Abstract
Many population studies on invasive plant pathogens are undertaken without knowing the center of origin of the pathogen. Most leaf pathogens of Eucalyptus originate in Australia and consequently with indigenous populations available, and it is possible to study the pathways of invasion. Teratosphaeria suttonii is a commonly occurring leaf pathogen of Eucalyptus species, naturally distributed in tropical and subtropical regions of eastern Australia where it is regarded as a minor pathogen infecting older leaves; however, repeated infections, especially in exotic plantations, can result in severe defoliation and tree deaths. Nine polymorphic microsatellite markers were used to assess the genetic structure of 11 populations of T. suttonii of which four where from within its native range in eastern Australia and the remaining seven from exotic Eucalyptus plantations. Indigenous populations exhibited high allele and haplotype diversity, predominantly clonal reproduction, high population differentiation, and low gene flow. The diversity of the invasive populations varied widely, but in general, the younger the plantation industry in a country or region, the lower the diversity of T. suttonii. Historical gene flow was from Australia, and while self‐recruitment was dominant in all populations, there was evidence for contemporary gene flow, with South Africa being the most common source and Uruguay the most common sink population. This points distinctly to human activities underlying long‐distance spread of this pathogen, and it highlights lessons to be learned regarding quarantine.
Keywords: Forest biosecurity, gene flow, microsatellite markers, plantation forestry, Teratosphaeria suttonii
Introduction
Eucalyptus species, commercially propagated in plantations cover approximately 20 million ha, become the dominant source of hardwood fiber globally. The majority of these plantations are in the Southern Hemisphere and Asia with only 600 thousand ha planted within areas of endemism in Australia (Gavran 2013). Many pathogens have been inadvertently introduced from Australia into these exotic plantations, and in some cases, they have caused very serious problems (Wingfield et al. 2008). Over time, increasing numbers of pathogens have been introduced into these plantations of non‐native Eucalyptus spp., requiring considerable resources to be allocated for the selection and screening of tolerant hybrids and clones (Wingfield et al. 2008).
Of the Eucalyptus leaf pathogens, there are approximately 150 species of Mycosphaerellaceae and Teratospheriaceae associated with exotic eucalypts, with most of the devastating being members of the Teratospheriaceae having Kirramyces‐like spores, namely T. destructans, T. viscidus, T. eucalypti, T. pseudoeucalypti, and T. suttonii, all of which cause leaf blights, as opposed to the less damaging leaf spot diseases (Hunter et al. 2011). They can cause serious damage in both nurseries and plantations reducing the growth and form of trees. When environmental conditions are conducive for infection and where susceptible planting stock has been deployed, repeated infections can lead to the death of large numbers of trees. However, despite their actual and potential importance, little is known regarding the global movement of these pathogens.
Population genetics studies of pathogens can make it possible to determine the origin of an introduced pathogen (Robert et al. 2012; Ali et al. 2014). In the case of most Eucalyptus leaf pathogens, the origin is known to be Australia. Thus, by sampling across the natural range in Australia and then assessing the genetic structure of introduced populations, pathways of introduction can be determined (McDonald and Linde 2002; Dlugosch and Parker 2008). Given the occurrence of these pathogens in Eucalyptus plantations outside the native range of the trees, it is clear that there have been multiple introductions either directly from Australia or from intermediary countries. It is therefore also reasonable to expect the level of diversity of the introduced pathogen populations to reflect the age of plantation industries where the tree hosts are deployed commercially.
Teratosphaeria suttonii (=Phaeoseptoria epicoccoides, Phaeophloespora epicoccoides=Kirramyces epicoccoides) is a well‐known pathogen of Eucalyptus foliage (Hunter et al. 2011). The asexual state of the fungus is one of the most prevalent leaf pathogens of Eucalyptus spp. grown as non‐natives in plantations in the tropics and Southern Hemisphere (Heather 1965; Walker et al. 1992; Burgess et al. 2006; Carnegie 2007a). The only evidence for the existence of a sexual state for the fungus is based on collections made in Queensland in Australia (Crous 1998). T. suttonii is known to be indigenous to eastern Australia (Hansford 1957; Heather 1965; Walker et al. 1992) and possibly introduced into Western Australia (Jackson et al. 2008).
The fungus has undergone several name changes (Crous et al. 2009), and variation in ITS sequence data has led several authors to propose that T. suttonii represents a species complex. However, the application of a combination of multiple gene phylogeny and morphological assessment for collections from various parts of the world led to the recent conclusion that T. suttonii is a single but morphologically variable species (Taole et al. 2012).
In natural forests and temperate plantations, T. suttonii is considered to be a relatively unimportant pathogen infecting old or stressed leaves (Knipscheer et al. 1990). However, in the subtropics, this pathogen can cause significant damage both in Eucalyptus plantations in Australia (Andjic et al. 2010; Taole et al. 2012) and plantations of exotic Eucalyptus spp. in other parts of the world including South Africa (Nichol et al. 1992a), South America (Pérez et al. 2009), China (Burgess et al. 2006), Japan, Indonesia, Philippines, New Zealand (Crous et al. 1998; Old et al. 2003), Vietnam (Pegg et al. 2003), and Zambia (Chungu et al. 2010). In general, stress favors infection by the pathogen (Nichol et al. 1992b; Carnegie 2007b) and this is presumably why it occurs predominantly on mature rather than on than the juvenile leaves of host trees. However, when inoculum levels are high and conditions are conducive for infection, T suttonii can infect juvenile foliage causing repeated tip dieback. Infection by this fungus reduces the surface area of leaves available for photosynthesis, resulting in defoliation, reduced growth, and in some cases death of trees (Knipscheer et al. 1990; Carnegie 2007a).
In Australia, T suttonii appears regularly in plantation disease surveys, but it is also commonly observed in natural Eucalyptus stands where it has a very wide host range. The production of a sexual state is extremely rare, but abundant sporulation of the asexual morph in long spore tendrils ensures rapid multiplication of infective propagules under favorable conditions. Dissemination is predominantly by rain splash, and thus, dispersal over long distances can occur only through anthropogenic activities. Such activities include plantation forestry, and because T. suttonii has a wide host range, it can spread naturally from plantations to adjacent natural forests.
In this study, microsatellite markers developed for T. suttonii (Aksoy et al. 2013) were used to determine its population structure and genetic diversity within its natural range in Australia as well as for introduced populations. Specifically, the data were used to confirm the origin of this pathogen in Australia and to establish the most likely migration history for its global movement.
Materials and Methods
Sampling and isolations
A total of 200 conidial cultures (Table 1) of T. suttonii were obtained from lesions on Eucalyptus leaves collected in Australia from different parts of Queensland (QLD, four locations), New South Wales (NSW, nine locations), and Western Australia (WA, one location) (Table 1). Additionally, 206 conidial cultures (Table 1) of T. suttonii were obtained from lesions on Eucalyptus leaves collected from plantations in Uruguay (URY), the United States of America (USA), China (CHN), Indonesia (IDN), Vietnam (VTN), and a nursery in South Africa (ZAF). At each site, a single leaf was sampled from 30 to 40 trees. Variation in the number of isolates available per site is a reflection of the isolation success. Isolates were acquired from diseased Eucalyptus leaves with typical T. suttonii symptoms as previously described (Taole et al. 2012). All isolates used in this study have been deposited in the culture collection (CMW) of the Forestry and Agricultural Biotechnology Institute (FABI) housed at University of Pretoria.
Table 1.
Teratosphaeria suttonii isolates used in this study
| Origin | Coded | No. of isolates | Collector | Collection year |
|---|---|---|---|---|
| Australia; Davies Creek, north QLD | FNQ | 27 | MM Taole & K Taylor | 2010 |
| Australia; Koumala, central QLD | CQLD | 46 | MM Taole & K Taylor | 2010 |
| Australia, Miriam Vale, central QLD | CQLD | 8 | V Andjic | 2009 |
| Australia; Imbil, south QLD | SQLD | 47 | MM Taole & K Taylor | 2010 |
| Australia; Wedding Bells, NSW | NSW | 37 | AJ Carnegie | 2010 |
| Australia; Emu Creek, NSW | NSW | 3 | AJ Carnegie | 2010 |
| Australia; Kew, NSW | NSW | 6 | V Andjic | 2009 |
| Australia; Kimbel, NSW | NSW | 4 | V Andjic | 2009 |
| Australia; Neaves, NSW | NSW | 2 | V Andjic | 2009 |
| Australia; Tunglebung, NSW | NSW | 3 | MM Taole & K Taylor | 2010 |
| Australia; Morrow, NSW | NSW | 3 | MM Taole & K Taylor | 2010 |
| Australia; Myrtle Creek, NSW | NSW | 3 | AJ Carnegie | 2010 |
| Australia; Garas, NSW | NSW | 2 | MM Taole & K Taylor | 2010 |
| Australia; Manjimup, WA | WA | 11 | T Burgess | 2007 |
| Uruguay; La Negra | URY | 38 | MJ Wingfield | 2009 |
| Uruguay; Caldras | URY | 23 | MJ Wingfield | 2009 |
| United States of America; Florida | USA | 18 | MJ Wingfield | 2007 |
| China; Fujian Province | CHN | 32 | MJ Wingfield & TI Burgess | 2004 |
| Indonesia; Aek Nauli, Sumatra | IDN | 22 | PA Barber | 2004 |
| Vietnam; Dai Lai, Vinh Phuc Province | VTN | 17 | TI Burgess | 2006 |
| South Africa; Pretoria, Gauteng | ZAF | 36 | MM Taole | 2007 |
DNA extraction and amplification of microsatellite loci
DNA was extracted from all cultures as previously described (Taole et al. 2012). For each isolate, 9 SSR loci were amplified in SSR‐PCRs using primer pairs specifically designed for T. suttonii (Aksoy et al. 2013). The resulting PCR products were separated on an ABI Prism DNA sequencer (Applied Biosystems, Foster City, CA). Estimates of allele sizes for the DNA fragments were determined by comparison with the internal size standard GENSCAN LIZ 600 (Applied Biosystems) and with the software GENMAPPER version 3.0 (Applied Biosystems).
Microsatellite haplotypes
Alleles at each locus were provided with an alphabetical letter, and the letters representing the different alleles together constituted the multilocus haplotypes (MLHs) for each isolate. Identical haplotypes within a population were regarded as clones. Based on the lack of genetic differentiation among populations from the same region, isolates from multiple locations for NSW were pooled; likewise, the two collections from central QLD and the two collections from Uruguay were pooled. Statistical analyses were carried out on clone‐corrected populations coded as FNQ, C‐QLD, S‐QLD, NSW, WA, URY, USA, CHN. IDN, VTN, ZAF (Table 1). When clone corrected, there were only three MLHs from WA and this population was excluded from some analyses.
Microsatellite analysis
Population diversity
For each clone‐corrected population, the frequency of alleles at each locus was calculated (Table S1). Gene diversity (H) was determined using the program POPGENE version 1.31 (Yeh et al. 1999). Population‐specific allelic richness (Ar) was calculated using a generalized rarefaction approach as implemented in the program ADZE (Szpiech et al. 2008). The hypothesis of random mating within populations was tested using the Index of Association IA and rBarD assessed using the program MULTILOCUS version 1.3 (Agapow and Burt 2000). In association tests, if rBarD is significantly different from zero, alleles are associated and there is a significant deviation from random mating (i.e., predominantly clonal). AMOVA was performed in GeneAlEx 6.5 (Peakall and Smouse 2012) to examine the distribution of variation among all clone‐corrected populations (PhiPT) except WA. Population pairwise PhiPT (genetic diversity among populations) was also estimated in GeneAIEX.
Geneflow
The software package BAYESASS v 3.0 (Wilson and Rannala 2003) was used to obtain an estimate of the magnitude and direction of gene flow between pairs of populations. This program uses Bayesian statistics to estimate posterior probability distribution of the proportion of migrants from one population to another assuming genetic equilibrium. MCMC chains were run using 5,000,000 iterations and burn‐in period of 500,000.
Genotype clustering
Assignment of isolates was performed in STRUCTURE v. 2.3.4 (Pritchard et al. 2000) using the clone‐corrected MLHs from each population. STRUCTURE estimates the probability of the haplotypes being distributed into K number of clusters (K = 1–10). The probability of admixture model without prior population information was applied assuming correlated allele frequencies. Each model was simulated 15 times with a burn‐in period of 20,000 iterations and run length of 100,000 MCMC repeats. Prior parameters were set to a nonadmixture model of ancestry and independent allele frequencies. The optimal value for K was inferred using the L(K) (Pritchard et al., 2000) and ΔK method of Evanno et al. (2005) implemented in STRUCTURE HARVESTER (Earl and van Holdt 2011). The genetic distances among K structure clusters were computed by applying Saitou and Nei's neighbor‐joining method (Saitou and Nei 1987) to the matrix of allele‐frequency divergence among clusters. The tree was estimated using the program NEIGHBOR and the plot produced using DRAWTREE (Felsenstein 2005), both of which are part of the STRUCTURE platform.
Haplotype networks
In order to gain a visual representation of how the MLHs from the disparate populations are linked, haplotype networks were constructed after the aggregation of similar MLHs based on Nei's unbiased genetic distance. Two separate networks were constructed as follows: (1) Australia alone or (2) all Australian MLHs were combined and then compared with those from elsewhere in the world. Based on the resultant NJ tree, tips were collapsed and similar MLHs grouped for the purpose of haplotype network analysis (to reduce the complexity of the network). This resulted in 41 groups for Australia and 71 for complete analysis. Relationships between MLHs were inferred via a minimum‐spinning network calculated using NETWORK 4.6.1.2 (http://www.fluxus-engineering.com), and the resultant networks were manually edited.
Results
Population diversity
A total of 83 alleles were detected in the entire T. suttonii data set of 406 isolates across the nine microsatellite loci tested (Table S1). All loci were polymorphic with a range of 2–13 alleles. With the exception of WA that had only 1 polymorphic locus, all other populations had between 5 and 8 polymorphic loci. Among the introduced populations, IDN and CHN had the most alleles, followed by VTN, ZAF, and USA, while URY had the lowest number of alleles (Table 2). Of the 83 alleles, all except 4 were found in Australia. Overall, allelic richness was higher for the Australian populations (except WA). All Australian populations other than WA contained private alleles; the only introduced population having private alleles was IDN (Table 2). Again with the exception of WA, gene diversity (H) was universally high in Australia. For introduced populations, IDN was comparable to Australia while gene diversity of other populations was somewhat lower (Table 2). Locus TE1 returned many null alleles and was excluded from further analyses. Of the total genetic diversity, 64% was attributed to individual differences within populations and 36% was due to population differences between populations (Table S2). The population pairwise PhiPT values indicated that there were significant differences between all pairs of populations except URY and ZAF (Table S3).
Table 2.
Population diversity parameters for the populations of Teratosphaeria suttonii
| Parameters | FNQ | CQLD | SQLD | NSW | WA | AUS | URY | USA | ZAF | VTN | IDN | CHN |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ni | 27 | 54 | 47 | 61 | 11 | 200 | 61 | 18 | 36 | 19 | 32 | 40 |
| Nh | 15 | 39 | 44 | 43 | 3 | 144 | 5 | 13 | 16 | 13 | 32 | 28 |
| Na | 2.75 | 4.88 | 4.13 | 4.63 | 1.13 | 8.13 | 2.25 | 2.00 | 2.25 | 2.13 | 3.75 | 3.00 |
| Ne | 2.08 ± 0.34 | 2.73 ± 0.36 | 2.41 ± 0.38 | 3.22 ± 0.62 | 1.1 ± 0.10 | 4.21 ± 0.7 | 1.86 ± 0.32 | 1.63 ± 0.2 | 1.52 ± 0.18 | 1.68 ± 0.16 | 2.32 ± 0.28 | 1.65 ± 0.23 |
| Nua | 2 | 6 | 4 | 2 | 1 | 28 | 0 | 0 | 0 | 0 | 2 | 0 |
| %P | 87.5 | 87.5 | 100 | 87.5 | 12.5 | 100 | 62.5 | 75 | 75 | 100 | 100 | 87.5 |
| P | 7 | 7 | 8 | 7 | 1 | 8 | 5 | 5 | 6 | 8 | 8 | 7 |
| Ar | 2.35 ± 0.33 | 3.91 ± 0.54 | 3.34 ± 0.41 | 3.72 ± 0.60 | 1.10 ± 0.10 | 6.82 ± 0.88 | 1.72 ± 0.23 | 1.79 ± 0.25 | 1.88 ± 0.31 | 1.89 ± 0.12 | 2.48 ± 0.62 | 2.36 ± 0.29 |
| Pa | 0.25 ± 0.16 | 0.75 ± 0.41 | 0.50 ± 0.27 | 0.25 ± 0.16 | 0.13 ± 0.13 | 3.5 ± 0.96 | Absent | Absent | Absent | Absent | 0.25 ± 0.16 | Absent |
| H | 0.44 ± 0.08 | 0.56 ± 0.09 | 0.49 ± 0.09 | 0.59 ± 0.09 | 0.06 ± 0.56 | 0.67 ± 0.09 | 0.34 ± 0.11 | 0.32 ± 0.08 | 0.27 ± 0.08 | 0.36 ± 0.06 | 0.52 ± 0.07 | 0.32 ± 0.08 |
| rBarD | 0.138 | 0.234 | 0.025 | 0.087 | na | 0.079 | 0.289 | 0.257 | −0.032 | −0.014 | 0.05 | 0.095 |
| P‐value for rBarD | <0.001 | <0.001 | 0.022 | <0.001 | na | <0.001 | 0.043 | <0.001 | 0.848 | 0.692 | <0.001 | 0.002 |
Ni: total number of isolates (nonclone corrected); Nh; number of haplotypes; Na: mean number of alleles; Ne: mean number of effective alleles ± standard error; Nua: number of unique alleles; %P: Percentage polymorphic loci; P: number of polymorphic loci (eight loci considered); Ar: allelic richness; mean ± standard error; Pa: private alleles; mean ± standard error; H: Gene diversity (Nei 1973).
The index of association (rBarD) was significantly different from zero for all populations except VTN and ZAF. The hypothesis of random mating could thus be rejected for these populations (Table 2). The association of alleles indicated predominantly asexual reproduction.
Gene Flow
Based on assessment of contemporary gene flow in BAYESASS, self‐recruitment dominated with proportions of over 0.9 for most populations (Table S4, red diagonal). In Australia, gene flow was observed from all eastern Australian populations toward Western Australia (Table S5). Within eastern Australia, there was limited evidence for gene flow between regions, with the exception of FNQ that appears to have received migrants from other eastern Australian populations (Table S5). The establishment of a new plantation industry is associated with the import of germplasm either as seeds, seedlings, or tissue culture, and it is during this phase that new pathogens can be introduced. We thus disallowed scenarios that would include migration from a younger to an older population. There was evidence of limited contemporary gene flow from native Australian populations (especially CQLD and NSW) to URY, USA, and VTN. Among exotic plantations, there appeared to be three‐way gene flow between USA, VTN, and URY populations (Table S4, Fig. 1). IDN and CHN were the source of gene flow to URY, VTN, and USA, while ZAF was a source of contemporary gene flow to all other introduced populations, in particular URY (Table S4, Fig. 1).
Figure 1.
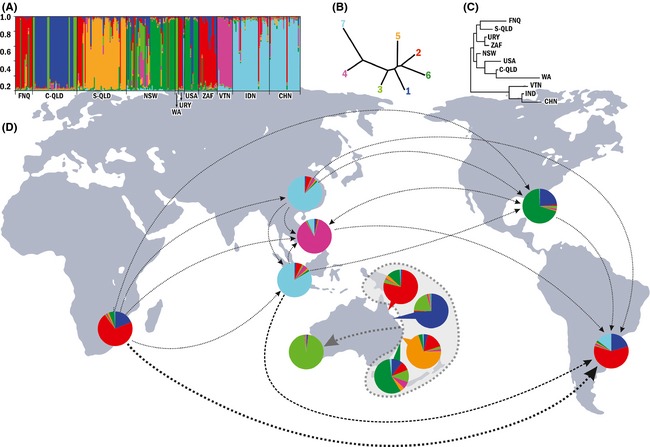
Assignment of and gene flow between populations of Teratosphaeria suttonii. (A) Genetic clustering (k = 7) of the 11 populations using eight microsatellite markers. Each bar represents an individual MLH divided populations and the colors code for the proportion (out of 1) of membership to each cluster. (B) Neighbor‐joining phenogram based on Nei's genetic distance showing the relationship between the seven clusters. (C) Neighbor‐joining phenogram based on Nei's genetic distance showing the relationship between the 11 populations. (D) Pie charts placed depicting the proportion of membership of each population to each cluster with lines depicting the relative intensity of gene flow between introduced populations (see Table S4) and arrows indicating the direction.
Clustering and genetic distance
Structure analysis for all MLHs showed that Ln P(X/K) produced a peak at K = 7 and this was deemed the optimal number of clusters to explain the genetic structure in the data set (Figs S1 and S2). Within the native range of T. suttonii in eastern Australia, clusters predominantly corresponded to the geographic origin of the isolates, with most MLHs in each population residing in the same structure group (Table S6, Fig. 1); most MLHs from FNQ resided in cluster 2 (red), C‐QLD in clusters 1 (blue) and 3 (lime green), S‐QLD in cluster 5 (orange), and NSW in cluster 6 (forest green). MLHs from the introduced population in Western Australia were in cluster 3 (lime green). For the introduced populations, clusters 1, 2, and 7 were represented in URY population, clusters 1 and 6 in USA population, and clusters 1 and 2 in ZAF population. Clusters 4 (pink) and 7 (aqua) were poorly represented among Australian populations, but dominated in Asia.
The NJ phenogram indicated that clusters 4 and 7 were related to each other, but more distant to the other clusters (Fig. 1B). Populations from Asia (VTN, IDN, and CHN) were more closely related to each other than to other populations (Fig. 1C).
Haplotype networks
Within Australia, haplotypes were for the most part clustered with geographic location (Fig. 2). The haplotypes from the introduced population in WA were found in the network linked with isolates from C‐QLD. MLHs from the NSW populations were scattered throughout the network (Fig. S3). The network of global population MLHs clearly depicted the representation and domination of Australian MLHs throughout. MLHs from CHN and IDN fell in the center of the network linked to Australian MLHs on both sides. MLHs from USA and VTN fell at the end of branches. MLHs from URY clustered in the network with some MLHs from ZAF.
Figure 2.
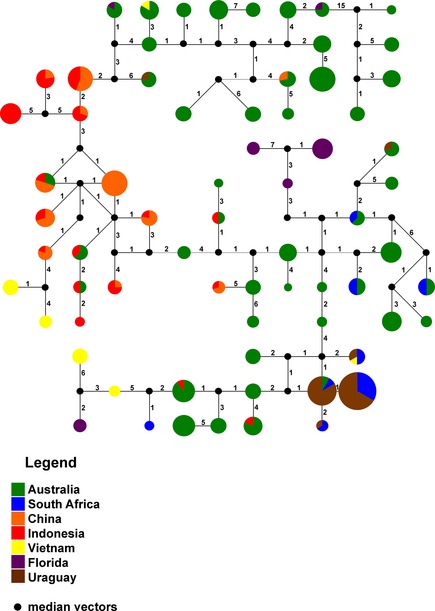
A network showing relationships between the global populations of Teratosphaeria suttonii. Each circle represents cluster of related haplotypes, the size of the circle corresponds to the number of MLHs in each cluster; the colors correspond to geographic regions as given in the legend. The numbers on the branches represent the number of mutations.
Discussion
Indigenous populations of the Eucalyptus leaf pathogen T. suttonii were characterized by high allele and haplotype diversity, low gene flow, and geographic structure. Most introduced populations also exhibited high levels of diversity. While sampling between indigenous and introduced populations was balanced with approximately 200 isolates from each, 95% of the 83 alleles obtained from eight microsatellite loci were recovered from indigenous Australia populations, with 34% of the alleles being recovered only from Australia. The allele recovery and relative proportions of these alleles differed greatly within and between the introduced populations resulting in significant population differentiation.
Teratosphaeria suttonii in Australia
Due to its widespread occurrence, wide host range among indigenous Eucalyptus spp., and low impact, T. suttonii is assumed to be indigenous to eastern Australia. This study provided experimental evidence to support this assumption. Accordingly, microsatellite data displayed strong geographic isolation between populations and a different structure cluster dominated in each region sampled, indicating independent evolution of populations sustained by restricted gene flow between the populations (McDonald and Linde 2002). This is the expected scenario for fungi within their natural range as has, for example, been shown for the pine‐infecting fungus Diplodia scrobiculata populations in North America (Burgess et al. 2004) and the banana fungal pathogen Mycosphaerella fijiensis (Robert et al. 2012). Based on structure and distance analyses, some admixture was, however, observed. This most probably indicates that the pathogen in different localities was initially introduced from the same source. This could have been as a consequence of a common source of seedlings for plantation development in subtropical Australia in the first decade of the 21st century (Andjic et al. 2010; Whyte et al. 2011).
Sampling in all indigenous populations was at the level of one isolate per tree, and yet, the number of MLHs was smaller than the total number of isolates, indicating the presence of clones in these populations. Because T. suttonii sporulates profusely on infected leaves releasing asexual spores into the environment, it is not surprising that we observed association of alleles in all indigenous populations. This supports the currently accepted dogma of predominantly asexual reproduction for this species.
When T. suttonii was first discovered in Western Australia, it was proposed that this was due to a recent introduction (Jackson et al. 2008). This hypothesis was proven to be correct in the present study with only three genetically similar MLHs being recovered from WA, and these were almost identical to haplotypes recovered from central Queensland. While WA is separated from the eastern Australia by a 6000 km of desert, a very effective natural geographic barrier (Bowler 1976), this barrier has clearly been breached. In early 2000s, there was a rapid expansion of the Eucalyptus globulus plantation industry in WA, and all seedlings could not be grown locally, so many were sourced from nurseries in eastern Australia. While, nursery material was apparently disease free, the use of fungicides could have masked asymptomatic infections. Thus, during this phase of rapid plantation expansion, many new Teratospheriaceae and Mycosphaerellaceae were unwittingly introduced to WA (Jackson et al. 2008) and this is clearly also true for T. suttonii.
Infection routes of Teratosphaeria suttonii in exotic plantations
The diversity of T. suttonii recovered from most exotic Eucalyptus plantations was moderate to high, although with the exception of Indonesia, it was lower than that of indigenous Australian populations. However, even for exotic populations with high diversity, there were few unique alleles. Population differentiation was significantly different between most pairwise comparisons of populations, and again, this was reflected in high self‐recruitment and low gene flow between populations. Population differentiation was not significant between URY and USA and URY with ZAF (but not USA and ZAF). Thus, for these populations, we would expect high gene flow and there was evidence for both flow between USA and URY and from ZAF to URY. However, populations of isolates from URY and USA were assigned to different structure clusters. This is possible because while they had the same alleles they were in different relative proportions.
The invasion history of Phytophthora ramorum in California was recently reconstructed by combining field epidemiology data (i.e., known age of infestation) with genetic microsatellite data (Croucher et al. 2013). In their study, unidirectional minimal infection routes were determined by disallowing models that included migration from younger to older populations. Following this approach, we have considered knowledge of quarantine regulations, age of plantation industry, and the first reports of T. suttonii in different regions to interpret gene flow between populations. Eucalyptus spp. were planted worldwide soon after colonization of Australia (200 ya) as ornamentals, for firewood, and charcoal, and they have naturalized in many places. Because T. suttonii has a very broad host range and early plant movement occurred prior to an understanding of the importance of biosecurity, it is likely that this species was an early and common hitchhiker and indeed T. suttonii occurs virtually wherever Eucalyptus spp. are planted as non‐natives (Taole et al. 2012). Consequently, historical gene flow would have been out of Australia.
Contemporary gene flow provides a different scenario; in the present era of known biosecurity threats in forestry (Liebhold et al. 2012; Wingfield et al. 2013; Eschen et al. 2014) and with numerous serious outbreaks due to introduced pathogens (Britton and Liebhold 2013), many countries are highly vigilant regarding quarantine procedures. Of these countries, Australia has some of the most rigorous import restrictions on many plant products including Eucalyptus germplasm while many of the other countries have imported germplasm during the establishment of eucalypt plantation industry. Thus, for determining the most likely migration scenarios (minimum infection routes), we interpret the data from the BAYESASS analysis under the following provisos; firstly, contemporary gene flow to Australia is excluded, and secondly, gene flow is only considered from older to newer populations. Effectively, the age of the populations, based on the first reports in the countries examined, equates to the age of the plantation industry; with the oldest industries in South Africa and Indonesia and the newest in Uruguay. Unsurprisingly, Uruguay seems to have acquired T. suttonii from a variety of different sources, and of all the exotic populations tested, South Africa is the most common source population.
In all population and haplotype clustering, T. suttonii MLHs obtained from Asia, in particular China and Indonesia, were more closely related to each other than to the other populations considered. The implication here is that there has been a substantial exchange of seeds and vegetative material among these countries, in particular among plantation forestry companies that own land in several different countries and as suggested by Wingfield et al. (2008). This has been clearly demonstrated for the related Eucalyptus pathogens T. destructans (Andjic et al. 2011) and T. nubilosa (Hunter et al. 2008; Pérez et al. 2012).
Fate of introduced populations
Geographically isolated exotic populations of T. suttonii that are genetically undifferentiated suggest the introduction was recent (Dlugosch and Parker 2008). Plantation forestry in Western Australia has expanded from a few thousand hectare in the mid 1980s to over 300,000 ha today. It is a young industry, and the plantations underwent regular tree health surveys. T. suttonii was introduced into the region in early 2000s (Jackson et al. 2008), as is supported here by very low diversity. Similarly, the plantation industry in Uruguay is relatively young and gene and haplotype diversity is low (from 61 isolates, five MLHs). In both these cases, there is evidence for a small founder populations and the observed diversity matches the scenario depicted in Figure S4a. Similar low diversity has also been observed for known introduced populations of other Teratosphaeria species (Hunter et al. 2008; Andjic et al. 2011; Pérez et al. 2012).
The relatively high levels of genetic diversity observed for the other introduced populations could be a consequence of multiple introductions of the pathogen from numerous sources within Australia or from other exotic plantations where the pathogen has become established (Fig. S4b,c). Assortment of alleles as a result of selection, genetic drift, and bottlenecks among these populations could have resulted in the loss of some alleles and or increased frequency of other alleles leading to apparent differentiation among populations. The high diversity observed in the populations from South Africa and Vietnam could also be a consequence of recombination, as the hypothesis of random mating could not be rejected for these populations (Fig. S4f). However, the observed diversity could also be a consequence repeated introductions for diverse sources. In well‐established fungal populations, in the absence of sexual reproduction, apparent diversity can also be increased by the stepwise accumulation of mutations at microsatellite loci, resulting in new haplotypes that are only slightly different to the original haplotype (Fig. S4d). However, there was very limited evidence for mutation in the studied populations with only a small number of unique alleles observed in one of the introduced populations (Indonesia).
The Asian populations cluster together in two main structure groups and in the haplotype network. Of the 83 alleles recognized in this study, only four are absent from Australia and these are all found in Asia. However, while structure clusters 4 and 7 were rare in Australia, some isolates from NSW did align with these groups and in the haplotype analysis there were Australian MLH intermixed with the Asian MLH. There are two complementary explanations. Firstly, there were regions in Australia that were not sampled (southern NSW, for example) and this could have been the source of the founder population in Asia. Secondly, mutation and subsequent drift in Asian populations have resulted in the current population structure.
Although Eucalyptus spp. have been planted worldwide as ornamentals for over 100 years, the plantation industries in the countries represented in this study have all experienced different periods of rapid expansion. Of these countries, South Africa has the longest record of intensive Eucalyptus planting dating back to the early 1900s, expanded most rapidly in the 1970s with the development of a paper/pulp industry. The other countries, or areas in the case of WA, have developed more recently commencing in 1990s, linked strongly to a growing worldwide demand for wood fiber. Within Asia, rapid plantation developement commenced first in Sumatra, Indonesia and a little later in Vietnam and China. During this period of rapid expansion, germplasm in the form of seeds and seedlings would have been imported from Australia as well as from other countries with well‐established Eucalyptus breeding and improvement programs (Liebhold et al. 2012; Wingfield et al. 2013). We believe the introduction of new genotypes of T. suttonii would have occurred during this phase.
Conclusion
Teratosphaeria suttonii is indigenous to eastern Australia where populations are characterized by asexual reproduction and high levels of genetic diversity and differentiation, that is, attributed to geographic location. Diversity in introduced populations differs depending upon age of plantation industry, with younger industries in Western Australia and Uruguay exhibiting low diversity. Because most alleles observed in introduced populations are present in Australia, mutation rates in new populations appear to be quite low. However, assortment of alleles as a result of selective forces (and perhaps sexual recombination) has resulted in new MLHs and population differentiation. While the original source of invasions was Australia, the data emerging from this study clearly show the role of contemporary gene flow from intermediary countries. Importantly, it emphasizes the need for substantial vigilance when moving Eucalyptus germplasm between regions and countries.
Conflict of Interest
None declared.
Supporting information
Figure S1. Optimum number of clusters (K = 7) established from LnK and DeltaK values obtained from STRUCTURE analysis.
Figure S2. Comparison of genetic clustering among the 11 populations at k = 2, k = 5, k = 7 and k = 10
Figure S3. Haplotype network for Australian MLHs of Teratosphaeria suttonii.
Figure S4 Some of the assumed possible models through which alleles and genotypes can be introduced into or acquired by pathogen populations over time.
Table S1. Distribution of alleles in clone corrected populations of Teratosphaeria suttonii. Australian populations are given first by region and then combined (AUS).
Table S2. Analysis of molecular variance (AMOVA) for 10 populations (WA excluded). P‐value estimates are based on 999 permutations; df = degrees of freedom, and MS = mean squared deviations.
Table S3. Pairwise comparison of population differentiation (PhiPT) among clone corrected populations of Teratosphaeria suttonii (lower diagonal) with probability values based on 999 permutations (upper diagonal).
Table S4. Bayesian assessment of migration within and among sampling locations of Teratosphaeria suttoniae.
Table S5. Bayesian assessment of migration within and among sampling locations of Teratosphaeria suttoniae in Australia.
Table S6. Assignment of Teratospheria suttonii populations into the seven clusters recognised in STRUCTURE. Numbers in brackets indicate the number of MLH in each population assigned to each cluster.
Acknowledgments
We are grateful to forestry companies colleagues in many parts of the world and too numerous to mention individually for providing us with the opportunity to collect isolates of T. suttonii without which this study could not have been undertaken. Funding for this study was provided by members of the Tree Protection Co‐operative Programme (TPCP) and the THRIP initiative of the Department of Trade and Industry, South Africa, and the CRC Forestry through Murdoch University in Australia.
Ecology and Evolution 2015; 5(18): 4210–4220
References
- Agapow, P.‐M. , and Burt A.. 2000. Multilocus 1.2: Department of Biology, Imperial College, Silwood Park, Ascot, Berks, SL5 7PY.
- Aksoy, S. , Almeida‐Val V. M. F., Azevedo V. C. R., Baucom R., Bazaga P., Beheregaray L. B.., … and Paula‐Silva M. N.. 2013. Permanent genetic resources added to molecular ecology resources database 1 October 2012–30 November 2012. Mol. Ecol. Resour. 13:341–343. [DOI] [PubMed] [Google Scholar]
- Ali, S. , Gladieux P., Leconte M., Gautier A., Justesen A. F., Hovmøller M. S., et al. 2014. Origin, migration routes and worldwide population genetic structure of the wheat yellow rust pathogen Puccinia striiformis f. sp. tritici. PLoS Pathog. 10:e1003903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andjic, V. , Pegg G. S., Carnegie A. J., Callister A., Hardy G. E. St. J., and Burgess T. I.. 2010. Phylogeographic study reveals new cryptic species Teratosphaeria pseudoeucalypti responsible for leaf blight of Eucalyptus in subtropical and tropical Australia. Plant. Pathol. 59:900–912. [Google Scholar]
- Andjic, V. , Dell B., Barber P. A., Hardy G. E. St. J., Wingfield M. J., and Burgess T. I.. 2011. Plants for Planting; evidence for the movement of serious forest pathogen, Teratosphaeria destructans on infected germplasm. Eur. J. Plant Pathol. 131:49–58. [Google Scholar]
- Bowler, J. M. 1976. Aridity in Australia: age, origins and expression in aeolian landforms and sediments. Earth Sci. Rev. 12:279–310. [Google Scholar]
- Britton, K. O. , and Liebhold A. M.. 2013. One world, many pathogens!. New Phytol. 197:9–10. [DOI] [PubMed] [Google Scholar]
- Burgess, T. I. , Gordon T. R., Wingfield M. J., and Wingfield B. D.. 2004. Geographic isolation of Diplodia scrobiculata and its association with native Pinus radiata . Mycol. Res. 108:1399–1406. [DOI] [PubMed] [Google Scholar]
- Burgess, T. I. , Andjic V., Hardy G. E. S. J., Dell B., and Xu D.. 2006. First report of Phaeophleospora destructans in China. J. Trop. Forest Sci. 18:144–146. [Google Scholar]
- Carnegie, A. J. 2007a. Forest health condition in New South Wales, Australia, 1996–2005. I. Fungi recorded from eucalypt plantations during forest health surveys. Australas. Plant Pathol. 36:213–224. [Google Scholar]
- Carnegie, A. J. 2007b. Forest health condition in New South Wales, Australia, 1996–2005. II. Fungal damage recorded in eucalypt plantations during forest health surveys and their management. Australas. Plant Pathol. 36:225–239. [Google Scholar]
- Chungu, D. , Muimba‐Kankolongo A., Wingfield M. J., and Roux J.. 2010. Identification of fungal pathogens occurring in eucalypt and pine plantations in Zambia by comparing DNA sequences. Forestry 83:507–515. [Google Scholar]
- Croucher, P. J. , Mascheretti S., and Garbelotto M.. 2013. Combining field epidemiological information and genetic data to comprehensively reconstruct the invasion history and the microevolution of the sudden oak death agent Phytophthora ramorum (Stramenopila: Oomycetes) in California. Biol. Invasions, 15:2281–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous, P. W. 1998. Mycosphaerella spp. and their anamorphs associated with leaf spot diseases of Eucalyptus. APS Press, St Paul, MN. [Google Scholar]
- Crous, P. W. , Wingfield M. J., Mohammed C., and Yuan Z. Q.. 1998. New foliar pathogens of Eucalyptus From Australia and Indonesia. Mycol. Res. 102:527–532. [Google Scholar]
- Crous, P. W. , Summerell B. A., Carnegie A. J., Wingfield M. J., Hunter G. C., Burgess T. I.. 2009. Unravelling Mycosphaerella: do you believe in genera? Persoonia 23:99–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlugosch, K. M. , and Parker I. M.. 2008. Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol. Ecol. 17:431–449. [DOI] [PubMed] [Google Scholar]
- Earl, D. A. , and van Holdt B. M.. 2011. Structure harvester: a website and program for visualizing structure output and implementing the Evanno method. Conserv. Genet. Resour. 4:359–361. [Google Scholar]
- Eschen, R. , Holmes T., Smith D., Roques A., Santini A., and Kenis M.. 2014. Likelihood of establishment of tree pests and diseases based on their worldwide occurrence as determined by hierarchical cluster analysis. For. Ecol. Manage. 315:103–111. [Google Scholar]
- Evanno, G. , Regnaut S., and Goudet J.. 2005. Detecting the number of clusters of individuals using the software STRUCTURE : a simulation study. Mol. Ecol. 14:2611–2620. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. 2005. PHYLIP (Phylogeny Inference Package) version 3.6. Department of Genome Sciences, University of Washington, Seattle. Distributed by the author. [Google Scholar]
- Gavran, M. 2013. Australian plantation statistics 2013 update: ABARES technical report 13.3, Canberra.
- Hansford, C. G. 1957. Australian fungi. IV. New records and revisions (continued). Proc. Linnean Soc. NSW 82:209–229. [Google Scholar]
- Heather, W. A. 1965. Some aspects of the ecology and pathology of Phaeoseptoria eucalypti (Hansf.) Walker, on some species of the genus Eucalyptus. Australian National University.
- Hunter, G. C. , van der Merwe N. A., Burgess T. I., Carnegie A. J., Wingfield B. D., Crous P. W., et al. 2008. Global movement and population biology of Mycosphaerella nubilosa infecting leaves of cold‐tolerant Eucalyptus globulus and E. nitens . Plant. Pathol. 57:235–242. [Google Scholar]
- Hunter, G. C. , Crous P. W., Carnegie A. J., Burgess T. I., and Wingfield M. J.. 2011. Mycosphaerella and Teratosphaeria diseases of Eucalyptus: Easily confused and with serious consequences. Fungal Divers. 50:145–166. [Google Scholar]
- Jackson, S. L. , Maxwell A., Burgess T. I., Dell B., and Hardy G. E. S.. 2008. Incidence and new records of Mycosphaerella species within a Eucalyptus globulus plantation in Western Australia. For. Ecol. Manage. 255:3931–3937. [Google Scholar]
- Knipscheer, N. S. , Wingfield M. J., and Swart W. J.. 1990. Phaeoseptoria leaf spot of Eucalyptus in South Africa. S Afr. For. J., 154:56–59. [Google Scholar]
- Liebhold, A. M. , Brockerhoff E. G., Garrett L. J., Parke J. L., and Britton K. O.. 2012. Live plant imports: the major pathway for forest insect and pathogen invasions of the US. Front. Ecol. Environ., 10:135–143. [Google Scholar]
- McDonald, B. A. , and Linde C.. 2002. Pathogen population genetics, evolutionary potential and durable resistance. Annu. Rev. Phytopathol. 40:349–379. [DOI] [PubMed] [Google Scholar]
- Nichol, N. S. , Wingfield M. J., and Swart W. J.. 1992a. Differences in susceptibility of Eucalyptus species to Phaeoseptoria eucalypti . Eur. J. For. Pathol. 22:418–423. [Google Scholar]
- Nei, M. 1973. Analysis of gene diversity in subdivided populations. Proc. Natl Acad. Sci. USA 70:3321–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichol, N. S. , Wingfield M. J., and Swart W. J.. 1992b. The effect of site preparation and fertilization on the severity of Phaeoseptoria eucalypti on Eucalyptus species. Eur. J. For. Pathol. 22:424–431. [Google Scholar]
- Old, K. M. , Wingfield M. J., and Yuan Z. Q.. 2003. A manual of diseases of Eucalypts in South‐East Asia. Center for International Forestry Research, Bogor, Indonesia. [Google Scholar]
- Peakall, R. , and Smouse P. E.. 2012. GenAlEx 6.5: genetic analysis in Excel. population genetic software for teaching and research‐an update. Bioinformatics 28:2537–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg, G. S. , Brown B. N., and Ivory M.. 2003. Eucalypt diseases in hardwood plantations in Queensland. Hardwood Queensland Report no. 16. Forestry Research, Agency for Food and Fibre Sciences, DPI. 30 p.
- Pérez, C. A. , Wingfield M. J., Altier N. A., and Blanchette R. A.. 2009. Mycosphaerellaceae and Teratosphaeriaceae associated with Eucalyptus leaf diseases and stem cankers in Uruguay. Forest Pathol. 39:349–360. [Google Scholar]
- Pérez, G. , Slippers B., Wingfield M. J., Wingfield B. D., Carnegie A. J., and Burgess T. I.. 2012. Cryptic species, native populations and biological invasions by a eucalypt forest pathogen. Mol. Ecol. 21:4452–4471. [DOI] [PubMed] [Google Scholar]
- Pritchard, J. K. , Stephens M., and Donnelly P.. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert, S. , Ravigne V., Zapater M. F., Abadie C., and Carlier J.. 2012. Contrasting introduction scenarios among continents in the worldwide invasion of the banana fungal pathogen Mycosphaerella fijiensis . Mol. Ecol. 21:1098–1114. [DOI] [PubMed] [Google Scholar]
- Saitou, N. , and Nei M.. 1987. The neighbor‐joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425. [DOI] [PubMed] [Google Scholar]
- Sajid, A. , Gladieux P., Leconte M., et al. 2014. Origin, migration routes and worldwide population genetic structure of the wheat yellow rust pathogen Puccinia striiformis f. sp. tritici. PLoS Pathog. 10:e1003903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpiech, Z. A. , Jakobsson M., and Rosenberg N. A.. 2008. ADZE: a rarefaction approach for counting alleles private to combinations of populations. Bioinformatics 24:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taole, M. , Burgess T. I., Gryzenhout M., Wingfield M. J., and Wingfield B. D.. 2012. DNA sequence incongruence and inconsistent morphology obscure species boundaries in the Teratosphaeria suttonii species complex. Mycoscience 53:270–282. [Google Scholar]
- Walker, J. , Sutton B. C., and Pascoe I. G.. 1992. Phaeoseptoria eucalypti and similar fungi on Eucalyptus with description of Kirramyces new genus (Coelomycetes). Mycol. Res. 96:911–924. [Google Scholar]
- Whyte, G. , Howard K., Hardy G. E. S. J., and Burgess T. I.. 2011. Foliar pests and pathogens of Eucalyptus dunnii plantations in southern Queensland. Aust. For. 74:161–169. [Google Scholar]
- Wilson, G. A. , and Rannala B.. 2003. Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163:1177–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield, B. D. , Slippers B., Hurley B., Coutinho T. A., Wingfield B. D., and Roux J.. 2008. Eucalypt pests and diseases: growing threats to plantation productivity. South. For. 70:139–144, 70, 139‐144. [Google Scholar]
- Wingfield, M. J. , Roux J., Slippers B., Hurley B. P., Garnas J., Myburg A. A., and Wingfield B. D.. 2013. Established and new technologies reduce increasing pest and pathogen threats to Eucalypt plantations. For. Ecol. Manage. 301:35–42. [Google Scholar]
- Yeh, F. C. , Yang R.‐C., and Boyle T.. 1999. POPGENE version 1.31. Microsoft Windows based free ware for population genetic analysis. Alberta.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Optimum number of clusters (K = 7) established from LnK and DeltaK values obtained from STRUCTURE analysis.
Figure S2. Comparison of genetic clustering among the 11 populations at k = 2, k = 5, k = 7 and k = 10
Figure S3. Haplotype network for Australian MLHs of Teratosphaeria suttonii.
Figure S4 Some of the assumed possible models through which alleles and genotypes can be introduced into or acquired by pathogen populations over time.
Table S1. Distribution of alleles in clone corrected populations of Teratosphaeria suttonii. Australian populations are given first by region and then combined (AUS).
Table S2. Analysis of molecular variance (AMOVA) for 10 populations (WA excluded). P‐value estimates are based on 999 permutations; df = degrees of freedom, and MS = mean squared deviations.
Table S3. Pairwise comparison of population differentiation (PhiPT) among clone corrected populations of Teratosphaeria suttonii (lower diagonal) with probability values based on 999 permutations (upper diagonal).
Table S4. Bayesian assessment of migration within and among sampling locations of Teratosphaeria suttoniae.
Table S5. Bayesian assessment of migration within and among sampling locations of Teratosphaeria suttoniae in Australia.
Table S6. Assignment of Teratospheria suttonii populations into the seven clusters recognised in STRUCTURE. Numbers in brackets indicate the number of MLH in each population assigned to each cluster.


