Highlights
-
•
An alpaca-derived γ-H2AX nanobody was generated.
-
•
γ-H2AX chromobody was able to bind and precipitate phosphorylated H2AX peptide.
-
•
γ-H2AX chromobody could be produced in bacterial as well as mammalian cells.
-
•
Alternative epitope recognition by γ-H2AX chromobody was induced by ectopic XRCC1.
-
•
Accessibility of γ-H2AX chromobody was hindered by MDC1 masking in vivo.
Abbreviations: VHH, variable domain of heavy-chain antibody; H2AX, histone H2AX; MDC1, mediator of DNA damage checkpoint-1; MEF, mouse embryonic fibroblast; GFP, green fluorescent protein; RFP, red fluorescent protein; HEK293, human embryonic kidney 293 cells; siRNA, short interfering RNA; XRCC1, X-ray repair cross-complementing protein 1; CKM, casein kinase 2 mutant; ELISA, enzyme linked immunosorbent assay; KLH, keyhole limpet hemocyanin; FRAP, fluorescence recovery after photobleaching
Keywords: Chromobodies, DNA repair, Post-translational modifications, Live cell microscopy, Alpaca heavy chain antibodies, Laser microirradiation
Abstract
Post-translational modifications are difficult to visualize in living cells and are conveniently analyzed using antibodies. Single-chain antibody fragments derived from alpacas and called nanobodies can be expressed and bind to the target antigenic sites in living cells. As a proof of concept, we generated and characterized nanobodies against the commonly used biomarker for DNA double strand breaks γ-H2AX. In vitro and in vivo characterization showed the specificity of the γ-H2AX nanobody. Mammalian cells were transfected with fluorescent fusions called chromobodies and DNA breaks induced by laser microirradiation. We found that alternative epitope recognition and masking of the epitope in living cells compromised the chromobody function. These pitfalls should be considered in the future development and screening of intracellular antibody biomarkers.
1. Introduction
Damage to the DNA is a common event occurring in the life of a cell due to exogenous factors like viruses, carcinogens, different types of radiation, other environmental factors and endogenous factors. These damages may lead to mutations in DNA, causing cancer and other harmful effects to the cell. Hence, the study of cellular DNA damage is very important and requires extensive data on the modification, localization and interaction of cellular components. Phosphorylation on serine 139 of histone H2AX (γ-H2AX) is associated with DNA double strand breaks (DSBs) [1]. Its amplification and signalling function in recruiting other repair factors to the site of damage and its removal after damage repair is completed, makes γ-H2AX the most sensitive biomarker used to study DNA DSBs. Existing methods are based on antibody staining and their application is not suitable for live cells. Though microinjection or bead loading of antibodies is possible for live cell analyses [2], it is not commonly used due to antibody aggregation and toxicity problems as well as nuclear accessibility. Fluorescent fusion proteins allow live cell kinetic measurements but in general cannot be used to distinguish post-translationally modified derivatives of ectopically expressed proteins, albeit, to a certain extent, GFP fused 53BP1 has been used as a surrogate marker for γ-H2AX foci [3]. Recent reports have demonstrated the isolation of the cDNAs coding for the variable regions of the IgG heavy and light chains from hybridoma cell lines and the feasibility of their expression in mammalian cells as fluorescent fusion intracellular antibodies [4]. The need for the isolation of both matched cDNAs (light and heavy chain variable fragments) and to select the appropriate linker sequence to create a functioning fusion of the heavy and light chain variable fragments together with a fluorescent protein, makes this a very difficult and challenging approach.
Here, we introduce a novel technology with the aim of studying DNA double strand breaks based on recognition of post-translational modifications of H2AX in living cells. Sera of llamas and camels were found to have both conventional heterotetrameric and also heavy chain only antibodies. These heavy chain antibodies possess a single variable domain (VHH) and two constant domains CH2 and CH3 [5]. The antigen binding domains of heavy chain antibodies (VHH), known as nanobodies, are of small size (between 12 and 15 kDa) and retain fully functional antigen-binding capacity. These domains can be chemically conjugated to fluorophores or genetically fused with fluorescent tags and subsequently expressed in living cells. Such fluorescently labeled intrabodies, which are called chromobodies, can be used to detect and trace proteins as well as other cellular components in vivo [6,7]. The paratope of nanobodies often adopts a convex-shaped conformation, which allows it to access differently shaped epitopes easier than the flat paratope of conventional antibodies. Nanobodies are very stable, monomeric and their highly soluble nature in aqueous solutions makes them easy to purify on a large scale in heterologous expression systems [8–10]. Their antigen binding affinities can reach subnanomolar range [11] and they possess high stability over a wide range of temperatures and show a very good resistance to chemical and thermal denaturation compared to their conventional counterparts [12–14]. Based on the choice of the epitope, chromobodies can be used to detect endogenous factors and dynamic events occurring in the living cell. In principle, any antigenic property including post-translational modifications or non-protein structures can be detected.
To expand our present understanding of DNA DSBs, we developed a γ-H2AX nanobody/chromobody that can specifically detect histone H2AX phosphorylated on serine 139 and characterize its binding in vitro and in vivo.
2. Material and methods
2.1. Quick outline for the production of chromobodies
-
(1)
An alpaca (Llama pacos) was immunized with γ-H2AX peptide coupled to KLH (keyhole limpet hemocyanin) (Fig. 1A). About two months later, serum was isolated by centrifugation of blood (2000 rpm, 10 min and 4 °C).
Immunizations of alpacas for the purpose of generating antibodies were approved by the Government of Upper Bavaria, according to the animal experimentation law, permit number 55.2.-154-2532.6-9-06.
-
(2)
To test for an immune response, an ELISA test was performed on the serum. 96-well plates (Maxisorp, Thermo Scientific GmbH, Schwerte, North Rhine-Westphalen, Germany) were coated with 1 μg of the antigen and the serum was added in serial dilutions. Bound alpaca antibodies were further detected with HRP-conjugated anti-alpaca IgG antibody (Bethyl Laboratories Inc, Montgomery, Alabama, USA).
-
(3)
Upon positive ELISA test, B cells were isolated with a Ficoll gradient using UNI-SEPMAXI (Novamed Ltd., Jerusalem, Israel).
-
(4)
From the B cells, RNA was extracted with the TRIzol reagent (Life Technologies, Carlsbad, California, USA) according to the manufacturer’s protocol.
-
(5)
From this RNA, complementary DNA (cDNA) was generated using the First-Strand cDNA Synthesis Kit (GE Healthcare, Uppsala, Sweden) according to the manufacturer’s protocol.
-
(6)
VHHs were amplified by three sequential PCR reactions. cDNA was used as the DNA template for the first PCR. For the PCR reactions, the following primers were used:
1st PCR:
Forward primer CALL001:
5′-GTC CTG GCT GCT CTT CTA CA A GG-3′
Reverse primer CALL002:
5′-GGT ACG TGC TGT TGA ACT GTT CC-3′;
2nd PCR:
Forward primer SM017:
5′-CCA GCC GGC CAT GGC TCA GGT GCA GCT GGT GGA GTC TGG-3′
Reverse primer SM018:
5′-CCA GCC GGC CAT GGC TGA TGT GCA GCT GGT GGA GTC TGG-3′;
3rd PCR:
Forward primer A4short:
5′-CAT GCC ATG ACT CGC GGC CAC GCC GGC CAT GGC-3′
Reverse primer 38:
5′-GGA CTA GTG CGG CCG CTG GAG ACG GTG ACC TGG GT-3′.
-
(7)
The amplified product and the plasmid vector pHEN4 were digested with NotI and NcoI restriction enzymes, thus producing compatible overhangs to ligate.
-
(8)
Electro-competent TG1 cells (Agilent Technologies GmbH & Co.KG, Waldbronn, Baden-Wuerttemberg, Germany) were used to generate VHH libraries. They were transformed by electroporation with the ligation preparations performed according to the manufacturer’s protocol.
-
(9)
The transformed TG1 cells were incubated with hyperphage (Progen Biotechnik GmbH, Heidelberg, Baden-Wuerttemberg, Germany). The phage particles presenting the VHH library on their tips were collected.
-
(10)
Solid phase panning is a conventional method to enrich for phages containing the antibody fragments from the whole library. Initially immunotubes were coated with 10 μg of the antigen at 4 °C. Phage particles were added to them and incubated for 1.5 h at room temperature.
-
(11)
The bound phages were eluted with 0.1 M triethylamine over four rounds of panning and used for reinfection of TG1 cells, which were then used for the subsequent panning round.
Fig. 1.
Schematic representation of alpaca derived γ-H2AX specific VHHs generation and biochemical in vitro and in vivo characterization. (A) In the top is shown the alignment of histone H2A variants depicting the unique C-terminal peptide sequence phosphorylated upon DNA damage and used for immunization. Following it, the steps of γ-H2AX specific VHH generation are summarized. (For details, see materials and methods.) (B) In the dot blot assay, γ-H2AX-chromobody (clones 3 and 4; FITC conjugated) was allowed to bind to increasing concentrations of γ-H2AX peptide-KLH and non-phosphorylated control peptide. (C) In the western blot experiments, different amounts of HeLa cell lysates treated or not with neocarcinostatin were loaded and the blot was probed with γ-H2AX-chromobody (clones 3 and 4; FITC conjugated) as well as the commercial γ-H2AX antibody. (D) Selected clones were used for immunoprecipitation experiments. Cells expressing the selected γ-H2AX-chromobody (clones 3 and 4) tagged with GFP or GFP alone were treated with neocarcinostatin. After cell lysis, the extract was incubated with the GFP-binder protein coupled to Sepharose beads [18]. The bound fraction and equivalent input cell lysate control were analyzed by western blot with anti-GFP and anti-γ-H2AX antibodies.
2.2. Phage ELISA
Phage ELISA was used to measure the binding and confirm the specificity to the antigen of the phages selected in the panning method described above. Initially 1 μg of antigen was coated onto 96 well plates. After blocking with 3% milk in PBS, phage particles were added to the plates coated with antigen and incubated at room temperature for 2 h. After washing multiple times with PBST (PBS with 0.05% Tween20), bound phages were detected by standard ELISA procedures using a horseradish peroxidase-labeled anti-M13 monoclonal antibody (GE Healthcare, Uppsala, Sweden).
2.3. Dot blot assay
Dot blot analysis was performed to validate the specificity of the VHH (nanobody) to the phospho epitope. Firstly 2 μg of peptide was spotted onto nitrocellulose membrane and incubated with FITC labeled VHH. The latter was generated via N-hydroxysuccinimide (NHS) based conjugation according to the manufacturer’s protocol (Thermo Scientific GmbH, Schwerte, North Rhine-Westphalen, Germany) and free fluorescent dyes separated using PD-10 desalting columns (GE Healthcare, Uppsala, Sweden). The binding signals were obtained by scanning with a Typhoon Scanner (excitation 480 ± 20 nm, emission: 520 ± 20 nm, GE Healthcare, Uppsala, Sweden) and normalized against the background. Quantification of the signals was performed with the ImageQuant software.
2.4. Mammalian expression plasmids
γ-H2AX-VHH (clones 3 and 4) was cloned in frame into the pEGFP-N1 vector (Clontech Laboratories Inc, Mountain View, California, USA) using BglII/HindIII restriction sites to generate γ-H2AX chromobody mammalian expression plasmid.
To obtain the RFP-XRCC1 full-length construct, human XRCC1 was cloned by amplifying XRCC1 from cDNA using the following primers: XRCC1 forward 5′ AA ACCGGT ATGCCGGAGATCCGCCTCC 3′ (HpaI), XRCC1 reverse 5′ AA GCTAGC GGCTTGCGGCACCACCCC 3′ (NheI), and cloned into the pmRFP-C1 backbone. To obtain the RFP-XRCC1-NTD construct, the sequence after the BRCT1 domain (amino acid number 357–578) was excised by XmaI digestion followed by self ligation. The phospho-mutant RFP-XRCC1-CKM construct was generated from the XRCC1 (CKM) construct described in [15] (obtained from Keith Caldecott; University of Sussex, UK). The CKM mutant region of the XRCC1 (CKM) was excised with XmaI and XhoI and was used to replace the corresponding part within the pmRFP-XRCC1 wild type.
Human MDC1-GFP plasmid was described in [16] (obtained from Roland Kanaar; Erasmus MC University, Rotterdam, The Netherlands).
2.5. Cell culture and transfection
HeLa, HEK293, H2AX knock out and wild-type mouse embryonic fibroblasts (MEFs; obtained from A. Nussenzweig, National Cancer Institute, USA) [17] were used. Cells were cultured at 37 °C, 5% CO2 in DMEM containing 50 μg/ml gentamicin supplemented with 10% fetal calf serum. For passaging, cells were washed with 2 ml PBS, and 1 ml trypsin/EDTA was added and distributed over the cell layer. Once the cells were detached, 9 ml of DMEM with serum were added to stop the enzymatic action of trypsin.
For live cell analysis, cells were seeded 24 h before transfection in a μ-8 well dish (ibidi GmbH, Munich, Bavaria, Germany). For fixed cell analysis, cells were grown on glass cover slips. Polyethylenimine (PEI, Sigma–Aldrich, Steinheim, North Rhine-Westphalia, Germany) with pH 7 was used to transfect the MEF and HEK293 cells and PEI with pH 10 was used for HeLa cells and cells were irradiated, imaged or pelleted one day after transfection. Hiperfect (Qiagen GmbH, Hilden, North Rhine-Westphalia, Germany) was used for the transfection of siRNAs according to the manufacturer’s instructions.
2.6. Immunoprecipitation and western blotting
For immunoprecipitation, HEK293 cells transfected with plasmids coding for γ-H2AX-3 chromobody or GFP alone, were treated with 50 μg neocarcinostatin to generate DSBs. Cell pellets were resuspended in lysis buffer (20 mM Tris/Cl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% NP-40) plus 1 mM PMSF, 1X protease inhibitor mix (Serva Electrophoresis GmbH, Heidelberg, Baden-Wuerttemberg, Germany), 0.5 μg/μl DNase, and 2.5 mM MgCl2 and centrifuged. Then, 50 μl slurry of NHS-activated Sepharose beads covalently coupled with a GFP binding nanobody, as detailed in [18], were equilibrated by washing three times with dilution buffer (20 mM Tris/Cl pH 7.5, 150 mM NaCl, 0.5 mM EDTA). HEK293 cell lysate was added to the equilibrated beads and incubated for four hours at 4 °C. The samples were loaded in an SDS–PAGE followed by western blot analysis.
Proteins separated on SDS–PAGE gels were transferred to a nitrocellulose membrane by semi-dry blotting at 240 mA. The membrane was incubated for one hour with 3% milk powder in Tris buffered saline with 0.075% Tween-20 (TBST) at room temperature. Primary antibodies anti-GFP (1:1000, Roche Diagnostics, Mannheim, Baden-Wuerttemberg, Germany) and anti γ-H2AX (1:1000, clone JBW301, Upstate, Millipore-Merck, Darmstadt, Hessen, Germany) were incubated in 3% milk-TBST at 4 °C overnight. Secondary antibodies (1:1000) were diluted in 3% milk-TBST and incubated for 1 h at room temperature. The membrane was washed with TBST and treated with ECL solution and bound proteins were detected with the Typhoon scanner.
2.7. Western blot
To test the usefulness of the γ-H2AX chromobody compared to the commercial γ-H2AX antibody for western blotting applications, HeLa cells incubated or not with neocarcinostatin (50 ng/ml) were harvested in PBS, snap-frozen in liquid nitrogen and stored at −20 °C. Cell pellets were homogenized in 200 μl RIPA buffer (10 mM Tris/Cl pH 7.5, 150 mM NaCl, 0.1% SDS, 1% Triton X-100, 1% deoxycholate, 5 mM EDTA, 1 μg/ml DNaseI, 2.5 mM MgCl2, 2 mM PMSF, 1X protease inhibitor mix M (Serva)) by repeated pipetting for 40 min on ice. After centrifugation (10 min at 18,000×g) protein concentrations of the supernatants were determined by BCA Protein Assay (Pierce). Lysates were analyzed on SDS–PAGE and transferred onto nitrocellulose membrane as described above. For detection of γ-H2AX, western blots were incubated with FITC labeled γ-H2AX nanobodies (0.2 mg/ml) diluted in 1X PBS, 0.1% BSA for 12 h at 4 °C. Blots were washed 3 times with 1X PBS, 0.05% Tween-20 and detected with the Typhoon scanner.
2.8. Microscopy
Live cell experiments were performed using confocal microscopy. Confocal images were collected using an UltraVIEW VoX spinning disc system (Perkin Elmer) on a Nikon Ti microscope equipped with an oil immersion Plan-Apochromat 60x/1.49 numerical aperture (NA) objective lens (pixel size in XY 120.45 nm/pixel) in a live-cell microscopy chamber (ACU control, Olympus) with a temperature of 37 °C, 5% CO2, and 60% humidity.
For fixed cell analysis, fluorescent images were obtained on a Zeiss Axiovert 200 inverted microscope equipped with a Plan-Apochromat 63x/1.4 NA oil immersion objective lens (pixel size in XY 104 nm/pixel) and a CCD camera AxioCam MRm.
2.9. Irradiation and FRAP experiments
Microirradiation of cells was carried out with a 405 nm diode laser set to maximum power at 100% transmission. Preselected spots of ∼1 μm in diameter within the nucleus were microirradiated for 1200 ms resulting in 1 mJ energy. Before and after microirradiation confocal image series were recorded.
For FRAP analysis, previously microirradiated spots were photobleached with 488 nm laser set to 100% for 600 ms. Similar bleach regions were selected either at the sites of repair or in the nucleoplasm (control) and the kinetics of recovery were monitored.
easyFRAP, a MatLab stand-alone application, was used to visualize the data, normalize the raw recovery curves and curve fitting [19]. Raw intensities of microirradiated area (ROI1), the whole nucleus (ROI2), and a background area (ROI3) as well as the corresponding time-points were calculated using a custom made ImageJ macro [20]. For quantifying the accumulation kinetics, the mean intensity of the microirradiated region was divided by mean intensity of the cell and both the intensities are corrected for the background using the easyFRAP software.
The FRAP fluorescence intensity curves were normalized by setting the first post-bleach value to zero. The normalized curves were subjected to double term curve fitting. The mobile fraction and t-half values were extracted from the double exponential fitting using easyFRAP.
For fixed cell experiments the cells were irradiated prior to fixation with a X-ray tube (GE isovolt titan) set to 90 kV and 19 mA filtered by a 2 mm aluminum sheet. The dose rate was controlled by DIADOS T11003 diagnostic dosimeter and a dose of 2 Gy was achieved by varying the distance and duration of the irradiation.
2.10. Knockdown experiments
For knockdown experiments, siRNA with the sequence of the sense strand 5′-CCAGAAATCTTTATGAATAAA-3′ targeting human MDC1 was used, and universal siRNA (Invitrogen, Karlsruhe, Baden-Wuerttemberg, Germany) was used as a negative control. Hiperfect (see above) was used as a cationic lipid cell transfection reagent. Human cells (HEK 293) were seeded at a density of 4 × 104 cells/well in a μ-8 well dish. 2 μl of Hiperfect transfection reagent was added along with 20 nM MDC1 siRNA in a μ-8 well dish immediately after seeding the cells. After 24 h, the media was removed and fresh media was added. After 48 h the transfection of γ-H2AX-3 chromobody was done using PEI pH 7. After 72 h, the cells were rinsed with fresh media and the microirradiation experiments were performed.
2.11. Immunofluorescence
Cells were grown on glass coverslips, fixed in 3.7% formaldehyde for 10 min at room temperature (RT), and permeabilized for 20 min at room temperature in 0.5% Triton X-100/PBS. Immunofluorescence staining was performed by diluting the antibodies in 4% BSA/PBS for 1 h at room temperature (primary antibodies) and for 45 min at room temperature (secondary antibodies). The following primary antibodies were used: mouse anti-γ-H2AX (1:300, clone JBW301, Millipore GmbH, Schwalbach, Hessen, Germany), anti-MDC1 (1:100, Sigma–Aldrich Inc, Steinheim, North Rhine-Westphalia, Germany). For detection, the cells were stained with donkey anti-mouse IgG Alexa 488 (Jackson Immuno Research, Suffolk, UK) and goat anti-mouse IgG Cy3 (Jackson Immuno Research, Suffolk, UK). Nuclear DNA was visualized with 4,6-diamidino-2-phenylindole (DAPI) (Sigma–Aldrich GmbH, Munich, Bavaria, Germany). Cells were mounted in Vectashield antifade (Vector Laboratories Inc).
3. Results and discussion
3.1. Development of γ-H2AX specific VHHs
The outline in Fig. 1A summarizes the main steps of the nanobody generation process, which are explained in more detail in the methods. Alpacas were immunized with specific peptides following a seven-week period, RNA from lymphocytes was extracted and retrotranscribed to DNA, and the sequences corresponding to the VHH domains were amplified with specific primers for cloning into a phage display vector. This library was then subjected to phage display using plates coated with the antigens (peptides). Two subsequent cycles of biopanning revealed an enrichment of three unique VHH sequences, which were positively tested for antigen binding in a phage ELISA. The binding ability of VHH to γ-H2AX was subsequently tested by a panel of in vitro and in vivo methods.
The selected VHHs were cloned with a C-terminal 6×His-tag, expressed in Escherichia coli and purified using immobilized metal ion affinity chromatography followed by size exclusion chromatography [18]. Purified VHH domains were chemically labeled with an organic dye (FITC) and used for dot blot and western blot assays. To this end the peptides were spotted at different concentrations on the membrane, and incubated with γ-H2AX-3 and 4 chromobodies (clones 3 and 4) confirming its phospho epitope specificity (Fig. 1B). Different concentration of cell lysates extracted from HeLa cells treated or not with neocarcinostatin (NCS) to generate γ-H2AX as well as untreated E. coli cells were used for western blot and the membrane was subsequently probed with γ-H2AX-3 and 4 chromobodies (Fig. 1C). The signals obtained with the chromobodies were specific to damaged cells but showed more background than the commercial anti-γ-H2AX antibody.
3.2. γ-H2AX chromobody interacts with and precipitates the phosphorylated histone
To test the binding specificity in cellulo, we generated a mammalian expression construct comprising the antigen-binding domain of the γ-H2AX-VHH (clones 3 and 4) in fusion with GFP (γ-H2AX-3 and 4 chromobodies). Human HEK293 cells were transfected with the γ-H2AX-3 and 4 chromobodies or GFP alone as negative control. To induce DNA damage, cells were treated with neocarcinostatin to generate DSBs and ensuing γ-H2AX formation. Subsequently, the cellular interaction of γ-H2AX and the γ-H2AX-3 and 4 chromobodies was analyzed by immunoprecipitation experiments. To this end, the respective cell lysates were incubated with the GFP-binding nanobody coupled to Sepharose [18] and the input as well the bound fractions were analyzed by SDS–PAGE followed by immunoblotting. The results showed substantial binding of the γ-H2AX-3 and 4 chromobodies to γ-H2AX, while no γ-H2AX was detectable in the bound fraction of GFP (Fig. 1D).
From the in vitro and in vivo results, the γ-H2AX-3 chromobody showed better binding. Hence, we decided to use this clone for all the subsequent live cell experiments.
3.3. In situ live cell characterization of γ-H2AX chromobody
The γ-H2AX-3 chromobody was next tested for its ability to mark DNA double strand breaks in living cells (Fig. 2). Here, we wanted to mark the phosphorylation event that occurs on the histone H2AX upon DSBs. Initially, cells expressing the γ-H2AX-3 chromobody were irradiated with the 405 nm laser, which has been shown to induce different types of DNA damage including DSBs [21]. No recruitment of γ-H2AX-3 chromobody was observed at the sites of DNA damage (Fig. 2B and D). Hence, XRCC1 (X-ray cross complementing protein 1), a loading protein [22] involved in base excision and single strand break repair in mammalian cells, was co-transfected with the γ-H2AX-3 chromobody to confirm the induction of DNA damage. Recruitment of γ-H2AX-3 chromobody was observed in the presence of the XRCC1 at the microirradiated sites (Fig. 2C and D).
Fig. 2.
γ-H2AX chromobody recruitment to DNA damage sites in living cells. (A) Schematic representation of the experimental strategy. In (B) and (C) HeLa cells were transfected with γ-H2AX-3 chromobody alone or with mRFP-XRCC1 and were microirradiated with a 405 nm laser. Confocal microscopy time series images were acquired 24 h post transfection before and after irradiation. H2AX wild type (E) and knockout (F) MEF cells were transfected with the γ-H2AX-3 chromobody and mRFP-XRCC1. The γ-H2AX-3 chromobody recruitment to microirradiated sites was measured. Scale bar represents 5 μm. (D) and (G) Kinetics of the recruitment of the γ-H2AX-3 chromobody in the presence and absence of XRCC1 is shown for the indicated cell lines. (H) Schematic illustration of FRAP experiments performed on the preselected microirradiated spots. (I-J) Recovery curves of γ-H2AX-3 chromobody in damaged and undamaged sites (± micro IR, respectively) in the presence (J) or absence (I) of XRCC1 overexpression and the corresponding t-half values, as indicated on the left hand side. In both the recruitment and recovery kinetics curves, mean values were plotted and the error bar (shaded and lined) denotes standard deviation.
To check if XRCC1 overexpression aids the binding of γ-H2AX-3 chromobody to the target or provides additional binding sites, we next used H2AX knock out and wildtype MEF cells. Unexpectedly, we observed the recruitment of the chromobody in the presence of XRCC1 in the mutant cells (Fig. 2E–G). The latter suggested either lack of specificity in vivo, which is not corroborated by the immunoprecipitation results (Fig. 1D) or alternative epitope recognition by the chromobody.
Based on these live-cell findings two important questions arose. Why is the γ-H2AX-3 chromobody exclusively recruited when co-expressed with the XRCC1? Why is the γ-H2AX-3 chromobody not recruited to sites of double strand breaks in living cells?
To measure whether XRCC1 influences binding affinity of the γ-H2AX-3 chromobody, we performed FRAP experiments in the presence or absence of XRCC1 (Fig. 2H–J). From the fluorescence intensity recovery curves, the half time (t-half) of the γ-H2AX-3 chromobody at DNA damage sites was calculated. Whereas there was no difference between the mobility of γ-H2AX-3 chromobody at microirradiated sites versus the nucleoplasmic pool, the overexpression of XRCC1 lead to a 1.5–1.7 times slower recovery and correspondingly higher t-half values. This implies that XRCC1 directly or indirectly increases the binding of the γ-H2AX-3 chromobody to microirradiated sites.
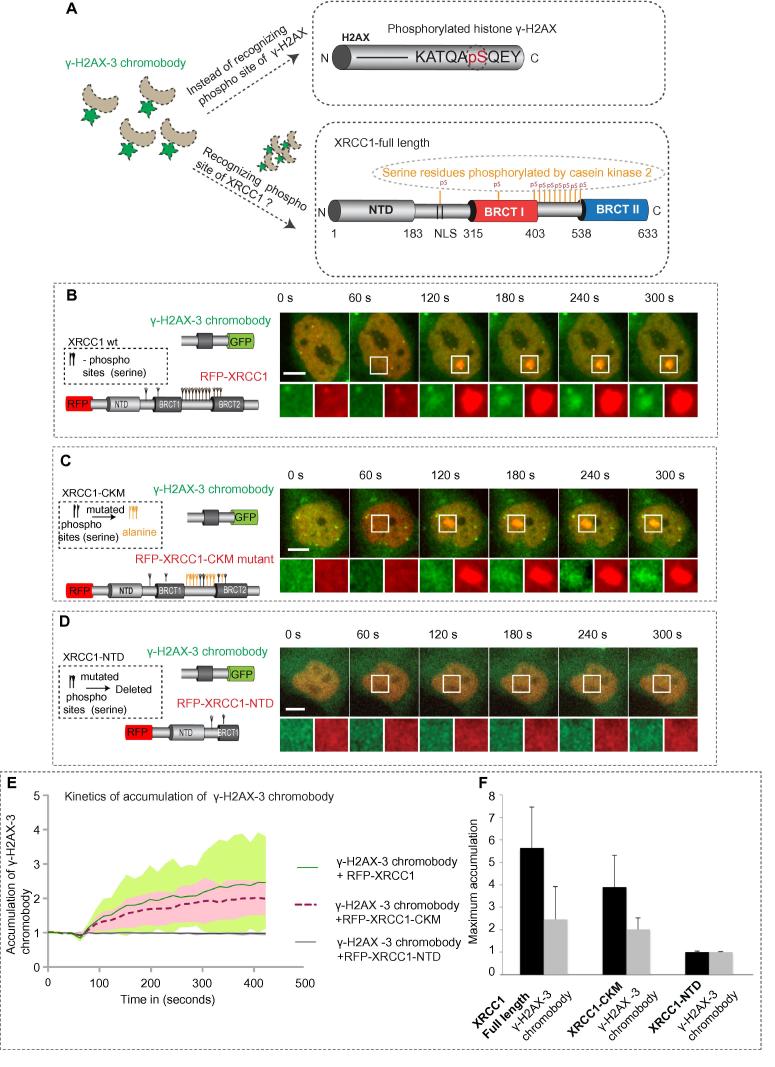
3.4. Alternative epitope recognition of γ-H2AX chromobody
To determine why the chromobody is recruited to sites of DNA damage in the presence of the overexpressed XRCC1, mutants of XRCC1 were developed (Fig. 3). XRCC1 has been found to have multiple phosphorylation sites containing serine residues (Fig. 3A), which are modified by casein kinase 2, a highly conserved protein serine/threonine kinase. Hence, we used multi-phospho site point mutants as well as a deletion mutant containing only the N-terminal domain excluding most of the phosphorylation sites. These mutants were compared with the full length XRCC1 (Fig. 3) to test for alternative phospho epitope recognition. The XRCC1 CKM mutant retaining some of the phosphorylation sites showed only a mild difference in the recruitment kinetics of the γ-H2AX-3 chromobody, when compared to XRCC1 full length. The XRCC1-NTD mutant, on the other hand, could no longer support γ-H2AX-3 chromobody recruitment. This was further substantiated by comparing the maximal accumulation of both proteins (Fig. 3F). Hence, it could be concluded that the chromobody’s recruitment is proportional to the XRCC1 accumulation and XRCC1 recruitment is slightly affected by the phosphosite mutation and completely affected by the deletion mutation. This indicates that either alternative XRCC1 phospho-epitope recognition allows the chromobody to accumulate at damage sites or a phospho-protein whose accumulation is dependent on XRCC1.
Fig. 3.
Efficiency of alternative epitope recognition. (A) Rationale for the chromobody’s alternative phospho epitope recognition. (B-D) HeLa cells cotransfected with the indicated plasmids were microirradiated with a 405 nm laser. Confocal microscopy time series images were acquired 24 h post transfection before and after irradiation. Scale bar represents 5 μm. (E) Kinetics of recruitment of the γ-H2AX-3 chromobody in the presence of the different XRCC1 proteins are shown. Shaded error bars represent standard deviation. (F) Maximum accumulation of the γ-H2AX-3 chromobody is provided along with the maximum accumulation of the XRCC1 mutants. The error bar represents standard deviation.
3.5. Unmasking γ-H2AX epitope in living cells
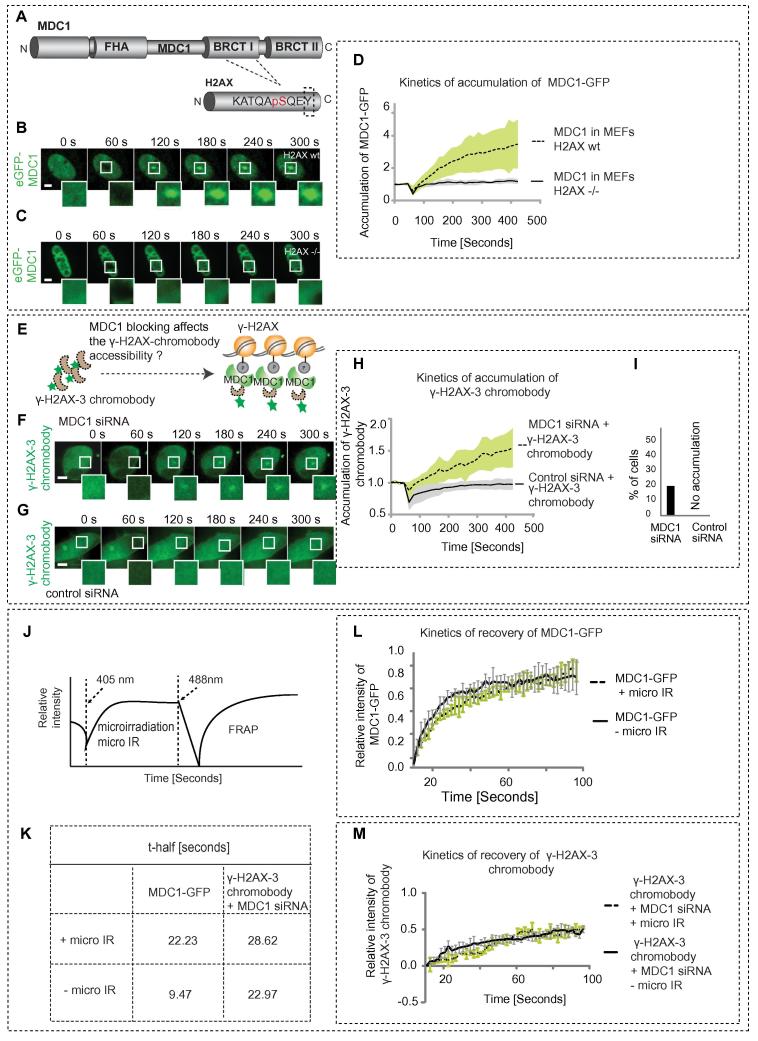
As the immunoprecipitation experiments demonstrated that the γ-H2AX-3 chromobody recognized the γ-H2AX in cell lysates, the question was how this was apparently not the case in living cells. We hypothesized that cellular proteins may mask the epitope. The most likely candidate is MDC1 (mediator of DNA damage checkpoint protein 1). MDC1 recognizes and binds the tyrosine 142 residue of the phosphorylated histone H2AX (Fig. 4A–D) [23]. MDC1 being a ∼250 kDa large protein could, by binding to tyrosine 142, mask the serine at position 139. The binding of MDC1 is known to facilitate the loading of repair proteins and initiate the DNA repair process [24]. Hence MDC1 knock down experiments were performed to check if this allows for epitope recognition by the γ-H2AX-3 chromobody. HEK 293 cells were transfected with MDC1 siRNA and the knockdown of MDC1 was controlled by immunostaining the cells. At 72 h, the siRNA treated cells were microirradiated with a 405 nm laser as before. Mild recruitment of the γ-H2AX-3 chromobody could be observed at the irradiated sites in MDC1 siRNA treated cells but not in the control siRNA cells (Fig. 4E-I). Notably, this occurs in the absence of ectopically expressed XRCC1. This confirms that MDC1 binding in living cells competitively inhibits likely by steric hindrance the binding of the γ-H2AX specific nanobody. Additionally, MDC1 knock down lead also to a mild decrease in the γ-H2AX foci numbers per se, which indicates that MDC1 reinforces γ-H2AX formation [25], maybe by facilitating spreading of the H2AX phosphorylation along the chromatin surrounding the DSB [2]. The lower amount of γ-H2AX upon MDC1 knock down explains the modest maximum amount of γ-H2AX-3 chromobody accumulation by contributing to the reduction of its epitope.
Fig. 4.
Epitope unmasking by knocking down MDC1. (A) MDC1 binds to the tyrosine at the 142nd residue of phosphorylated H2AX. H2AX wild type (B) or knockout (C) MEFs, were transfected with the constructs indicated and microirradiated with 405 nm laser 24 h post transfection. Recruitment to damage sites was measured before and after damage. (D) Kinetics of recruitment of MDC1 in the presence and absence of H2AX is shown. (E) Rationale for epitope masking by MDC1. In (F) and (G) MDC1 siRNA (or control siRNA) plus the indicated constructs were transfected into HEK293 cells and cells were microirradiated as described in detail in the methods. The scale bar represents 5 μm. (H) Kinetics of recruitment of the γ-H2AX-3 chromobody in cells transfected with MDC1 siRNA and control siRNA. (I) The percentage of cells that showed γ-H2AX-3 chromobody recruitment with MDC1 siRNA and control siRNA is shown. (J) Schematic illustration of FRAP experiments performed on the preselected microirradiated spots. (L) and (K) Recovery curves of MDC1-GFP expressed in HeLa cells at damaged and undamaged sites (± micro IR, respectively) and the corresponding t-half values. (M) and (K) Recovery curves of γ-H2AX-3 chromobody expressed in HEK293 cells upon MDC1 knockdown in damaged and undamaged sites (± micro IR, respectively) and corresponding t-half values. In all the recruitment and recovery kinetics curves, mean values were plotted and the error bars (shaded and lined) denote standard deviation.
To further test the effect of MDC1 on γ-H2AX-3 chromobody binding, we performed FRAP experiments (Fig. 4J–M) in the presence or absence of MDC1 (siRNA knockdown). MDC1 mobility itself at microirradiated sites was 2.3 times slower than within the unbound nucleoplasmic pool (Fig. 4K and L). The absence of MDC1 lead to a 1.2 times increase in the t-half of the γ-H2AX-3 chromobody at microirradiated sites (Fig. 4K and M). The latter agrees with the hypothesis that MDC1 presence blocks the access of the γ-H2AX-3 chromobody to its phospho-histone binding site.
4. Conclusions
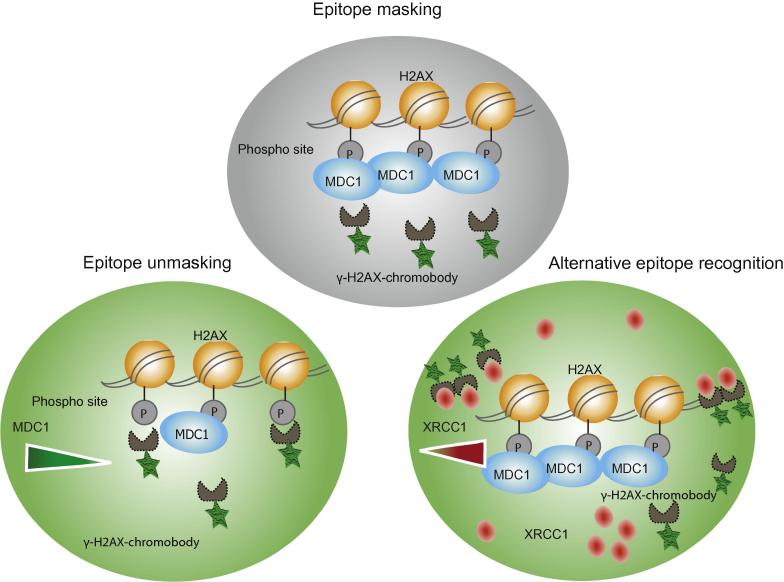
The main objective of this work was the generation and characterization of γ-H2AX specific VHHs to be used in vitro and in living cells. The generated VHHs were found to be functional in vitro as well as in vivo. In addition, the functionality in living cells was assessed by the localization of the fluorescently tagged γ-H2AX-VHH-3 (chromobody) at sites of DNA damage. The ability to identify native proteins within the intact cellular context is the ultimate test for the application of VHHs in living cells. However, during in vivo characterization we found that alternative epitope recognition and epitope masking limit their in vivo applications. These two pitfalls must be considered in the future development and screening of live-cell biomarkers and are summarized in Fig. 5.
Fig. 5.
Model for γ-H2AX chromobody recruitment upon epitope unmasking and alternative epitope recognition.
Conflict of interest
UR and KS are shareholders of the company ChromoTek GmbH, which develops and markets research reagents based on the chromobody technology.
Author contributions
MCC and HL conceived and supervised the study; MR, OM, KS, UL, FDH, AR performed experiments and analyzed the data; MR and MCC wrote the manuscript; all authors commented on the manuscript.
Acknowledgments
We thank Anne Lehmkuhl for excellent technical assistance.
We are indebted to Andre Nussenzweig, Keith Caldecott and Roland Kanaar for the generous gift of biological materials.
This work was supported by grants of the German Research Council to MCC (DFG GRK 1657 TP1B) and to HL (DFG LE 721/13).
Contributor Information
Heinrich Leonhardt, Email: h.leonhardt@lmu.de.
M. Cristina Cardoso, Email: cardoso@bio.tu-darmstadt.de.
References
- 1.Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 2.Stasevich T.J., Hayashi-Takanaka Y., Sato Y., Maehara K., Ohkawa Y., Sakata-Sogawa K., Tokunaga M., Nagase T., Nozaki N., McNally J.G., Kimura H. Regulation of RNA polymerase II activation by histone acetylation in single living cells. Nature. 2014;516:272–275. doi: 10.1038/nature13714. [DOI] [PubMed] [Google Scholar]
- 3.Asaithamby A., Chen D.J. Cellular responses to DNA double-strand breaks after low-dose gamma-irradiation. Nucleic Acids Res. 2009;37:3912–3923. doi: 10.1093/nar/gkp237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sato Y., Mukai M., Ueda J., Muraki M., Stasevich T.J., Horikoshi N., Kujirai T., Kita H., Kimura T., Hira S., Okada Y., Hayashi-Takanaka Y., Obuse C., Kurumizaka H., Kawahara A., Yamagata K., Nozaki N., Kimura H. Genetically encoded system to track histone modification in vivo. Sci. Rep. 2013;3:2436. doi: 10.1038/srep02436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamers-Casterman C., Atarhouch T., Muyldermans S., Robinson G., Hamers C., Songa E.B., Bendahman N., Hamers R. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 6.Zolghadr K., Gregor J., Leonhardt H., Rothbauer U. Case study on live cell apoptosis-assay using lamin-chromobody cell-lines for high-content analysis. Methods Mol. Biol. 2012;911:569–575. doi: 10.1007/978-1-61779-968-6_36. [DOI] [PubMed] [Google Scholar]
- 7.Rothbauer U., Zolghadr K., Tillib S., Nowak D., Schermelleh L., Gahl A., Backmann N., Conrath K., Muyldermans S., Cardoso M.C., Leonhardt H. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat. Methods. 2006;3:887–889. doi: 10.1038/nmeth953. [DOI] [PubMed] [Google Scholar]
- 8.Salema V., Fernandez L.A. High yield purification of nanobodies from the periplasm of E. coli as fusions with the maltose binding protein. Protein Expr. Purif. 2013;91:42–48. doi: 10.1016/j.pep.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Frenken L.G., van der Linden R.H., Hermans P.W., Bos J.W., Ruuls R.C., de Geus B., Verrips C.T. Isolation of antigen specific llama VHH antibody fragments and their high level secretion by Saccharomyces cerevisiae. J. Biotechnol. 2000;78:11–21. doi: 10.1016/s0168-1656(99)00228-x. [DOI] [PubMed] [Google Scholar]
- 10.Conrath K.E., Lauwereys M., Galleni M., Matagne A., Frere J.M., Kinne J., Wyns L., Muyldermans S. Beta-lactamase inhibitors derived from single-domain antibody fragments elicited in the camelidae. Antimicrob. Agents Chemother. 2001;45:2807–2812. doi: 10.1128/AAC.45.10.2807-2812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muyldermans S., Lauwereys M. Unique single-domain antigen binding fragments derived from naturally occurring camel heavy-chain antibodies. J. Mol. Recognit. 1999;12:131–140. doi: 10.1002/(SICI)1099-1352(199903/04)12:2<131::AID-JMR454>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 12.Dumoulin M., Conrath K., Van Meirhaeghe A., Meersman F., Heremans K., Frenken L.G., Muyldermans S., Wyns L., Matagne A. Single-domain antibody fragments with high conformational stability. Protein Sci. 2002;11:500–515. doi: 10.1110/ps.34602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Linden R.H., Frenken L.G., de Geus B., Harmsen M.M., Ruuls R.C., Stok W., de Ron L., Wilson S., Davis P., Verrips C.T. Comparison of physical chemical properties of llama VHH antibody fragments and mouse monoclonal antibodies. Biochim. Biophys. Acta. 1999;1431:37–46. doi: 10.1016/s0167-4838(99)00030-8. [DOI] [PubMed] [Google Scholar]
- 14.Arbabi M., Ghahroudi A., Desmyter L., Wyns R., Hamers S. Muyldermans, Selection and identification of single domain antibody fragments from camel heavy-chain antibodies. FEBS Lett. 1997;414:521–526. doi: 10.1016/s0014-5793(97)01062-4. [DOI] [PubMed] [Google Scholar]
- 15.Loizou J.I., El-Khamisy S.F., Zlatanou A., Moore D.J., Chan D.W., Qin J., Sarno S., Meggio F., Pinna L.A., Caldecott K.W. The protein kinase CK2 facilitates repair of chromosomal DNA single-strand breaks. Cell. 2004;117:17–28. doi: 10.1016/s0092-8674(04)00206-5. [DOI] [PubMed] [Google Scholar]
- 16.Hable V., Drexler G.A., Bruning T., Burgdorf C., Greubel C., Derer A., Seel J., Strickfaden H., Cremer T., Friedl A.A., Dollinger G. Recruitment kinetics of DNA repair proteins Mdc1 and Rad52 but not 53BP1 depend on damage complexity. Plos One. 2012;7 doi: 10.1371/journal.pone.0041943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernandez-Capetillo O., Liebe B., Scherthan H., Nussenzweig A. H2AX regulates meiotic telomere clustering. J. Cell. Biol. 2003;163:15–20. doi: 10.1083/jcb.200305124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rothbauer U., Zolghadr K., Muyldermans S., Schepers A., Cardoso M.C., Leonhardt H. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell. Proteomics. 2008;7:282–289. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Rapsomaniki M.A., Kotsantis P., Symeonidou I.E., Giakoumakis N.N., Taraviras S., Lygerou Z. easyfrap: an interactive, easy-to-use tool for qualitative and quantitative analysis of FRAP data. Bioinformatics. 2012;28:1800–1801. doi: 10.1093/bioinformatics/bts241. [DOI] [PubMed] [Google Scholar]
- 20.Kind B., Muster B., Staroske W., Herce H.D., Sachse R., Rapp A., Schmidt F., Koss S., Cardoso M.C., Lee-Kirsch M.A. Altered spatio-temporal dynamics of RNase H2 complex assembly at replication and repair sites in Aicardi-Goutieres syndrome. Hum. Mol. Genet. 2014;23:5950–5960. doi: 10.1093/hmg/ddu319. [DOI] [PubMed] [Google Scholar]
- 21.B. Muster, DNA Repair and Chromatin, Online-Edition: http://tuprints.ulb.tu-darmstadt.de/id/eprint/3989, TU Darmstadt, 2014.
- 22.Mortusewicz O., Leonhardt H. XRCC1 and PCNA are loading platforms with distinct kinetic properties and different capacities to respond to multiple DNA lesions. BMC Mol. Biol. 2007;8:81. doi: 10.1186/1471-2199-8-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stucki M., Clapperton J.A., Mohammad D., Yaffe M.B., Smerdon S.J., Jackson S.P. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 24.Lou Z., Chen B.P., Asaithamby A., Minter-Dykhouse K., Chen D.J., Chen J. MDC1 regulates DNA-PK autophosphorylation in response to DNA damage. J. Biol. Chem. 2004;279:46359–46362. doi: 10.1074/jbc.C400375200. [DOI] [PubMed] [Google Scholar]
- 25.Jungmichel S., Stucki M. MDC1: The art of keeping things in focus. Chromosoma. 2010;119:337–349. doi: 10.1007/s00412-010-0266-9. [DOI] [PubMed] [Google Scholar]