Highlights
-
•
Amyloid precursor protein (APP) dimerizes more than its C-terminal fragments in cells.
-
•
Mutations of membrane GXXXG motifs affect Aβ production but not APP dimerization.
-
•
Deletion of the APP intracellular domain increases APP dimerization.
Abbreviations: Aβ, β-amyloid peptide; AD, Alzheimer’s disease; APP, amyloid precursor protein; AICD, APP intracellular domain; sAPPα, soluble APPα; sAPPβ, soluble APPβ; CHO, chinese hamster ovary; CTF, C-terminal fragment; DAPT, N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester; DTT, dithiothreitol; ECL, enzymatic chemi-luminescence; ECLIA, electro-chemiluminescence immuno-assay; FBS, fetal bovine serum; FRET, fluorescence resonance energy transfer; KPI, Kunitz-type protease inhibitor; NSAIDs, nonsteroidal anti-inflammatory drugs; PBS, phosphate buffered saline; PS1/PS2, presenilin1/presenilin2; RLU, relative light unit; SP, signal peptide; TM, transmembrane; YFP, yellow fluorescent protein
Keywords: Alzheimer disease, APP, Dimerization, GXXXG motifs, Amyloid beta peptide, Split luciferase
Abstract
Alzheimer’s disease (AD) is a neurodegenerative disease that causes progressive loss of cognitive functions, leading to dementia. Two types of lesions are found in AD brains: neurofibrillary tangles and senile plaques. The latter are composed mainly of the β-amyloid peptide (Aβ) generated by amyloidogenic processing of the amyloid precursor protein (APP). Several studies have suggested that dimerization of APP is closely linked to Aβ production. Nevertheless, the mechanisms controlling APP dimerization and their role in APP function are not known. Here we used a new luciferase complementation assay to analyze APP dimerization and unravel the involvement of its three major domains: the ectodomain, the transmembrane domain and the intracellular domain. Our results indicate that within cells full-length APP dimerizes more than its α and β C-terminal fragments, confirming the pivotal role of the ectodomain in this process. Dimerization of the APP transmembrane (TM) domain has been reported to regulate processing at the γ-cleavage site. We show that both non-familial and familial AD mutations in the TM GXXXG motifs strongly modulate Aβ production, but do not consistently change dimerization of the C-terminal fragments. Finally, we found for the first time that removal of intracellular domain strongly increases APP dimerization. Increased APP dimerization is linked to increased non-amyloidogenic processing.
1. Introduction
The amyloid precursor protein (APP) is a ubiquitously expressed type 1 transmembrane protein [1,2]. APP undergoes proteolysis via two distinct pathways known as the amyloidogenic and the non-amyloidogenic pathways. APP processing is initiated by the shedding of the large ectodomain by either an α-secretase (non-amyloidogenic pathway) or the β-secretase BACE1 (amyloidogenic pathway). APP β-cleavage generates a membrane-anchored β C-terminal fragment (βCTF or C99), which is further cleaved by the γ-secretase complex to generate the Aβ peptides. The 40 and 42 amino acids Aβ isoforms (Aβ40 and Aβ42, respectively) are the major constituents of the senile plaques, a typical lesion found in the brain of patients with Alzheimer’s disease (AD) [4]. Mutations responsible for inherited or familial AD cases (FAD) are located in the APP or presenilin genes (PS1 and PS2). The presenilin proteins are the catalytic subunits of the γ-secretase. AD mutations typically result in an increased Aβ42/Aβ40 ratio [2]. Imbalanced production of Aβ, along with its aggregation and accumulation in the brain, may therefore play a critical role in the onset and progression of AD [5].
Although Aβ involvement in the pathology has been extensively studied over the past decades, our knowledge of the physiological function of APP and the cellular mechanism regulating its processing remain remarkably incomplete. APP belongs with its two paralogs APLP1 and APLP2 to the APP-like protein family. Unique features in each member of the family could account for its specialized and specific function [6]. Ten APP isoforms generated by alternative splicing of the APP transcript have been identified [3]. The major ones (APP695, APP751 and APP770) differ in their extracellular domain by a Kunitz-type protease inhibitor (KPI) domain present in non-neuronal isoforms (APP751 and APP770), but absent in the neuronal one (APP695).
APP has been proposed to mediate dendritic spine arrangement, neural cell migration and synapse formation, including neuromuscular junctions formation [7] that could underlie the neuromuscular phenotype observed in APP knock-out mice [8]. The possible physiological role of APP can be related to its structural properties. APP resembles a transmembrane receptor with an extracellular region displaying features of a cell surface receptor or an adhesion molecule [9,10]. These different functional regions include copper binding and growth factor-like domains and are required for homo- and heterophilic interactions [11,12]. For instance, APP has been shown to interact with Notch [11,13], another major γ-secretase substrate, and even with the Aβ peptide generated by its processing [14]. APP dimerization is therefore likely to play a pivotal role in its processing and function. Recent studies have indicated that APP dimerization involves both the E1 and KPI regions of the ectodomain [11,15–17] and GXXXG motifs of the transmembrane (TM) domain [18,19]. GXXXG motifs are structural determinants favoring close apposition of TM helices and formation of TM dimers [20,21,38]. The TM region of APP contains three consecutive GXXXG motifs. The FAD A21G mutation (Aβ numbering) known as the Flemish mutation [24] extends the GXXXG interface by adding a fourth GXXXG motif and triggers Aβ production. This strongly suggests that TM interactions are involved in pathophysiological processing of APP [19,23,25,26,33]. It is of particular interest to clearly establish the relation between APP dimerization and its processing, especially its cleavage at the γ site. Studies on APP dimerization have largely been carried out using biochemical approaches (crosslinking, co-immunoprecipitation) [27] focused on TM domain interactions in reconstituted micelles or membrane bilayers [22] or have used purified peptides for structural approaches [27–30]. Very few studies have addressed APP dimerization in living cells. Although split fluorescent proteins assays [13,31,32] have revealed a positive role of the KPI domain in APP dimerization, the role of TM dimerization has appeared much more controversial. It has even been suggested that the TM domain plays only a marginal role in full-length APP dimerization [9,27].
Indeed, the extent of APP and CTFs dimerization in living cells is poorly known. There is no information about the respective contribution of its 3 major domains, and especially of its intracellular domain in this process. The link between APP dimerization and processing is controversial [27,36]. Here we use a new dynamic and highly sensitive split protein assay, the split luciferase assay [37] to define the role and the contribution of the different APP regions to dimerization and clarify the correlation between its dimerization and processing. Our major findings are that full-length APP forms more dimers than APP β and αCTFs. Mutations in the GXXXG motifs, including FAD mutants (Flemish), do not consistently alter dimerization. Strikingly, deletion of intracellular domain strongly favors dimerization. Finally, we found that the extent of dimerization is not correlated to Aβ production, but that increased dimerization observed with APP lacking its intracellular region is linked to increased non-amyloidogenic processing.
2. Material and methods
2.1. Chemicals and reagents
Restriction enzymes, Taq DNA polymerase, all culture media, penicillin-streptomycin solution and Lipofectamine® transfection reagent, Nu-Page® Novex® 4–12% Bis-Tris gels and buffers were from Life Technology Corporation (Carlsbad, CA). Fetal bovine serum (FBS) for culture media was purchased from Thermo Scientific (Rockford, IL). Transfection reagent Trans-IT2020 was from Mirus Bio Corporation (Madison, WI). Analytical grade solvents, salts and poly-l-lysine were from Sigma-Aldrich (St Louis, MO). N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine-butyl ester (DAPT) was from Calbiochem (Camarillo, CA). Protease inhibitor cocktail was purchased from Roche (Basel, Switzerland). BCA protein assay kit was from Pierce (Rockford, IL, USA). Nitrocellulose membranes were obtained from GE Healthcare (Fairfield, CT). ECL reagents were obtained from Perkin Elmer Inc. (Waltham, MA). Gaussia luciferase substrate Coelenterazine native was purchased from Prolume® Ltd. (Pinetop, AZ). The luciferase cell lysis buffer was from New England Biolabs (Ipswich, MA). The following primary antibodies were used: anti-amyloid β antibody, clone W0-2 (EMD Millipore, Billerica, MA), anti-amyloid precursor protein, C-terminal antibody (Sigma-Aldrich, St Louis, MO), anti-GLuc antibody (New England Biolabs, Ipswich, MA). Fluorescent nucleic acid stain DAPI was obtained from Sigma–Aldrich (St Louis, MO). Secondary antibodies coupled to HRP were obtained from Amersham Bioscience (Uppsala, Sweden) and fluorescent secondary antibodies coupled to Alexa fluorochromes were from Life Technology Corporation (Carlsbad, CA). Fluorescent mounting medium was from DAKO (Agilent Technologies, Santa Clara, CA, USA).
2.2. Cells lines and cell culture
Chinese hamster ovary (CHO) cell lines were grown in Ham’s F-12 medium. The medium was supplemented with 10% of fetal bovine serum (FBS) and penicillin–streptomycin solution (10 units-10 μg). All cell cultures were maintained at 37 °C in a humidified atmosphere (5% CO2).
2.3. Plasmids, site-directed mutagenesis and cloning
GCN4 leucine zipper split-luciferase constructs Zip-hGLuc1 and Zip-hGLuc2 in pcDNA3.1 vectors were obtained from the group of S.W. Michnick [37]. All the constructs expressing APP and APP fragments fused to humanized Gaussia luciferase (hGluc) halves were obtained by PCR amplification of APP sequences encoded by expression vectors previously described [19] with forward and reverse primers harboring the NotI and ClaI restriction sites, respectively. PCR products were digested and further inserted in the NotI/ClaI restrictions sites of the Zip-hGLuc1 and Zip-hGLuc2 constructs, removing the GCN4 leucine zipper sequence of the backbone. All constructs were verified by full sequencing (Macrogen Europe, Amsterdam, The Netherlands). C83 mutants were obtained by Quick-change site-specific mutagenesis (Stratagene, La Jolla, CA) as previously described [31].
2.4. Cell transfection and treatment
CHO cells were transfected with Lipofectamin reagent 24 h after seeding following manufacturer’s instructions. Plasmids expressing the split-luciferase proteins were cotransfected in a 1:1 ratio. The control plasmid (Mock) was the corresponding empty vector. MEF cells (PS+/+ and PS−/−) were transfected using Trans-IT2020 according to the manufacturer’s instructions. CHO cells were treated with DAPT for 15 h at a final concentration of 1 μM. 48 h after transfection, medium was collected, treated with protease inhibitors cocktail (Roche) and stored at 20 °C for ECLIA assay. Cells were harvested and lysed in Luciferase Cell Lysis Buffer (New England Biolab) and pelleted by quick centrifugation at 4 °C for 1 min. Protein concentrations of cell lysates were measured by the BCA protein assay kit (Pierce). Cell lysates were further used for Gaussia luciferase assay and Western blotting.
2.5. Gaussia luciferase assay
Samples were aliquoted in 5 ml polystyrene round-bottom tubes at a final concentration of 10 μg of proteins in 20 μl in Luciferase Cell Lysis Buffer. Native coelenterazine was reconstituted as a stock solution of 1 mg/ml in methanol (stored frozen), diluted 30 min prior reading in DMEM without phenol red and used at a final concentration of 20 μM. 50 μl of coelenterazine was added to tubes and luminescence directly measured on a Sirius Luminometer (Berthold, Pforzheim, Germany).
2.6. Western blotting
Ten μg of protein of cell lysates were heated for 10 min at 70 °C in loading buffer (Luciferase Cell Lysis Buffer with 0.5 M DTT and staining Nupage blue™), loaded and separated onto 4–12% Nupage™ bis-Tris gel, and then transferred for 2 h at 30 V onto nitrocellulose membranes. Ponceau Red staining was used to check gel loading and transfer accuracy. After blocking with 5% non-fat milk in PBS, membranes were incubated overnight at 4 °C with one of the primary antibodies: anti-amyloid β antibody, clone W0-2 (1/2500), anti-amyloid precursor protein, C-terminal antibody (1/2000), anti-hGLuc antibody (1/2000). Membranes were washed with PBS-Tween (0.005%) and incubated with the secondary antibodies anti-mouse (1:10,000) or anti-rabbit (1:10,000) coupled to peroxidase prior to ECL detection (GE Healthcare). Signals were quantified with a Gel Doc 2000 imaging system coupled to a Quantity One™ software (Bio-Rad).
2.7. Immunocytochemistry
Cells were seeded on coverslips previously incubated with poly-l-lysine (10 mg/ml). Prior to staining, cells were rinsed twice with Opti-MEM® (Life Technology Corporation) and fixed with 4% paraformaldehyde (PFA) for 15 min. After 3 washes in PBS, cells were permeabilized with PBS1X/0.3% Triton100X for 30 min and blocked in PBS1X/fetal bovine serum 5%/0.1% Triton100X for 30 min. Primary antibodies were prepared in the blocking solution and incubated O/N at 4 °C. Primary antibodies used were anti-human amyloid β antibody, clone W0-2 (1:2500), anti-amyloid precursor protein, C-terminal antibody (1:2000), anti-hGLuc antibody (1:2000). After 3 washes in PBS, cells were incubated with secondary antibodies (goat anti-mouse Alexa 465 and goat anti-rabbit Alexa 488 and 465, 1:500 in blocking solution) and DAPI (1:2000) for 1 h at 4 °C. After 3 washes in PBS, cells were stored in PBS-azide 0.1% at 4 °C or mounted with fluorescent mounting medium for coverslips. Pictures were acquired with an Evos fluorescence microscope (Advanced Microscopy Group, Mill Creek, Washington, USA) or with an Olympus Fluoview confocal microscope (Olympus America Inc., Center Valley, Pennsylvania, USA).
2.8. Aβ and sAPP measurements
Aβ38, Aβ40 and Aβ42 peptides were quantified in the cell medium as previously described [39] by the Aβ multiplex electro-chemiluminescence immunoassay (ECLIA) (Meso Scale Discovery, Gaithersburg, MD). sAPPα and β were quantified using a sAPPα/sAPPβ multiplex ECLIA (Meso Scale Discovery). Cells were conditioned in serum-free medium for 16 h before harvesting. Cell medium was then collected and Aβ or sAPP were quantified according to the manufacturer’s instructions. Aβ produced from split-luciferase constructs was quantified with the human Aβ specific 6E10 multiplex assay.
2.9. Statistical analysis
The number of samples (n) in each experimental condition is indicated in figure legends. The data were analyzed using GraphPad Prism software by analysis of variance (ANOVA) followed by unpaired t test (2 experimental conditions) or by Bonferroni’s Multiple Comparison tests (more than 2 experimental conditions).
3. Results
3.1. APP and APP C-terminal fragments dimerization in cells analyzed by luciferase complementation assays
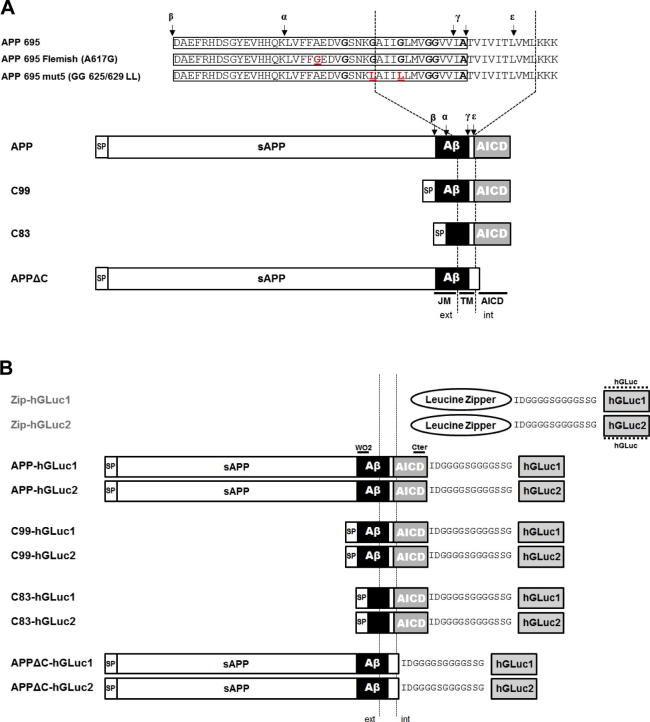
To analyze human APP dimerization and the contribution of its extracellular, juxtamembrane/transmembrane (JM/TM) and intracellular (AICD) domains to this process, we generated vectors expressing the full-length and truncated APP proteins (Fig. 1A) fused to complementary humanized Gaussia luciferase (hGluc) fragments referred to as hGluc1 and hGluc2 corresponding to N-terminal and C-terminal moieties, respectively. C99 and C83 correspond to β- and α-secretase cleavage products fused to the APP signal peptide. APPΔC corresponds to the APP protein truncated after the KKKYQ sequence at the TM/intracellular interface. To analyze the role of membrane GXXXG motifs in dimerization, we generated 2 mutants modifying the interactive properties of the GXXXG interface [19,24]. The first mutation corresponds to the FAD Flemish mutation (A617G, APP695 numbering or A21G, Aβ numbering). It extends the GXXXG interface and helical structure of the surrounding residues [29]. The double GG625/629LL mutant (GG29/33LL, Aβ numbering) hereafter referred as mutant 5 (mut5) carries glycine to leucine mutations of the central GXXXG motif. These mutations have been previously reported to affect interaction of APP TM helices and strongly impair amyloidogenic processing [19]. As a positive control, we used yeast transcription factor GCN4 leucine zipper fusion proteins (Zip-hGLuc1 and Zip-hGLuc2) [37]. The leucine zipper of GCN4 is a strong dimerization domain. All the constructs generated for this study are represented in Fig. 1A and B.
Fig. 1.
Schematic representation of the different APP split-luciferase constructs. (A) Schematic representation of the different human APP and APP C-terminal fragments generated for fusion to the humanized Gaussia luciferase moieties (hGLuc). APPΔC corresponds to APP695 deleted from its intracellular C-terminal domain (stop after the KKKQY intracellular sequence). C99 and C83 correspond to the APP β and α C-terminal fragments, respectively. All the N-terminally truncated CTFs are fused to the APP signal peptide (SP). Abbreviations are as follows: TM, transmembrane; JM, juxtamembrane; AICD, APP intracellular domain; ext, extracellular; int, intracellular. The positions of Flemish and mutant 5 (mut5) mutations are underlined and amino acid substitutions are in red. The cleavage sites of α (α)-, β (β)- and γ (γ and ε)-secretases are indicated by arrows. (B) Schematic representation of APP constructs fused to hGLuc moieties (hGLuc1 and hGLuc2). The epitopes of the human-specific W0-2 antibody, the APP C-terminal and hGLuc antibodies are indicated.
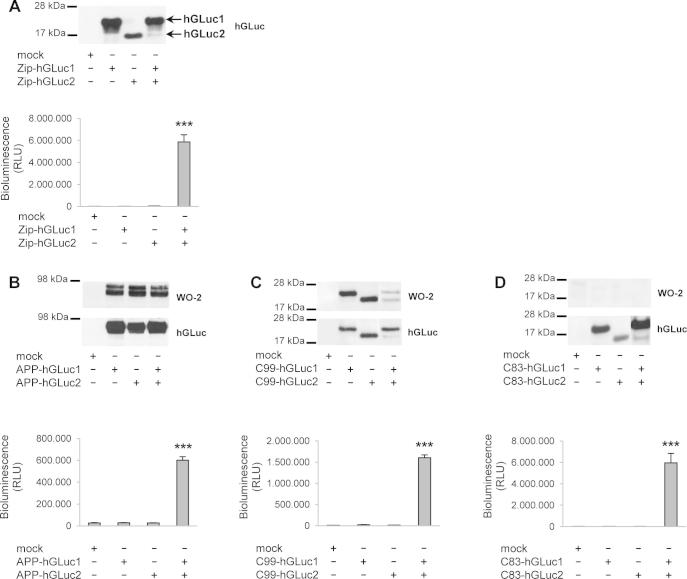
We first validated the luciferase complementation approach as a tool to measure protein dimerization in living CHO cells by measuring bioluminescence upon transfection with the GCN4 leucine zipper hGLuc constructs. Western blotting of cell lysates indicated that Zip-hGLuc1 and Zip-hGLuc2 were detected as 17 and 18 kDa bands recognized by the polyclonal anti-hGLuc antibody (Fig. 2A). Transfection of Zip-hGLuc1 and Zip-hGLuc2 alone did not generate any bioluminescent signal whereas co-transfection of both generated high levels of luciferase activity (Fig. 2A). Co-expression of the hGLuc1 and hGLuc2 moieties alone (not fused to any APP protein sequence) did not generate luciferase activity (data not shown). This clearly validated the luciferase complementation approach as a very sensitive tool to measure dimerization in cells.
Fig. 2.
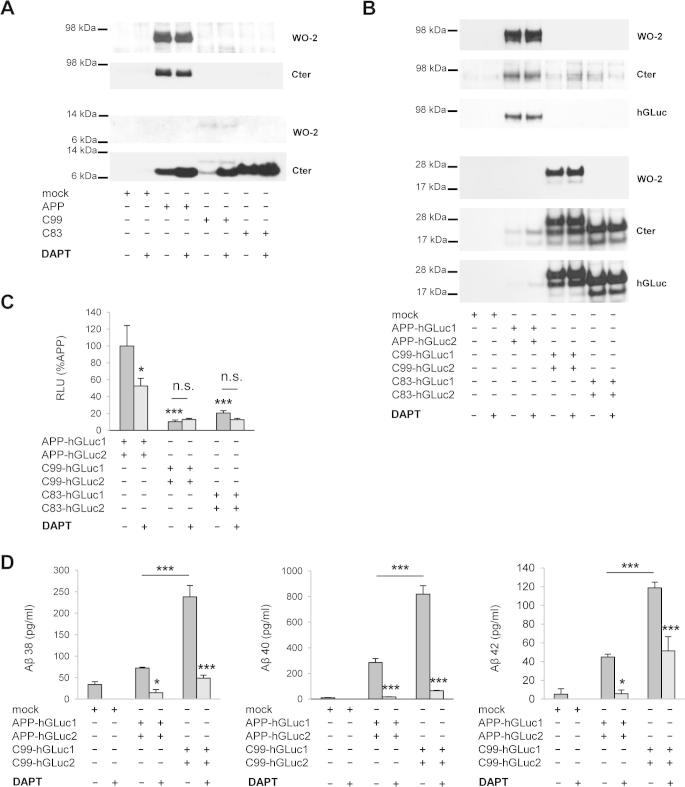
Dimerization of APP and APP C-terminal fragments in living cells measured by the split-luciferase complementation assay. (A) Validation of the luciferase complementation assay for measuring protein dimerization in CHO cells. Cells were transfected with the control empty vector (mock) or the GCN4 leucine zipper–coding sequences fused to hGLuc moieties (Zip-hGluc1 and 2). Expression of the fusion proteins was checked in cell lysates by Western blotting with the hGLuc antibody (top). Luciferase activity (bioluminescence) was measured and expressed as RLU (bottom). Values (means ± SEM) are representative of 3 independent experiments (n = 4 in each experiment). ***p < 0.0001, as compared to control (mock). APP-hGLuc1 and 2 (B), C99-hGLuc1 and 2 (C) or C83-hGLuc1 and 2 proteins (D) were transfected in CHO cells. Protein expression was monitored in cell lysates by Western blotting with the W0-2 and hGluc antibodies (top panels). Luciferase activity was measured and expressed as RLU (bottom). Values (means ± SEM) are representative of 3 independent experiments (n = 4 in each experiment). ***p < 0.0001, as compared to control (mock).
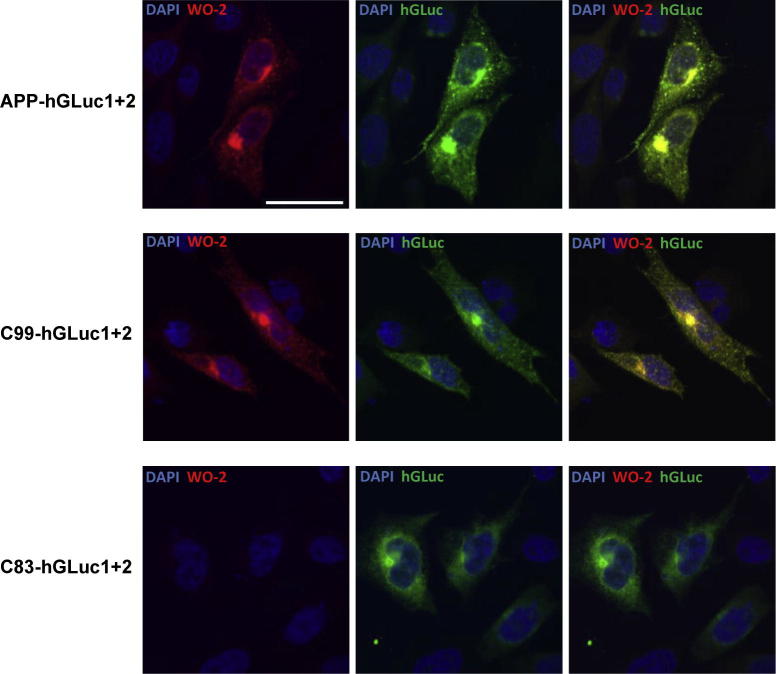
We next compared dimerization of the APP, C99 and C83 constructs (Fig. 2B–D). Immunoblots performed with the W0-2 antibody, the hGLuc antibody or the APP C-terminal antibody (Cter) showed that all constructs were expressed at readily detectable levels. Immunolabelling of the double transfected cells indicated a vesicular localization of the APP fusion proteins (Fig. 3), particularly in the perinuclear region. APP hGLuc fusion proteins had a subcellular distribution consistent with a post-ER/cis-Golgi localization previously reported for endogenous APP and APP fragments, but also for fusion constructs used in split protein assays [27,31,40]. Signals of antibodies directed against human APP epitopes (W0-2) or hGLuc epitopes completely co-localized, excluding the hypothesis that spurious cleavage or degradation products could be responsible for bioluminescent signals.
Fig. 3.
Localization of split-luciferase constructs in CHO cells. Cells were co-transfected with the two split-luciferase constructs expressing either APP C99 or C83. Nuclei were stained with DAPI, and APP fusion constructs were stained by the W0-2 and/or hGLuc antibodies. Scale bar: 5 μm.
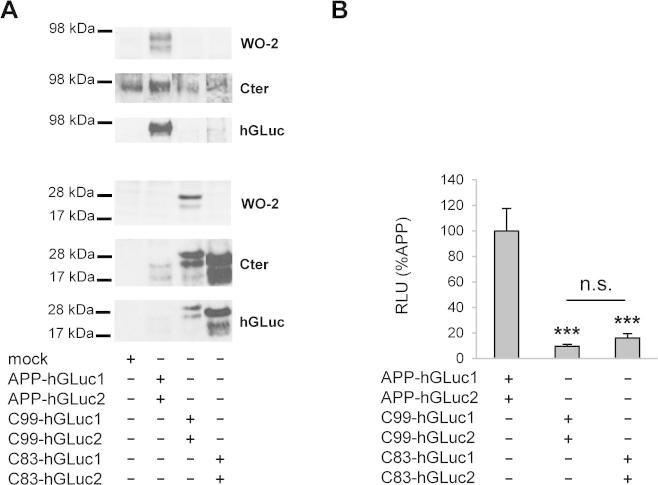
After normalization to expression levels of the fusion proteins (Fig. 4A), our results indicated that APP dimerizes significantly more (∼5 times) than its α and β C-terminal fragments (Fig. 4B). Similar results were obtained with all the antibodies used for cell lysate quantifications (W0-2, Cter or hGLuc). These observations suggest that C-terminal fragments of APP (β or αCTFs) form dimers to a smaller extent than full-length APP. We further investigated this hypothesis by treating transfected cells with DAPT, a γ-secretase allosteric inhibitor, inducing the accumulation of γ-secretase substrates in cells. DAPT effects were consistent with those observed on non-tagged human APP in the same cells (Fig. 5A) [19]. DAPT treatment resulted mainly in the accumulation of αCTFs (Fig. 5A and B). Bioluminescence measured after DAPT treatment indicated a small -although significant- decrease in APP dimerization, but importantly no increase in αCTF or βCTF dimerization was measured under the same conditions (Fig. 5C). Similar results were observed in PSdKO mouse embryonic fibroblast (MEFs) that are devoid of γ-secretase activity (data not shown). Together, these data indicate that APP CTFs dimerize much less than full-length APP. Their accumulation by a γ-secretase inhibitor did not increase dimerization, suggesting that only a pool of CTFs are forming dimers, which are not accessible to cleavage by γ-secretase. DAPT treatment efficiently inhibited the γ-cleavage of APP split luciferase constructs, as evidenced by the strong inhibition of Aβ release (Fig. 5D).
Fig. 4.
Comparison of APP and APP C-terminal fragment dimerization. Cells were transfected with the control empty vector (mock), the APP-hGLuc1 and 2, C99-hGLuc1 and 2 or C83-hGLuc1 and 2 constructs. (A) Protein expression was monitored in cell lysates by Western blotting with the W0-2, Cter or hGLuc antibodies. (B) Luciferase activity was measured and expressed as RLU normalized to APP (set to 100%). Values (means ± SEM) are representative of 3 independent experiments (n = 4 in each experiment). ***p < 0.001, n.s. (non-significant), as compared to APP-hGLuc1 and 2.
Fig. 5.
Effects of gamma-secretase inhibition on APP C99 and C83 dimerization. CHO cells transfected with the empty vector (mock) or the luciferase constructs expressing either APP, C99 and C83 or APP-hGLuc1 and 2, C99-hGLuc1 and 2 and C83-hGLuc1 and 2. Cells were treated with DAPT 1 μM for 18 h. (A) Expression of the non-tagged proteins and effect of the treatment on metabolism were checked in cell lysates by western blotting with APP specific antibodies (W0-2 and Cter). (B) Expression of the hGLuc fusion proteins was checked in cell lysates by Western blotting with the hGLuc antibody and APP specific antibodies (W0-2 and Cter). (C) Luciferase activity measured was expressed as RLU normalized to APP-hGLuc1 and 2 (set to 100%). Values (means ± SEM) are representative of 3 independent experiments (n = 4 in each experiment). *p < 0.05, ***p < 0.001, n.s. (non-significant), as compared to APP-hGLuc1 and 2. (D) Gamma-secretase inhibition was confirmed by monitoring Aβ38, Aβ40 and Aβ42 production by ECLIA in the culture medium of cells expressing APP-hGLuc1 and 2 or C99-hGLuc1 and 2. Results are given as Aβ levels in pg/ml. Values (means ± SEM) are representative of 3 independent experiments (n = 4 in each experiment). *p < 0.05 and ***p < 0.001, as compared to non-treated cells.
3.2. GXXXG motifs are not critical for CTFs dimerization
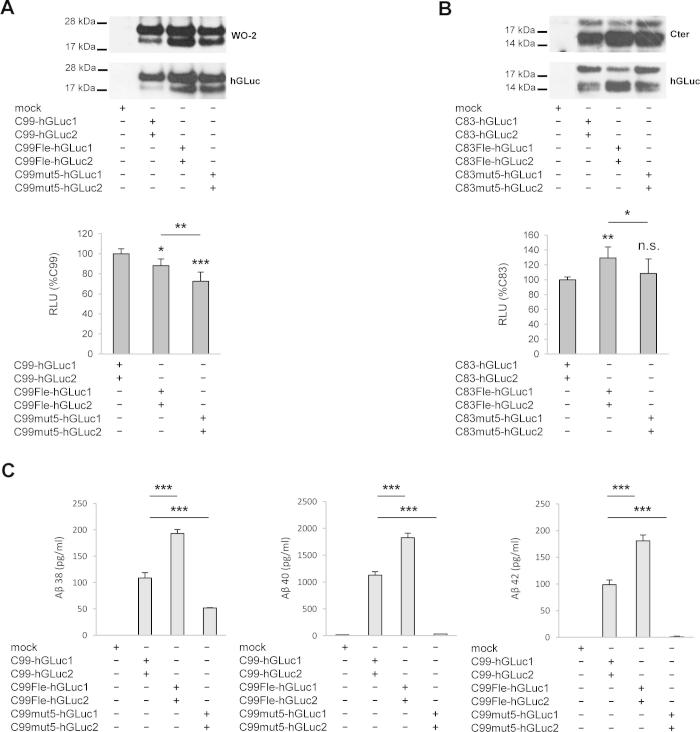
Our recent work showed that mutation of the GXXXG motifs modifies TM interactions of the APP CTFs [19]. To determine whether GXXXG motifs are involved in CTFs homo-dimerization by a quantitative approach, we compared dimerization of C99 (corresponding to the βCTF of APP), C99 with the Flemish mutation or C99 mut5 (Fig. 1A). Both C99 mutants exhibited the same dimerization profile as wild-type C99 (Fig. 6A). Similar experiments were carried out with C83 constructs, corresponding to the APP αCTF. C99 and C83 constructs had similar expression profiles (Fig. 6B). Both the Flemish mutant and mut5 showed a slight but significant decrease in dimerization of C99. For C83, dimerization of the Flemish mutant was slightly but significantly increased, whereas dimerization of mut5 was not affected. Mutation of the GXXXG motifs was reported to dramatically impact Aβ production (Fig. 6C). We previously showed [19,29] that the Flemish FAD mutation increased Aβ production whereas mutant 5 strongly decreased it. Our data (Fig. 6) indicate that mutations of GXXXG motifs strongly affect APP γ-cleavage without a corresponding change in dimerization of the APP CTFs.
Fig. 6.
Involvement of GXXXG motifs in CTF dimerization and Aβ production. CHO cells were transfected with C99-hGLuc1 and 2 or C83-hGLuc1 and 2 and their GXXXG Flemish (Fle) and mutant 5 (mut5) corresponding mutants. (A) Cells transfected with the control empty vector (mock), the C99-hGLuc1 and 2, C99Fle-hGLuc1 and 2 or C99mut5-hGLuc1 and 2 proteins. Protein expression was monitored in cell lysates by Western blotting with the W0-2 or hGluc antibodies (top panels). Luciferase activity was measured and expressed as RLU normalized to non-mutated C99 (bottom). Values (means ± SEM) are representative of 5 independent experiments (n = 4 in each experiment). *p < 0.05, **p < 0.01 and ***p < 0.001, as compared to C99-hGLuc1 and 2. (B) Cells transfected with the control empty vector (mock), the C83-hGLuc1 and 2, C83Fle-hGLuc1 and 2 or C83mut5-hGLuc1 and 2 proteins. Protein expression was monitored in cell lysates by Western blotting with the Cter or hGLuc antibodies (top panels). Luciferase activity was measured and expressed as RLU normalized to non-mutated C83 (bottom). Values (means ± SEM) are representative of 3 independent experiments (n = 4 in each experiment). *p < 0.05, **p < 0.01 and n.s. (non significant), as compared to C83-hGLuc1 and 2. (C) Aβ 38, 40 and 42 production for C99-hGLuc1 and 2, C99Fle-hGLuc1 and 2 or C99mut5-hGLuc1 and 2 was measured by ECLIA in the culture media and given in pg/ml. Values (means ± SEM) are representative of 3 independent experiment (n = 4 in each experiment). ***p < 0.001, as compared to non-mutated C99.
3.3. APP C-terminal region regulates APP dimerization and non-amyloidogenic processing
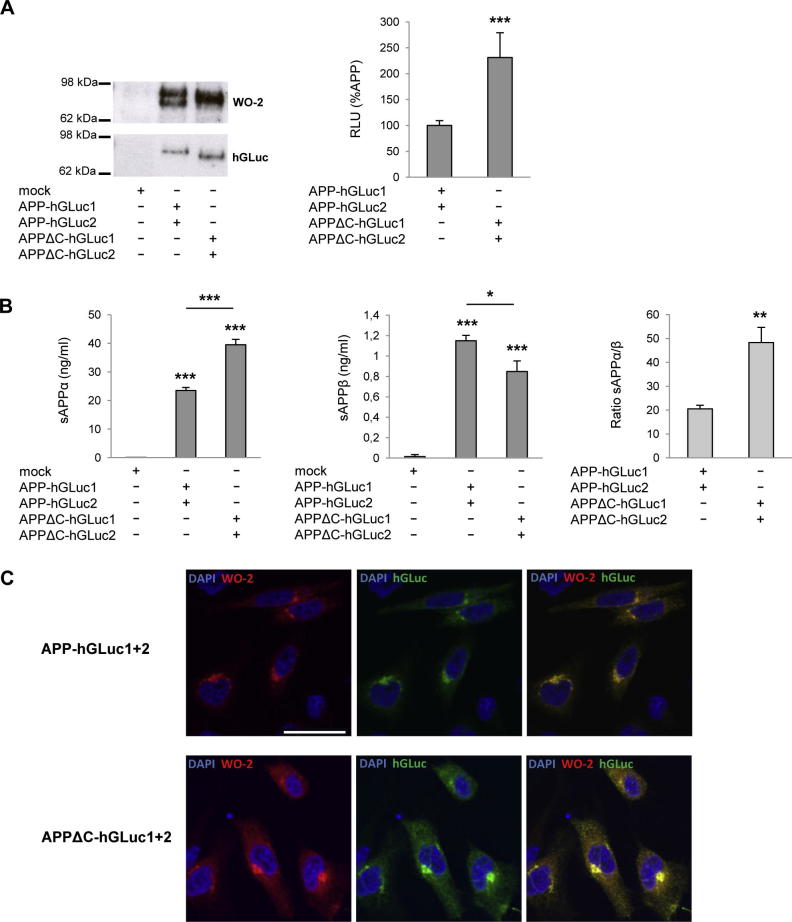
We finally examined the role of the APP intracellular C-terminal domain in dimerization. We generated APPΔC split-luciferase constructs, corresponding to APP deleted from its intracellular region (Fig. 1). Indeed, much attention has been given so far to the contribution of the extracellular and TM domains to APP dimerization, but nothing is known about the role of its intracellular region. Both APP and APPΔC were recognized by the W0-2 and hGLuc antibodies, and both constructs were expressed at similar levels (Fig. 7A). Immunostaining (Fig. 7C) indicated that APP and APPΔC displayed similar subcellular distribution profiles. Results in Fig. 7A clearly showed that APPΔC formed more dimers than APP in cells. Expression of C99 constructs identically lacking their intracellular domains (C55) led to the same increased dimerization (data not shown). To know whether this drastic change in dimerization is related to APP metabolism, we measured sAPPα and sAPPβ levels produced by those cells. Soluble APPα and sAPPβ production are indicators of the ectodomain shedding occurring as a first step of the non-amyloidogenic and amyloidogenic processing, respectively. Cells expressing APPΔC-hGLuc showed increased sAPPα levels and reduced sAPPβ production. The ratio between sAPPα and sAPPβ significantly increased, indicating that APPΔC metabolism is shifted towards non-amyloidogenic processing.
Fig. 7.
Influence of the intracellular in APP dimerization. (A) Protein expression was monitored in cell lysates by Western blotting with the W0-2 or hGluc antibodies (left panel). Luciferase activity was measured and expressed as RLU normalized to APP (set to 100%, right panel). Values (means ± SEM) are representative of 2 independent experiment (n = 4 in each experiment). ***p < 0.001, as compared to APP-hGLuc1 and 2. (B) sAPPα and β production of APP-hGLuc1 and 2 and APPΔC-hGLuc1 and 2 constructs were monitored by ECLIA in the culture media of cells and are given in ng/ml. *p < 0.05 and ***p < 0.001, as compared to non-transfected cells or as indicated. Ratio of sAPPα on sAPPβ produced was calculated in the same experiments. **p < 0.01, as compared to APP-hGLuc1 and 2 (C) Immunostaining of cells co-expressing APP or APPΔC constructs of either. Nuclei were stained with DAPI, and APP fusion constructs were stained by the W0-2 and/or hGLuc antibodies. Scale bar: 5 μm.
4. Discussion
Dimerization appears to be a key modulator of APP processing and function. APP shares many features with cell adhesion molecules and TM receptors [9]. APP homo- and hetero-association are involved in trans–cellular interactions [11]. Homodimerization was proposed to be directly correlated to amyloidogenic processing and Aβ production. We and others found that disruption of TM interaction motifs leads to impaired Aβ production [18,19], suggesting that the formation of dimeric amyloidogenic CTFs is a control mechanism of APP γ-cleavage. However, whether dimerization promotes or inhibits dimerization itself has been a matter of debate [25,26]. Recent studies indicated that, in contrast to Aβ production, dimerization is not altered in FAD mutants [27], challenging the hypothesis that dimerization is involved in amyloidogenic processing. Indeed, the extent of APP and APP CTFs dimerization in cells, its relation to amyloidogenic processing and the contribution of the different APP domains in this process remain unclear and poorly known.
APP homo- and/or hetero-dimerization have been reported so far using different approaches, including co-immunopreciptation and non-denaturing electrophoresis, FRET-based assays and split-protein assays [19,27,41]. We recently used the bimolecular fluorescence complementation (BiFC) method, or split-YFP, to analyze dimerization of APP695 and APP751 isoforms [17,31]. This method has gained popularity in the field to study protein–protein interactions in living cells [13,42]. Its major advantage is to allow direct visualization of protein interactions without the use of biochemical reagents or antibodies that might themselves modify the dimerization properties of the proteins investigated. The major drawbacks of BiFC are that the fluorescent protein halves are prone to self-assembly independent of a protein–protein interaction event, and that the split position promotes irreversible self-interaction of the two non-fluorescent fragments, which may result in the detection of false positive signals [43]. Here we used the split Gaussia luciferase assay, a more recent, quantitative and dynamic complementation method to circumvent drawbacks of previous split protein approaches [37]. The humanized Gaussia princeps luciferase (hGLuc) can generate over 100-fold higher bioluminescent signal than other luciferases and is the smallest known coelenterazine-using luciferase, making it an ideal candidate for protein complementation assays.
APP and the various APP C-terminal fragments fused to the split-protein used in our study were readily expressed in CHO cells and detectable by Western blotting by different antibodies, thus allowing normalization of the bioluminescence to the bioluminescent protein content. This is an important asset when compared to the split YFP approach [31], in which quantitative comparison of protein dimerization is more difficult to achieve. We also checked that the co-expression of the hGLuc halves alone do not generate a bioluminescent signal (data not shown), excluding the possibility that the luciferase protein halves self-assemble independently of a protein–protein interaction event, a critical possible bias in split protein assays [43]. Grafting hGLuc halves at the C-terminus does not impair APP subcellular localization or processing. The split luciferase assays we developed appeared therefore as a very sensitive and reliable tool to decipher the mechanisms of APP dimerization in cells.
We first observed that full length APP dimerizes more than its C-terminal fragments. At similar expression levels, the extent of APP dimerization is at least 5 times higher than for C99 or C83. C99 and C83 seem to have only poor and restricted dimerization properties [30]. Their accumulation upon cell treatment with a γ-secretase inhibitor (DAPT) does not increase the bioluminescent signal. This observation strongly argues that CTF dimers are not substrates of the γ-secretase. Under the same conditions (DAPT treatment), APP dimerization was slightly but significantly reduced although cellular levels of the APP protein were unchanged. One possible interpretation to this unexpected observation is that the accumulation of CTFs (endogenous or hGLuc tagged) upon DAPT treatment could interfere with full-length APP dimerization. This seems to be indeed the case since co-transfection of APP-hGLuc and C83-hGLuc formed dimers to a lesser extent than APP homodimers (see Supplementary Fig. S1).
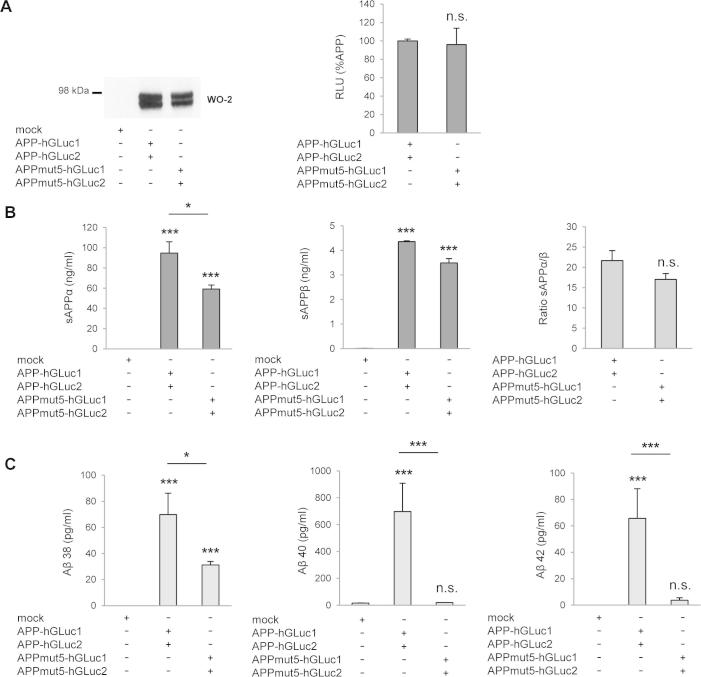
Previous studies have shown that APP dimers can be readily observed in cells and that dimers are formed in the early secretory pathway [31]. Our results are in line with these observations, and confirm that determinants playing a major role in APP dimerization (E1 and KPI domains) are located in its extracellular region [15,31]. Dimerization driven by ectodomain regions appears as a key mechanism for APP trafficking to the cell surface [15,16]. Much more attention has been recently given to dimerization of the APP JM/TM regions and the role of these regions in amyloidogenic processing and Aβ production. APP contains 3 in-register GXXXG motifs that start in the extracellular JM sequence and continue into the TM domain. These motifs are well known to stabilize TM protein interactions by close apposition of TM α-helices [44,45]. Structural approaches have established that APP JM/TM sequences interact through GXXXG motifs interfaces [28–30]. Mutations of these motifs strongly modulate their interaction and impair Aβ production [18,19]. We found in a previous study [17] that mutation of GXXXG motifs did not affect dimerization of full length APP. We confirmed by the split luciferase system that mutation in a critical GXXXG motif (mut5) has no effect on full-length APP dimerization (see Supplementary Fig. S2). In addition, GXXXG mutations did no modify the sAPPα/sAPPβ ratio, excluding the hypothesis that they indirectly contribute to the production of Aβ by facilitating the shedding of the of APP ectodomain through the α- or β-secretase pathways. This strongly suggested that GXXXG motifs might play a critical role in APP CTF dimerization, after the removal of the bulky ectodomain by α- or β-secretase. We directly addressed this point by mutating the GXXXG motifs in C99 and C83 (corresponding to β and αCTF, respectively) and measuring the effect on dimerization by the split luciferase assay. We analyzed 2 different mutants: mut5 with mutations of the central GXXXG motif (GG29/33LL) disrupting the GXXXG interface and the FAD Flemish mutant (A21G) adding a fourth in register GXXXG motif extending the GXXXG interface. These mutations had only a very moderate effect on C83 and C99 dimerization. No consistent effect was measured of C83 mut5 and A21G constructs. Decreased dimerization was measured for both mutated C99 constructs, although the effect was moderate. These observations first indicate that C99 and C83 dimerization might be different, due to determinants located between the α- and β-cleavage sites. Contrary to what was previously suggested, mutation of GXXXG motifs do not trigger the formation of stable TM dimers through the GXXXA motif located downstream [19]. Importantly, there was no direct relationship between the effects of the mutation on dimerization and Aβ production. The Flemish mutation significantly increased Aβ release, whereas mut5 blocked it. This is consistent with very recent results showing an independent relationship between APP dimerization and γ-secretase processivity [26,27,36]. Rather than changing dimerization, mutation of glycine residues in the GXXXG motifs might induce a conformational change increasing the fitness of the substrate for γ-secretase activity, and thus Aβ production [29]. In fact, the GXXXG motif has been shown to interact with NSAIDs and cholesterol, playing the role of a cholesterol sensor pocket [30,35]. JM/TM regions are sensitive to the membrane lipid context and are targeted by drugs acting as γ-secretase modulators, but probably independent of their dimerization properties. It would be of interest to evaluate with the split luciferase assay how these compounds impact dimerization, if GXXXG mutants show different sensitivities, and how they can be related to their modulatory effect on Aβ production.
One major finding in this study is that deletion of APP intracellular domain importantly increased its dimerization. So far, the intracellular domain has never been shown to influence APP dimerization. We built AICD split luciferase constructs and found that AICD does not dimerize by itself (data not shown). One possible explanation could be that the interaction of the APP intracellular region with proteins like G0 protein or Fe65 [46,47] favors dimerization. Indeed, APP has been shown to interact with adaptor proteins like Fe65 by its YENPTY intracellular motif [48,49] and this association could impair or compete with dimerization. When we expressed Fe65 together with APP in CHO cells we did not modify APP dimerization measured by split-luciferase (see Supplementary Fig. S3). This does not exclude the possibility that other unknown interacting protein modulate dimerization. An alternative hypothesis could be that dimerization is related to trafficking, which in turn impacts processing. APPΔC is directed to the secretory pathway but cannot be internalized properly due to loss of the NPTY internalization motif [31,50]. At this stage, we cannot define whether increased dimerization of APPΔC is the cause or a consequence of the reduced endocytosis, but we can propose that the amount of APPΔC stays high in cells because they fail to be endocytosed in compartments where they are further processed. This would in turn favor its non-amyloidogenic processing, known to occur close to the cell surface. This is consistent with our observations, indicating that increased dimerization of APPΔC is linked to increased cleavage by α-secretase, inducing a shift towards non-amyloidogenic pathway. Interestingly, compounds that have been shown to destabilize dimerization of the APP ectodomain indeed regulate cleavage by α-secretase [34]. However, the question of whether α- secretase cleaves APP dimers, or whether dimerization favors trafficking to compartments where α-cleavage occurs, remains totally open.
5. Conclusion
We used the recent split luciferase assay to study APP dimerization in cells. In contrast to what was suggested in previous studies, we found that the APP CTFs (C99 and C83) dimerized at a low level when compared to full-length APP. Mutation of the TM interaction GXXXG motifs did not significantly affect CTF dimerization. Combining dimerization studies and measurement of Aβ production, we found that there is no direct relationship between APP dimerization and γ-cleavage. In fact, major determinants controlling APP dimerization appear to be its intracellular domain. Increasing APP dimerization favors its non-amyloidogenic processing. This brings new and important insights on the different regions driving APP dimerization, and the relationship between dimerization and processing.
Contributions
M.D. and P.K.C. designed research; M.D., L.E.H. and S.S. conducted experiments, M.D. and P.K-C. analyzed data and wrote the paper with fundamental input of S.O.S., S.C., I.D., and J-N.O.
Acknowledgements
This work was supported by a grant of the Belgian F.N.R.S FRIA (Fonds National pour la Recherche Scientifique) to M.D., Foundation for Research on Alzheimer’s disease (P.K-C.), by the Interuniversity Attraction Poles Programme-Belgian Sate-Belgian Science Policy (IAP-P7/16 and IAP-P7/13) to J-N.O. and P.K-C, and by the NIH (AG027317) to S.O.S. We are grateful to J-F. Paradis and S.W. Michnick (UMontreal) for the leucine zipper split-luciferase plasmids. We greatly acknowledge B. Tasiaux for her excellent technical support.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.fob.2015.09.002.
Appendix A. Supplementary data
Supplementary Fig. 1.
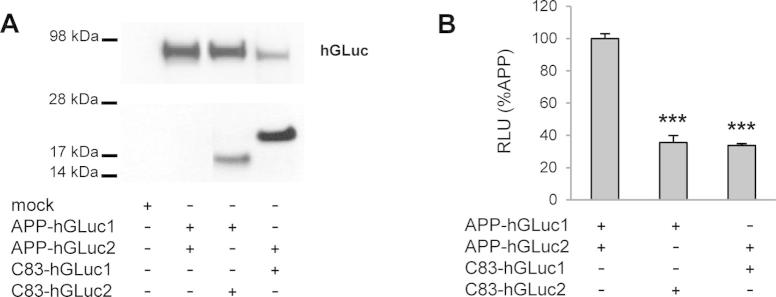
Dimerization of APP and C83 heterodimers.
Supplementary Fig. 2.
Dimerization and metabolism of APP GXXXG mutant 5.
Supplementary Fig. 3.
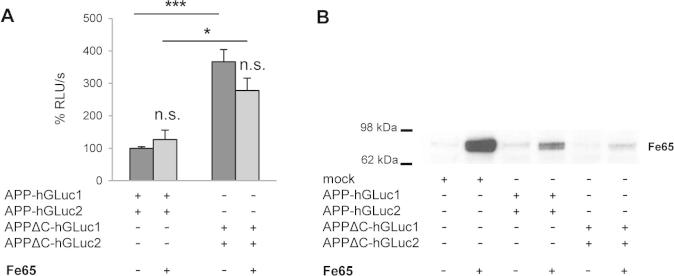
Influence of Fe65 overexpression on APP dimerization.
References
- 1.Kang J., Lemaire H.G., Unterbeck A., Salbaum J.M., Masters C.L., Grzeschik K.H., Multhaup G., Beyreuther K., Muller-Hill B. The precursor of Alzheimer’s disease amyloid A4 protein resembles a cell-surface receptor. Nature. 1987;325:733–736. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- 2.Selkoe D.J. Alzheimer disease: mechanistic understanding predicts novel therapies. Ann. Intern. Med. 2004;140:627–638. doi: 10.7326/0003-4819-140-8-200404200-00047. [DOI] [PubMed] [Google Scholar]
- 3.Octave J.N. The amyloid peptide and its precursor in Alzheimer’s disease. Rev. Neurosci. 1995;6:287–316. [PubMed] [Google Scholar]
- 4.Glenner G.G., Wong C.W. Alzheimer’s disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem. Biophys. Res. Commun. 1984;120:885–890. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- 5.Hardy J., Selkoe D.J. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 6.Shariati S.A., De S.B. Redundancy and divergence in the amyloid precursor protein family. FEBS Lett. 2013;587:2036–2045. doi: 10.1016/j.febslet.2013.05.026. [DOI] [PubMed] [Google Scholar]
- 7.Weyer S.W., Klevanski M., Delekate A., Voikar V., Aydin D., Hick M., Filippov M., Drost N., Schaller K.L., Saar M., Vogt M.A., Gass P., Samanta A., Jaschke A., Korte M., Wolfer D.P., Caldwell J.H., Muller U.C. APP and APLP2 are essential at PNS and CNS synapses for transmission, spatial learning and LTP. EMBO J. 2011;30:2266–2280. doi: 10.1038/emboj.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klevanski M., Saar M., Baumkotter F., Weyer S.W., Kins S., Muller U.C. Differential role of APP and APLPs for neuromuscular synaptic morphology and function. Mol. Cell Neurosci. 2014;61:201–210. doi: 10.1016/j.mcn.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Khalifa N.B., Van H.J., Tasiaux B., Huysseune S., Smith S.O., Constantinescu S.N., Octave J.N., Kienlen-Campard P. What is the role of amyloid precursor protein dimerization? Cell Adh. Migr. 2010;4:268–272. doi: 10.4161/cam.4.2.11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coulson E.J., Paliga K., Beyreuther K., Masters C.L. What the evolution of the amyloid protein precursor supergene family tells us about its function. Neurochem. Int. 2000;36:175–184. doi: 10.1016/s0197-0186(99)00125-4. [DOI] [PubMed] [Google Scholar]
- 11.Soba P., Eggert S., Wagner K., Zentgraf H., Siehl K., Kreger S., Lower A., Langer A., Merdes G., Paro R., Masters C.L., Muller U., Kins S., Beyreuther K. Homo- and heterodimerization of APP family members promotes intercellular adhesion. EMBO J. 2005;24:3624–3634. doi: 10.1038/sj.emboj.7600824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaden D., Voigt P., Munter L.M., Bobowski K.D., Schaefer M., Multhaup G. Subcellular localization and dimerization of APLP1 are strikingly different from APP and APLP2. J. Cell Sci. 2009;122:368–377. doi: 10.1242/jcs.034058. [DOI] [PubMed] [Google Scholar]
- 13.Chen C.D., Oh S.Y., Hinman J.D., Abraham C.R. Visualization of APP dimerization and APP-Notch2 heterodimerization in living cells using bimolecular fluorescence complementation. J. Neurochem. 2006;97:30–43. doi: 10.1111/j.1471-4159.2006.03705.x. [DOI] [PubMed] [Google Scholar]
- 14.Shaked G.M., Kummer M.P., Lu D.C., Galvan V., Bredesen D.E., Koo E.H. Abeta induces cell death by direct interaction with its cognate extracellular domain on APP (APP 597–624) FASEB J. 2006;20:1254–1256. doi: 10.1096/fj.05-5032fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaden D., Munter L.M., Joshi M., Treiber C., Weise C., Bethge T., Voigt P., Schaefer M., Beyermann M., Reif B., Multhaup G. Homophilic interactions of the amyloid precursor protein (APP) ectodomain are regulated by the loop region and affect beta-secretase cleavage of APP. J. Biol. Chem. 2008;283:7271–7279. doi: 10.1074/jbc.M708046200. [DOI] [PubMed] [Google Scholar]
- 16.Isbert S., Wagner K., Eggert S., Schweitzer A., Multhaup G., Weggen S., Kins S., Pietrzik C.U. APP dimer formation is initiated in the endoplasmic reticulum and differs between APP isoforms. Cell Mol. Life Sci. 2012;69:1353–1375. doi: 10.1007/s00018-011-0882-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ben K.N., Tyteca D., Courtoy P.J., Renauld J.C., Constantinescu S.N., Octave J.N., Kienlen-Campard P. Contribution of Kunitz protease inhibitor and transmembrane domains to amyloid precursor protein homodimerization. Neurodegener. Dis. 2012;10:92–95. doi: 10.1159/000335225. [DOI] [PubMed] [Google Scholar]
- 18.Munter L.M., Voigt P., Harmeier A., Kaden D., Gottschalk K.E., Weise C., Pipkorn R., Schaefer M., Langosch D., Multhaup G. GxxxG motifs within the amyloid precursor protein transmembrane sequence are critical for the etiology of Abeta42. EMBO J. 2007;26:1702–1712. doi: 10.1038/sj.emboj.7601616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kienlen-Campard P., Tasiaux B., Van H.J., Li M., Huysseune S., Sato T., Fei J.Z., Aimoto S., Courtoy P.J., Smith S.O., Constantinescu S.N., Octave J.N. Amyloidogenic processing but not amyloid precursor protein (APP) intracellular C-terminal domain production requires a precisely oriented APP dimer assembled by transmembrane GXXXG motifs. J. Biol. Chem. 2008;283:7733–7744. doi: 10.1074/jbc.M707142200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russ W.P., Engelman D.M. The GxxxG motif: a framework for transmembrane helix-helix association. J. Mol. Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 21.Brosig B., Langosch D. The dimerization motif of the glycophorin A transmembrane segment in membranes: importance of glycine residues. Protein Sci. 1998;7:1052–1056. doi: 10.1002/pro.5560070423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorman P.M., Kim S., Guo M., Melnyk R.A., McLaurin J., Fraser P.E., Bowie J.U., Chakrabartty A. Dimerization of the transmembrane domain of amyloid precursor proteins and familial Alzheimer’s disease mutants. BMC Neurosci. 2008;9:17. doi: 10.1186/1471-2202-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munter L.M., Botev A., Richter L., Hildebrand P.W., Althoff V., Weise C., Kaden D., Multhaup G. Aberrant amyloid precursor protein (APP) processing in hereditary forms of Alzheimer disease caused by APP familial Alzheimer disease mutations can be rescued by mutations in the APP GxxxG motif. J. Biol. Chem. 2010;285:21636–21643. doi: 10.1074/jbc.M109.088005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hendriks L., van Duijn C.M., Cras P., Cruts M., Van H.W., van H.F., Warren A., McInnis M.G., Antonarakis S.E., Martin J.J. Presenile dementia and cerebral haemorrhage linked to a mutation at codon 692 of the beta-amyloid precursor protein gene. Nat. Genet. 1992;1:218–221. doi: 10.1038/ng0692-218. [DOI] [PubMed] [Google Scholar]
- 25.Scheuermann S., Hambsch B., Hesse L., Stumm J., Schmidt C., Beher D., Bayer T.A., Beyreuther K., Multhaup G. Homodimerization of amyloid precursor protein and its implication in the amyloidogenic pathway of Alzheimer’s disease. J. Biol. Chem. 2001;276:33923–33929. doi: 10.1074/jbc.M105410200. [DOI] [PubMed] [Google Scholar]
- 26.Eggert S., Midthune B., Cottrell B., Koo E.H. Induced dimerization of the amyloid precursor protein leads to decreased amyloid-beta protein production. J. Biol. Chem. 2009;284:28943–28952. doi: 10.1074/jbc.M109.038646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.So P.P., Khodr C.E., Chen C.D., Abraham C.R. Comparable dimerization found in wildtype and familial Alzheimer’s disease amyloid precursor protein mutants. Am. J. Neurodegener. Dis. 2013;2:15–28. [PMC free article] [PubMed] [Google Scholar]
- 28.Sato T., Tang T.C., Reubins G., Fei J.Z., Fujimoto T., Kienlen-Campard P., Constantinescu S.N., Octave J.N., Aimoto S., Smith S.O. A helix-to-coil transition at the epsilon-cut site in the transmembrane dimer of the amyloid precursor protein is required for proteolysis. Proc. Natl. Acad. Sci. USA. 2009;106:1421–1426. doi: 10.1073/pnas.0812261106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang T.C., Hu Y., Kienlen-Campard P., El H.L., Decock M., Van H.J., Fu Z., Octave J.N., Constantinescu S.N., Smith S.O. Conformational changes induced by the A21G Flemish mutation in the amyloid precursor protein lead to increased Abeta production. Structure. 2014;22:387–396. doi: 10.1016/j.str.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song Y., Hustedt E.J., Brandon S., Sanders C.R. Competition between homodimerization and cholesterol binding to the C99 domain of the amyloid precursor protein. Biochemistry. 2013;52:5051–5064. doi: 10.1021/bi400735x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ben K.N., Tyteca D., Marinangeli C., Depuydt M., Collet J.F., Courtoy P.J., Renauld J.C., Constantinescu S., Octave J.N., Kienlen-Campard P. Structural features of the KPI domain control APP dimerization, trafficking, and processing. FASEB J. 2012;26:855–867. doi: 10.1096/fj.11-190207. [DOI] [PubMed] [Google Scholar]
- 32.So P.P., Zeldich E., Seyb K.I., Huang M.M., Concannon J.B., King G.D., Chen C.D., Cuny G.D., Glicksman M.A., Abraham C.R. Lowering of amyloid beta peptide production with a small molecule inhibitor of amyloid-beta precursor protein dimerization. Am. J. Neurodegener. Dis. 2012;1:75–87. [PMC free article] [PubMed] [Google Scholar]
- 33.Asada-Utsugi M., Uemura K., Noda Y., Kuzuya A., Maesako M., Ando K., Kubota M., Watanabe K., Takahashi M., Kihara T., Shimohama S., Takahashi R., Berezovska O., Kinoshita A. N-cadherin enhances APP dimerization at the extracellular domain and modulates Abeta production. J. Neurochem. 2011;119:354–363. doi: 10.1111/j.1471-4159.2011.07364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Libeu C.A., Descamps O., Zhang Q., John V., Bredesen D.E. Altering APP proteolysis: increasing sAPPalpha production by targeting dimerization of the APP ectodomain. PLoS One. 2012;7:e40027. doi: 10.1371/journal.pone.0040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richter L., Munter L.M., Ness J., Hildebrand P.W., Dasari M., Unterreitmeier S., Bulic B., Beyermann M., Gust R., Reif B., Weggen S., Langosch D., Multhaup G. Amyloid beta 42 peptide (Abeta42)-lowering compounds directly bind to Abeta and interfere with amyloid precursor protein (APP) transmembrane dimerization. Proc. Natl. Acad. Sci. USA. 2010;107:14597–14602. doi: 10.1073/pnas.1003026107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jung J.I., Premraj S., Cruz P.E., Ladd T.B., Kwak Y., Koo E.H., Felsenstein K.M., Golde T.E., Ran Y. Independent relationship between amyloid precursor protein (APP) dimerization and gamma-secretase processivity. PLoS One. 2014;9:e111553. doi: 10.1371/journal.pone.0111553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Remy I., Michnick S.W. A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nat. Methods. 2006;3:977–979. doi: 10.1038/nmeth979. [DOI] [PubMed] [Google Scholar]
- 38.Marinangeli C., Tasiaux B., Opsomer R., Hage S., Sodero A.O., Dewachter I., Octave J.N., Smith S.O., Constantinescu S.N., Kienlen-Campard P. Presenilin transmembrane domain 8 conserved AXXXAXXXG motifs are required for the activity of the gamma-secretase complex. J. Biol. Chem. 2015;290:7169–7184. doi: 10.1074/jbc.M114.601286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hage S., Stanga S., Marinangeli C., Octave J.N., Dewachter I., Quetin-Leclercq J., Kienlen-Campard P. Characterization of Pterocarpus erinaceus kino extract and its gamma-secretase inhibitory properties. J. Ethnopharmacol. 2015;163:192–202. doi: 10.1016/j.jep.2015.01.028. [DOI] [PubMed] [Google Scholar]
- 40.Annaert W.G., Levesque L., Craessaerts K., Dierinck I., Snellings G., Westaway D., George-Hyslop P.S., Cordell B., Fraser P., De S.B. Presenilin 1 controls gamma-secretase processing of amyloid precursor protein in pre-golgi compartments of hippocampal neurons. J. Cell Biol. 1999;147:277–294. doi: 10.1083/jcb.147.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Devauges V., Marquer C., Lecart S., Cossec J.C., Potier M.C., Fort E., Suhling K., Leveque-Fort S. Homodimerization of amyloid precursor protein at the plasma membrane: a homoFRET study by time-resolved fluorescence anisotropy imaging. PLoS One. 2012;7:e44434. doi: 10.1371/journal.pone.0044434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kerppola T.K. Complementary methods for studies of protein interactions in living cells. Nat. Methods. 2006;3:969–971. doi: 10.1038/nmeth1206-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horstman A., Tonaco I.A., Boutilier K., Immink R.G. A cautionary note on the use of split-YFP/BiFC in plant protein-protein interaction studies. Int. J. Mol. Sci. 2014;15:9628–9643. doi: 10.3390/ijms15069628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith S.O., Song D., Shekar S., Groesbeek M., Ziliox M., Aimoto S. Structure of the transmembrane dimer interface of glycophorin A in membrane bilayers. Biochemistry. 2001;40:6553–6558. doi: 10.1021/bi010357v. [DOI] [PubMed] [Google Scholar]
- 45.Lemmon M.A., Flanagan J.M., Treutlein H.R., Zhang J., Engelman D.M. Sequence specificity in the dimerization of transmembrane alpha-helices. Biochemistry. 1992;31:12719–12725. doi: 10.1021/bi00166a002. [DOI] [PubMed] [Google Scholar]
- 46.Nishimoto I., Okamoto T., Matsuura Y., Takahashi S., Okamoto T., Murayama Y., Ogata E. Alzheimer amyloid protein precursor complexes with brain GTP-binding protein G(o) Nature. 1993;362:75–79. doi: 10.1038/362075a0. [DOI] [PubMed] [Google Scholar]
- 47.Fiore F., Zambrano N., Minopoli G., Donini V., Duilio A., Russo T. The regions of the Fe65 protein homologous to the phosphotyrosine interaction/phosphotyrosine binding domain of Shc bind the intracellular domain of the Alzheimer’s amyloid precursor protein. J. Biol. Chem. 1995;270:30853–30856. doi: 10.1074/jbc.270.52.30853. [DOI] [PubMed] [Google Scholar]
- 48.Guenette S.Y., Chen J., Jondro P.D., Tanzi R.E. Association of a novel human FE65-like protein with the cytoplasmic domain of the beta-amyloid precursor protein. Proc. Natl. Acad. Sci. USA. 1996;93:10832–10837. doi: 10.1073/pnas.93.20.10832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Borg J.P., Ooi J., Levy E., Margolis B. The phosphotyrosine interaction domains of X11 and FE65 bind to distinct sites on the YENPTY motif of amyloid precursor protein. Mol. Cell Biol. 1996;16:6229–6241. doi: 10.1128/mcb.16.11.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Selkoe D.J., Yamazaki T., Citron M., Podlisny M.B., Koo E.H., Teplow D.B., Haass C. The role of APP processing and trafficking pathways in the formation of amyloid beta-protein. Ann. N. Y. Acad. Sci. 1996;777:57–64. doi: 10.1111/j.1749-6632.1996.tb34401.x. [DOI] [PubMed] [Google Scholar]