Abstract
Yeast Bro1 and Rim20 belong to a family of proteins which possess a common architecture of Bro1 and V domains. Alix and His domain protein tyrosine phosphatase (HD-PTP), mammalian Bro1 family proteins, bind YP(X)nL (n = 1 to 3) motifs in their target proteins through their V domains. In Alix, the Phe residue, which is located in the hydrophobic groove of the V domain, is critical for binding to the YP(X)nL motif. Although the overall sequences are not highly conserved between mammalian and yeast V domains, we show that the conserved Phe residue in the yeast Bro1 V domain is important for binding to its YP(X)nL-containing target protein, Rfu1. Furthermore, we show that Rim20 binds to its target protein Rim101 through the interaction between the V domain of Rim20 and the YPIKL motif of Rim101. The mutation of either the critical Phe residue in the Rim20 V domain or the YPIKL motif of Rim101 affected the Rim20-mediated processing of Rim101. These results suggest that the interactions between V domains and YP(X)nL motif-containing proteins are conserved from yeast to mammalian cells. Moreover, the specificities of each V domain to their target protein suggest that unidentified elements determine the binding specificity.
INTRODUCTION
Yeast Bro1 belongs to a family of related proteins that share a architecture comprising an N-terminal Bro1 homology domain and a following V domain (Fig. 1A). Bro1/Vps31 was originally isolated as one of the vacuolar protein-targeting mutants and later classified as class E vps mutants (1, 2). Bro1 is reported to function as an accessory factor for the endosomal sorting complex required for transport (ESCRT) apparatus in the multivesicular body (MVB) pathway (3, 4). The ESCRT apparatus, which comprises four complexes (ESCRT-0, -I, -II, and -III), is responsible for the sorting of ubiquitinated membrane proteins into MVBs for degradation in the lysosome/vacuole (5). Bro1 is directed to endosomes by the association of the Bro1 domain with ESCRT-III subunit Snf7 (6) and was reported to regulate the membrane scission activity of ESCRT-III (7). Moreover, Bro1 binds to the deubiquitinating enzyme Doa4 through its C-terminal region, recruits Doa4 to endosomes, and activates Doa4 (8, 9). Doa4 plays a role in the recovery of ubiquitins from ubiquitinated cargoes just prior to the invagination of the cargo protein-enriched membranes; therefore, it maintains cellular ubiquitin homeostasis in yeast (10). Intriguingly, we revealed that Bro1 also binds to Rfu1 (a regulator for free ubiquitin chains) through its V domain and recruits Rfu1 to endosomes (11). Rfu1 also has a function to maintain ubiquitin homeostasis by inhibiting Doa4 activity (12). Bro1 has an additional region called the Pro-rich region (PRR), which was reported to bind Rsp5, a major ubiquitin ligase for ubiquitinating cargo proteins (13).
FIG 1.
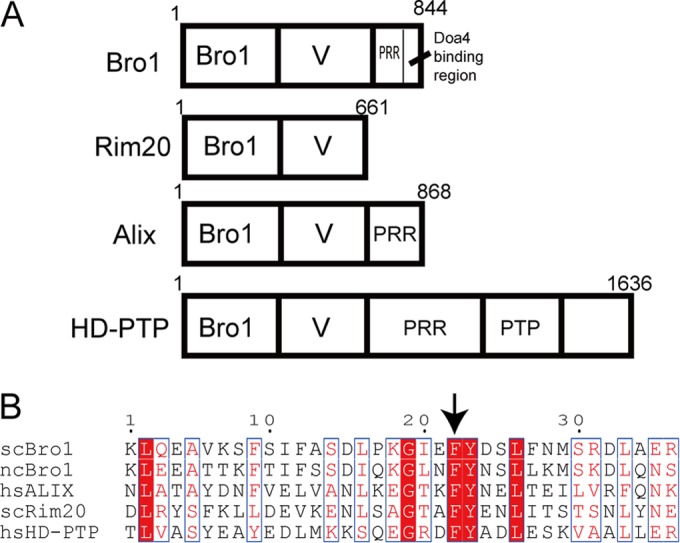
Bro1 family proteins. (A) Schematic organization of Bro1, Rim20, Alix, and HD-PTP. The Bro1, V domain, and PRR (proline-rich region) are indicated. PTP, phosphatase domain. (B) Conservation of putative YP(X)nL binding regions of the V domains of Saccharomyces cerevisiae (sc) Bro1, Naumovozyma castellii (nc) Bro1, Saccharomyces cerevisiae Rim20, human (hs) Alix, and human HD-PTP. Alignments of scBro1 versus ncBro1, scRim20 versus Alix, and HD-PTP versus Alix were generated by Clustal W. Alignments of nsBro1 and hsAlix were made by structural comparisons using DaliLite. These alignments were then assembled. The arrow indicates the critical Phe residue in the Alix V domain and the corresponding Phe in other Bro1 family proteins.
Rim20, another Bro1 family protein in yeast, functions in the pH-responsive pathway (14, 15). The pathway has been intensively studied in the fungus Aspergillus nidulans and the yeast Saccharomyces cerevisiae (16). In this pathway, Rim101, a transcription factor, is processed through the proteolytic removal of its C-terminal region in response to alkaline pH. The processed Rim101 then regulates the expression of alkali-responsive genes, resulting in the adaptation to alkaline conditions (17). During this activation process, Rim20 is required for the proteolytic cleavage of Rim101 along with other factors such as Rim13, Rim9, Rim21, Dfg16, Rim8, and several ESCRT-I, -II, and -III factors (18–20). Rim20 appears to function as an adaptor by directly binding to Rim101 and several ESCRT components, such as Snf7 (18, 19). Recently, it was reported that the events of the Rim101 pathway, after alkaline conditions, occurred on the plasma membrane (21, 22).
The mammalian Bro1 homolog, apoptosis-linked gene 2 interacting protein X (Alix), functions in ESCRT-mediated budding of enveloped viruses and membrane abscission in cytokinesis (3, 5). During the process of virus budding, cellular ESCRT machineries are hijacked by the viruses to facilitate their release from the cell membrane. Like Bro1, Alix has three main domains, Bro1, V, and PRR. The PRR of Alix was shown to directly bind multiple proteins such as Tsg101 (yeast Vps23) or CEP55 (23, 24). In addition, PRR keeps Alix in an autoinhibited conformation (25, 26). Although Alix has not been reported to function in the sorting of ubiquitinated cargoes such as the epidermal growth factor (EGF) receptor, His domain protein tyrosine phosphatase (HD-PTP), another member of Bro1 family proteins is required for EGF receptor sorting to the MVB (27).
The Alix V domain is about 320 amino acids (aa) long, forming the structure of two trihelical bundles taking the shape of the letter V. It has been studied extensively for its interaction with YP(X)nL motif-containing viral and cellular proteins (28, 29). The Alix V domain binds to the YP(X)nL motif-containing late domains of retroviruses such as HIV-1, equine infectious anemia virus (EIAV), and Ebola virus and appears to play an important role in virus budding (30, 31). A hydrophobic pocket on the second arm of Alix V was identified as a region for binding to the YP(X)nL motif peptide (28, 29). Particularly, the Phe residue in the pocket plays a critical role in the interaction with the YP(X)nL motif, and F676D is an inactivation mutation of Alix V in binding. As for cellular proteins, Alix V was shown to bind to the YPX(3)L motif of the G-protein-coupled receptor protease-activated receptor 1 (PAR1) to mediate the ubiquitin-independent sorting of PAR1 (32). In yeast, the Bro1 and Rim20 V domains were shown to have a structure very similar to that of the Alix V domain, although they have a low sequence similarity (33). Recently, the V domains of Alix, HD-PTP, Bro1, and Rim20 were shown to bind to ubiquitins, particularly to K63-linked ubiquitin chains (33–35), leading to the proposal that V domains are ubiquitin receptors. The ubiquitin binding regions within the V domains were reported to be different from the YP(X)nL binding region.
Because amino acid sequences are not highly conserved between Alix and yeast V domains (11 to 13% amino acid identity for the Alix and Bro1 V domains [see Fig. S1 in the supplemental material), the interaction of the yeast V domain with a YP(X)nL motif-containing protein has been overlooked (33). Recently, we showed a direct interaction between a region containing the YPEL motif of Rfu1 and the V domain of Bro1 (11). In this study, we observed that a region containing a critical Phe residue that is reported to bind to the YP(X)nL motif of the target proteins in Alix is relatively conserved in the V domains of Alix, HD-PTP, Bro1, and Rim20 (Fig. 1). Therefore, we tested whether the yeast V domain's interaction with YP(X)nL motif-containing target proteins could be analogous to that of mammalian V domains. We examined the Bro1 V-Rfu1 and Rim20 V-Rim101 interactions by focusing on the conserved Phe residue in the V domains of Bro1 and Rim20.
MATERIALS AND METHODS
Media.
Yeast strains were grown in YPAD medium (1% yeast extract, 2% Bacto Peptone, 2% glucose, and 0.002% adenine), in synthetic complete medium (SD) (0.67% yeast nitrogen base and 2% glucose supplemented with amino acids), or in synthetic Casamino medium (SC) (0.67% yeast nitrogen base, 2% glucose, and 0.5% Casamino Acids). If necessary, tryptophan, uracil, or adenine was added. For microscopy studies, 0.02% adenine was added.
Yeast strains and plasmids.
Lists of the yeast strains and plasmids used in this study are provided in Tables S1 and S2, respectively, in the supplemental material. Plasmid pGST2-Alix(360–702) was obtained from Addgene. Plasmid expressing N-terminally myc-tagged Rim20 under the control of a RIM20 promoter was created as follows. Two kinds of DNA fragments, F and B, were amplified using a RIM20 plasmid as a template and two sets of primers, (i) RIM20-up875-BamHI (AATTAGGATCCACGTTGTATATTTTCAATCTGGAAAGTAA) and RIM20-BtsI-AS (GTTCACTCATGTCACACTGCCTGGATCTCC) and (ii) RIM20-BtsI-Myc-sense (AATTGCAGTGTGACATGGAACAAAAGCTTATTTCTGAAGAAGACTTGATGAGTGAACTGCTTGCCATTCC) and RIM20-Down-XhoI-AS (AATTCTCGAGCTGTTGTCTAAAGGCGAAACTACGATGAAG), respectively. The obtained F and B fragments were cut with BamHI-BtsI and BtsI-XhoI, respectively. The two fragments were ligated to the BamHI-XhoI vector portion of pRS315.
Immunoblotting.
Preparation of whole-cell extracts and immunoblot analysis were performed as previously described (36). In Western blotting, blots were incubated with a mouse anti-green fluorescent protein (anti-GFP) monoclonal antibody (Roche), an anti-hemagglutinin (anti-HA) antibody (HA.11; Covance), or an anti-yeast phosphoglycerate kinase (anti-PGK) antibody (Molecular Probes, Eugene, OR), followed by horseradish peroxidase (HRP)-conjugated anti-mouse IgG (NA931V, Amersham) and then visualized using ECL-plus reagent (Amersham). To detect glutathione S-transferase (GST), an HRP-conjugated anti-GST antibody (Wako) was used. A rabbit anti-yeast Bro1 antibody was described previously (11).
Recombinant protein purification.
Maltose binding protein (MBP)-Rfu1 and MBP fusions of the Rfu1 mutants were purified as previously described (12). Recombinant GST, GST-Bro1, or the various GST-Bro1 mutants were purified using glutathione chromatography as recommended by the manufacturer (GE Healthcare). Recombinant proteins were eluted with 20 mM glutathione, 50 mM Tris-HCl (pH 8.0), and 2 mM dithiothreitol (DTT), dialyzed against 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, and 10% glycerol, and then stored at −80°C.
In vitro binding between various MBP-fused proteins and GST-fused proteins.
Binding experiments were performed as previously described (11).
Microscopy.
FM4-64 (Molecular Probes, Inc.) staining was performed as previously described (37). Cells were imaged at room temperature using a confocal microscope (LSM780; Carl Zeiss) equipped with an αPlan-Apochromat 100× oil objective lens. Images were processed using the LSM image browser, and the brightness and contrast were adjusted using Adobe Photoshop CS4.
Detection of HA-Rim101.
Logarithmically growing cells in SC-Ura Leu or SC-Ura (pH 4.0) medium were harvested by centrifugation and resuspended in the same medium, SC-Ura Leu or SC-Ura (pH 8.0). After incubating for 20 min at 30°C, trichloroacetic acid (TCA) was added to make a final concentration of 6%, and the mixture was kept on ice for 20 min. Cells collected by centrifugation were suspended in a urea buffer (50 mM Tris-HCl [pH 7.5], 5 mM EDTA, 6 M urea, 1% SDS, 50 mM NaF, 1 mM phenylmethylsulfonyl fluoride) and were disrupted by vortexing with glass beads. The obtained cell lysates were cleared by centrifugation, and the total protein concentrations were determined using the DC protein assay (Bio-Rad, Richmond, CA). The cell lysates were incubated with Laemmli SDS sample buffer at 65°C for 15 min and were subjected to SDS-PAGE and Western blotting. To detect HA-tagged Rim101 and Myc-tagged Rim20, membranes were blocked with 1% skim milk and then immunoblotted with an anti-HA antibody (12CA5) or an anti-myc antibody (9E10), respectively. The membranes were then treated with an anti-mouse IgG secondary antibody conjugated with horseradish peroxidase (NA931; GE Healthcare) and developed with ECL Prime (GE Healthcare). To detect actin, an anti-actin monoclonal antibody (C4; ICN, Aurora, OH) and IRDye-conjugated anti-mouse IgG antibody (Rockland, Gilbertsville, PA) were used. The signals were detected by using the infrared imaging system Odyssey (LiCor, Lincoln, NE).
RESULTS
Effect of Phe687 of Bro1 on the interaction with Rfu1.
We noticed that the Phe residue and the following Tyr as well as several neighboring residues were conserved around this region in the Bro1 and Rim20 V domains of S. cerevisiae and the Bro1 V domain of Naumovozyma castellii, whose crystal structure was resolved (33) (Fig. 1B). This suggests that this region in the V domains of Bro1 and Rim20 may have functions similar to those of the Alix V domain.
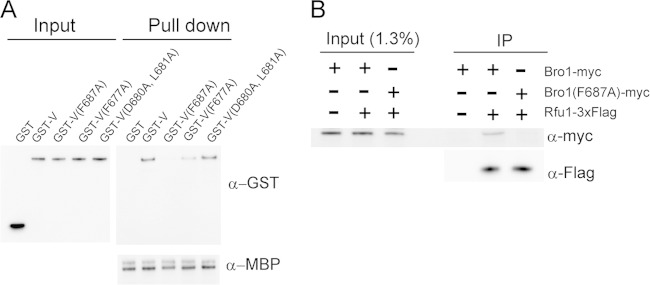
In Bro1, Phe687 is the corresponding Phe residue. First, we examined the effect of the Phe 687 mutation in the Bro1 V domain for the binding of Rfu1 in vitro (Fig. 2A). In previous work, we found that recombinant MBP-Rfu1 specifically bound to the recombinant GST-fused Bro1 V domain (11). We observed that the binding activity of GST-Bro1 V(F687A) to MBP-Rfu1 was drastically reduced (Fig. 2A). Additionally, we examined the binding abilities of mutants whose mutations were located close to F687: F677A, D680A, and L681A. The binding of Bro1 V(F677A) was reduced moderately but not as much as that of F687A Bro1 V, and D680A L681A double mutations had no effects.
FIG 2.
Impaired binding of Bro1(F687A) to Rfu1. (A) Impaired binding of Bro1 V(F687A) to MBP-Rfu1 in vitro. MBP or MBP-Rfu1 was mixed with GST, GST-Bro1 V, or the indicated GST-Bro1 V mutants, and the proteins were isolated with amylose resin. Samples were examined by immunoblot analysis using anti-GST (top panel) and anti-MBP (bottom panel) antibodies. (B) Impaired association of Bro1(F687A) with Rfu1-3 × Flag in vivo. Lysates of Δbro1cells or Δbro1 RFU1-3xF cells harboring a plasmid expressing C-terminally myc-tagged Bro1 or Bro1(F687A) were immunoprecipitated with anti-Flag. The resulting immune complexes (IP) were analyzed by immunoblotting using anti-Flag and anti-myc.
Next, we investigated whether the Bro1 F687 residue functioned in the interaction with Rfu1 in vivo. Immunoprecipitation analysis, using lysates from cells expressing Rfu1-3 × Flag plus myc-tagged Bro1 or myc-tagged Bro1(F687A), was performed using anti-Flag. Myc-tagged Bro1, but not myc-tagged Bro1(F687A), was specifically precipitated with Rfu1-3 × Flag (Fig. 2B). These results indicated that Bro1 Phe687 played a critical role in the Bro1-Rfu1 interaction both in vitro and in vivo.
The Rfu1 localization at endosomes largely depends on Bro1; Rfu1-GFP is mainly diffusive in the Δbro1 mutant, and the Rfu1 mutant in which the YPEL motif was changed to AAEL showed impaired endosomal localization (11). We therefore examined the effect of the F687A mutation on the localization of Rfu1-GFP fusions in yeast. First, we found that Bro1-GFP and Bro1(F687A)-GFP were similarly observed, mainly at the class E compartments in Δvps4 Δbro1 cells (data not shown). Next, the localization of Rfu1-GFP expressed under control of the RFU1 promoter was examined in Δrfu1 Δvps4 Δbro1 cells expressing either a wild-type or F687A Bro1. Rfu1-GFP fluorescence was present at foci that overlapped with FM4-64-stained class E compartments in Bro1-expressing Δvps4 Δrfu1 cells (Fig. 3A and B). In contrast, the localization of Rfu1-GFP at class E compartments was reduced in Bro1(F687A)-expressing cells. The accumulation of Rfu1-GFP was slightly reduced, probably due to its impaired binding to Bro1(F687A) because a reduction in the accumulation of Rfu1 was previously observed in the Δbro1 mutant (Fig. 3C) (11).
FIG 3.
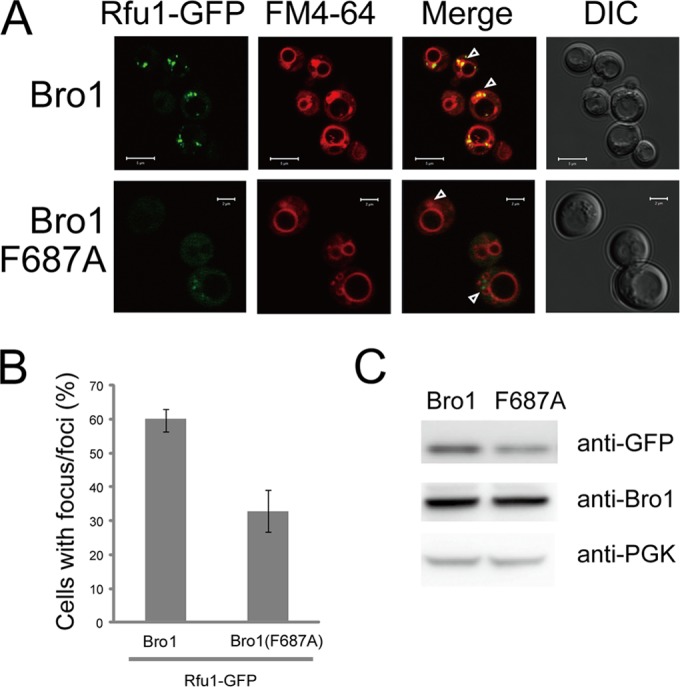
Impaired endosome localization of Rfu1-GFP in Bro1(F687A)-expressing cells. (A) GFP and FM4-64 fluorescence and differential interference contrast (DIC) microscopy of Rfu1-GFP in Δvps4 Δrfu1 Δbro1 cells expressing Bro1 or Bro1(F687A). Arrowheads indicate the class E compartments. Scale bars, 5 μm for upper panels and 2 μm for lower panels. (B) Quantification of Rfu1-GFP foci in panel A. Cells containing GFP foci around the vacuolar membrane were counted (n = 50 cells in each experiment), and mean values from three independent experiments are shown. Standard errors (SE) are shown. (C) Rfu1-GFP expression as determined by the anti-GFP immunoblot analysis in panel A. Anti-GFP (top), anti-Bro1 (middle), and anti-phosphoglycerate kinase (anti-PGK) (bottom) (a control for protein loading) immunoblots are shown.
Rim20 and Rim101 interaction through the V domain of Rim20 and the YPKIL motif in Rim101.
Next, we looked for different interactions between V domains and YP(X)nL motif-containing proteins. Rim20, another V-domain-containing protein, is required for Rim101p processing by direct binding to Rim101 (14, 38). Xu and Mitchell showed that the C-terminal region containing PEST-like sequences in Rim101 was sufficient for binding to Rim20 (14). Within the C-terminal region of Rim101, there is a YPKIL motif close to the C-terminal end that matches with the consensus YP(X)nL (n = 1 to 3) motif (39). This motif is located downstream of the cleavage site of Rim101. In addition, approximately the C-terminal half (aa 353 to 661) of Rim20, which corresponds to its V domain, was reported to bind to Rim101 (14).
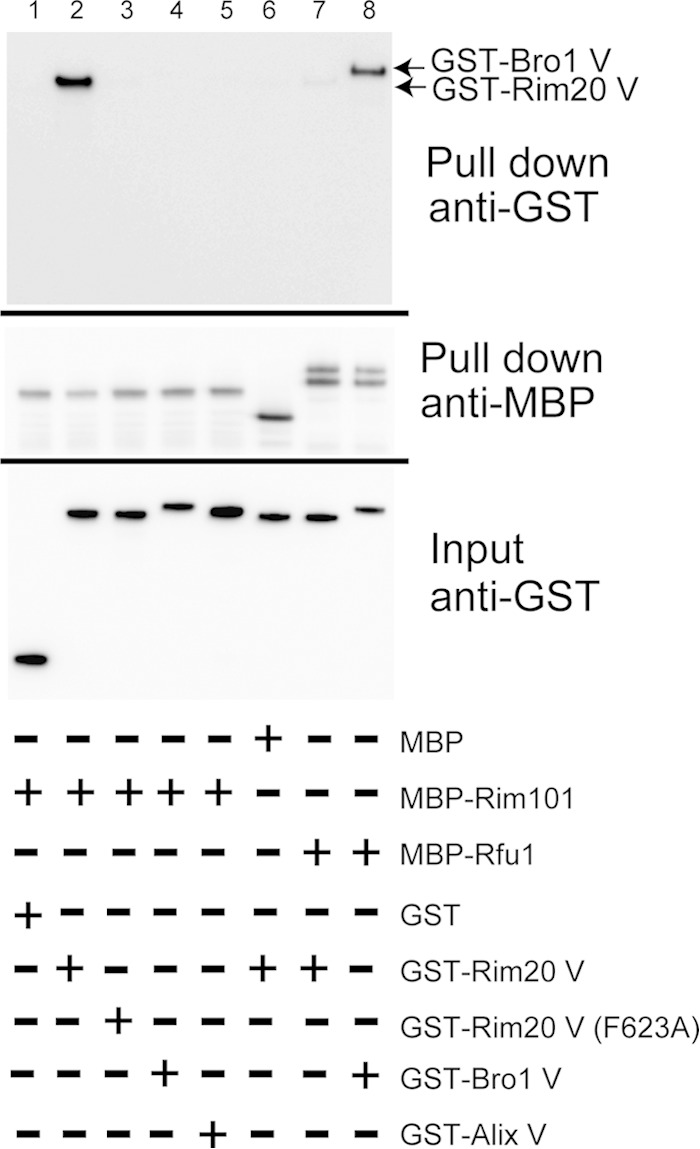
To test whether the Rim101-Rim20 interaction was mediated by Rim20's V domain and Rim101's YPKIL motif, we assessed the interaction in in vitro binding experiments. We made recombinant MBP-Rim101-C, a fusion of MBP with the 125 aa of the C-terminal region of Rim101, and checked whether MBP-Rim101-C bound to recombinant GST-Rim20 V (aa 330 to 661) in vitro (Fig. 4). As expected, we observed an efficient interaction between the two proteins (Fig. 4). When the conserved Rim20 Phe623 was replaced by Ala in GST-Rim20 V, the binding ability was significantly lost (Fig. 4). Moreover, we observed that the interaction between GST-Rim20V and MBP-Rim101-C(Y620A, P621A), in which the YPKIL motif was changed to AAKIL, was significantly lost (Fig. 4). These results suggest that the interaction between Rim20 and Rim101 is mediated by the Rim20 V domain and the YPKIL motif of Rim101 and that the conserved Phe in Rim20V is critical for the interaction.
FIG 4.
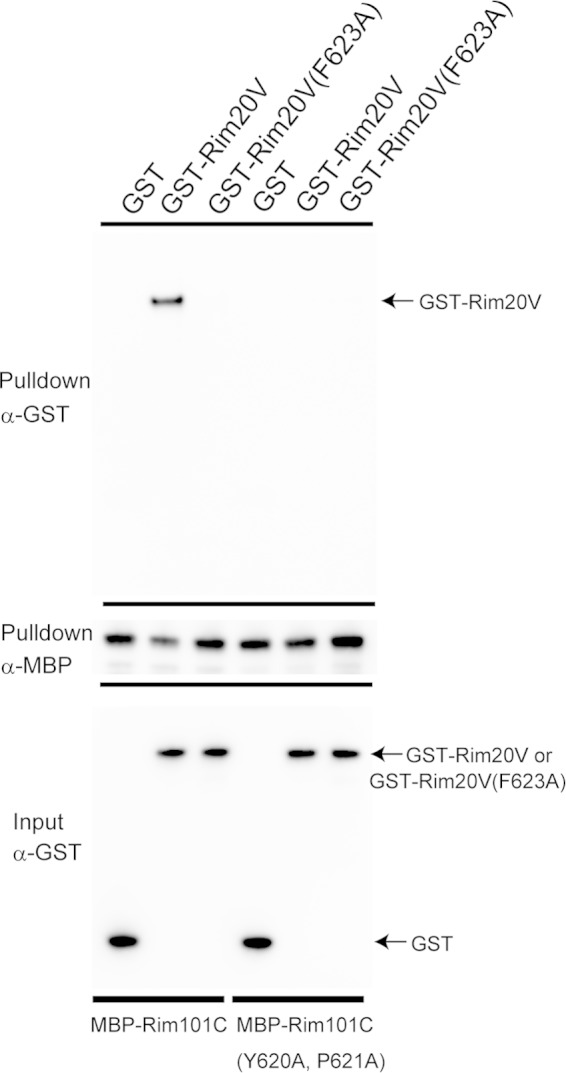
Binding of the Rim20 V domain to the Rim101 YP(X)nL motif in vitro. MBP, MBP-Rim101-C, or MBP-Rim101-C(Y620A, P621A) was incubated with GST, GST-Rim20 V, or GST-Rim20-V(F623A), and the proteins were isolated with amylose resin. GST, GST-Rim20V, and GST-Rim20V(F623A) are indicated by arrows. An anti-GST immunoblot for pulldown samples (top), an anti-MBP immunoblot for pulldown samples (middle), and an anti-GST immunoblot for input (bottom) are shown.
Next, we investigated the role of Phe623 in the Rim20 V domain by examining the processing of Rim101 in vivo (Fig. 5). N-terminally myc-tagged or nontagged Rim20 or Rim20(F623A) was expressed in the Δrim20 mutant together with HA-tagged Rim101. Under acidic conditions (pH 4), the intact full length of Rim101 is a major form, whereas under alkaline conditions (pH 8), Rim101 undergoes proteolytic processing that removes the C-terminal region of Rim101 (15). As previously reported (14), the processing was defective in the Δrim20 mutant. When wild-type Rim20 or myc-tagged Rim20 was introduced into the Δrim20 mutant, Rim101 processing became normal. In contrast, Rim101 processing was defective in the Δrim20 mutant expressing Rim20(F623A) or myc-tagged Rim20(F623A). We observed that the amino acid change of F623A did not affect the stability of myc-Rim20(F623A), and its level was similar to that of myc-Rim20 (Fig. 5A).
FIG 5.
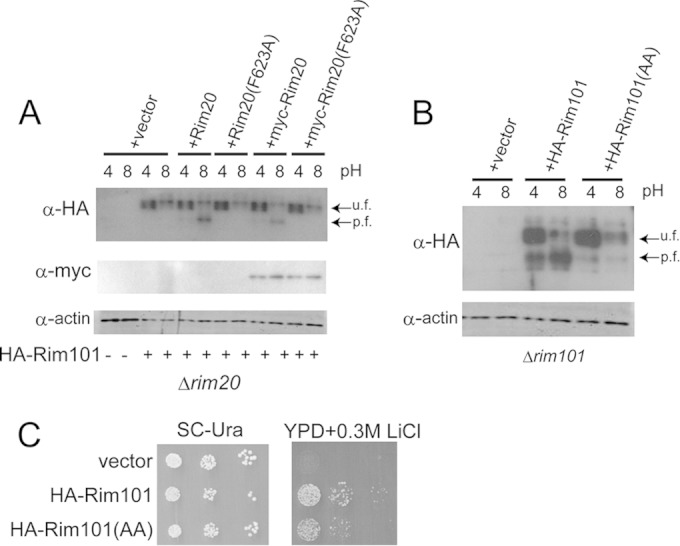
Effects of Rim101 or Rim20 mutation on HA-Rim101 processing. (A) Effects of Rim20(F623A) mutation on HA-Rim101 processing. HA-tagged Rim101 was expressed in Δrim20 cells harboring a vector or expressing Rim20, Rim20(F623A), myc-tagged Rim20, or myc-tagged Rim20(F623A) at the indicated pH of 4 or 8. Processed (p.f.) and unprocessed (u.f.) forms of HA-Rim101 are indicated. An anti-HA immunoblot (top), an anti-myc immunoblot for myc-tagged Rim20 or Rim20(F623A) (middle), and an antiactin blot, used as a loading control (bottom), are shown. (B) Effect of HA-Rim101(AAKIL) mutation on processing. Δrim101 cells harboring a vector or plasmids expressing HA-Rim101 or HA-Rim101(AAKIL) were tested. (C) Li sensitivity. Cells were diluted and spotted on SC-Ura plates and YPD containing 0.3 M LiCl and incubated for 3 days.
In addition, we investigated the effect of the YPKIL mutation of Rim101 on its processing by expressing HA-Rim101 or HA-Rim101(Y620A, P621A) in Δrim101 cells, and we observed that proteolytic processing was defective in cells expressing Rim101(Y620A, P621A) (Fig. 5B). Because the mutants with mutations in the Rim101 pathway show sensitivity to LiCl-containing medium (20, 40), LiCl sensitivity was examined (Fig. 5C). Cells expressing Rim101(Y620A, P621A) showed a marginal but significant sensitivity, indicating that the active form of Rim101 was not efficiently produced from Rim101(Y620A, P621A). These results indicated that the interaction between the Rim20 V domain and the YPKIL motif of Rim101 was important for their biological function.
Specificity of the V domain-YP(X)nL interaction.
The V domains of Bro1 and Rim20 are structurally similar; however, their physiological roles have been reported to be different. It was reported that the Δbro1 mutant showed normal Rim101 processing (14), and the involvement of Rim20 in MVB sorting has not been reported. We therefore suspected that there were specificities for their interactions. To test the idea, we examined whether Rfu1 bound to the Rim20 V domain or whether Rim101 bound to Bro1 V (Fig. 6). We observed that under the conditions that MBP-Rfu1 bound to Bro1 V, MBP-Rfu1 did not bind Rim20 V. Similarly, MBP-Rim101-C bound to Rim20 V but did not bind to Bro1 V or Alix V. These results suggest that there are more unidentified determinants for the specific interaction between V domains and YP(X)nL motifs.
FIG 6.
V domain specificity against YP(X)nL motif-containing proteins. MBP, MBP-Rfu1, and MBP-Rim101-C were incubated with GST, GST-Bro1V, GST-Rim20 V, and GST-Alix V, and the proteins were isolated with amylose resin. GST-tagged samples were examined by immunoblot analysis using anti-MBP and anti-GST antibodies. Top, anti-GST immunoblot for pulldown samples. GST-Rim20 V and GST-Bro1V in pulldown samples are indicated by arrows. Middle, anti-MBP immunoblot for pulldown samples. Bottom, anti-GST immunoblot for input.
DISCUSSION
In this study, we showed that the conserved Phe residue in the V domains of yeast Bro1 and Rim20 plays an important role in binding to the YP(X)nL motif of their target proteins, Rfu1 and Rim101. The results suggest that the yeast Bro1 and Rim20 V domains bind to their target proteins in a way similar to that of mammalian Alix V, indicating that V domains are YP(X)nL motif binding domains from yeast to mammals. Therefore, results from yeast Bro1 family V domain studies not only will contribute to our understanding of the cellular events in yeast but also may be informative in our understanding of the interactions between mammalian Bro1 family proteins and their YP(X)nL-containing target proteins such as virus proteins.
In addition, we showed that there are specificities of each V domain with its target protein; the C-terminal region of Rim101 specifically binds to the Rim20 V domain but not to the Bro1 and Alix V domains. Likewise, Rfu1 binds only to Bro1 V and not to Rim20 V domains in our in vitro assay. These results suggest that there must be more unidentified sequence or structural determinants of the interaction between V domains and their cognate YP(X)nL motif-containing partners (e.g., particular sequence or structures). Indeed, there are many proteins that possess YP(X)nL motifs in a cell, but only a subset of them seem to bind to the V domains. For example, the YPFL motif of Doa4 does not bind to the Bro1 V domain; instead, this motif binds to the C-terminal region of Bro1 (8). Although we do not have any hints for the determinants, an intensive mutagenesis approach may give us a clue to understand the nature of the specificity. In any case, a structural analysis of the Bro1 V domain YPEL peptide or the Rim20 V domain YPKIL peptide will be needed to define the precise mechanism of the interaction. Moreover, finding more Bro1 V domain binding proteins and comparing their sequences or structures with those of Rfu1 or Alix V binding proteins may give us some ideas of the specificities.
Bro1 is required for Rfu1 to function at endosomes (11). Rfu1 is involved in ubiquitin homeostasis, because in Δrfu1 cells, monomer ubiquitin is increased and unanchored ubiquitin chains or small ubiquitin species decreased (11, 12). We expected that Δbro1 cells expressing Bro1(F687A) would show a ubiquitin profile similar to that of the Δrfu1 mutant, but they did not show obvious aberrant profiles (data not shown). The reason was unknown, but it may be that the residual binding of Rfu1 to Bro1(F687A) in vivo is enough to support ubiquitin homeostasis, although other possibilities cannot be excluded.
Alix has been reported to have a flexible structure (25, 26). The PRR was reported to fold back and inhibit V domain binding to viral proteins. The V domain appears to take a closed conformation in the presence of the PRR and an open conformation in the absence of the PRR. Alix was also reported to have a dimer structure via its V domain (41). Moreover, binding of ubiquitins to Alix V was shown to induce oligomerization of the V domain (35). Thus, conformational change of Bro1 family proteins as a whole as well as their V domain seem to be regulated in complex ways in a cell, and Bro1 family studies using tractable yeasts would be suitable to reveal such complex mechanisms.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by JSPS KAKENHI grants 26440070 (to Y. Kimura) and 25291042 (to T. Maeda).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00091-15.
REFERENCES
- 1.Robinson JS, Klionsky DJ, Banta LM, Emr SD. 1988. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol 8:4936–4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. 1992. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol Biol Cell 3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bissig C, Gruenberg J. 2014. ALIX and the multivesicular endosome: ALIX in Wonderland. Trends Cell Biol 24:19–25. doi: 10.1016/j.tcb.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 4.Odorizzi G, Katzmann DJ, Babst M, Audhya A, Emr SD. 2003. Bro1 is an endosome-associated protein that functions in the MVB pathway in Saccharomyces cerevisiae. J Cell Sci 116:1893–1903. doi: 10.1242/jcs.00395. [DOI] [PubMed] [Google Scholar]
- 5.Henne WM, Buchkovich NJ, Emr SD. 2011. The ESCRT pathway. Dev Cell 21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 6.Kim J, Sitaraman S, Hierro A, Beach BM, Odorizzi G, Hurley JH. 2005. Structural basis for endosomal targeting by the Bro1 domain. Dev Cell 8:937–947. doi: 10.1016/j.devcel.2005.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wemmer M, Azmi I, West M, Davies B, Katzmann D, Odorizzi G. 2011. Bro1 binding to Snf7 regulates ESCRT-III membrane scission activity in yeast. J Cell Biol 192:295–306. doi: 10.1083/jcb.201007018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richter C, West M, Odorizzi G. 2007. Dual mechanisms specify Doa4-mediated deubiquitination at multivesicular bodies. EMBO J 26:2454–2464. doi: 10.1038/sj.emboj.7601692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luhtala N, Odorizzi G. 2004. Bro1 coordinates deubiquitination in the multivesicular body pathway by recruiting Doa4 to endosomes. J Cell Biol 166:717–729. doi: 10.1083/jcb.200403139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swaminathan S, Amerik AY, Hochstrasser M. 1999. The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol Biol Cell 10:2583–2594. doi: 10.1091/mbc.10.8.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimura Y, Kawawaki J, Kakiyama Y, Shimoda A, Tanaka K. 2014. The ESCRT-III adaptor protein Bro1 controls functions of regulator for free ubiquitin chains 1 (Rfu1) in ubiquitin homeostasis. J Biol Chem 289:21760–21769. doi: 10.1074/jbc.M114.550871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimura Y, Yashiroda H, Kudo T, Koitabashi S, Murata S, Kakizuka A, Tanaka K. 2009. An inhibitor of a deubiquitinating enzyme regulates ubiquitin homeostasis. Cell 137:549–559. doi: 10.1016/j.cell.2009.02.028. [DOI] [PubMed] [Google Scholar]
- 13.Nikko E, Andre B. 2007. Split-ubiquitin two-hybrid assay to analyze protein-protein interactions at the endosome: application to Saccharomyces cerevisiae Bro1 interacting with ESCRT complexes, the Doa4 ubiquitin hydrolase, and the Rsp5 ubiquitin ligase. Eukaryot Cell 6:1266–1277. doi: 10.1128/EC.00024-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu W, Mitchell AP. 2001. Yeast PalA/AIP1/Alix homolog Rim20p associates with a PEST-like region and is required for its proteolytic cleavage. J Bacteriol 183:6917–6923. doi: 10.1128/JB.183.23.6917-6923.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W, Mitchell AP. 1997. Proteolytic activation of Rim1p, a positive regulator of yeast sporulation and invasive growth. Genetics 145:63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Penalva MA, Lucena-Agell D, Arst HN Jr. 2014. Liaison alcaline: Pals entice non-endosomal ESCRTs to the plasma membrane for pH signaling. Curr Opin Microbiol 22:49–59. doi: 10.1016/j.mib.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Maeda T. 2012. The signaling mechanism of ambient pH sensing and adaptation in yeast and fungi. FEBS J 279:1407–1413. doi: 10.1111/j.1742-4658.2012.08548.x. [DOI] [PubMed] [Google Scholar]
- 18.Boysen JH, Mitchell AP. 2006. Control of Bro1-domain protein Rim20 localization by external pH, ESCRT machinery, and the Saccharomyces cerevisiae Rim101 pathway. Mol Biol Cell 17:1344–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu W, Smith FJ Jr, Subaran R, Mitchell AP. 2004. Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans. Mol Biol Cell 15:5528–5537. doi: 10.1091/mbc.E04-08-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayashi M, Fukuzawa T, Sorimachi H, Maeda T. 2005. Constitutive activation of the pH-responsive Rim101 pathway in yeast mutants defective in late steps of the MVB/ESCRT pathway. Mol Cell Biol 25:9478–9490. doi: 10.1128/MCB.25.21.9478-9490.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Obara K, Kihara A. 2014. Signaling events of the Rim101 pathway occur at the plasma membrane in a ubiquitination-dependent manner. Mol Cell Biol 34:3525–3534. doi: 10.1128/MCB.00408-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galindo A, Calcagno-Pizarelli AM, Arst HN Jr, Penalva MA. 2012. An ordered pathway for the assembly of fungal ESCRT-containing ambient pH signalling complexes at the plasma membrane. J Cell Sci 125:1784–1795. doi: 10.1242/jcs.098897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Schwedler UK, Stuchell M, Muller B, Ward DM, Chung HY, Morita E, Wang HE, Davis T, He GP, Cimbora DM, Scott A, Krausslich HG, Kaplan J, Morham SG, Sundquist WI. 2003. The protein network of HIV budding. Cell 114:701–713. doi: 10.1016/S0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 24.Morita E, Sandrin V, Chung HY, Morham SG, Gygi SP, Rodesch CK, Sundquist WI. 2007. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J 26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou X, Si J, Corvera J, Gallick GE, Kuang J. 2010. Decoding the intrinsic mechanism that prohibits ALIX interaction with ESCRT and viral proteins. Biochem J 432:525–534. doi: 10.1042/BJ20100862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhai Q, Landesman MB, Chung HY, Dierkers A, Jeffries CM, Trewhella J, Hill CP, Sundquist WI. 2011. Activation of the retroviral budding factor ALIX. J Virol 85:9222–9226. doi: 10.1128/JVI.02653-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doyotte A, Mironov A, McKenzie E, Woodman P. 2008. The Bro1-related protein HD-PTP/PTPN23 is required for endosomal cargo sorting and multivesicular body morphogenesis. Proc Natl Acad Sci U S A 105:6308–6313. doi: 10.1073/pnas.0707601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fisher RD, Chung HY, Zhai Q, Robinson H, Sundquist WI, Hill CP. 2007. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell 128:841–852. doi: 10.1016/j.cell.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 29.Lee S, Joshi A, Nagashima K, Freed EO, Hurley JH. 2007. Structural basis for viral late-domain binding to Alix. Nat Struct Mol Biol 14:194–199. doi: 10.1038/nsmb1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han Z, Madara JJ, Liu Y, Liu W, Ruthel G, Freedman BD, Harty RN. 18 March 2015. ALIX rescues budding of a double PTAP/PPEY L-domain deletion mutant of Ebola VP40: a role for ALIX in Ebola virus egress. J Infect Dis doi: 10.1093/infdis/jiu838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Votteler J, Sundquist WI. 2013. Virus budding and the ESCRT pathway. Cell Host Microbe 14:232–241. doi: 10.1016/j.chom.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dores MR, Chen B, Lin H, Soh UJ, Paing MM, Montagne WA, Meerloo T, Trejo J. 2012. ALIX binds a YPX(3)L motif of the GPCR PAR1 and mediates ubiquitin-independent ESCRT-III/MVB sorting. J Cell Biol 197:407–419. doi: 10.1083/jcb.201110031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pashkova N, Gakhar L, Winistorfer SC, Sunshine AB, Rich M, Dunham MJ, Yu L, Piper RC. 2013. The yeast Alix homolog Bro1 functions as a ubiquitin receptor for protein sorting into multivesicular endosomes. Dev Cell 25:520–533. doi: 10.1016/j.devcel.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dowlatshahi DP, Sandrin V, Vivona S, Shaler TA, Kaiser SE, Melandri F, Sundquist WI, Kopito RR. 2012. ALIX Is a Lys63-specific polyubiquitin binding protein that functions in retrovirus budding. Dev Cell 23:1247–1254. doi: 10.1016/j.devcel.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keren-Kaplan T, Attali I, Estrin M, Kuo LS, Farkash E, Jerabek-Willemsen M, Blutraich N, Artzi S, Peri A, Freed EO, Wolfson HJ, Prag G. 2013. Structure-based in silico identification of ubiquitin-binding domains provides insights into the ALIX-V:ubiquitin complex and retrovirus budding. EMBO J 32:538–551. doi: 10.1038/emboj.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimura Y, Koitabashi S, Kakizuka A, Fujita T. 2001. Initial process of polyglutamine aggregate formation in vivo. Genes Cells 6:887–897. doi: 10.1046/j.1365-2443.2001.00472.x. [DOI] [PubMed] [Google Scholar]
- 37.Vida TA, Emr SD. 1995. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol 128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A 98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vincent O, Rainbow L, Tilburn J, Arst HN Jr, Penalva MA. 2003. YPXL/I is a protein interaction motif recognized by aspergillus PalA and its human homologue, AIP1/Alix. Mol Cell Biol 23:1647–1655. doi: 10.1128/MCB.23.5.1647-1655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lamb TM, Xu W, Diamond A, Mitchell AP. 2001. Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J Biol Chem 276:1850–1856. doi: 10.1074/jbc.M008381200. [DOI] [PubMed] [Google Scholar]
- 41.Pires R, Hartlieb B, Signor L, Schoehn G, Lata S, Roessle M, Moriscot C, Popov S, Hinz A, Jamin M, Boyer V, Sadoul R, Forest E, Svergun DI, Gottlinger HG, Weissenhorn W. 2009. A crescent-shaped ALIX dimer targets ESCRT-III CHMP4 filaments. Structure 17:843–856. doi: 10.1016/j.str.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.