Abstract
Saussurea lappa has been reported to possess anti-atopic properties. In this study, we have confirmed the S. lappa’s anti-atopic properties in Nc/Nga mice and investigated the candidate gene related with its properties using microarray. We determined the target gene using real time PCR in in vitro experiment. S. lappa showed the significant reduction in atoptic dermatitis (AD) score and immunoglobulin E compared with the AD induced Nc/Nga mice. In the results of microarray using back skin obtained from animals, we found that S. lappa’s properties are closely associated with cytokine-cytokine receptor interaction and the JAK-STAT signaling pathway. Consistent with the microarray data, real-time RT-PCR confirmed these modulation at the mRNA level in skin tissues from S. lappa-treated mice. Among these genes, PI3Kca and IL20Rβ were significantly downregulated by S. lappa treatment in Nc/Nga mouse model. In in vitro experiment using HaCaT cells, we found that the S. lappa components, including alantolactone, caryophyllene, costic acid, costunolide and dehydrocostus lactone significantly decreased the expression of PI3Kca but not IL20Rβ in vitro. Therefore, our study suggests that PI3Kca-related signaling is closely related with the protective effects of S. lappa against the development of atopic-dermatitis.
Keywords: atopic dermatitis, microarray, Nc/Nga mouse model, PI3Kca, Saussurea lappa
INTRODUCTION
Saussurea lappa has traditionally been used in Asian countries as an ingredient in the treatment of abdominal pain and tenesmus, and an analgesic, digestive aid, aphrodisiac, and diuretic. Many authors have reported that S. lappa exhibits biological effects such as antiulcer, anticancer, and hepatoprotective effects (Byambaragchaa et al., 2014; Chen et al., 1995; Kim et al., 2014; Yoshikawa et al., 1993). We have recently reported anti-AD effects of S. lappa as shown in both in vitro and in vivo assays (Lim et al., 2014). S. lappa decreased the immunoglobulin E, cytokines and chemokines with the reduction in histopatholgical features of atopic dermatitis (AD) lesion. However, there is no study on identifying an active component of S. lappa exhibiting curative effects against AD and its mechanism of actions.
AD is a chronic skin disease involving skin barrier dysfunction and cutaneous inflammatory hypersensitivity, and has a strong genetic basis (Cookson, 2001). Various studies have indicated that AD has a complex etiology that involves the activation of multiple immunological and inflammatory mechanisms (Leung and Bieber, 2003; Novak et al., 2003). Currently, many researchers have been investigating target genes that can apply to treat AD using a various experiments (Choi et al., 2014; Choy et al., 2012; Lu et al., 2009; Zhang et al., 2014). Therefore, candidate for treating AD need to identify target molecule exerting its curative effects throughout a gene analysis.
Microarray analysis is a molecular technique that enables the parallel analysis of expression by a very large number of genes encompassing a significant fraction of the human genome. This method is both qualitative and quantitative because it can detect changes in the expression levels in treated cells based on comparisons with control samples (Kim et al., 2006; Wang et al., 2006; Yu et al., 2011). The use of microarray analysis may be helpful in the development of more advanced therapies for the treatment of AD using naturally derived products.
In the present study, we used microarray analysis to evaluate the systemic biological activities of S. lappa extract in a house dust mite-induced AD model in Nc/Nga mice. We found that of S. lappa extract reduced the expression of genes related to allergy metabolism in Nc/Nga mice. The results of our evaluation and the possible mechanisms to explain these results are discussed herein.
MATERIALS AND METHODS
Reagents and chemicals
Biostir-AD®, which is an ointment that contains house dust mite (Dermatophagoides farinae) extract, was purchased from Biostir Inc. (Japan). Reference compounds, costunolide, dehydrocostus lactone, alantolactone, costic acid, and trans- caryophyllene, were purchased from ChemFaces (China). The purities of the five components were ≥ 98.0% by HPLC analysis.
Plant materials and preparations 70% methanol extract
The S. lappa material used in this experiment was purchased from HMAX (Korea) in 2009. A specimen (2009-KIOM62) has been deposited in the K-herb Research Center, Korea Institute of Oriental Medicine. Dried S. lappa (100 g) was extracted three times with 70% methanol (1 L) by refluxing for 90 min. The extract was filtered, evaporated to dryness, and freeze-dried using a freeze dryer (PVTFD100R, Ilshin Lab Co., Korea). The yield was 28.6%.
Experimental animals
Specific pathogen-free male Nc/Nga mice (eight weeks old) were purchased from Central Laboratory Animal Inc. (Korea). The animals were maintained in an air-conditioned room and maintained at 24 ± 2°C with 55 ± 15% humidity. The animals were housed one per cage and were allowed sterilized tap water and standard rodent chow (Samyang Feed Co, Korea) ad libitum. All experimental procedures were carried out in accordance with the NIH Guidelines for the Care and Use of Laboratory Animals and were approved by Korea Institute of Oriental Medicine Institutional Animal Care and Use Committee (Approval number: #12-047).
Induction of AD in Nc/Nga mice
AD-like skin lesions were induced in 10-week-old male Nc/Nga mice using Biostir-AD, as described by the manufacturer (Lee et al., 2010). The animals were housed in conventional conditions. Briefly, the upper back was shaved, and 200 μl of 4% (w/v) sodium dodecyl sulfate was applied to the shaved dorsal skin and both surfaces of each ear to induce barrier disruption. Two hours after shaving, 50 mg of Biostir-AD was applied topically twice per week for six weeks. Prednisolone and S. lappa were dissolved in distilled water and administrated by oral gavage 6 h after Biostir-AD application for six weeks. The mice (n = 30) were allocated to five groups (n = 6/group): (A) normal control group that received distilled water (DW, p.o.); (B) Biostir group that received Biostir-AD (topical application) and DW (p.o); (C) prednisolone group that received Biostir-AD (topical application) and prednisolone (3 mg/kg, p.o.); S. lappa groups that received Biostir-AD (topical application) and S. lappa (100 or 300 mg/kg, p.o.). Prednisolone was used as a positive control and was a recommended medication to patient with AD. The mice were sacrificed under anesthesia via intraperitoneal injection of pentobarbital sodium (Entobar Inj., Hanlim Pharm. Co., Ltd., Korea) on the day of the experiment. The incidence of atopic dermatitis was determined by gross and histopathological examination for back skin and ear and the level of IgE in plasma (Supplementary Fig.1).
RNA preparation
RNA was isolated from dorsal skin tissue using TRIzol reagent (Invitrogen, USA) using the following procedure. Initially, 1 ml of TRIzol reagent was added to cells grown in culture dishes. After 5 min at room temperature, 0.2 ml of chloroform was added for each sample of reagent, and the tubes were shaken vigorously by hand for 15 s and then incubated at room temperature for 3 min. Next, the mixture was centrifuged at 14,000 rpm for 15 min at 4°C, after which the resulting upper aqueous phase (400 μl) was transferred to a fresh tube into which 0.5 ml of 2-propanol was also added. After incubation for 10 min at 4°C, the mixture was centrifuged again at 14,000 rpm for 10 min at 4°C. After separation, the supernatant was removed, washed with 1 ml of 75% ethanol and centrifuged again at 10,000 rpm for 5 min at 4°C. The resulting RNA pellet was then dried briefly, and the purified RNA was dissolved in diethyl pyrocarbonate (DEPC)-distilled water. The RNA was cleaned up using the Rneasy Mini kit (Qiangen, Germany) in accordance with the manufacturer’s instructions. The quality and quantity of total RNA were measured with an ExperionTM system (Bio-Rad, USA).
Labeling and purification
Total RNA was amplified and purified using and Ambion Illumina RNA amplification kit (Ambion, USA) to yield biotinylated cRNA according to the manufacturer’s instructions. Briefly, 550 ng of total RNA was reverse-transcribed to cDNA using a T7 oligo (dT) primer. Second-strand cDNA was synthesized, transcribed in vitro, and labeled with biotin-NTP. After purification, the cRNA was quantified using an ND-1000 Spectrophotometer (NanoDrop Technologies Inc., USA).
Hybridization and data export
Illumina Mouse Ref-8 expression BeadChip (P/N BD-25-203, Illumina Inc., Ambion) arrays were used in this study. Seven hundred fifty nanograms of labeled cRNA samples were hybridized to each mouse Ref-8 expression bead array for 16–18 h at 58°C according to the manufacturer’s instructions. The array signal was detected using Amersham Fluorolink streptavidin- Cy3 (GE Healthcare Bio-Sciences, USA) following the bead array reader confocal scanner according to the manufacturer’s instructions. Array data export processing and analysis were performed using Illumina GenomeStudio v2009.2 (Gene Expression Module v1.5.4).
Data analysis
The BeadStudio (version 3.0) was used to evaluate the expression signals generated by the Illumina Mouse Ref-8 expression BeadChip array. Global scaling normalization was then performed, and the normalized data were log-transformed using base 2. Next, the fold change and Welch’s t-test were applied to select the differentially expressed genes (DEGs) using a fold change threshold of 2-fold and a p < 0.05 to indicate significance. The 2-fold DEGs were clustered using GenPlexTM v3.0 software (ISTECH Inc., Korea) using hierarchical clustering with Pearson correlation as a similarity measure and complete linkage as the linkage method. Gene ontology classification was obtained from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database.
Real-time RT-PCR analysis
To verify the microarray results, real-time RT-PCR analysis was performed for selected genes using an Applied Biosystems 7300 Real-time PCR system and the SYBR green fluorescence quantification system (Applied Biosystems, USA) to quantify the amplicons. cDNA was synthesized using 100 ng of RNA in a reverse transcription reaction. The PCR conditions were 50 cycles of 95°C (30 s), 53°C (30 s), and a standard denaturation curve. The primer sequences are listed in the 5′ to 3′ orientation in Supplementary Table 1. The PCR conditions for each target were optimized according to the primer concentration, the absence of primer dimer formation, and the efficiency of amplification of both the target genes and the housekeeping control gene. PCR reactions mixture comprised 1 μl of cDNA and 9.5 μl of PCR master mix, which contained 2× SYBR Green, 10 pmole each of the forward and reverse primer, and 4.5 μl of DEPC-distilled water in a final volume of 15 μl. To normalize the cDNA content of the samples, we used the comparative threshold (CT) cycle method, which includes normalization of the number of target gene copies vs. the endogenous reference gene, β-actin. The CT is defined as the fractional cycle number at which the fluorescence generated by cleavage of the probe passes a fixed threshold baseline when amplification of the PCR products is first detected.
Cell culture
Human keratinocyte HaCaT cells were obtained from CLS Cell Lines Service GmbH (Eppelheim, Germany). The cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco Inc., USA), supplemented with 10% heat-inactivated fetal bovine serum (Gibco Inc.), penicillin (100 U/ml), and streptomycin (100 μg/ml), in a 5% CO2 incubator at 37°C.
Statistical analyses
The data are expressed as the mean ± SEM. The data were analyzed by one-way analysis of variance and Dunnett’s multiple-comparisons test. Results with a P value < 0.05 were considered significant.
RESULTS
Gene expression profile of S. lappa extract in a house dust mite-induced AD Nc/Nga mouse model
Gene expression profiles were significantly up or downregulated in the S. lappa extract group compared with the Biostir group. A total of 4255 genes that were differentially expressed from about 24,000 genes detected in the experimental group. A hierarchical clustering algorithm was used to group the genes based on their similar expression patterns. Gene ontology annotation was achieved using the KEGG database and the genes were placed into 173 biological pathways (Supplementary Table 2): MAPK signaling pathway (48 genes), neuroactive ligand-receptor interaction (31 genes), glycan structures biosynthesis 1 (29 genes), cytokine-cytokine receptor interaction (29 genes), calcium signaling pathway (26 genes), Wnt signaling pathway (25 genes), regulation of actin cytoskeleton (25 genes), oxidative phosphorylation (25 genes), JAK-STAT signaling pathway (24 genes), focal adhesion (23 genes), tight junction (22 genes), purine metabolism (23 genes), cell cycle (21 genes), leukocyte transendothelial migration (20 genes), and cell adhesion molecules (20 genes) (Fig. 1). Among the 173 biological pathways that were up or downregulated, we selected five upregulated and six downregulated allergy-related pathways. The functionally annotated upregulated (40) and downregulated (45) genes are listed in Supplementary Tables 3 and 4, which show comparisons of the expression levels for a variety of genes between the Biostir and S. lappa group.
Fig. 1.
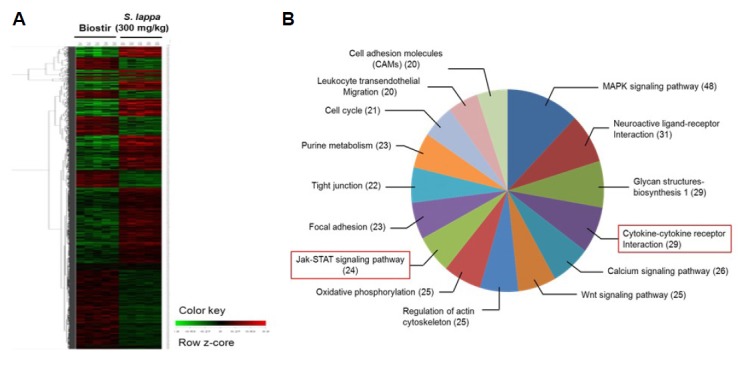
Gene ontology assignment of differentially expressed genes. Clustergram of up- and down-regulated genes in skin tissue from Nc/Nga mice. Microarray data from control (untreated cells) and experimental groups (300 mg/kg S. lappa-treated group) were combined and clustered. (A) Each gene is represented by a single row of clustered boxes and each group is represented by a single column. (B) Gene ontology classification based on biological processes.
Validation of selected genes using real-time RT-PCR
To confirm the effects of S. lappa extract on AD metabolism genes at the mRNA level, real-time RT-PCR was conducted for 10 selected genes involved in cytokine-cytokine receptor interaction and the JAK-STAT signaling pathway. Consistent with the results of the microarray analysis, S. lappa extract increased the expression of kinase insert domain protein receptor (KDR), interleukin 13 receptor alpha 2 (IL13Rα2), interferon alpha 1 (IFNα1), thymic stromal lymphopoietin (TSLP), and chemokine (C-X-C motif) ligand 11 (CXCL11) in the Biostir group (Fig. 2). In addition, S. lappa extract suppressed the expression of chemokine (C-C motif) receptor 1 (CCR1), interleukin 2 receptor beta (IL2Rβ), phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3Kca), and interleukin 20 receptor beta (IL20Rβ) in the Biostir group (Fig. 3). Among the downregulated genes, S. lappa significantly suppressed PI3Kca and IL20Rβ. The relative expression levels of each gene were normalized relative to the expression of β-actin, a well-known housekeeping gene.
Fig. 2.
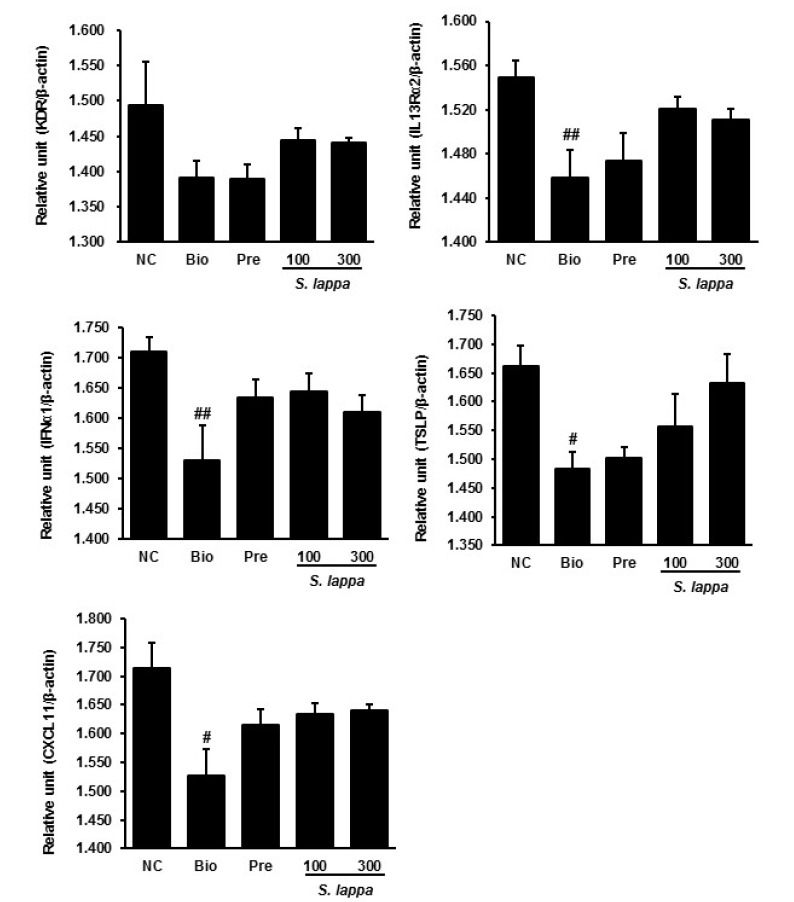
Validation of selected up-regulated genes using real-time qPCR in skin tissue from Nc/Nga mice. The results are normalized as a ratio of each specific mRNA signal (KDR, IL13Rα2, IFNα1, TSLP, and CXCL11) to the β-actin gene signal within the same sample and the values expressed. Data are presented as mean ± SD (n = 3). #p < 0.05 and ##p < 0.01 vs. normal control.
Fig. 3.
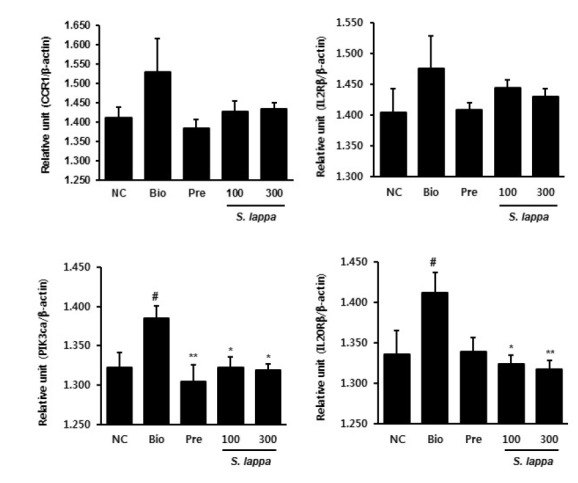
Validation of selected down-regulated genes using real-time qPCR in skin tissue from Nc/Nga mice. The results are normalized as a ratio of each specific mRNA signal (CCR1, IL2Rβ, PI3Kca, and IL20Rβ) to the β-actin gene signal within the same sample and the values expressed. Data are presented as mean ± SD (n = 3). #p < 0.05 vs. normal control. *p < 0.05 and **p < 0.01 vs. Biostir-treated group.
Effects of S. lappa and its components on validation of gene expression
To examine the downregulatory effects of PI3Kca, and IL20Rβ in S. lappa-treated in HaCaT cells, the cells were treated with tumor necrosis factor alpha (TNF-α) and IFN-γ in the absence or presence of S. lappa extract. As shown in Figure 3, S. lappa had no significant effect on PI3Kca or IL20Rβ expression in TNF-α/IFN-γ stimulated HaCaT cells. However, five components of S. lappa suppressed PI3Kca mRNA expression in TNF-α/IFN-γ-stimulated HaCaT cells. Among the five components of S. lappa, costic acid and dehydrocostus lactone suppressed PI3Kca mRNA expression in a dose-dependent manner (Fig. 4). By contrast, no inhibitory effect of these components on IL20Rβ expression was observed in TNF-α/IFN-γ-stimulated HaCaT cells (Fig. 5).
Fig. 4.
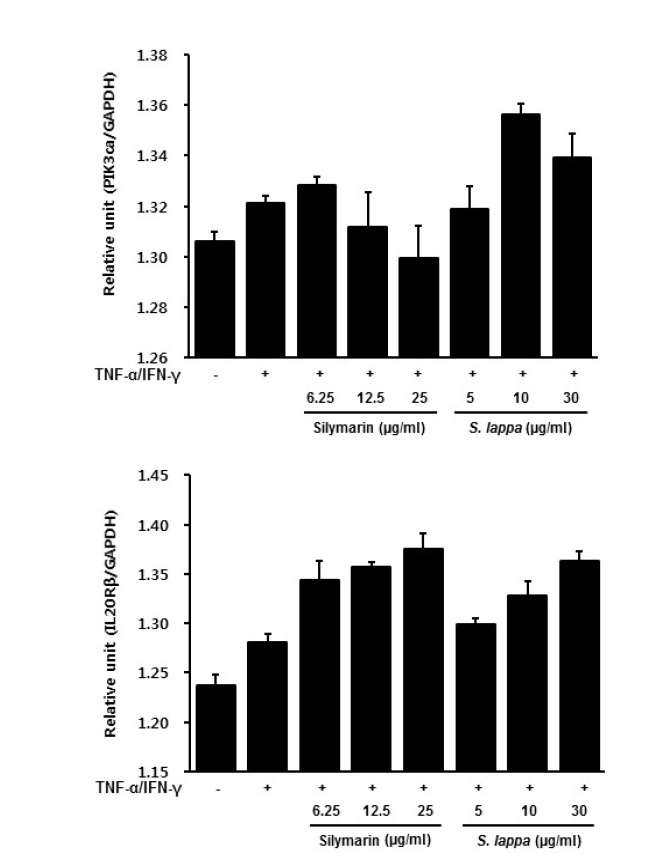
Effects of S. lappa on gene expression determined using real-time RT-PCR in TNF-α and IFN-γ-treated HaCaT cells. The results are normalized for each specific mRNA signal to the GAPDH gene signal within the same sample and the values expressed. (A) PI3Kca and (B) IL20Rβ.
Fig. 5.
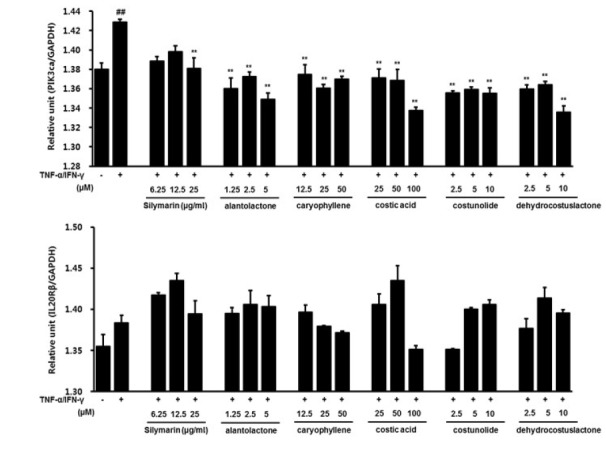
Effects of five components of S. lappa on the validation of gene expression using real-time RT-PCR in HaCaT cells. The results are normalized as a ratio of the (A) PI3Kca and (B) IL20Rβ mRNA signal to the GAPDH gene signal within the same sample, and the values expressed. Values are expressed as mean ± SEM of three independent experiments. ##P < 0.01 vs. vehicle control cells; **P < 0.01 vs. TI-treated cells.
DISCUSSION
Genome-wide association studies are a hypothesis-free way to discover disease-associated microarray (Barnes, 2010). This avoids the limitations of using candidate genes, while retaining the advantages of the case-control approach. Genome sequencing has identified large numbers of genetic variants that can be read on microarrays (Lindblad-Toh et al., 2005). This allows identification of microarrays that are expressed more frequently in affected individuals than in control subjects. The disease-associated microarray marker regions of the genome identified in this study may be involved in the pathogenesis of AD. In the present study, we used microarray analysis to elucidate the underlying biological effects in S. lappa extract-treated Nc/Nga mice. The genes regulated by S. lappa extract in Nc/Nga mice were classified using the KEGG database. Interestingly, we detected genes related to allergy-related molecular mechanisms including cytokine-cytokine receptor interaction, JAK-STAT signaling, T-cell receptor signaling, Toll-like receptor signaling, and the mTOR signaling pathways. To confirm these findings, we selected genes related to cytokine-cytokine receptor interactions (CCR1, IL2Rβ, TSLP, KDR, and CXCL11) and JAK-STAT signaling (PI3Kca, IL20Rβ, IFNα1, TSLP, and IL13Rα2).
Several researchers have demonstrated the role of cytokine-cytokine receptor interactions and JAK-STAT signaling in development of AD. TSLP, an interleukin 7-like cytokine, is known to trigger dendritic cell-mediated Th2 inflammatory responses and highly expresses in activated mast cells and skin lesion of AD, which is triggers allergic inflammation (Isaksen et al., 2002; Liu et al., 2006; Sebastian et al., 2008). KDR is a protein-coding gene that is also known as vascular endothelial growth factor receptor 2 (VEGFR-2), the gene that encodes the VEGFR. Activation of the VEGFR leads to activation of endothelial nitric oxide synthase, which also depends on the activation of KDR (Holmes et al., 2007). KDR significantly up-regulated by activated protein C (Tanimoto et al., 2002). CXCR3 is activated by the three IFN-γ-inducible chemokines of the CXC family: CXCL9, CXCL10, and CXCL11 (Lu et al., 1999). The three ligands for CXCR3 are responsible for the recruitment of immune cells at sites of infection and inflammation (Lasagni et al., 2003; Lu et al., 1999). By contrast, interferon alpha1 (INF-α1) has immunoregulatory functions in autoimmune inflammatory diseases and is an important component of the innate immune system in a number of autoimmune or inflammatory diseases including AD (Borden et al., 2007; Theofilopoulos et al., 2005). IL-13 is associated with multiple diseases such as asthma and allergy (Heinzmann et al., 2000). Two IL-13 receptors have been identified, IL-13Rα1 and IL-13Rv2. IL-13Rα2 has been implicated in allergy, bronchial asthma, atopy, and esophageal diseases (Heinzmann et al., 2000; Zuo et al., 2010). In addition, IL2Rβ is associated with a greater predisposition to type I diabetes, rheumatoid arthritis, and multiple sclerosis (Burchill et al., 2007). The association between the IL2Rβ genes and multiple autoimmune diseases suggests a common mechanism in their pathogenesis (Burchill et al., 2007; Matesanz et al., 2001). In this study, we found that S. lappa extract upregulated the genes related to AD metabolism in skin samples from Nc/Nga mice. S. lappa extract caused overexpression of TSLP, KDR, CXCL11, IFNα1, and IL13Rα2 genes in house dust mite-treated mice. These results suggest that S. lappa extract has protective effects on AD metabolism.
PI3K, the most important member of the PI3K complex, comprises a heterodimer with a p85 regulatory subunit and a p110 catalytic subunit (PI3Kca). PI3Kca is one of the most important downstream regulators of multiple receptor kinase families, which are involved in many fundamental cellular processes, including proliferation, cell survival, motility, and cell growth (Bader et al., 2005; Engelman et al., 2006). The importance of PI3K p110 in various functions of leukocytes such as B cells, T cells, NK cells, myeloid cells, macrophages, keratinocytes, and mast cells is well documented (Fung-Leung, 2011). Previous studies have demonstrated that blockade of p110 activity significantly inhibits allergic inflammation, which suggests an important role for PI3K p110 in allergy (Nashed et al., 2007). The IL-20 cytokine subunit, IL20Rβ is highly expressed in skin lesions (Sa et al., 2007) and induces STAT3 phosphorylation (Dumoutier et al., 2001). Previous studies have shown that STAT3 phosphorylation in epidermal keratinocytes is implicated in the development of psoriasis (Sano et al., 2005). In the present study, the effects of S. lappa extract on genes involved in cytokine-cytokine receptor interaction and JAK-STAT signaling were confirmed by real-time RT-PCR. We found that S. lappa extract significantly decreased the mRNA levels of PI3Kca and IL20Rβ in a dose-dependent manner in skin tissue from Nc/Nga mice. Although S. lappa extract did not affect PI3Kca expression, some of its components significantly reduced PI3Kca mRNA expression in TNF-α/IFN-γ-treated Ha-CaT cells. Difference between S. lappa extract and its components was considered to be due to biological condition of in vivo and in vitro. These results indicate that S. lappa include another components exerting elevation of PI3Kca expression. The drugs may change its character by biological conditions including gastric acid and digestive enzymes in the body. Therefore, we considered that PI3Kca mRNA expression in vivo significantly decreased because components of S. lappa exhibiting PI3Kca expression eliminate or change by biological system in the body.
Overall, our genomic analysis data demonstrate that S. lappa can regulate the expression of genes related to cytokine-cytokine receptor interactions and JAK-STAT signaling in a house dust mite-induced Nc/Nga mouse model of AD. Real-time PCR analysis confirmed that PI3Kca and IL20Rβ expression were significantly downregulated at the mRNA level by S. lappa treatment in Nc/Nga mouse model. We also found that the S. lappa components alantolactone, caryophyllene, costic acid, costunolide and dehydrocostus lactone significantly decreased the expression of PI3Kca but not IL20Rβ in vitro. Our study suggests that S. lappa extract may be a potential candidate for the treatment of AD by targeting PI3Kca-related signaling.
Acknowledgments
This work was supported by a grant from the Korea Institute of Oriental Medicine (No. K14030).
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Bader A.G., Kang S., Zhao L., Vogt P.K. Oncogenic PI3K deregulates transcription and translation. Nat. Rev. Cancer. 2005;5:921–929. doi: 10.1038/nrc1753. [DOI] [PubMed] [Google Scholar]
- Barnes K.C. An update on the genetics of atopic dermatitis: scratching the surface in 2009. J. Allergy Clin. Immunol. 2010;125:16–29.e11. doi: 10.1016/j.jaci.2009.11.008. –11; quiz 30-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden E.C., Sen G.C., Uze G., Silverman R.H., Ransohoff R.M., Foster G.R., Stark G.R. Interferons at age 50: past, current and future impact on biomedicine. Nat. Rev. Drug Discov. 2007;6:975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchill M.A., Yang J., Vang K.B., Farrar M.A. Interleukin-2 receptor signaling in regulatory T cell development and homeostasis. Immunol. Lett. 2007;114:1–8. doi: 10.1016/j.imlet.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byambaragchaa M., Dela Cruz J., Kh A., Hwang S.G. Anticancer potential of an ethanol extract of saussurea involucrata against hepatic cancer cells in vitro. Asian Pac. J. Cancer Prev. 2014;15:7527–7532. doi: 10.7314/apjcp.2014.15.18.7527. [DOI] [PubMed] [Google Scholar]
- Chen H.C., Chou C.K., Lee S.D., Wang J.C., Yeh S.F. Active compounds from Saussurea lappa Clarks that suppress hepatitis B virus surface antigen gene expression in human hepatoma cells. Antiviral Res. 1995;27:99–109. doi: 10.1016/0166-3542(94)00083-k. [DOI] [PubMed] [Google Scholar]
- Choi J.S., Roh J.Y., Lee J.R. Clinical availability of component-resolved diagnosis using microarray technology in atopic dermatitis. Ann. Dermatol. 2014;26:437–446. doi: 10.5021/ad.2014.26.4.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy D.F., Hsu D.K., Seshasayee D., Fung M.A., Modrusan Z., Martin Z., Liu F.T., Arron J.R. Comparative transcriptomic analyses of atopic dermatitis and psoriasis reveal shared neutrophilic inflammation. J. Allergy Clin. Immunol. 2012;130:1335–1343. doi: 10.1016/j.jaci.2012.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson W.O. The genetics of atopic dermatitis: strategies, candidate genes, and genome screens. J. Am. Acad. Dermatol. 2001;45:S7–9. doi: 10.1067/mjd.2001.117026. [DOI] [PubMed] [Google Scholar]
- Dumoutier L., Leemans C., Lejeune D., Kotenko S.V., Renauld J.C. Cutting edge: STAT activation by IL-19, IL-20 and mda-7 through IL-20 receptor complexes of two types. J. Immunol. 2001;167:3545–3549. doi: 10.4049/jimmunol.167.7.3545. [DOI] [PubMed] [Google Scholar]
- Engelman J.A., Luo J., Cantley L.C. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- Fung-Leung W.P. Phosphoinositide 3-kinase delta (PI3Kdelta) in leukocyte signaling and function. Cell. Signal. 2011;23:603–608. doi: 10.1016/j.cellsig.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Heinzmann A., Mao X.Q., Akaiwa M., Kreomer R.T., Gao P.S., Ohshima K., Umeshita R., Abe Y., Braun S., Yamashita T., et al. Genetic variants of IL-13 signalling and human asthma and atopy. Hum. Mol. Genet. 2000;9:549–559. doi: 10.1093/hmg/9.4.549. [DOI] [PubMed] [Google Scholar]
- Holmes K., Roberts O.L., Thomas A.M., Cross M.J. Vascular endothelial growth factor receptor-2: structure, function, intracellular signalling and therapeutic inhibition. Cell. Signal. 2007;19:2003–2012. doi: 10.1016/j.cellsig.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Isaksen D.E., Baumann H., Zhou B., Nivollet S., Farr A.G., Levin S.D., Ziegler S.F. Uncoupling of proliferation and Stat5 activation in thymic stromal lymphopoietin-mediated signal transduction. J. Immunol. 2002;168:3288–3294. doi: 10.4049/jimmunol.168.7.3288. [DOI] [PubMed] [Google Scholar]
- Kim C.S., Sohn S.H., Jeon S.K., Kim K.N., Ryu J.J., Kim M.K. Effect of various implant coatings on biological responses in MG63 using cDNA microarray. J. Oral Rehabil. 2006;33:368–379. doi: 10.1111/j.1365-2842.2005.01553.x. [DOI] [PubMed] [Google Scholar]
- Kim H.R., Kim J.M., Kim M.S., Hwang J.K., Park Y.J., Yang S.H., Kim H.J., Ryu D.G., Lee D.S., Oh H., et al. Saussurea lappa extract suppresses TPA-induced cell invasion via inhibition of NF-kappaB-dependent MMP-9 expression in MCF-7 breast cancer cells. BMC Complement. Altern. Med. 2014;14:170. doi: 10.1186/1472-6882-14-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasagni L., Francalanci M., Annunziato F., Lazzeri E., Giannini S., Cosmi L., Sagrinati C., Mazzinghi B., Orlando C., Maggi E., et al. An alternatively spliced variant of CXCR3 mediates the inhibition of endothelial cell growth induced by IP-10, Mig, and I-TAC, and acts as functional receptor for platelet factor 4. J. Exp. Med. 2003;197:1537–1549. doi: 10.1084/jem.20021897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.K., Ha H., Lee H.Y., Park S.J., Jeong S.L., Choi Y.J., Shin H.K. Inhibitory effects of heartwood extracts of Broussonetia kazinoki Sieb on the development of atopic dermatitis in NC/Nga mice. Biosci. Biotechnol. Biochem. 2010;74:1802–1806. doi: 10.1271/bbb.100138. [DOI] [PubMed] [Google Scholar]
- Leung D.Y., Bieber T. Atopic dermatitis. Lancet. 2003;361:151–160. doi: 10.1016/S0140-6736(03)12193-9. [DOI] [PubMed] [Google Scholar]
- Lim H.S., Ha H., Lee M.Y., Jin S.E., Jeong S.J., Jeon W.Y., Shin N.R., Sok D.E., Shin H.K. Saussurea lappa alleviates inflammatory chemokine production in HaCaT cells and house dust mite-induced atopic-like dermatitis in Nc/Nga mice. Food Chem. Toxicol. 2014;63:212–220. doi: 10.1016/j.fct.2013.10.050. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., 3rd, Zody M.C., et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Liu L.Y., Mathur S.K., Sedgwick J.B., Jarjour N.N., Busse W.W., Kelly E.A. Human airway and peripheral blood eosinophils enhance Th1 and Th2 cytokine secretion. Allergy. 2006;61:589–597. doi: 10.1111/j.1398-9995.2006.01060.x. [DOI] [PubMed] [Google Scholar]
- Lu B., Humbles A., Bota D., Gerard C., Moser B., Soler D., Luster A.D., Gerard N.P. Structure and function of the murine chemokine receptor CXCR3. Eur. J. Immunol. 1999;29:3804–3812. doi: 10.1002/(SICI)1521-4141(199911)29:11<3804::AID-IMMU3804>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Lu Z.R., Kim W.S., Cho I.H., Park D., Bhak J., Shi L., Zhou H.W., Lee D.Y., Park Y.D., Yang J.M., et al. DNA microarray analyses and interactomic predictions for atopic dermatitis. J. Dermatol. Sci. 2009;55:123–125. doi: 10.1016/j.jdermsci.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Matesanz F., Fedetz M., Collado-Romero M., Fernandez O., Guerrero M., Delgado C., Alcina A. Allelic expression and interleukin-2 polymorphisms in multiple sclerosis. J. Neuroimmunol. 2001;119:101–105. doi: 10.1016/s0165-5728(01)00354-x. [DOI] [PubMed] [Google Scholar]
- Nashed B.F., Zhang T., Al-Alwan M., Srinivasan G., Halayko A.J., Okkenhaug K., Vanhaesebroeck B., Hayglass K.T., Marshall A.J. Role of the phosphoinositide 3-kinase p110delta in generation of type 2 cytokine responses and allergic airway inflammation. Eur. J. Immunol. 2007;37:416–424. doi: 10.1002/eji.200636401. [DOI] [PubMed] [Google Scholar]
- Novak N., Tepel C., Koch S., Brix K., Bieber T., Kraft S. Evidence for a differential expression of the FcepsilonRIgamma chain in dendritic cells of atopic and nonatopic donors. J. Clin. Invest. 2003;111:1047–1056. doi: 10.1172/JCI15932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sa S.M., Valdez P.A., Wu J., Jung K., Zhong F., Hall L., Kasman I., Winer J., Modrusan Z., Danilenko D.M., et al. The effects of IL-20 subfamily cytokines on reconstituted human epidermis suggest potential roles in cutaneous innate defense and pathogenic adaptive immunity in psoriasis. J. Immunol. 2007;178:2229–2240. doi: 10.4049/jimmunol.178.4.2229. [DOI] [PubMed] [Google Scholar]
- Sano S., Chan K.S., Carbajal S., Clifford J., Peavey M., Kiguchi K., Itami S., Nickoloff B.J., DiGiovanni J. Stat3 links activated keratinocytes and immunocytes required for development of psoriasis in a novel transgenic mouse model. Nat. Med. 2005;11:43–49. doi: 10.1038/nm1162. [DOI] [PubMed] [Google Scholar]
- Sebastian K., Borowski A., Kuepper M., Friedrich K. Signal transduction around thymic stromal lymphopoietin (TSLP) in atopic asthma. Cell Commun. Signal. 2008;6:5. doi: 10.1186/1478-811X-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto T., Jin Z.G., Berk B.C. Transactivation of vascular endothelial growth factor (VEGF) receptor Flk-1/KDR is involved in sphingosine 1-phosphate-stimulated phosphorylation of Akt and endothelial nitric-oxide synthase (eNOS) J. Biol. Chem. 2002;277:42997–43001. doi: 10.1074/jbc.M204764200. [DOI] [PubMed] [Google Scholar]
- Theofilopoulos A.N., Baccala R., Beutler B., Kono D.H. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu. Rev. Immunol. 2005;23:307–336. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- Wang Y., Barbacioru C., Hyland F., Xiao W., Hunkapiller K.L., Blake J., Chan F., Gonzalez C., Zhang L., Samaha R.R. Large scale real-time PCR validation on gene expression measurements from two commercial long-oligonucleotide microarrays. BMC Genomics. 2006;7:59. doi: 10.1186/1471-2164-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M., Hatakeyama S., Inoue Y., Yamahara J. Saussureamines A, B, C, D, and E, new anti-ulcer principles from Chinese Saussureae Radix. Chem. Pharm. Bull. 1993;41:214–216. doi: 10.1248/cpb.41.214. [DOI] [PubMed] [Google Scholar]
- Yu W.S., Jeong S.J., Kim J.H., Lee H.J., Song H.S., Kim M.S., Ko E., Lee H.J., Khil J.H., et al. The genome-wide expression profile of 1,2,3,4,6-penta-O-galloyl-β-D-glucose-treated MDA-MB-231 breast cancer cells: molecular target on cancer metabolism. Mol. Cells. 2011;32:123–132. doi: 10.1007/s10059-011-2254-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.K., Yang Y., Bai S.R., Zhang G.Z., Liu T.H., Zhou Z., Wang C.M., Tang L.J., Wang J., He S.X. Screening for key genes associated with atopic dermatitis with DNA microarrays. Mol. Med. Rep. 2014;9:1049–1055. doi: 10.3892/mmr.2014.1908. [DOI] [PubMed] [Google Scholar]
- Zuo L., Fulkerson P.C., Finkelman F.D., Mingler M., Fischetti C.A., Blanchard C., Rothenberg M.E. IL-13 induces esophageal remodeling and gene expression by an eosinophil-independent, IL-13R alpha 2-inhibited pathway. J. Immunol. 2010;185:660–669. doi: 10.4049/jimmunol.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


