Abstract
Local adaptation is often obvious when gene flow is impeded, such as observed at large spatial scales and across strong ecological contrasts. However, it becomes less certain at small scales such as between adjacent populations or across weak ecological contrasts, when gene flow is strong. While studies on genomic adaptation tend to focus on the former, less is known about the genomic targets of natural selection in the latter situation. In this study, we investigate genomic adaptation in populations of the three‐spined stickleback Gasterosteus aculeatus L. across a small‐scale ecological transition with salinities ranging from brackish to fresh. Adaptation to salinity has been repeatedly demonstrated in this species. A genome scan based on 87 microsatellite markers revealed only few signatures of selection, likely owing to the constraints that homogenizing gene flow puts on adaptive divergence. However, the detected loci appear repeatedly as targets of selection in similar studies of genomic adaptation in the three‐spined stickleback. We conclude that the signature of genomic selection in the face of strong gene flow is weak, yet detectable. We argue that the range of studies of genomic divergence should be extended to include more systems characterized by limited geographical and ecological isolation, which is often a realistic setting in nature.
Keywords: Adaptation, candidate gene, evolution, fish, genome scan, population genomics, selection
Introduction
Recent questions on the mechanisms of evolutionary biology revolve around the genomic architecture of species and the effects that processes such as selection, drift, mutation, and gene flow have on the genome. Changes in the environment can now be linked to the genetic signatures of these processes and hence promote the understanding of the genetic basis of ecological adaptation. This allows us to understand the various components of the mechanism of adaptation such as which genes are involved, how they are distributed in genome, and how often the same genes lead to certain adaptations both in the laboratory (Becks et al. 2012; see Barrick and Lenski 2013 for other examples) and in nature (Mitchell‐Olds and Schmitt 2006; Yang et al. 2011; Jones et al. 2012a; Orsini et al. 2012). Additionally, we want to understand which processes are essential for adaptation to succeed. One way to find genes that are important for the process of adaptation is to identify genetic signatures of adaptation in natural populations. These “genome scans” identify genes that are under selection across contrasting environments and might thus be involved in the transition from one ecological extreme to the other. When interpreted with care (de Villemereuil et al. 2014), this approach has shown to be a promising way to find links between genotype, phenotype, and fitness in natural populations (Storz 2005).
Genome scans have so far generated two major insights. First, selection may act on many parts of the genome (Nosil 2009; Hohenlohe et al. 2010; Feder et al. 2012; Jones et al. 2012a,b; Strasburg et al. 2012; Arnegard et al. 2014; Seehausen et al. 2014). Nosil et al. (2009) estimated that 5–10% of the genome is affected by natural selection. Based on a genomewide analysis of selection, Hohenlohe et al. (2012) showed that the genome is much more structured and dynamic than expected from theory. More studies have now contributed to these topics and show that the areas affected by natural selection are typically patchily distributed (“hotspots”) across the genome (Voight et al. 2006; Papa et al. 2008; Hohenlohe et al. 2010). Genomic inversions, deletions, repetitive elements, and other structural changes facilitate the rise of these islands of divergence, causing a physical barrier to crossing over and thus adding to linkage disequilibrium (Hohenlohe et al. 2010). On the other hand, most cases of adaptation in plants involve many genomic areas with a wide distribution across the genome rather than clustered hotspots (Strasburg et al. 2012).
A second common finding of genome scans is that organisms may show parallel as well as nonparallel genetic responses to environmental change. For example, Atlantic salmon (Salmo salar) populations that undergo a parallel change in environment show nonparallel adaptive divergence at the genomic level (Perrier et al. 2013). In cases of parallel phenotypic evolution in the three‐spined stickleback (Gasterosteus aculeatus L.), common (Hohenlohe et al. 2010; Shimada et al. 2011; Jones et al. 2012b) as well as unique (Mäkinen et al. 2008; Jones et al. 2012b; Roesti et al. 2012) genomic regions are targeted by selection. This has been confirmed in experiments with stick insects (Timema cristinae) (Soria‐Carrasco et al. 2014) and in genome scans of lake white fish (Coregonus sp.) (Bernatchez et al. 2010). In contrast, parallel phenotypic evolution in other species, such as the nine‐spined stickleback (Pungitius pungitius) (Shikano et al. 2010a) or the rough periwinkle Littorina saxatilis (Butlin et al. 2014), was mainly characterized by nonparallel genomic signatures of selection.
Adaptive divergence is thought to proceed as a balance between divergent selection and homogenizing gene flow (Levene 1953; Hagen 1967; Endler 1973; Bell 1982) and hence may reach various stages. This has long been recognized at the phenotypic level (Tregenza 2002; Moore et al. 2007; Hendry 2009; Schluter 2009). More recently, theoretical and empirical studies have improved our understanding of the genomic architecture at various stages of adaptation as well (Pinho and Hey 2010; Yeaman and Otto 2011; Yeaman and Whitlock 2011). Nevertheless, the focus of genome scans is often on systems where it is reasonable to assume that population divergence has a strong adaptive component. To do so, genome scans often target populations at large spatial scales and across strong ecological contrasts (Hohenlohe et al. 2010; Poncet et al. 2010; Stapley et al. 2010; Zulliger et al. 2013). While this has generated great insight into the genomic basis of adaptation and speciation, knowledge might be biased toward stages where gene flow may be largely impeded, and where adaptation is already largely achieved. Gene flow modifies the response to selection by modulating the distribution of the genes that underlie ecologically relevant traits (Slatkin 1987). The study of adaptation with gene flow, especially in study systems where genetic divergence is far from complete such as across a small‐scale ecological transitions or between highly connected populations, is therefore crucial to understand how populations diverge from different ecological optima (Hansen et al. 2002; Storz 2005; Nielsen et al. 2009; Coscia et al. 2011; DeFaveri et al. 2013; Vandamme et al. 2014).
The three‐spined stickleback represents an excellent model for the study of adaptive divergence, as phenotypic responses to several ecological changes are frequent and well documented (McPhail 1994; Foster et al. 1998; McKinnon and Rundle 2002; Boughman 2007). A diverse number of populations can be found that occupy various stages of divergence from panmixia to complete and irreversible reproductive isolation (Hendry et al. 2009). This provides an excellent framework to investigate the population divergence at the phenotypic and genomic level. The genome scans that have been applied to the three‐spined stickleback have contrasted marine–freshwater (Mäkinen et al. 2008; Hohenlohe et al. 2010; DeFaveri et al. 2011; Shimada et al. 2011; Jones et al. 2012a,b; DeFaveri and Merilä 2013), lake–stream (Deagle et al. 2012; Roesti et al. 2012), and benthic–limnetic (Olafsdottir and Snorrason 2009; Jones et al. 2012a; Lucek et al. 2014) population pairs, as well as populations from clean versus polluted water (Lind and Grahn 2011). A common finding is that several genes or gene regions are repeatedly selected across populations and locations, although population‐specific regions do appear as well (Hohenlohe et al. 2010; DeFaveri et al. 2011; Shimada et al. 2011; Jones et al. 2012b). Others have found that most regions under selection were highly specific to the location under study (Mäkinen et al. 2008; Deagle et al. 2012; Roesti et al. 2012).
In this study, we investigate genomic adaptation in three‐spined stickleback populations from the Belgian–Dutch lowlands. Populations in this area differ in various morphological traits, which are often correlated with salinity and distance to the coast (Heuts 1947; Raeymaekers et al. 2005, 2007, 2012; Van Dongen et al. 2009). At the same time, gene flow between these populations is moderate to strong due to high connectivity (Raeymaekers et al. 2014). This has the advantage that we can study adaptation with ongoing gene flow, which is often a realistic setting in nature. All sites are part of an interconnected landscape of canals and streams along an ecological transition (i.e., discrete habitats with salinities ranging between brackish and freshwater) at a much smaller geographical scale than other genome scan studies across salinity gradients in the three‐spined stickleback (Table 1).
Table 1.
Comparison of microsatellite‐based genome scan studies in three‐spined sticklebacks across freshwater–brackish/saltwater gradients, including spatial scale (from regional to global), percentage of conservative outliers, F ST, F ST at the Eda locus, and F ST at the ATP1A1 locus
In order to explore the adaptive changes across this transition, we screened two populations from each end of the transition for genomic signatures of selection. We hypothesized that (1) the outcome of small‐scale adaptation along ecological transitions may vary at the genomic level across populations and (2) adaptation at the genomic level may be influenced by gene flow. Populations were screened for 87 microsatellite markers, of which 41 are linked to genes with a range of ecologically relevant functions (Shikano et al. 2010b; Shimada et al. 2011). We expect that the relatively high gene flow among these populations might constrain local adaptation, despite obvious differences in phenotype across these extremes. We determine outlier loci and compare the results to previous studies in three‐spined sticklebacks across similar ecological contrasts.
Materials and Methods
Study area
Three‐spined sticklebacks (Gasterosteus aculeatus L.; Gasterosteidae) from the coastal lowlands (polder) of Belgium and the Netherlands (Fig. 1) reside in ponds, ditches, streams, estuaries, or polder creeks. They have an anadromous or landlocked life style (Heuts 1947; Wootton 1976; Raeymaekers et al. 2005, 2007). The polder and surrounding areas contain diked brackish and freshwater habitats of Holocene origin with varying levels of connectivity to adjacent estuaries and the open sea. Populations that live in close proximity to the sea (<10 km) reside in brackish water, of which the salinity is influenced by rainfall and water management. On a scale of less than 50 km further inland, salinity drops to freshwater levels (Raeymaekers et al. 2014). Lateral plate number, an important ecological trait, also decreases with distance to the coast, with population averages ranging between 5 and 20 (Heuts 1947; Raeymaekers et al. 2014). Populations bordering the North Sea and the Baltic are typically polymorphic for lateral plate number (Heuts 1947; Raeymaekers et al. 2014), so higher or lower population averages are rare. This range is therefore representative for the phenotypic extremes we can find in this part of the stickleback's distribution range.
Figure 1.
Map with the four sampling locations of three‐spined sticklebacks in northwestern Belgium and the southwestern Netherlands. River network and major towns are mapped.
Field sampling
Field sampling was conducted in spring 2009 in parallel with a multiyear study by Raeymaekers et al. (2014), describing the distribution of lateral plate number in the study area. Two brackish water creeks (L01 and L02) and two freshwater ditches (L12 and U01), representing the sites with the most extreme values for salinity, were selected (Table 2). Thirty fish from each population were collected with dip nets, immediately anaesthetized with MS222 (2 g·l−1) and frozen on dry ice. In the laboratory, sticklebacks were thawed on ice, measured (standard length (SL); ±0.1 cm), weighed (±0.01 g), photographed, and fin‐clipped for a DNA sample.
Table 2.
Characteristics of the four‐three‐spined stickleback populations used in this study. DTC: distance to coast; MPN: mean plate number; Eda L: frequency of Eda low‐plated allele; AR: allelic richness; H e: expected heterozygosity; F IS: inbreeding coefficient, with confidence interval (CI 95%)
| Code | Coordinates (N, E) | Habitat | DTC (km) | Salinity (psu) | MPN | EdaL | AR | H e | F IS (CI 95%) |
|---|---|---|---|---|---|---|---|---|---|
| L01 | 51°21′10.66, 3°26′01.83 | Creek | 3.94 | 2.04 | 17.4 | 0.44 | 9.47 | 0.69 | 0.032 (−0.017–0.033) |
| L02 | 51°21′56.33, 3°31′11.09 | Creek | 4.30 | 1.83 | 16.8 | 0.51 | 9.06 | 0.69 | 0.095 (0.048–0.096) |
| L12 | 51°10′29.03, 3°28′10.45 | Stream | 22.84 | 0.32 | 12.7 | 0.74 | 6.80 | 0.61 | 0.044 (−0.012–0.050) |
| U01 | 51°02′48.40, 3°33′01.60 | Ditch | 36.20 | 0.52 | 7.0 | 0.93 | 6.51 | 0.57 | 0.039 (−0.010–0.040) |
Morphological measurements
In line with the previous studies on phenotypic divergence between the stickleback populations of the Belgian–Dutch lowlands (Heuts 1947; Raeymaekers et al. 2005, 2007, 2012; Van Dongen et al. 2009), we investigated variation at six morphological traits. The left side of each fish was photographed from a standard angle, and a ruler was placed in each photograph for scaling. Dorsal spine length, pelvic spine length, and pelvic plate length were measured from pictures using the software tpsDig 1.37 (Rohlf 2002). A subsample of the fish was rinsed with water for 72 h, bleached for 4 h (1% KOH bleach solution), and stained with alizarin red (Taylor and Dyke 1985). Stained fish were used to determine the number of lateral plates on the right side of the fish. The right part of the gills was then dissected, and the number and length of the large gill rakers were quantified under a dissection scope.
Marker selection
A set of 110 microsatellite markers was selected, including a range of putatively neutral markers, to set a proper neutral F ST background. In our study area, there is a salinity cline, but various other factors may covary with this cline. Therefore, we included 41 markers that are known to be linked to functional genes in a range of ecologically relevant functions such as salinity, growth, and immunity (Shikano et al. 2010b; Shimada et al. 2011). Several of these genes have been found to be under selection in the three‐spined stickleback or nine‐spined stickleback of other salinity transitions (Hohenlohe et al. 2010; Shikano et al. 2010a; DeFaveri et al. 2011; Shimada et al. 2011; Jones et al. 2012a,b). This allows us to compare these systems to some extent with ours and pinpoint parallel changes across systems. The marker closely linked to the Eda gene (Stn380), a major effect gene underlying variation in plate number (Colosimo et al. 2005), was included as a reference gene that is often under selection in freshwater–saltwater comparisons (Raeymaekers et al. 2007; Mäkinen et al. 2008; DeFaveri et al. 2011; DeFaveri and Merilä 2013).
DNA extraction and genotyping
Genomic DNA was extracted from fin clips using a proteinase K digestion step and the Nucleospin 96 Tissue DNA Extraction kit (Macherey‐Nagel). Individuals were genotyped at 110 microsatellite loci that were arranged in 21 multiplexes of 4–8 markers at a time, with EST‐based markers and markers within or near genes with relevant functions included (Table S1). Loci were amplified with the Qiagen® Multiplex PCR Kit (Qiagen, Venlo, the Netherlands). The 10 μL PCR cocktail contained 1–100 ng genomic DNA, 2 pmol each of forward and reverse primers, 1× Qiagen Multiplex PCR master mix, 0.5× Q‐solution, and RNase‐free water. The reaction consisted of an initial activation step of 15 min at 95°C, followed by 30 cycles of 30 s at 94°C, 90 s at 53°C and 60 s at 72°C. A final elongation step of 5 min at 60°C was performed. Allele sizes were determined by means of an internal ET ROX 550 size standard (Amersham Biosciences, Uppsala, Sweden). Polymerase chain products were visualized using a MegaBace 1000 automated sequencer (Amersham Biosciences). Alleles were scored with the Fragment Profiler v1.2 software (Amersham Biosciences), using visual scoring and manual corrections. Marker Stn380, linked to the Eda gene, was scored separately to determine the frequency of the “low‐plated” allele in each population (Table 2).
Data analysis
Phenotypic differentiation
For each trait, analysis of variance (ANOVA) was performed to test for statistical differences between the four populations. For traits that depend on size, standard length was included as a covariate.
Genetic diversity and genetic differentiation
Genotypes were checked for scoring errors attributable to stutter products, large allele dropout, or null alleles, using micro‐checker v2.3 (van Oosterhout et al. 2004). Estimates of allelic richness, genetic diversity (H e, H o), and global and pairwise F ST, with a significance calculated with 1000 bootstraps over loci, were calculated using the GENETIX v4.05.02 software (Belkhir et al. 1996).
Genomic signatures of selection
We conducted global outlier tests to find outliers across all populations and used pairwise comparisons to check whether the outliers found by global tests were due to habitat differences. Loci that are under directional selection are expected to have lower intrapopulation variability and larger interpopulation variability than neutral loci. Loci under directional selection can thus be traced by patterns in heterozygosity, differences in F ST values, or a combination of the two. Evaluation of several outlier detection methods has shown that these methods differ in number of false positives and false negatives (Narum and Hess 2011). Four methods were therefore compared: LOSITAN (Antao et al. 2008), the outlier detection method implemented in Arlequin v3.5.2.3 using hierarchical clustering (Excoffier and Lischer 2010), BayeScan v2.01 (Foll and Gaggiotti 2008), and lnRH (Kauer et al. 2003). The first three methods were used to determine global outliers. Additionally, we did pairwise comparisons of all populations using LOSITAN and lnRH to detect specific signatures of selection in freshwater–brackish water, freshwater–freshwater, and brackish water–brackish water population pairs.
The four methods are based on different underlying assumptions. LOSITAN is based on an island model that uses a coalescent F ST‐outlier method based on the distribution of F ST as a function of the heterozygosity. We used the function that first establishes a neutral F ST baseline by removing putative markers under selection outside the 95% interval with 105 simulations. The infinite allele model was used with a 95% and 99% confidence interval. We ran 105 simulations as recommended by Antao et al. (2008). The outlier detection software implemented in Arlequin uses the same island model, but adds on the option for hierarchical clustering. In the presence of strong hierarchical population structure, it reduces false positives by a hierarchical analysis of genetic differentiation (Excoffier et al. 2009). We clustered populations according to two scenarios: in two groups (brackish water populations versus freshwater populations) and three groups (brackish water populations versus each freshwater population separately). The first scenario simulates a common descent of the two freshwater populations from the marine population, while the second one simulates a separate split of the two freshwater populations from the marine population. We used the standard settings of 20,000 simulations for each run and 100 demes per group. The method executed by BayeScan uses a logistic regression model which explains the observed pattern of diversity by dividing it in a locus‐ and a population‐specific component (Beaumont and Balding 2004). One benefit of this method is that it allows for different migration rates and different effect sizes and thus can be used for scenarios that deviate from the island model. We conducted 10 pilot runs of 5000 iterations, followed by an additional 150,000 iterations and a burn‐in of 50,000 iterations. Outliers were appointed based on 90%, 95%, and 99% posterior probabilities. Finally, the lnRH method is designed especially for microsatellite markers and determines the reduction in heterozygosity. This method is based on the assumption that microsatellites linked to a gene under selection will show reduced levels of diversity between two populations. After standardization of the lnRH estimates with a mean of zero and a standard deviation of one, we determined outliers at 95% and 99% levels. We subtracted outliers that were found in pairwise comparisons within the same habitat type, to limit the number of false positives.
All tests based on simulations were executed three times to test for robustness of the results. Only outliers that were detected at least twice with each method were scored as a putative outlier. We detected outlier loci that are under balancing and directional selection. However, interpreting loci under balancing selection is difficult, as there are still limitations for the identification of loci under balancing selection (Hansen et al. 2010; Narum and Hess 2011). Therefore, we only discuss the loci under directional or positive selection. Loci under balancing selection are provided in Supplementary Table S2.
Results
Phenotypic differentiation
Statistical analysis of variance (ANOVA) confirmed the presence of significant differences between the four populations for standard length, lateral plate number, pelvic plate and spine length, and large gill raker length (Fig. 2). In particular, the brackish water populations (L01 and L02) were larger in size than the freshwater populations (L12 and U01; F 3,155 = 75.6; P < 0.0001) and had more lateral plates (F 3,147 = 10.4; P < 0.0001), longer pelvic spines (especially population L01; F 3,93 = 5.51; P = 0.0016), longer pelvic plates (F 3,93 = 8.60; P < 0.0001), and larger gill rakers (F 3,93 = 19.8; P < 0.0001).
Figure 2.
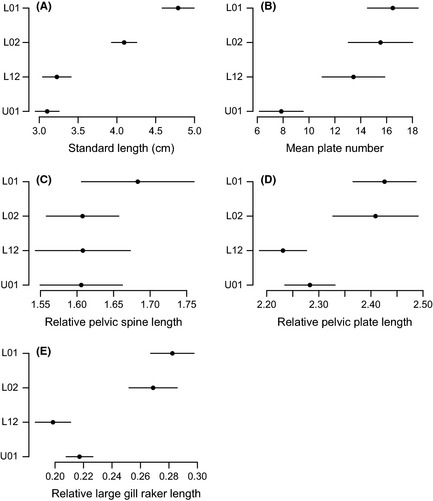
Average values for (A) standard length, (B) plate number, (C) relative pelvic spine length, (D) relative pelvic plate length, and (E) relative length of the second large gill raker in two brackish water populations (L01, L02) and two freshwater populations (L12 and U01) of the three‐spined stickleback. Horizontal bars represent 95% confidence intervals.
Genetic diversity and genetic differentiation
After genotyping of the individuals with 110 microsatellite markers, we selected 87 markers with good amplification quality to perform the data analysis. A total of 1116 alleles were observed in four populations of 26–28 individuals each, with an average of 13 alleles per locus and a range from 2 to 34. Observed heterozygosity ranged from 0.022 to 0.95 across loci. Expected heterozygosity ranged from 0.05 to 0.92 across loci and from 0.57 to 0.69 across populations (Table 2). Allelic richness among populations varied from 6.51 to 9.47, with the freshwater populations being less diverse than the brackish water populations. Eight loci were possibly affected by null alleles or stutters. We therefore performed all final analyses with and without these loci and specify when results differ. Differentiation among populations was moderate with the global F ST value being 0.059. Pairwise F ST values were significant, except between the two brackish water populations (Table 3).
Table 3.
Pairwise F ST values below the diagonal and associated P‐value above the diagonal. For site codes, see Table 2
| Code | L01 | L02 | L12 | U01 |
|---|---|---|---|---|
| L01 | – | 0.600 | <0.010 | <0.010 |
| L02 | 0.005 | – | <0.010 | <0.010 |
| L12 | 0.054 | 0.056 | – | <0.010 |
| U01 | 0.094 | 0.097 | 0.047 | – |
Genomic signatures of selection
Six of the 87 loci were assigned as outliers in at least one of the methods, with the different methods identifying 4, 2, 2, and 4 outliers for LOSITAN, BayeScan, lnRH, and Arlequin, respectively (Table 4). The methodologically similar tests performed by LOSITAN and Arlequin with hierarchical clustering resulted in the same set of outliers. In contrast, only two outliers were shared across methodologically different outlier detection methods; our conservative measure of outliers hence amounts to 2.3% of the total number of loci. This value is low in comparison with other genome scan studies of three‐spined sticklebacks (Table 1).
Table 4.
Global outlier loci that are putatively under directional selection detected in four populations of the three‐spined stickleback. The table includes the locus considered, its F ST value, and either the q value for BayeScan or the P‐value for lnRH, LOSITAN, and Arlequin
| Locus | F ST | BayeScan | lnRH | LOSITAN | Arlequin (2 groups) | Arlequin (3 groups) |
|---|---|---|---|---|---|---|
| ATP1A1 | 0.149 | * | * | * | ||
| HSPA14 | 0.159 | * | * | * | ||
| Ppgm44 | 0.120 | † | *** | |||
| Stn74 | 0.083 | * | ||||
| Stn177 | 0.189 | ** | ** | ** | ||
| Stn46 | 0.199 | †† | ** | ** | ** | |
| Stn380 (Eda) | 0.118 |
Significance is marked as †q < 0.1, and ††q < 0.01 or as *P < 0.05, **P < 0.01, and ***P < 0.001. Marker Stn380, linked to the Eda gene, was added as a reference.
The two outliers for directional selection that appear consistently across methodologically different tests are marker Stn46 (identified by Arlequin/LOSITAN and BayeScan) and marker Ppgm44 (identified by lnRH and BayeScan). Stn46 has been previously associated with the Rho guanine nucleotide exchange factor 9 (Table S1). It was also assigned as an outlier in one pairwise freshwater versus brackish water comparison. Ppgm44 is a marker that is linked to the gene myostatin2, which is associated with growth (Table S1). It was also assigned as an outlier in all four pairwise freshwater versus brackish water comparisons. Among the four other outliers that were detected (Table 4), one is associated with osmoregulation, namely an alpha subunit of the Na+/K+ ATPase (ATP1A1). Other outliers are either linked to functions such as thermal response (HSPA14) or were assumed to be neutral. Locus Stn34 was also assigned as an outlier, but as null alleles were detected at this locus, it was excluded from Table 4. The Eda gene, with frequencies of the low‐plated allele ranging between 0.44 and 0.51 in the brackish populations and 0.74 and 0.93 in the freshwater populations (Table 2), did not show up as an outlier in the analyses (Table 4).
Discussion
We found a limited number of loci showing a signature of selection among three‐spined stickleback populations inhabiting the coastal Dutch–Belgian lowlands. Six outliers were found when all outlier detection methods were considered and only two of those were shared among methodologically different tests, despite differentiation among populations in ecology and phenotype. Not only the number but also the proportion of outliers was low as compared to other studies (Table 1). The function of one of the outlier loci could be directly related to salinity, the most obvious ecological gradient in our study area. We here discuss possible explanations for the limited amount of outliers, the putative function of the genes linked to outliers, and the consequences for adaptive divergence in the face of gene flow.
The presence of a limited number and proportion of outliers might be attributed to a number of technical aspects. First, we used relatively few markers. However, among the 87 markers selected, 41 markers are linked to ecologically relevant functions, of which several have been shown to be under selection in systems with similar salinity clines (Mäkinen et al. 2008; Shimada et al. 2011; DeFaveri et al. 2013). Second, analyzing relatively few populations may have two drawbacks: (1) It might increase the risk of false negatives due to the limited number of individuals sampled, and (2) it might increase the difficulty in separating the signal of selection from the geographical and historical signal (Bierne et al. 2011, 2013). The small overlap in outlier loci among detection methods might be a symptom of this. However, in a larger study across 14 populations (2320 individuals) describing the distribution of lateral plate number and the underlying Eda gene in our study area (Raeymaekers et al. 2014), the signature of selection at the Eda gene was neither significant. This bi‐allelic gene is experiencing selection across various other salinity transitions in the three‐spined stickleback (Raeymaekers et al. 2007; Mäkinen et al. 2008; DeFaveri et al. 2011; DeFaveri and Merilä 2013). This suggests that increasing sample size does not necessarily enhance the detectability of selection. We therefore do not expect that technical issues are a major explanation for our findings.
An alternative explanation for the low proportion of outlier loci is that selection might be weak, owing to weaker environmental contrasts in our study area (i.e., brackish to freshwater) compared to other studies (Table 1). For instance, Raeymaekers et al. (2014) found that shifts in Eda allele frequencies from one generation to the next were associated with salinity, but that at the landscape level, salinity did not correlate with Eda allele frequencies. This suggests that selection is still acting, but might be too weak to contribute to local adaptation and leave a signature of selection at this gene. In addition, strong gene flow might confound the effect of selection by mixing adapted and nonadapted alleles in the respective populations. Gene flow being moderate to high, we expect this to be another explanation for why we find so few consistent outlier loci compared to other studies. Accordingly, Raeymaekers et al. (2014) found that the spatial distribution of the Eda allele frequency correlated with distance to the coast, a proxy for population connectivity. Recurrent contact between freshwater and estuarine or marine populations might lead to the exchange of maladaptive alleles, but at the same time, it has been argued that gene flow might cause an opposite effect: efficient flow of advantageous loci (Schluter and Conte 2009; Hohenlohe et al. 2012; Bell and Aguirre 2013; Raeymaekers et al. 2014), thus facilitating adaptation from standing variation. Whether gene flow in this case fuels or constrains adaptation is hard to say, but the signal of selection might be more difficult to pick up in genome scans due to mixing of the genomic background. We therefore expect that the low number of outlier loci is rather explained by either weak selection, confounding effects of gene flow, or a combination of both and are thus inherent to the system we study.
Genes that have been found in previous studies to be under selection relate to biological functions such as bone formation, osmoregulation, growth, thermal response, maturation, pigmentation, scent detection, spiggin production, and morphology (Hohenlohe et al. 2010; DeFaveri et al. 2011; Shimada et al. 2011; Jones et al. 2012b). Genes that appeared as outliers in this study include markers linked to osmoregulation (ATP1A1), thermal response (HSPA14), and growth (myostatin2). The marker linked to ATP1A1 has been identified as outlier gene in multiple stickleback saltwater–freshwater transitions worldwide (Jones et al. 2006, 2012a,b; McCairns and Bernatchez 2010; DeFaveri et al. 2011; Shimada et al. 2011). A major outlier in our study was marker Stn46, which is linked to the gene for Rho guanine nucleotide exchange factor 9. It is a member of the gene family coding for rho proteins, a subfamily of the guanine nucleotide exchange factors (GEFs), which are multidomain proteins involved in the activation of small GTPases (Rossman et al. 2005). The Rho family is involved in relaying signals from cell‐surface receptors to the actin cytoskeleton and elsewhere (Dvorsky and Ahmadian 2004). Its function can be associated with juvenile growth; in zebrafish, it has been specifically linked to angiogenesis (Garnaas et al. 2008) and striated muscle and neural development (Raeker et al. 2010). Interestingly, locus Stn46 was also under selection in populations of the nine‐spined stickleback (Shikano et al. 2010a), indicating that the gene might be involved in local adaptation in multiple species. Another major outlier in our study was the marker linked to myostatin2. This gene, a member of the transforming growth factor‐beta (TGF‐beta) family, is known to function as a negative regulator of skeletal muscle development and growth in mammals (Walsh and Celeste 2005) and teleost fish (Radaelli et al. 2003). Blocking the expression of myostatin in zebrafish has led to the development of a giant phenotype (Acosta et al. 2005), but myostatin is produced in many other tissues than skeletal muscles and is expected to influence many more functions (Radaelli et al. 2003). Understanding why these specific genes are selected requires further study due to the broad range of functions these genes might have.
The low number of outlier loci that we find contrasts with the differentiation across populations in several morphological traits. Local differentiation in the number of lateral plates, for instance, has been shown to significantly exceed the level of neutral differentiation in our study area (Raeymaekers et al. 2014). Plate number and other morphological traits such as spine length and gill raker length have reasonably high heritability values exceeding 40% (Schluter 1996; Peichel et al. 2001; Berner et al. 2014), suggesting that phenotypes are largely determined by genetic rather than by plastic effects. Yet, theory predicts that only functional loci with a relatively large effect size under strong divergent selection will be able to surpass gene flow (Via 2009; Yeaman and Whitlock 2011). The discrepancy between phenotypic and genomic signatures of selection might therefore become particularly strong for traits that involve many genes of small effect. Arnegard et al. (2014) have another explanation for the relatively large phenotypic divergence. They show that niche differentiation in sticklebacks, even in early stages of differentiation, can involve many different genes and that gene flow between divergent niche‐adapted populations has a bigger impact on the phenotype than just the traits that are directly targeted by selection. It may be attributed to incompatibilities in hybrids that harbor a mix of genes of differentially adapted genotypes. These effects imply that phenotypic changes might not always be adaptive. McCairns and Bernatchez (2010) found indications that freshwater populations might suffer from a loss of plasticity and it might be that epigenetic effects further enhance this discrepancy. For instance, Chaturvedi et al. (2014) found that regulation by miRNAs might make a significant contribution to freshwater adaptation in stickleback populations.
Conclusion
We find that weak selection, high levels of gene flow, or a combination of both can limit the number of outliers in genome scans. Although genome scans targeting populations across strong environmental contrasts are possibly more effective for pinpointing genes that are involved in adaptation, the genes identified by these studies do not necessarily play an important role at every stage of divergence. Many of the genes involved in saltwater–freshwater transitions might be site specific or might not be involved when gene flow is constantly mixing the gene pool. The genes that we do find are likely to be those with a major effect size and thus an underrepresentation of the total number of genes involved. In addition, phenotypic adaptation is not necessarily genetic, but might be facilitated by plastic and epigenetic effects. It remains a challenging task to find which genes and how many are truly involved in local adaptation. We here showed that even with ample gene flow and across weak ecological contrasts, interesting insights on the repeatability of genomic signatures of selection can be obtained.
Data Accessibility
Genotypes are available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.hm926.
Conflict of Interest
None declared.
Supporting information
Table S1. Characterisation of microsatellite loci of the three‐spined stickleback, including name, type of marker, gene assignment and putative function.
Table S2. Global outlier loci that are putatively under balancing selection as detected with the methods of BayeScan, LOSITAN and Arlequin.
Acknowledgments
We would like to thank A. Chaturvedi, S. Geldof, B. Hellemans, L. Jaquemin, S. Ruyts, N. Thiercelin, I. Verdonck, and D. Verheyen for field support and technical assistance. We thank I. Coscia and J. Merilä for insightful and inspiring comments and the latter for collaboration on the whole project. We appreciated the insightful suggestions of two anonymous reviewers. Research was sponsored by the Research Foundation – Flanders (postdoctoral fellowship to J.A.M.R. and research project G.0142.03) and by the KU Leuven (GOA/2006/06 “Evolutionary responses to strong selection pressures and their ecological consequences”; PF/2010/07 “Eco‐ and socio‐evolutionary dynamics”) and facilitated by the research community W0.037.10N “Eco‐evolutionary dynamics in natural and anthropogenic communities” sponsored by the Research Foundation – Flanders. The study received additional support from Academy of Finland grant (129662, 134728 and 218343) to Juha Merilä.
Ecology and Evolution 2015; 5(18): 4174–4186
References
- Acosta, J. , Carpio Y., Borroto I., González O., and Estrada M. P.. 2005. Myostatin gene silenced by RNAi show a zebrafish giant phenotype. J. Biotechnol. 119:324–331. [DOI] [PubMed] [Google Scholar]
- Antao, T. , Lopes A., Lopes R., Beja‐Pereira A., and Luikart G.. 2008. LOSITAN: a workbench to detect molecular adaptation based on a Fst‐outlier method. BMC Bioinformatics 9:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnegard, M. E. , McGee M. D., Matthews B., Marchinko K. B., Conte G. L., Kabir S., et al. 2014. Genetics of ecological divergence during speciation. Nature 511:307–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick, J. E. , and Lenski R. E.. 2013. Genome dynamics during experimental evolution. Nat. Rev. Genet. 14:827–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont, M. A. , and Balding D. J.. 2004. Identifying adaptive genetic divergence among populations from genome scans. Mol. Ecol. 13:969–980. [DOI] [PubMed] [Google Scholar]
- Becks, L. , Ellner S. P., Jones L. E., and Hairston N. G.. 2012. The functional genomics of an eco‐evolutionary feedback loop: linking gene expression, trait evolution, and community dynamics. Ecol. Lett. 15:492–501. [DOI] [PubMed] [Google Scholar]
- Belkhir, K. , Borsa P., Chikhi L., Raufaste N., and Bonhomme F. 1996. GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations, Pp. 1996–2004. Laboratoire génome, populations, interactions, CNRS UMR 5000, Université de Montpellier II, Montpellier, France. [Google Scholar]
- Bell, M. A. 1982. Differentiation of adjacent stream populations of threespine sticklebacks. Evolution 36:189–199. [DOI] [PubMed] [Google Scholar]
- Bell, M. A. , and Aguirre W. E.. 2013. Contemporary evolution, allelic recycling, and adaptive radiation of the threespine stickleback. Evol. Ecol. Res. 15:377–411. [Google Scholar]
- Bernatchez, L. , Renaut S., Whiteley A. R., Derome N., Jeukens J., Landry L., et al. 2010. On the origin of species: insights from the ecological genomics of lake whitefish. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365:1783–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berner, D. , Moser D., Roesti M., Buescher H., and Salzburger W.. 2014. Genetic architecture of skeletal evolution in european lake and stream stickleback. Evolution 68:1792–1805. [DOI] [PubMed] [Google Scholar]
- Bierne, N. , Welch J., Loire E., Bonhomme F., and David P.. 2011. The coupling hypothesis: why genome scans may fail to map local adaptation genes. Mol. Ecol. 20:2044–2072. [DOI] [PubMed] [Google Scholar]
- Bierne, N. , Roze D., and Welch J. J.. 2013. Pervasive selection or is it…? why are FST outliers sometimes so frequent? Mol. Ecol. 22:2061–2064. [DOI] [PubMed] [Google Scholar]
- Boughman, J. W. 2007. Speciation in sticklebacks Pp. 83–126 in Östlund‐Nilsson S., Mayer I. and Huntingford F., eds. Biology of the three‐spined stickleback. CRC Press, Boca Raton, FL. [Google Scholar]
- Butlin, R. K. , Saura M., Charrier G., Jackson B., Andre C., Caballero A., et al. 2014. Parallel evolution of local adaptation and reproductive isolation in the face of gene flow. Evolution 68:935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi, A. , Raeymaekers J. A. M., and Volckaert F. A. M.. 2014. Computational identification of miRNAs, their targets and functions in three‐spined stickleback (Gasterosteus aculeatus). Mol. Ecol. Resour. 14:768–777. [DOI] [PubMed] [Google Scholar]
- Colosimo, P. F. , Hosemann K. E., Balabhadra S., G., Villarreal Jr. , Dickson M., Grimwood J., et al. 2005. Widespread parallel evolution in sticklebacks by repeated fixation of ectodysplasin alleles. Science 307:1928–1933. [DOI] [PubMed] [Google Scholar]
- Coscia, I. , Vogiatzi E., Kotoulas G., Tsigenopoulos C., and Mariani S.. 2011. Exploring neutral and adaptive processes in expanding populations of gilthead sea bream, Sparus aurata L., in the North‐East Atlantic. Heredity 108:537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deagle, B. E. , Jones F. C., Chan Y. F., Absher D. M., Kingsley D. M. and Reimchen T. E. 2012. Population genomics of parallel phenotypic evolution in stickleback across stream‐lake ecological transitions. Proc. R. Soc. Lond. B Biol. Sci. 279:1277–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFaveri, J. , and Merilä J.. 2013. Evidence for adaptive phenotypic differentiation in Baltic Sea sticklebacks. J. Evol. Biol. 26:1700–1715. [DOI] [PubMed] [Google Scholar]
- DeFaveri, J. , Shikano T., Shimada Y., Goto A., and Merilä J.. 2011. Global analysis of genes involved in freshwater adaptation in threespine stickleback (Gasterosteus aculeatus). Evolution 65:1800–1807. [DOI] [PubMed] [Google Scholar]
- DeFaveri, J. , Jonsson P. R., and Merilä J.. 2013. Heterogeneous genomic differentiation in marine threespine sticklebacks: adaptation along an environmental gradient. Evolution 67:2530–2546. [DOI] [PubMed] [Google Scholar]
- Dvorsky, R. , and Ahmadian M. R.. 2004. Always look on the bright site of Rho: structural implications for a conserved intermolecular interface. EMBO Rep. 5:1130–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler, J. A. 1973. Gene flow and population differentiation: studies of clines suggest that differentiation along environmental gradients may be independent of gene flow. Science 179:243–250. [DOI] [PubMed] [Google Scholar]
- Excoffier, L. , and Lischer H. E.. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10:564–567. [DOI] [PubMed] [Google Scholar]
- Excoffier, L. , Hofer T., and Foll M.. 2009. Detecting loci under selection in a hierarchically structured population. Heredity 103:285–298. [DOI] [PubMed] [Google Scholar]
- Feder, J. L. , Egan S. P., and Nosil P.. 2012. The genomics of speciation‐with‐gene‐flow. Trends Genet. 28:342–350. [DOI] [PubMed] [Google Scholar]
- Foll, M. , and Gaggiotti O.. 2008. A genome‐scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180:977–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, S. A. , Scott R. J., and Cresko W. A.. 1998. Nested biological variation and speciation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 353:207–218. [Google Scholar]
- Garnaas, M. K. , Moodie K. L., M‐l Liu., Samant G. V., Li K., Marx R., et al. 2008. Syx, a RhoA guanine exchange factor, is essential for angiogenesis in vivo. Circ. Res. 103:710–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen, D. W. 1967. Isolating mechanisms in threespine sticklebacks (Gasterosteus). J. Fish. Res. Board Can. 24:1637–1692. [Google Scholar]
- Hansen, M. M. , Ruzzante D. E., Nielsen E. E., Bekkevold D., and Mensberg K.‐L. D.. 2002. Long‐term effective population sizes, temporal stability of genetic composition and potential for local adaptation in anadromous brown trout (Salmo trutta) populations. Mol. Ecol. 11:2523–2535. [DOI] [PubMed] [Google Scholar]
- Hansen, M. M. , Meier K., and Mensberg K.‐L. D.. 2010. Identifying footprints of selection in stocked brown trout populations: a spatio‐temporal approach. Mol. Ecol. 19:1787–1800. [DOI] [PubMed] [Google Scholar]
- Hendry, A. P. 2009. Ecological speciation! Or the lack thereof? Can. J. Fish Aquat. Sci. 66:1383–1398. [Google Scholar]
- Hendry, A. P. , Bolnick D. I., Berner D., and Peichel C. L.. 2009. Along the speciation continuum in sticklebacks. J. Fish Biol. 75:2000–2036. [DOI] [PubMed] [Google Scholar]
- Heuts, M. J. 1947. The phenotypical variability of Gasterosteus aculeatus L. populations in Belgium. Verhandelingen van de Koninklijke Vlaamse Academie voor Wetenschappen, Letteren en Schone Kunsten van België 9:1–63. [Google Scholar]
- Hohenlohe, P. A. , Bassham S., Etter P. D., Stiffler N., Johnson E. A. and Cresko W. A. 2010. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genet. 6:e1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenlohe, P. A. , Bassham S., Currey M., and Cresko W. A.. 2012. Extensive linkage disequilibrium and parallel adaptive divergence across threespine stickleback genomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367:395–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, F. , Brown C., Pemberton J., and Braithwaite V.. 2006. Reproductive isolation in a threespine stickleback hybrid zone. J. Evol. Biol. 19:1531–1544. [DOI] [PubMed] [Google Scholar]
- Jones, F. C. , Chan Y. F., Schmutz J., Grimwood J., Brady S. D., Southwick A. M., et al. 2012a. A genome‐wide SNP genotyping array reveals patterns of global and repeated species‐pair divergence in sticklebacks. Curr. Biol. 22:83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, F. C. , Grabherr M. G., Chan Y. F., Russell P., Mauceli E., Johnson J., et al. 2012b. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484:55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauer, M. O. , Dieringer D., and Schlotterer C.. 2003. A microsatellite variability screen for positive selection associated with the “Out of Africa” habitat expansion of Drosophila melanogaster. Genetics 165:1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levene, H. 1953. Genetic equilibrium when more than one ecological niche is available. Am. Nat. 87:331–333. [Google Scholar]
- Lind, E. E. , and Grahn M.. 2011. Directional genetic selection by pulp mill effluent on multiple natural populations of three‐spined stickleback (Gasterosteus aculeatus). Ecotoxicology 20:503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucek, K. , Sivasundar A., Kristjánsson B. K., Skúlason S., and Seehausen O.. 2014. Quick divergence but slow convergence during ecotype formation in lake and stream stickleback pairs of variable age. J. Evol. Biol. 27:1878–1892. [DOI] [PubMed] [Google Scholar]
- Mäkinen, H. S. , Cano J. M., and Merilä J.. 2008. Identifying footprints of directional and balancing selection in marine and freshwater three‐spined stickleback (Gasterosteus aculeatus) populations. Mol. Ecol. 17:3565–3582. [DOI] [PubMed] [Google Scholar]
- McCairns, R. J. S. , and Bernatchez L.. 2010. Adaptive divergence between freshwater and marine sticklebacks: insights into the role of phenotypic plasticity from an integrated analysis of candidate gene expression. Evolution 64:1029–1047. [DOI] [PubMed] [Google Scholar]
- McKinnon, J. S. , and Rundle H. D.. 2002. Speciation in nature: the threespine stickleback model systems. Trends Ecol. Evol. 17:480–488. [Google Scholar]
- McPhail, J. 1994. Speciation and the evolution of reproductive isolation in the sticklebacks (Gasterosteus) of south‐western British Columbia Pp. 399–437 in Bell A. M. and Foster S., eds. The evolutionary biology of the threespine stickleback. Oxford University Press Inc., New York, NY. [Google Scholar]
- Mitchell‐Olds, T. , and Schmitt J.. 2006. Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature 441:947–952. [DOI] [PubMed] [Google Scholar]
- Moore, J.‐S. , Gow J. L., Taylor E. B., and Hendry A. P.. 2007. Quantifying the constraining influence of gene flow on adaptive divergence in the lake‐stream threespine stickleback System. Evolution 61:2015–2026. [DOI] [PubMed] [Google Scholar]
- Narum, S. R. , and Hess J. E.. 2011. Comparison of F‐ST outlier tests for SNP loci under selection. Mol. Ecol. Resour. 11:184–194. [DOI] [PubMed] [Google Scholar]
- Nielsen, E. , Hemmer‐Hansen J., Poulsen N., Loeschcke V., Moen T., Johansen T., et al. 2009. Genomic signatures of local directional selection in a high gene flow marine organism; the Atlantic cod (Gadus morhua). BMC Evol. Biol. 9:276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil, P. 2009. Adaptive population divergence in cryptic color‐pattern following a reduction in gene flow. Evolution 63:1902–1912. [DOI] [PubMed] [Google Scholar]
- Nosil, P. , Funk D. J., and Ortiz‐Barrientos D.. 2009. Divergent selection and heterogeneous genomic divergence. Mol. Ecol. 18:375–402. [DOI] [PubMed] [Google Scholar]
- Olafsdottir, G. A. , and Snorrason S. S.. 2009. Parallels, nonparallels, and plasticity in population differentiation of threespine stickleback within a lake. Biol. J. Linn. Soc. 98:803–813. [Google Scholar]
- van Oosterhout, C. , Hutchinson W. F., Wills D. P. M., and Shipley P.. 2004. Micro‐Checker: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 4:535–538. [Google Scholar]
- Orsini, L. , Spanier K. I., and De Meester L. U. C.. 2012. Genomic signature of natural and anthropogenic stress in wild populations of the waterflea Daphnia magna: validation in space, time and experimental evolution. Mol. Ecol. 21:2160–2175. [DOI] [PubMed] [Google Scholar]
- Papa, R. , Martin A., and Reed R. D.. 2008. Genomic hotspots of adaptation in butterfly wing pattern evolution. Curr. Opin. Genet. Dev. 18:559–564. [DOI] [PubMed] [Google Scholar]
- Peichel, C. L. , Nereng K. S., Ohgi K. A., Cole B. L. E., Colosimo P. F., Buerkle C. A., et al. 2001. The genetic architecture of divergence between threespine stickleback species. Nature 414:901–905. [DOI] [PubMed] [Google Scholar]
- Perrier, C. , Bourret V., Kent M. P., and Bernatchez L.. 2013. Parallel and nonparallel genome‐wide divergence among replicate population pairs of freshwater and anadromous Atlantic salmon. Mol. Ecol. 22:5577–5593. [DOI] [PubMed] [Google Scholar]
- Pinho, C. , and Hey J.. 2010. Divergence with gene flow: models and data. Annu. Rev. Ecol. Evol. Syst. 41:215–230. [Google Scholar]
- Poncet, B. N. , Herrmann D., Gugerli F., Taberlet P., Holdergger R., Gielly L., et al. 2010. Tracking genes of ecological relevance using a genome scan in two independent regional population samples of Arabis alpina . Mol. Ecol. 19:2896–2907. [DOI] [PubMed] [Google Scholar]
- Radaelli, G. , Rowlerson A., Mascarello F., Patruno M., and Funkenstein B.. 2003. Myostatin precursor is present in several tissues in teleost fish: a comparative immunolocalization study. Cell Tissue Res. 311:239–250. [DOI] [PubMed] [Google Scholar]
- Raeker, M. Ö. , Bieniek A. N., Ryan A. S., Tsai H., Zahn K. M. and Russell M. W.. 2010. Targeted deletion of the zebrafish obscurin A RhoGEF domain affects heart, skeletal muscle and brain development. Dev. Biol. 337:432–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raeymaekers, J. A. M. , Maes G. E., Audenaert E., and Volckaert F. A. M.. 2005. Detecting Holocene divergence in the anadromous‐freshwater three‐spined stickleback (Gasterosteus aculeatus) system. Mol. Ecol. 14:1001–1014. [DOI] [PubMed] [Google Scholar]
- Raeymaekers, J. A. M. , Van Houdt J. K. J., Larmuseau M. H. D., Geldof S., and Volckaert F. A. M.. 2007. Divergent selection as revealed by PST and QTL‐based F ST in three‐spined stickleback (Gasterosteus aculeatus) populations along a coastal‐inland gradient. Mol. Ecol. 16:891–905. [DOI] [PubMed] [Google Scholar]
- Raeymaekers, J. A. M. , Lens L. U. C., Van den Broeck F., Van Dongen S., and Volckaert F. A. M.. 2012. Quantifying population structure on short timescales. Mol. Ecol. 21:3458–3473. [DOI] [PubMed] [Google Scholar]
- Raeymaekers, J. A. M. , Konijnendijk N., Larmuseau M. H. D., Hellemans B., De Meester L. and Volckaert F. A. M.. 2014. A gene with major phenotypic effects as a target for selection vs. homogenizing gene flow. Mol. Ecol. 23:162–181. [DOI] [PubMed] [Google Scholar]
- Roesti, M. , Hendry A. P., Salzburger W., and Berner D.. 2012. Genome divergence during evolutionary diversification as revealed in replicate lake‐stream stickleback population pairs. Mol. Ecol. 21:2852–2862. [DOI] [PubMed] [Google Scholar]
- Rohlf, F. J. 2002. tpsDig, digitize landmarks and outlines. Department of Ecology and Evolution, State University of New York at Stony Brook, Stony Brook, NY. [Google Scholar]
- Rossman, K. L. , Der C. J., and Sondek J.. 2005. GEF means go: turning on RHO GTPases with guanine nucleotide‐exchange factors. Nat. Rev. Mol. Cell Biol. 6:167–180. [DOI] [PubMed] [Google Scholar]
- Schluter, D. 1996. Adaptive radiation along genetic lines of least resistance. Evolution, 176:6–1774. [DOI] [PubMed] [Google Scholar]
- Schluter, D. 2009. Evidence for Ecological Speciation and Its Alternative. Science 323:737–741. [DOI] [PubMed] [Google Scholar]
- Schluter, D. , and Conte G. L.. 2009. Genetics and ecological speciation. Proc. Natl Acad. Sci. USA 106:9955–9962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehausen, O. , Butlin R. K., Keller I., Wagner C. E., Boughman J. W., Hohenlohe P. A., et al. 2014. Genomics and the origin of species. Nat. Rev. Genet. 15:176–192. [DOI] [PubMed] [Google Scholar]
- Shikano, T. , Ramadevi J., and Merilä J.. 2010a. Identification of local‐ and habitat‐dependent selection: scanning functionally important genes in nine‐spined sticklebacks (Pungitius pungitius). Mol. Biol. Evol. 27:2775–2789. [DOI] [PubMed] [Google Scholar]
- Shikano, T. , Ramadevi J., Shimada Y., and Merilä J.. 2010b. Utility of sequenced genomes for microsatellite marker development in non‐model organisms: a case study of functionally important genes in nine‐spined sticklebacks (Pungitius pungitius). BMC Genom. 11:334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada, Y. , Shikano T., and Merilä J.. 2011. A high incidence of selection on physiologically important genes in the three‐spined stickleback, Gasterosteus aculeatus . Mol. Biol. Evol. 28:181–193. [DOI] [PubMed] [Google Scholar]
- Slatkin, M. 1987. Gene flow and the geographic structure of natural populations. Science 236:787–792. [DOI] [PubMed] [Google Scholar]
- Soria‐Carrasco, V. , Gompert Z., Comeault A. A., Farkas T. E., Parchman T. L., Johnston J. S., et al. 2014. Stick insect genomes reveal natural selection's role in parallel speciation. Science 344:738–742. [DOI] [PubMed] [Google Scholar]
- Stapley, J. , Reger J., Feulner P. G. D., Smadja C., Galindo J., Ekblom R., et al. 2010. Adaptation genomics: the next generation. Trends Ecol. Evol. 25:705–712. [DOI] [PubMed] [Google Scholar]
- Storz, J. F. 2005. Invited review: using genome scans of DNA polymorphism to infer adaptive population divergence. Mol. Ecol. 14:671–688. [DOI] [PubMed] [Google Scholar]
- Strasburg, J. L. , Sherman N. A., Wright K. M., Moyle L. C., Willis J. H. and Rieseberg L. H.. 2012. What can patterns of differentiation across plant genomes tell us about adaptation and speciation? Philos. Trans. R. Soc. Lond. B Biol. Sci. 367:364–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, W. R. , and Dyke G. C. V.. 1985. Revised procedures for staining and clearing small fishes and other vertebrates for bone and cartilage study. Cybium 9:107–119. [Google Scholar]
- Tregenza, T. 2002. Divergence and reproductive isolation in the early stages of speciation Pp. 291–300 in Etges W. J. and Noor M. A. F., eds. Genetics of mate choice: from sexual selection to sexual isolation. Springer Netherlands, Springer Science+Business Media B.V., Dordrecht. [PubMed] [Google Scholar]
- Van Dongen, S. , Lens L., Pape E., Volckaert F. A. M., and Raeymaekers J. A. M.. 2009. Evolutionary history shapes the association between developmental instability and population‐level genetic variation in three‐spined sticklebacks. J. Evol. Biol. 22:1695–1707. [DOI] [PubMed] [Google Scholar]
- Vandamme, S. G. , Maes G. E., Raeymaekers J. A. M., Cottenie K., Imsland A. K., Hellemans B., et al. 2014. Regional environmental pressure influences population differentiation in turbot (Scophthalmus maximus). Mol. Ecol. 23:618–636. [DOI] [PubMed] [Google Scholar]
- Via, S. 2009. Natural selection in action during speciation. Proc. Natl Acad. Sci. USA 106:9939–9946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Villemereuil, P. , Frichot É., Bazin É., François O., and Gaggiotti O. E.. 2014. Genome scan methods against more complex models: when and how much should we trust them? Mol. Ecol. 23:2006–2019. [DOI] [PubMed] [Google Scholar]
- Voight, B. F. , Kudaravalli S., Wen X., and Pritchard J. K.. 2006. A map of recent positive selection in the human genome. PLoS Biol. 4:e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, F. , and Celeste A.. 2005. Myostatin: a modulator of skeletal‐muscle stem cells. Biochem. Soc. Trans. 33:1513–1517. [DOI] [PubMed] [Google Scholar]
- Wootton, R. J. 1976. The biology of the sticklebacks. Academic Press, London. [Google Scholar]
- Yang, L. , Jelsbak L., Marvig R. L., Damkiæra S., Workmana C. T., Rau M. H., et al. 2011. Evolutionary dynamics of bacteria in a human host environment. Proc. Natl Acad. Sci. USA 108:7481–7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeaman, S. , and Otto S. P.. 2011. Establishment and maintenance of adaptive genetic divergence under migration, selection, and drift. Evolution 65:2123–2129. [DOI] [PubMed] [Google Scholar]
- Yeaman, S. , and Whitlock M. C.. 2011. The genetic architecture of adaptation under migration–selection balance. Evolution 65:1897–1911. [DOI] [PubMed] [Google Scholar]
- Zulliger, D. , Schnyder E., and Gugerli F.. 2013. Are adaptive loci transferable across genomes of related species? Outlier and environmental association analyses in Alpine Brassicaceae species. Mol. Ecol. 22:1626–1639. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Characterisation of microsatellite loci of the three‐spined stickleback, including name, type of marker, gene assignment and putative function.
Table S2. Global outlier loci that are putatively under balancing selection as detected with the methods of BayeScan, LOSITAN and Arlequin.
Data Availability Statement
Genotypes are available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.hm926.


