. & Furbank R. T. The control of single-celled cotton fiber elongation by developmentally reversible gating of plasmodesmata and coordinated expression of sucrose and K+ transporters and expansin. Plant Cell. 13, 47–60 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P. et al. Inheritance of long staple fiber quality traits of Gossypium barbadense in G. hirsutum background using CSILs. Theor Appl Genet. 124, 1415–1428 (2012). [DOI] [PubMed] [Google Scholar]
- Li F. et al. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat Biotechnol. 33, 524–530 (2015). [DOI] [PubMed] [Google Scholar]
- Zhang T. et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol. 33, 531–537 (2015). [DOI] [PubMed] [Google Scholar]
- Wang S. et al. Sequence-based ultra-dense genetic and physical maps reveal structural variations of allopolyploid cotton genomes. Genome Biol. 16, 108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome I. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 408, 796–815 (2000). [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing P. The map-based sequence of the rice genome. Nature. 436, 793–800 (2005). [DOI] [PubMed] [Google Scholar]
- Schmutz J. et al. Genome sequence of the palaeopolyploid soybean. Nature. 463, 178–183 (2010). [DOI] [PubMed] [Google Scholar]
- Brenchley R. et al. Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature. 491, 705–710 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page J. T. et al. Insights into the evolution of cotton diploids and polyploids from whole-genome re-sequencing. G3. 3, 1809–1818 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brubaker C. L., Paterson A. H. & Wendel J. F. Comparative genetic mapping of allotetraploid cotton and its diploid progenitors. Genome. 42, 184–203 (1999). [Google Scholar]
- Yu J. Z. et al. A high-density simple sequence repeat and single nucleotide polymorphism genetic map of the tetraploid cotton genome. G3. 2, 43–58 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystedt B. et al. The Norway spruce genome sequence and conifer genome evolution. Nature. 497, 579–584 (2013). [DOI] [PubMed] [Google Scholar]
- SanMiguel P. & Bennetzen J. L. Evidence that a Recent Increase in Maize Genome Size was Caused by the Massive Amplification of Intergene Retrotransposons. Annals of Botany. 82 (suppl 1), 37–44 (1998). [Google Scholar]
- Hawkins J. S., Proulx S. R., Rapp R. A. & Wendel J. F. Rapid D. N. A. loss as a counterbalance to genome expansion through retrotransposon proliferation in plants. P Natl Acad Sci USA. 106, 17811–17816 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou C., Lehti-Shiu M. D., Thibaud-Nissen F., Prakash T., Buell C. R. & Shiu S. H. Evolutionary and expression signatures of pseudogenes in Arabidopsis and rice. Plant Physiol. 151, 3–15 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo M. J. & Wendel J. F. Comparative evolutionary and developmental dynamics of the cotton (Gossypium hirsutum) fiber transcriptome. PLoS Genet. 10, e1004073 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walford S. A., Wu Y., Llewellyn D. J. & Dennis E. S. GhMYB25-like: a key factor in early cotton fibre development. Plant J. 65, 785–797 (2011). [DOI] [PubMed] [Google Scholar]
- Zhang M. et al. Spatiotemporal manipulation of auxin biosynthesis in cotton ovule epidermal cells enhances fiber yield and quality. Nat Biotechnol. 29, 453–458 (2011). [DOI] [PubMed] [Google Scholar]
- Shan C. M. et al. Control of cotton fibre elongation by a homeodomain transcription factor GhHOX3. Nat Commun. 5, 5519 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. J. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol. 58, 377–406 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M. Y., Fan M., Oh E. & Wang Z. Y. A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell. 24, 4917–4929 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. Y. et al. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell. 21, 3767–3780 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argout X. et al. The genome of Theobroma cacao. Nat Genet. 43, 101–108 (2011). [DOI] [PubMed] [Google Scholar]
- Gu Y. et al. Identification of a cellulose synthase-associated protein required for cellulose biosynthesis. P Natl Acad Sci USA. 107, 12866–12871 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane H. E., Doring A. & Persson S. The cell biology of cellulose synthesis. Annu Rev Plant Biol. 65, 69–94 (2014). [DOI] [PubMed] [Google Scholar]
- Gou J. Y., Wang L. J., Chen S. P., Hu W. L. & Chen X. Y. Gene expression and metabolite profiles of cotton fiber during cell elongation and secondary cell wall synthesis. Cell Res. 17, 422–434 (2007). [DOI] [PubMed] [Google Scholar]
- Pear J. R., Kawagoe Y., Schreckengost W. E. & Delmer D. P., Stalker DM. Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. P Natl Acad Sci USA. 93, 12637–12642 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor-Teeples M. et al. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature. 517, 571–575 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y. B. et al. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotechnol. 25, 1307–1313 (2007). [DOI] [PubMed] [Google Scholar]
- Tao X. Y., Xue X. Y., Huang Y. P., Chen X. Y. & Mao Y. B. Gossypol-enhanced P450 gene pool contributes to cotton bollworm tolerance to a pyrethroid insecticide. Mol Ecol. 21, 4371–4385 (2012). [DOI] [PubMed] [Google Scholar]
- Bohlmann J., Meyer-Gauen G. & Croteau R. Plant terpenoid synthases: molecular biology and phylogenetic analysis. P Natl Acad Sci USA. 95, 4126–4133 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. Y., Chen Y., Heinstein P. & Davisson V. J. Cloning, expression, and characterization of (+)-delta-cadinene synthase: a catalyst for cotton phytoalexin biosynthesis. Arch Biochem Biophys. 324, 255–266 (1995). [DOI] [PubMed] [Google Scholar]
- Comai L. et al. Phenotypic instability and rapid gene silencing in newly formed arabidopsis allotetraploids. Plant Cell. 12, 1551–1568 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B. & Wendel J. F. Non-Mendelian phenomenon in allopolyploid genome evolution. Curr Genomics. 3, 489–505 (2002). [Google Scholar]
- Adams K. L. & Wendel J. F. Novel patterns of gene expression in polyploid plants. Trends Genet. 21, 539–543 (2005). [DOI] [PubMed] [Google Scholar]
- Lomsadze A., Ter-Hovhannisyan V., Chernoff Y. O. & Borodovsky M. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 33, 6494–6506 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M. & Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 19 Suppl 2, ii215–225 (2003). [DOI] [PubMed] [Google Scholar]
- Salamov A. A. & Solovyev V. V. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 10, 516–522 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A., Gotz S., Garcia-Gomez J. M., Terol J., Talon M. & Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 21, 3674–3676 (2005). [DOI] [PubMed] [Google Scholar]
- Jin J., Zhang H., Kong L., Gao G. & Luo J. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 42, D1182–1187 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Kawashima S., Okuno Y. & Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 32, D277–280 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Stoeckert C. J. Jr. & Roos D. S. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Li J., Zhao X. Q., Wang J., Wong G. K. & Yu J. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics, proteomics & bioinformatics. 4, 259–263 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z. & Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 35, W265–268 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y. & Wessler S. R. MITE-Hunter: a program for discovering miniature inverted-repeat transposable elements from genomic sequences. Nucleic Acids Res. 38, e199 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T. et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 8, 973–982 (2007). [DOI] [PubMed] [Google Scholar]
- Zhang Z., Carriero N., Zheng D., Karro J., Harrison P. M. & Gerstein M. PseudoPipe: an automated pseudogene identification pipeline. Bioinformatics. 22, 1437–1439 (2006). [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M. & Salzberg S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Feng Z., Wang X., Wang X. & Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 26, 136–138 (2010). [DOI] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L. & Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 5, 621–628 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.