Abstract
Background
Protocadherin17 (PCDH17) is a tumor suppressor gene, and is frequently silenced by promoter methylation in human cancers, including clear cell renal cell carcinoma (ccRCC). However, the clinical significance of PCDH17 methylation in ccRCC remains largely unclear. The aim of the present study was to investigate the methylation status of PCDH17 in ccRCC and its potential relevance to clinicopathological parameters and prognosis.
Material/Methods
Methylation-specific PCR was used to examine the methylation status of PCDH17 in 191 ccRCC tumors and matched paired adjacent noncancerous tissues. Subsequently, the associations between PCDH17 methylation and clinicopathological parameters and prognosis of patients with ccRCC were analyzed.
Results
PCDH17 methylation occurred in 66.5% of ccRCC tumors, but in only 12.1% of adjacent noncancerous tissues. PCDH17 methylation is significantly correlated with advanced stage, higher grade, and lymph node metastasis in ccRCC. Moreover, it is an independent prognostic factor for progression-free survival and overall survival of patients with ccRCC.
Conclusions
PCDH17 methylation occurred more frequently and was associated with malignant clinicopathological characteristics and poor prognosis in ccRCC patients. Thus, PCDH17 methylation may be used as a novel biomarker to predict the prognosis of patients with ccRCC.
MeSH Keywords: Cadherins; Carcinoma, Renal Cell; Methylation
Background
Renal cell carcinoma (RCC) is a common genitourinary malignancy with high morbidity and mortality, with an estimated 63 920 new cases and 13 860 associated deaths in the USA in 2014 [1–5]. Unfortunately, the incidence of RCC has been rising steadily over the last 3 decades in the developed and developing countries [1–5]. Histopathologically, RCC is a heterogeneous disease, including several subtypes: clear cell subtype, papillary, chromophobe, and other rarer forms. However, the most common type of RCC is clear cell renal cell carcinoma (ccRCC), which accounts for more than 75% of tumors [1]. With the advance of diagnostic techniques, a considerable part of the disease can be found at the early stage and cured by surgery. However, some patients will experience recurrence and progression after surgery, and eventually die from RCC. RCC patients with similar tumor characteristics show heterogeneity in the course and outcome of the disease [6–8]. It is difficult to distinguish indolent disease from aggressive disease. Thus, novel predictors are needed to predict disease progression, therapeutic response and prognosis, in addition to traditional factors [6–8].
Recent advances in the understanding of ccRCC indicate that the initiation and progression of the disease is accumulated form genetic and epigenetic changes [7]. DNA methylation is the best studied epigenetic change; the methylation of cytosine residues within CpG dinucleotides can alter the transcription rate of a given gene and bring about transcriptional silencing [9–12]. Human cancers often demonstrate tumor suppressor silencing as a result of aberrant promoter hypermethylation, and aberrant DNA methylation may be a potential biomarker for diagnosis, prognosis, and treatment response [9–12]. Recent studies indicated that PCDH17 may function as a tumor suppressor gene in bladder cancer, prostate cancer, and gastric cancer. The PCDH17 gene is located on chromosome 13q21.2 in humans, and I often silenced by promoter methylation in some target CpG sites, such as −458, −731, and 870 in human cancers [13–16]. However, the clinical significance of PCDH17 methylation in ccRCC needs to be further elucidated.
In the current study, the promoter methylation of PCDH17 was measured in ccRCC tissues and adjacent non-cancerous tissues using methylation-specific PCR (MSP). MSP is a major technique for the detection of gene methylation and can provide specific and sensitive results; it is rapid, simple, and cost-effective and allows rapid examining multiple samples, which is convenient for clinical research [9–14]. Then we analyzed the correlation between PCDH17 methylation and clinicopathologic parameters, and evaluated its association with the outcome of patients with ccRCC in order to elucidate its clinical significance in this disease.
Material and Methods
Patients and tissue samples
A total of 191 ccRCC tumors and matched paired adjacent noncancerous tissues were collected from patients during radical nephrectomy, at the Third Hospital of Hebei Medical University, between May 2003 and May 2009. The tissue samples were immediately frozen in liquid nitrogen and stored at −80°C until used. These patients were all histologically confirmed ccRCC cases for the first time, and they did not receive any anti cancer treatment before surgery. Two senior pathologists assessed all the tissue samples considering tumor classification, grade and histological type in the same hospital. The TNM classification of all the tumors was evaluated according to the Union for International Cancer Control 2002 classification, and grading was assessed as previously described [17–21]. The patients were followed up at interval, and the time to disease progression was defined as the point at which patients had either a local recurrence or a synchronous/metachronous metastasis as detected by computerized tomography scan [21]. The 5-year overall survival data was also collected from all the patients. Clinical and pathological characteristics of the patients are summarized in Table 1. In addition, the study was approved by our local ethics committee (No. HMU20030365X) and informed consent was obtained from each patient.
Table 1.
The clinical and pathological parameters of patients with ccRCC.
| Features | Parameters | No. (%) |
|---|---|---|
| Sex | Male | 114 (59.7) |
| Female | 77 (40.3) | |
| Age (years) | <60 | 82 (42.9) |
| ≥60 | 109 (57.1) | |
| Pathological stage | T1 | 109 (57.1) |
| T2 | 57 (29.8) | |
| T3 | 25 (13.1) | |
| Lymph node metastasis | N0 | 182 (95.3) |
| N1 | 9 (4.7) | |
| Distant metastasis | M0 | 191 (100.0) |
| M1 | 0 (0.0) | |
| Fuhrman grade | I | 73 (38.2) |
| II | 57 (29.8) | |
| III | 45 (23.6) | |
| IV | 16 (8.4) | |
| Progression | Presence | 54 (28.3) |
| Absence | 137 (71.7) | |
| Total | 191 (100.0) |
DNA isolation, bisulfite conversion, and MSP
Genomic DNA was isolated from the frozen tissue samples using the DNeasy Tissue Kit (Qiagen, Valencia, CA) following the manufacturer’s instructions as previously described [12,18]. The bisulfite conversion of DNA was carried out using EpiTect Bisulfite Kit (Qiagen, Valencia, CA) according to the manufacture’s protocol as reported previously [12,18]. Methylation analysis of bisulfite-treated DNA was carried out using PCR assay, and the primers specific for methylated PCDH17 and unmethylated PCDH17 were used as previously reported [14,22]. The primers for the methylated sequences were: 5′-GATTATCGGGTGTCGTAGTTC-3′ (forward) and 5′-CCCTAACGCAACGTACGCG-3′ (reverse). The primers for the unmethylated sequences were: 5′-AGATTATTGGGTGTTGTAGTTT-3′ (forward) and 5′-AACCCTAACACAACATACACA-3′ (reverse). The PCR amplification was carried out as described previously [14,22]. In vitro methylated DNA and unmethylated DNA (New England Biolabs, Beverly, MA, USA) were used as methylation and unmethylation positive control. The MSP products were separated in 2% agarose gel, stained with ethidium bromide, and visualized under ultraviolet illumination for analysis. The product was defined as methylation positive when methylated allele was present in the methylated DNA lane or both in the methylated and unmethylated DNA lanes, and methylation negative when band was present only in the unmethylated DNA lane [14,22].
Quantitative Real-Time PCR (qRT-PCR) analysis of PCDH17 mRNA expression
PCDH17 mRNA expression was detected in 127 tumors with methylated PCDH17, 64 tumors with unmethylated PCDH17, and 168 adjacent noncancerous tissues with unmethylated PCDH17. In addition, PCDH17 mRNA expression in 68 adjacent noncancerous tissues with unmethylated PCDH17 was used as control. Total RNA was extracted from tissue samples by Trizol reagent (Invitrogen, Carlsbad, Calif, CA, USA). The relative PCDH17 mRNA expression was calculated using the comparative CT method, with GAPDH as the internal control. The primers for PCDH17: sense, 5′-CGGAGGTGATGTATCTCAAA-3′; antisense 5′-CAGGAGCCTTTGTTAGTGTC-3′.The primers for GAPDH as follow: sense: 5′-CGCTCTCTGCTCCTCCTGTTC-3′; antisense: 5′-ATCCGTTGACTCCGACCTTCA C-3′. The PCR conditions included a denaturation step of 95°C for 2 min, followed by 30 cycles for 94°C for 30 s, 50°C for 30 s, 72°C for 30 s, and a final elongation step of 72°C for 7 min. Each PCR was repeated at least 3 times to confirm the result.
Statistical analysis
Fisher’s exact test was used to assess the difference of PCDH17 methylation status between ccRCC tumors and matched adjacent noncancerous tissues. Chi-square test was used to assess the relationship between PCDH17 methylation and clinicopathologic features. Kaplan-Meier survival analysis and log-rank test were used to assess the differences of progression-free survival and 5-year overall survival between patients with methylated PCDH17 and unmethylated PCDH17. Univariate and Multivariate Cox proportional hazard model analysis was used to assess the prognostic value of PCDH17 methylation in ccRCC patients. The difference of PCDH17 mRNA expression among controls, tumors with methylated PCDH17 and unmethylated PCDH17 was analyzed by one-way ANOVA. A p value <0.05 was considered statistically significant. The statistical analysis was conducted using SAS version 8.0 (SAS Institute, Cary, N.C., USA) for windows.
Results
The methylation status of PCDH17 in ccRCC tumors and matched adjacent noncancerous tissues
The methylation status of PCDH17 was detected in 191 ccRCC tumors and matched adjacent noncancerous tissues using MSP. PCDH17 methylation was detected in 127 (66.5%) ccRCC tumors, but PCDH17 methylation was only found in 23 (12.1%) adjacent noncancerous tissues. Representative MSP results are shown in Figure 1. The methylation rate of PCDH17 in ccRCC tumors is higher than that in adjacent noncancerous tissues, and the difference is statistically significant (P<0.0001, Table 2).
Figure 1.
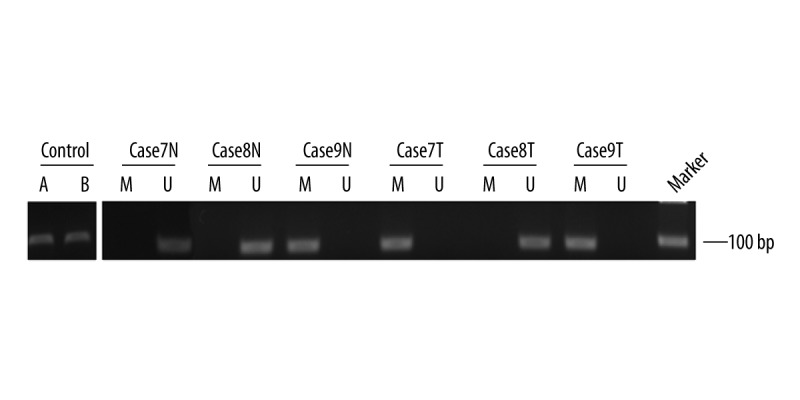
Representative MSP results of PCDH17 methylation in patients with ccRCC. A: methylation control; B: unmethylation control; M: methylated; U: unmethylated; N: adjacent noncancerous tissues; T: ccRCC tissues. Cases9N, Case7T and Case9T exhibited methylated PCDH17. Case7N, Case8N and Case8T exhibited unmethylated PCDH17.
Table 2.
PCDH17 methylation in ccRCC and matched adjacent noncancerous tissues.
| Group | M (%) | U (%) | P |
|---|---|---|---|
| ccRCC | 127 (66.5) | 64 (33.5) | <0.0001 |
| ANT | 23 (12.1) | 168 (87.9) |
M – methylation; U – unmethylation; ANT – adjacent noncancerous tissues.
Relationship between PCDH17 methylation and clinicopathological variables of patients with ccRCC
Subsequently, we linked the methylation status of PCDH17 in ccRCC tumors to clinicopathological features, in order to evaluate its clinical significance. The results indicated that PCDH17 methylation was significantly correlated with advanced stage (P=0.0478), higher grade (P=0.0449), lymph node metastasis (P=0.0302) and tumor progression (P=0.0002). However, the methylation of PCDH17 was not related to age or sex. These results are summarized in Table 3.
Table 3.
Relationship between PCDH17 methylation and clinicopathological variables of patients with ccRCC.
| Features | Variables | No. | M (%) | U (%) | P |
|---|---|---|---|---|---|
| Sex | Male | 114 | 71 (62.3) | 43 (37.7) | 0.1335 |
| Female | 77 | 56 (72.7) | 21 (27.3) | ||
| Age | <60 | 82 | 52 (63.4) | 30 (36.6) | 0.4345 |
| ≥60 | 109 | 75 (68.8) | 34 (31.2) | ||
| Pathological stage | T1 | 109 | 68 (62.4) | 61 (36.8) | 0.0478 |
| T2 | 57 | 37 (64.9) | 20 (35.1) | ||
| T3 | 25 | 22 (88.0) | 3 (12.0) | ||
| Fuhrman grade | I | 73 | 40 (54.8) | 33 (45.2) | 0.0449 |
| II | 57 | 40 (70.2) | 17 (29.8) | ||
| III | 45 | 34 (75.6) | 11 (24.4) | ||
| IV | 16 | 13 (81.3) | 3 (18.7) | ||
| Lymph node metastasis | N0 | 182 | 118 (64.8) | 64 (35.2) | 0.0302 |
| N1 | 9 | 9 (100.0) | 0 (0.0) | ||
| Progression | Presence | 54 | 47 (87.0) | 7 (13.0) | 0.0002 |
| Absence | 137 | 80 (58.4) | 57 (41.6) | ||
| Total | 191 | 127 (66.5) | 64 (33.5) |
M – methylation; U – unmethylation.
PCDH17 methylation to predict clinical outcome after radical nephrectomy
To examine if PCDH17 is a significant predictor of patients’ outcome after surgery, Kaplan-Meier curves and log-rank tests were performed. We found that patients with methylated PCDH17 had significantly shorter progression-free time and 5-year overall survival time than patients with unmethylated PCDH17. These findings are shown in Figures 2 and 3. To further determine the predictive value of PCDH17 methylation in ccRCC patients, the Univariate and Multivariate Cox proportional hazard model analysis was carried out, and the parameters that were significant risk factors in univariate analysis were entered into multivariate analysis. The results suggested that PCDH17 methylation is an independent prognostic factor for progression-free survival and 5-year overall survival of ccRCC patients. The results are shown in Tables 4 and 5.
Figure 2.
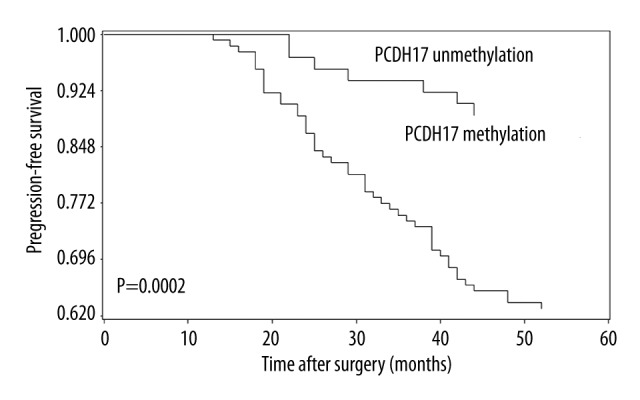
Correlation between PCDH17 methylation and progression-free survival in ccRCC patients. Patients with PCDH17 methylation showed significantly shorter progression-free survival than patients without (log-rank test, P=0.0002).
Figure 3.
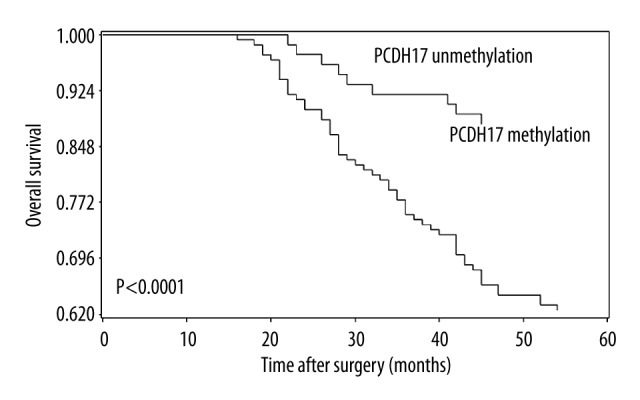
Correlation between PCDH17 methylation and 5-year overall survival in ccRCC patients. Patients with PCDH17 methylation showed significantly shorter 5-year overall survival than patients without (log-rank test, P < 0.0001).
Table 4.
The predictive value of PCDH17 methylation for the progression-free survival in ccRCC.
| Variable | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| Sex (Male vs. Female) | 1.325 | 0.805–2.316 | 0.3207 | |||
| Age (≥60 vs. <60) | 1.532 | 0.693–3.047 | 0.2031 | |||
| Pathological stage (T3 vs. T1/T2) | 2.968 | 1.853–9.335 | 0.0017 | 1.768 | 1.035–7.153 | 0.0136 |
| Fuhrman grade (III/IV vs. I/II) | 2.532 | 1.214–8.153 | 0.0075 | 1.541 | 0.793–6.832 | 0.0673 |
| Lymph node metastasis (N1 vs. N0) | 4.531 | 2.071–11.865 | <0.0001 | 3.652 | 1.872–9.461 | 0.0013 |
| PCDH17 methylation (M vs. U) | 3.973 | 1.795–8.792 | 0.0007 | 3.014 | 1.235–7.463 | 0.0028 |
HR – hazard ratio; M – methylated; U – unmethylated.
Table 5.
The predictive value of PCDH17 methylation for the five-year overall survival in ccRCC.
| Variable | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| Sex (Male vs. Female) | 1.133 | 0.765–2.068 | 0.3426 | |||
| Age (≥60 vs. <60) | 1.263 | 0.831–2.179 | 0.1683 | |||
| Pathological stage (T3 vs. T1/T2) | 2.476 | 1.128–7.336 | 0.0268 | 1.463 | 0.874–3.421 | 0.0769 |
| Fuhrman grade (III/IV vs. I/II) | 3.042 | 1.363–8.546 | 0.0043 | 2.127 | 1.093–5.862 | 0.0347 |
| Lymph node metastasis (N1 vs. N0) | 3.925 | 1.833–10.796 | <0.0001 | 2.563 | 1.248–7.552 | 0.0163 |
| PCDH17 methylation (M vs. U) | 3.673 | 1.615–7.435 | 0.0003 | 2.876 | 1.347–8.065 | 0.0097 |
HR – hazard ratio; M – methylated; U – unmethylated.
The relationship between PCDH17 methylation and mRNA expression
In order to certify that PCDH17 methylation was correlated with the down-regulation of its gene expression, quantitative real-time PCR was performed to detect the mRNA levels of PCDH17 in adjacent noncancerous tissues with unmethylated PCDH17, tumors with unmethylated PCDH17, and tumors with methylated PCDH17. We found that there was no difference between adjacent noncancerous tissues with unmethylated PCDH17 group and tumors with unmethylated PCDH17 group, but PCDH17 mRNA expression was significant lower in tumors with methylated PCDH17 group compared with the 2 groups with unmethylated PCDH17 mentioned above (P<0.05; Figure 4). The results indicate that PCDH17 methylation is associated with down-regulation of its expression.
Figure 4.
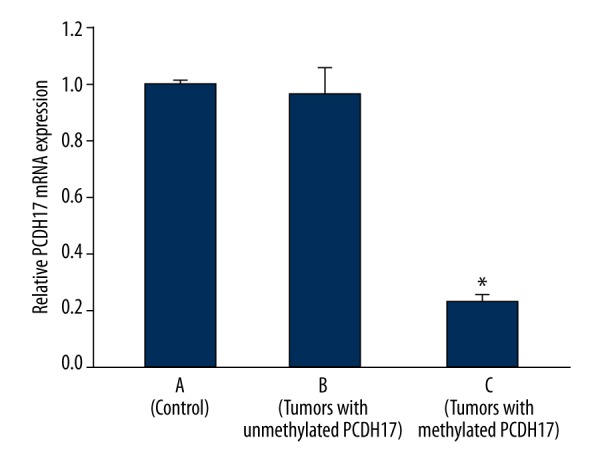
PCDH17 mRNA expression. A: 168 adjacent noncancerous tissues with unmethylated PCDH17; B: 64 tumors with unmethylated PCDH17; C: 127 tumors with methylated PCDH17; * P<0.05 (C vs. B or C vs. A).
Discussion
The protocadherins are a subgroup of the cadherin super-family, which plays crucial roles in cell-cell adhesion, proliferation, migration, invasion and signal transduction [15,23,24]. The protocadherin family can be divided into two groups: clustered protocadherins and non-clustered protocadherins, based on their genomic structure, and PCDH17 belongs to non-clustered group [15,23,24]. Recently, it has been reported that some non-clustered protocadherins act as tumor suppressor genes and were specifically silenced by promoter hypermethylation in many tumor tissues, including RCC [11–16]. In this study, we focused on the investigation of the relationship between PCDH17 methylation and clinicopathological parameters as well as prognosis of the patients with ccRCC, in order to elucidate its clinical significance. To our knowledge this is the first study to investigate the clinical significance of PCDH17 methylation in ccRCC.
Our study demonstrated that PCDH17 methylation occurred more frequently in ccRCC tissues compared to adjacent noncancerous tissues, and PCDH17 methylation is associated with down-regulation of its expression. Moreover, PCDH17 methylation not only exists in ccRCC tissues but also in adjacent noncancerous tissues, which implied that PCDH17 methylation is a critical trigger for ccRCC initiation. In addition, when we correlated the methylation of PCDH17 to clinicopathological features of ccRCC patients, we found that PCDH17 methylation associated with advanced stage, higher grade, lymph node metastasis and progression after surgery. Our findings agree with previous studies about PCDH17 methylation in bladder cancer, prostate cancer, gastric cancer and colorectal cancer [13,14,25–27]. These findings promoted us to more thoroughly investigate its clinical significance in ccRCC. Subsequently, we investigated the prognostic impact of PCDH17 methylation on the ccRCC patients. The progression-free survival and 5-year overall survival was analyzed by Kaplan-Meier curves and log-rank test. The result revealed that PCDH17 methylation was correlated with adverse progression-free survival and 5-year overall survival of patients with ccRCC. In addition, since parameters observed to have a prognostic impact by univariate analysis may covariate, PCDH17 methylation and those clinicopathologic parameters that were significant in univariate analysis were further investigated in multivariate analysis. Multivariate analysis revealed that PCDH17 methylation was an independent predictor of progression-free survival and 5-year overall survival of ccRCC patients. Taken together, our data revealed that PCDH17 methylation occurred more frequently in ccRCC tumors compared to noncancerous tissue; it was correlated with adverse clinicopathologic parameters and predicted poor outcome of ccRCC patients after surgery.
Aberrant DNA methylation is the best-studied epigenetic alteration in human tumors. DNA methylation as a potential biomarker for tumors is of particular interest because DNA can be collected conveniently from tissues and body fluids [28–30]. In addition, DNA methylation can be reversed by demethylation agents, and in the longer term may enable more individualized therapies [32,33]. To the best of our knowledge, this is the first study to demonstrate that PCDH17 methylation may be used as a potential prognostic marker in ccRCC. Consequently, PCDH17 methylation, if validated in future studies, could be used for selection of ccRCC patients for adjuvant treatment after surgery.
Conclusions
Our study indicated that PCDH17 methylation occurred more frequently and was associated with malignant clinicopathological characteristics and poor prognosis in ccRCC patients, suggesting that it could be used as a potential prognostic biomarker in this malignancy. Our study has some limitations. The current study is limited by a relatively small number of patients. We used MSP but not quantitative MSP to detect the methylation status of PCDH17 in ccRCC. In addition, we did not interrogate all of the CpG sites in the promoter, although the CpG sites used in our study are widely used in research. These may make our results less robust. To solve these problems, future studies with larger sample size using quantitative MSP to detect all the CpG sites in the promoter region may be needed to confirm our findings.
Footnotes
Conflicts of interest
None.
Source of support: This study was supported by Xuzhou Medical Talented Youth Project (No: 2014007) and Xuzhou Science and Technology Project (No: KC14SH015)
References
- 1.Cairns P. Renal cell carcinoma. Cancer Biomark. 2010;9(1–6):461–73. doi: 10.3233/CBM-2011-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmad ST, Arjumand W, Seth A, et al. Methylation of the APAF-1 and DAPK-1 promoter region correlates with progression of renal cell carcinoma in North Indian population. Tumour Biol. 2012;33(2):395–402. doi: 10.1007/s13277-011-0235-9. [DOI] [PubMed] [Google Scholar]
- 3.Keefe SM, Nathanson KL, Rathmell WK. The molecular biology of renal cell carcinoma. Semin Oncol. 2013;40(4):421–28. doi: 10.1053/j.seminoncol.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 4.Xiao W, Wang J, Li H, et al. Fibulin-1 is down-regulated through promoter hypermethylation and suppresses renal cell carcinoma progression. J Urol. 2013;190(1):291–301. doi: 10.1016/j.juro.2013.01.098. [DOI] [PubMed] [Google Scholar]
- 5.Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. Cancer J Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 6.Rogenhofer S, Kahl P, Holzapfel S, et al. Decreased levels of histone H3K9me1 indicate poor prognosis in patients with renal cell carcinoma. Anticancer Res. 2012;32(3):879–86. [PubMed] [Google Scholar]
- 7.Rydzanicz M, Wrzesiński T, Bluyssen HA, et al. Genomics and epigenomics of clear cell renal cell carcinoma: recent developments and potential applications. Cancer Lett. 2013;341(2):111–26. doi: 10.1016/j.canlet.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 8.Vasudev NS, Selby PJ, Banks RE. Renal cancer biomarkers: the promise of personalized care. BMC Med. 2012;10:112. doi: 10.1186/1741-7015-10-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van der Heijden AG. The role of methylation in urological tumours. Arch Esp Urol. 2013;66(5):432–39. [PubMed] [Google Scholar]
- 10.Ramakrishnan S, Pili R. Histone deacetylase inhibitors and epigenetic modifications as a novel strategy in renal cell carcinoma. Cancer J. 2013;19(4):333–40. doi: 10.1097/PPO.0b013e3182a09e07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin YL, Wang YL, Fu XL, et al. Aberrant methylation of PCDH8 is a potential prognostic biomarker for patients with clear cell renal cell carcinoma. Med Sci Monit. 2014;20:2380–85. doi: 10.12659/MSM.892433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin YL, Wang YL, Ma JG, et al. Clinical significance of protocadherin 8 (PCDH8) promoter methylation in non-muscle invasive bladder cancer. J Exp Clin Cancer Res. 2014;33:68. doi: 10.1186/s13046-014-0068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin YL, Xie PG, Wang L, et al. Aberrant methylation of protocadherin 17 and its clinical significance in patients with prostate cancer after radical prostatectomy. Med Sci Monit. 2014;20:1376–82. doi: 10.12659/MSM.891247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang XB, Lin YL, Li ZG, et al. Protocadherin 17 promoter methylation in tumour tissue from patients with bladder transitional cell carcinoma. J Int Med Res. 2014;42(2):292–99. doi: 10.1177/0300060513504364. [DOI] [PubMed] [Google Scholar]
- 15.Kim SY, Yasuda S, Tanaka H, et al. Non-clustered protocadherin. Cell Adh Migr. 2011;5(2):97–105. doi: 10.4161/cam.5.2.14374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costa VL, Henrique R, Danielsen SA, et al. TCF21 and PCDH17 methylation: An innovative panel of biomarkers for a simultaneous detection of urological cancers. Epigenetics. 2011;6(9):1120–30. doi: 10.4161/epi.6.9.16376. [DOI] [PubMed] [Google Scholar]
- 17.Pichler M, Hutterer GC, Chromecki TF, et al. Comparison of the 2002 and 2010 TNM classification systems regarding outcome prediction in clear cell and papillary renal cell carcinoma. Histopathology. 2013;62(2):237–46. doi: 10.1111/his.12001. [DOI] [PubMed] [Google Scholar]
- 18.Lin YL, Wang YL, Fu XL, et al. Aberrant methylation of PCDH8 is a potential prognostic biomarker for patients with clear cell renal cell carcinoma. Med Sci Monit. 2014;20:2380–85. doi: 10.12659/MSM.892433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lo CH, Huang WY, Chao HL, et al. Novel application of stereotactic ablative radiotherapy using CyberKnife® for early-stage renal cell carcinoma in patients with pre-existing chronic kidney disease: Initial clinical experiences. Oncol Lett. 2014;8(1):355–60. doi: 10.3892/ol.2014.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lessi F, Mazzanti CM, Tomei S, et al. VHL and HIF-1α: gene variations and prognosis in early-stage clear cell renal cell carcinoma. Med Oncol. 2014;31(3):840. doi: 10.1007/s12032-014-0840-8. [DOI] [PubMed] [Google Scholar]
- 21.Gebauer K, Peters I, Dubrowinskaja N, et al. Hsa-mir-124-3 CpG island methylation is associated with advanced tumours and disease recurrence of patients with clear cell renal cell carcinoma. Br J Cancer. 2013;108(1):131–38. doi: 10.1038/bjc.2012.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin YL, Xie PG, Wang L, et al. Aberrant methylation of protocadherin 17 and its clinical significance in patients with prostate cancer after radical prostatectomy. Med Sci Monit. 2014;20:1376–82. doi: 10.12659/MSM.891247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen WV, Maniatis T. Clustered protocadherins. Development. 2013;140(16):3297–302. doi: 10.1242/dev.090621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berx G, van Roy F. Involvement of members of the cadherin superfamily in cancer. Cold Spring Harb Perspect Biol. 2009;1(6):a003129. doi: 10.1101/cshperspect.a003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qu Y, Dang S, Hou P. Gene methylation in gastric cancer. Clin Chim Acta. 2013;424:53–65. doi: 10.1016/j.cca.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 26.Hu X, Sui X, Li L, et al. Protocadherin 17 acts as a tumour suppressor inducing tumour cell apoptosis and autophagy, and is frequently methylated in gastric and colorectal cancers. J Pathol. 2013;229(1):62–73. doi: 10.1002/path.4093. [DOI] [PubMed] [Google Scholar]
- 27.Yang Y, Liu J, Li X, et al. PCDH17 gene promoter demethylation and cell cycle arrest by genistein in gastric cancer. Histol Histopathol. 2012;27(2):217–24. doi: 10.14670/HH-27.217. [DOI] [PubMed] [Google Scholar]
- 28.Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 29.Iacobazzi V, Castegna A, Infantino V, et al. Mitochondrial DNA methylation as a next-generation biomarker and diagnostic tool. Mol Genet Metab. 2013;110(1–2):25–34. doi: 10.1016/j.ymgme.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 30.Gyparaki MT, Basdra EK, Papavassiliou AG. DNA methylation biomarkers as diagnostic and prognostic tools in colorectal cancer. J Mol Med (Berl) 2013;91(11):1249–56. doi: 10.1007/s00109-013-1088-z. [DOI] [PubMed] [Google Scholar]
- 31.Koukoura O, Spandidos DA, Daponte A, et al. DNA methylation profiles in ovarian cancer: implication in diagnosis and therapy (Review) Mol Med Rep. 2014;10(1):3–9. doi: 10.3892/mmr.2014.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarkar S, Goldgar S, Byler S, et al. Demethylation and re-expression of epigenetically silenced tumor suppressor genes: sensitization of cancer cells by combination therapy. Epigenomics. 2013;5(1):87–94. doi: 10.2217/epi.12.68. [DOI] [PubMed] [Google Scholar]
- 33.Teperek-Tkacz M, Pasque V, Gentsch G, et al. Epigenetic reprogramming: is deamination key to active DNA demethylation? Reproduction. 2011;142(5):621–32. doi: 10.1530/REP-11-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]


