Abstract
A chromosome territory is composed of chromosomal subdomains. The internal structure of chromosomal subdomains provides a structural framework for many genomic activities such as replication and DNA repair, and thus is key to determining the basis of their mechanisms. However, the internal structure and regulating proteins of a chromosomal subdomain remains elusive. Previously, we showed that the chromosome territory expanded after BAF53 knockdown. Because the integrity of chromosomal subdomains is a deciding factor of the volume of a chromosome territory, we examined here the effect of BAF53 knockdown on chromosomal subdomains. We found that BAF53 knockdown led to the disintegration of histone H2B-GFP-visualized chromosomal subdomains and BrdU-labeled replication foci. In addition, the size of DNA loops measured by the maximum fluorescent halo technique increased and became irregular after BAF53 knockdown, indicating DNA loops were released from the residual nuclear structure. These data can be accounted for by the model that BAF53 is prerequisite for maintaining the structural integrity of chromosomal subdomains.
Keywords: actin-related protein, BAF53, chromosomal subdomain, DNA loop, replication foci
INTRODUCTION
Each chromosome is encapsulated in its own chromosome territory (CT) within mammalian cell nuclei (Cremer and Cremer, 2010). The internal structure of CTs has been the subject of intensive studies, and there is compelling evidence supporting the idea that CTs are built up from chromosomal subdomains. Chromosomal subdomains are higher-order chromatin structures comprising several hundred kb to several Mb each and have been visualized as bead images with diameters of 300 to 800 nm in fluorescence in situ hybridization (FISH) studies of natural mammalian chromatin and artificial tandem array systems, such as the lac operator array and MMTV tandem array (Muller et al., 2001; 2004; Tumbar et al., 1999). Hi-C analysis has identified topological domains that are large, megabase-sized local chromatin interaction domains (Dixon et al., 2012). Topological domains are likely to correspond to chromosomal subdomains.
A chromosomal subdomain is also considered to be an independent structural and functional unit of replication. When labeled with a thymidine analog such as 5-bromodeoxyuridine (BrdU), replicated DNA is visualized as foci with a size of 400–800 nm diameter (Jackson and Pombo, 1998; O’Keefe et al., 1992). Replication foci appear to be discrete and stable structures because these foci persist throughout subsequent cell cycles (Kennedy et al., 2000; Leonhardt et al., 2000). Each replication focus contains an average of 1 Mbp of DNA. Taken together, it has been proposed that replication foci are another representation of chromosomal subdomains.
The internal structure of chromosomal subdomains has remained a matter of controversy. The multi-loop subcompartment model states that a series of ∼100-kb chromatin loops are clustered to form ∼1-Mb rosette-like multi-loop subcompartments (Munkel et al., 1999). Assuming that chromatin is compacted into a 30-nm fiber, the subcompartment turns out to be a ∼500-nm diameter compact structure in computer modeling, which is well consistent with the size of the subdomains demonstrated by FISH. On the other hand, EM images of thin sections of CHO and HeLa cells have demonstrated several features characteristic of fiber-like structure at a scale much smaller than the average diameter of ∼500 nm (Belmont and Bruce, 1994). Based on these observations, the chromonema fiber model has been proposed in which the chromatin constructs a series of progressive folds via intrafiber interactions, generating a series of thicker fibers: 60–80 nm and 100–130 nm in thickness. Recently, Hi-C data obtained for human cells turned out to be consistent with the fractal globule model on the scale of up to 10 Mb (Lieberman-Aiden et al., 2009). A fractal globule is characterized by a hierarchy of crumples that are formed spontaneously by polymer collapse. Polymer collapse in interphase chromatin fibers could be provided by nuclear matrix attachment regions and DNA-binding condensing/linking proteins like cohesin and CTCF (Ji et al., 2013; Rao et al., 2014). Therefore, chromosomal subdomains can be conceived as predominant crumples with sizes of approximately 1 Mb.
Interestingly, bead images of tandem array systems such as lac operator arrays and MMTV tandem arrays becoming elongated into a series of smaller beads in response to transactivation (Muller et al., 2001; Tumbar et al., 1999). A single bead image of transcriptionally active regions in natural chromosomes was elongated in response to the mode of transactivation as well (Muller et al., 2004). These results indicate the protein scaffold and/or molecular crosslinks that play a pivotal role in the formation of chromosomal subdomains could be reorganized responding to its environmental cue. For example, the outgrowth and retraction of protein scaffold may yield a separation of several clusters of ∼100-kb chromatin loops within a ∼1-Mb multi-loop subcompartment, or partial removal of molecular crosslinks that keep a crumple formed may lead to differentiation into smaller crumples (Mirny, 2011).
BAF53 is an actin-related protein (Arp) found in mammalian cell nuclei (Sung et al., 2001; Zhao et al., 1998). Previously, we showed that BAF53 is required for the maintenance of chromosome territories (Lee et al., 2007). When the expression of BAF53 was suppressed by short interfering RNA (siRNA) interference, chromosome territories were expanded by 2 to 2.4 fold. One likely cause of the expansion of chromosome territories is a release from constraints that limit free diffusion of chromatin fibers such as the linkage between DNA loops and the protein scaffold proposed in the multi-loop subcompartment model, or the molecular crosslinks according to the chromonema fiber model and fractal globule model. Regardless of the model being followed, a release from constraints that limit free diffusion of chromatin fibers within the chromosomal subdomains would inevitably result in disintegration of chromosomal subdomains. In this study, we examined this possibility, and found that BAF53 knockdown leads to the disintegration of chromosomal subdomains, as visualized by the expression of histone H2B-GFP. Chase experiments of BrdU-prelabeled replication foci revealed that BAF53 knockdown disintegrates replication foci structurally. Finally, DNA loops are released from the residual nuclear structure when BAF53 is depleted. Our findings suggest that the structural integrity of chromosomal subdomains can be disrupted when BAF53 is depleted.
MATERIALS AND METHODS
Cell culture and synchronization
NIH3T3 cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% calf serum with 100 U/ml penicillin and 100 μg/ml streptomycin at 37°C in a CO2 incubator. HeLa cells stably transfected with H2B-GFP (Kanda et al., 1998) were cultured in DMEM supplemented with 10% fetal bovine serum, 600 ug/ml G418, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C in a CO2 incubator. To synchronize the cell cycle at the G1/S border, NIH3T3 cells were incubated in complete medium with 5 mM thymidine for 12 h, washed and incubated in complete medium for 9 h, and then incubated in 5 mM thymidine again for 14 h.
Establishment of a HeLa H2B-GFP cell line
The H2B-GFP expression vector (Kanda et al., 1998) received from Addgene (Plasmid 11680) was transfected into HeLa cells using Lipofectamine. At 48 h after transfection, the cells were split into fresh medium containing 600 μg/ml G418 and then were cultured by feeding the selective medium every 3 days for 2 weeks until colonies could be identified. Twenty colonies were expanded, and we screened those clones that exhibited a high level of expression of H2B-GFP.
Formation of hypercondensed chromatin (HCC)
HCC formation was induced by incubating the cells in a hyper-osmolar medium. One ml of 20× phosphate-buffered saline (PBS; 2.8 M NaCl, 54 mM KCl, 130 mM Na2HPO4, and 30 mM KH2PO4 in H2O, pH adjusted to 7.4) was diluted with 19 ml of standard culturing medium (290 mOsm) to yield an osmolarity of 570 mOsm.
siRNA transfection and short hairpin RNA (shRNA) transduction
NIH3T3 cells were cultured for 24 h in complete medium without antibiotics, and the cells were incubated in serum-free medium containing 100 nM siRNA with siPORTamine (Applied Biosystems) for 20 min at room temperature. We constructed the expression vector of the shRNA-BAF53 using the BLOCK-iT™ Lentiviral RNAi Expression System (Invitrogen) and treated 1 × 105 NIH3T3 cells with 30% of the viral particles, which express shRNA-BAF53, to knockdown BAF53.
Immunoblot assay
Cells were lysed in 1× sodium dodecyl sulfate (SDS) sample buffer (62.5 mM Tris-Cl, pH 6.8, 10% glycerol, 2% SDS, 1% β–mercaptoethanol, and 0.01% bromophenol blue). The protein concentrations were determined using the BCA kit (PIERCE), and the samples were subjected to 10% or 15% SDS polyacrylamide gel electrophoresis. The proteins were transferred to polyvinylidene fluoride (PVDF) membranes (BIO-RAD) in 10% methanol in Tris-glycine for 90 min at room temperature; the membranes were then blocked in 5% nonfat dry milk in 0.1% Tween 20 in Tris-buffered saline (TBS), pH 7.6, for 1 h at room temperature. The membrane was incubated with primary antibody (BAF53, described in Lee et al., 2003; β-tubulin, #T4026, Sigma) overnight at 4°C and washed in 0.1% Tween 20 in TBS three times. After application of the secondary antibodies, the membrane was visualized by ECL (enhanced chemiluminescence).
Immunostaining
Fixed cells on a coverslip were incubated in 1.5 N HCl for 30 min to denature the DNA, and were washed with distilled water and neutralized in 0.1 M sodium borate for 5 min at room temperature. The cells were permeabilized in 0.5% PBS-T for 5 min, washed in 0.1% PBS-T briefly, and incubated in blocking solution (10% heat-inactivated calf serum and 0.5% gelatin in TBS) for 30 min. After a quick wash in 0.1% PBS-T, the primary antibody, anti-mouse BrdU (BD #555627), and then the secondary antibody, mouse FITC, were applied for 1 h each at room temperature. Next, 1 μg/ml Hoechst 33258 was used for counterstaining. Between these steps, we washed the cells three times in 0.1% PBS-T for 10 min each at room temperature.
DNA halo assay
Non-fixed nuclei on coverslips were incubated for 1 min in ice-cold NP40 buffer (0.5% NP40, 10 mM MgCl2, 0.5 mM CaCl2, 50 mM HEPES, pH 7.8) and sequentially dipped in 0.5 M, 1 M, 1.5 M, and 2 M NaCl containing 0.2 mM MgCl2 and 10 mM Tris-HCl (pH 7.4). To observe the DNA halo, the nuclei were incubated in 100 μg/ml ethidium bromide for 1 min in 2 M NaCl and exposed for 1 min to short-wave UV light.
BrdU chase
After 30 min or 6 h from releasing G1/S arrest, NIH3T3 cells were incorporated with 20 μM BrdU for 30 min. The cells labeled at early or late S phase were infected with lentivirus containing shRNA-BAF53 and then were cultured for 48 h before fixation. Fixed cells were immunostained with the anti-BrdU antibody.
RESULTS
BAF53 knockdown disintegrates chromosomal subdomains as visualized by the expression of histone H2B-GFP
We generated HeLa cells expressing histone H2B-GFP, which allows the visualization of chromosomal subdomains as bead images (Kanda et al., 1998). The midsection of a fixed nucleus reveals that bead images are evenly distributed throughout the nucleoplasm, nuclear periphery and perinucleolar regions, although individual beads cannot be clearly discerned from each other (Fig. 1). The less clear delineation of each bead image is probably due to out-of-focus light from the densely packed neighboring chromatin domains. To test whether BAF53 is required for the maintenance of chromosomal subdomains, we suppressed the expression of BAF53 by siRNA interference (Fig. 1A). We employed the same population of HeLa cells stably expressing histone H2B-GFP for the control and BAF53-knockdown groups, and the expression levels of H2B-GFP were in fact comparable for the both groups (Fig. 1B). After BAF53 knockdown, the nucleoplasm became more or less vacant of bead images, while the bead images at the nuclear periphery and perinucleolar regions became more conspicuous (Fig. 1C). To visualize this change more clearly, we assigned a series of threshold values to the images and highlighted pixels with gray values above the assigned thresholds in red. Pixels highlighted in red started to decrease with the low threshold, TH = 40, and decreased precipitously with the higher threshold, TH = 60, in BAF53 knockdown cells (Fig. 1D). By contrast, the number of red pixels decreased slightly with both thresholds in control cells. It is noteworthy that the red pixels that disappeared after BAF53 knockdown were preferentially located in the nucleoplasm, while the red pixels at the nuclear periphery remained relatively robust. Considering that the hallmark of bead images is repeated, stronger signals from locally overlapping segments of chromatin fibers arising from higher-order folding or clustering of chromatin loops, the decrease in the number of red pixels observed in BAF53 knockdown cells can be attributed to an unfolding of beads, leaving diffused fluorescence signals below the assigned thresholds. In addition, this finding suggests that beads in the nucleoplasm are preferentially unfolded after BAF53 knockdown.
Fig. 1.
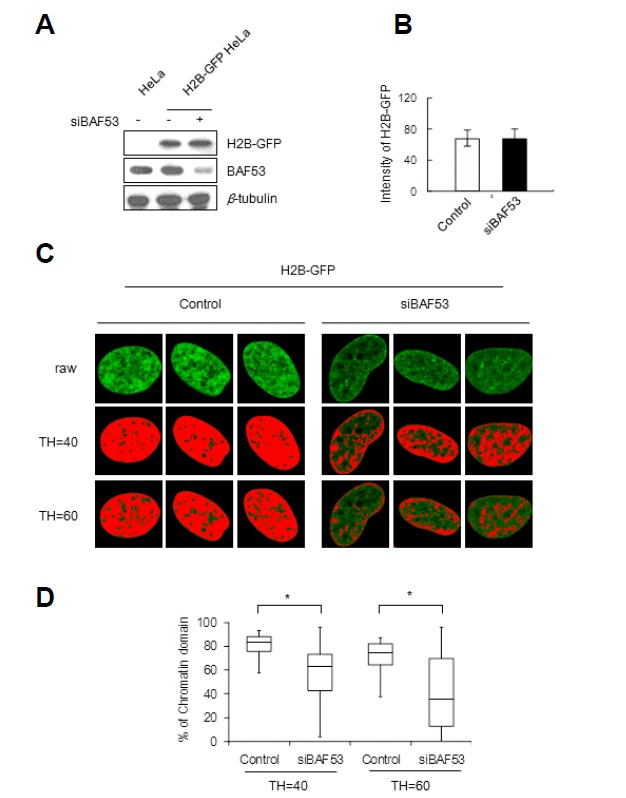
Disintegration of the chromosomal subdomains by BAF53 knockdown. HeLa cells stably expressing GFP-tagged histone H2B were transfected with siRNA-BAF53. (A) The expression levels of H2B-GFP and BAF53 were determined by immunoblotting. (B) The expression level of H2B-GFP of a cell was measured by integrating the signals from all of the pixels of the midsection image of a fixed nucleus revealing GFP-tagged histone H2B (t-test; P > 0.5). Error bars, mean ± SD (n = 10). (C) Representative images of the midsection of a fixed nucleus revealing GFP-tagged histone H2B. Pixels with gray values above the assigned thresholds (TH = 40 and TH = 60) are highlighted in red. (D) Distributions of the occupancies of red pixels in the control and BAF53-knockdown cell nuclei. Boxes represent median and quartile values, and bars define the 5th and 95th percentiles (n = 30). t-test; *P < 0.001.
It has been known that the placement of cells in hyper-osmolar medium leads to the formation of hypercondensed chromatin (HCC). This approach was employed to highlight chromosomal subdomains successfully (Albiez et al., 2006; Cremer et al., 2000). Under hyper-osmolar conditions, hypercondensation of chromatin allowed clear visualization of chromosomal subdomains of H2B-GFP-tagged chromatin (Fig. 2). Bead images with similar signal intensity and size were found throughout the nucleoplasm, nuclear periphery, and perinucleolar regions. Previously, it was shown that structural patterns of hypercondensed chromatin reflect a higher-order chromatin and interchromatin domain topology (Albiez et al., 2006). Since the protein scaffolds that play a pivotal role in the formation of chromosomal subdomains are expected to be a primary determinant of the higher-order chromatin and interchromatin domain topology, this result suggests that chromatin coalesces with the underlying protein scaffold into a highly condensed mass during the hypercondensation process, and this process is delimited by the domain structure. Placement of BAF53 knockdown cells into hyper-osmolar medium also led to hypercondensation of chromatin, resulting in bead images similar to those of the control cells. This finding implied that condensation of chromatin under hyper-osmolar conditions is apparently unaltered after BAF53 knockdown. Significantly, however, the interchromatin-lacunas space widened after BAF53 knockdown. This change is more clearly demonstrated by assigning thresholds and highlighting pixels with gray values below the assigned thresholds in blue. The widening of the interchromatin-lacunas space argues that the even distribution of the protein scaffold that contributes to the formation of hypercondensed chromatin bundles, as well as chromosomal subdomains, is at least in part distorted or disrupted by BAF53 knockdown.
Fig. 2.
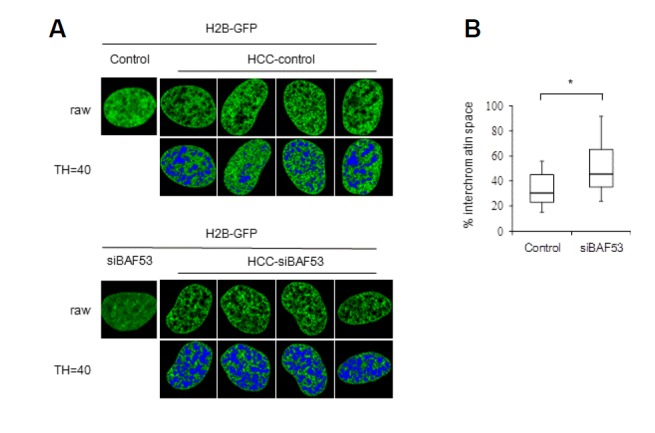
Expansion of the interchromatin compartment space by BAF53 knockdown. (A) Representative confocal midsections of the nuclei of HeLa cells (H2B-GFP) after treatment with the hyper-osmolar medium. Note that the formation of hypercondensed chromatin (HCC) is unchanged, but the interchromatin compartment space is expanded in BAF53-knockdown cell nuclei. Pixels with gray values below the assigned thresholds (TH = 40) are highlighted in blue. (B) Distributions of the occupancies of blue pixels in the control and BAF53-knockdown cell nuclei. Boxes represent median and quartile values, and bars define the 5th and 95th percentiles (n = 30). t-test; *P < 0.001.
Early S-phase replication foci are selectively disintegrated by BAF53 knockdown
Replication foci are stable structures that can persist throughout several cell cycles and have been proposed as other representations of chromosomal subdomains (Cremer et al., 2000; Kennedy et al., 2000; Leonhardt et al., 2000; Muck and Zink, 2009). When cells in which replication foci were pulse-labeled with BrdU undergo several cell cycles, BrdU-labeled chromosomes were segregated, and it allowed a clear visualization of BrdU-prelabeled replication foci. Therefore, this method provides another opportunity to examine the effect of BAF53 knockdown on chromosomal subdomains, as well. Another interesting feature of the formation of replication foci is its specific spatiotemporal order. Time course analysis of incorporated BrdU revealed that the formation of replication foci in NIH3T3 cells follows faithfully the specific spatiotemporal order that was demonstrated in CHO cells (O’Keefe et al., 1992). During early S-phase (up to 2 h into S-phase), replication foci were distributed at sites throughout the nucleoplasm, excluding the nucleolus and periphery of the nucleus. As S-phase progressed (mid S-phase, 4–6 h), replication foci at sites near perinucleolar and peripheral regions became predominant, and replication at the nucleoplasm decreased. In late S-phase (7 h), replication occurred at a few large regions that were presumably interior regions of heterochromatin.
NIH3T3 cells were labeled with BrdU at the early or late S-phase and were split into two groups (Fig. 3A). The control group was mock-treated, and the BAF53-knockdown group was infected with lentivirus containing shRNA-BAF53 (Fig. 3B). Cells were furthered incubated for 48 h and then fixed and immunostained with anti-BrdU antibody. The number of pixels with staining above the assigned threshold per cell was counted. BAF53 knockdown dramatically reduced the number of replication foci pre-labeled at early S-phase (Fig. 3C). Because the cells of the control group and BAF53-knockdown group were derived from the same cell population labeled with BrdU, the cells of the control group and BAF53-knockdown group should contain the same extent of BrdU. Considering this, this result strongly suggested that BAF53 knockdown leads to unfolding of BrdU-prelabeled stable units of chromosome structure, and this unfolding reduces the intensity of BrdU staining of replication foci below the assigned thresholds, which can no longer be recognized as foci.
Fig. 3.
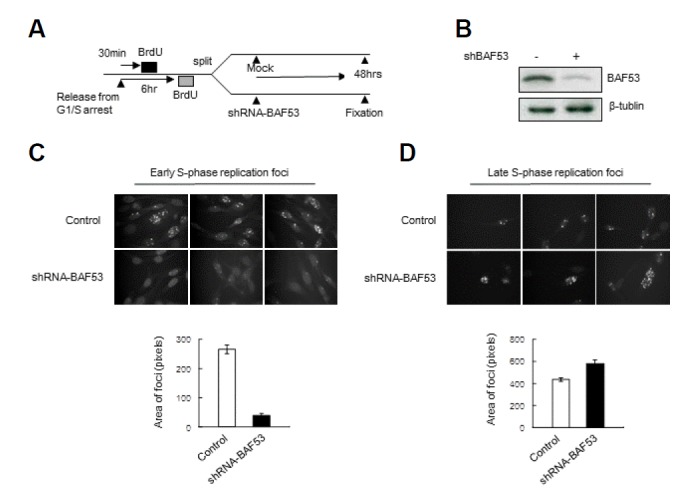
Early replication foci are selectively disintegrated by BAF53 knockdown. (A) The scheme of the experimental procedure. NIH3T3 cells were arrested at the G1/S border by double-thymidine blockade. After release from the G1/S arrest, the cells were pulse labeled for 20 min with BrdU at early or late S phase. Next, the cells were split into two groups—the control group was mock-treated, and the BAF53 knockdown group was infected with lentivirus containing shRNA-BAF53. At 48 h after the transduction, the cells were fixed and immunostained with anti-BrdU antibody. (B) The expression level of BAF53 at 48 h after the transduction was determined by immunoblotting. (C) Confocal midsections of the NIH3T3 cells labeled with BrdU at early S-phase. Pixels above the assigned thresholds were counted as replication foci in the cells. One hundred cells were counted for each experiment. Histograms show the number of replication foci in the control and BAF53-knockdown cell nuclei (n = 3, mean ± SD). t-test; P < 0.001. (D) Confocal midsections of the NIH3T3 cells labeled with BrdU at late S-phase. Pixels above the assigned thresholds were counted as replication foci in the cells. Thirty cells were counted for each experiment. The histograms show the number of replication foci in the control and BAF53-knockdown cell nuclei (n = 3, mean ± SD). t-test; P < 0.01.
Interestingly, BAF53 knockdown caused the opposite effect on the replication foci pre-labeled at late S-phase (Fig. 3D). It increased the number of replication foci. This increase seems to be due to the reduction of segregation of pre-labeled chromosome because BAF53 knockdown leads to cell cycle arrest (Lee et al., 2007). Taken together, these results indicate that BAF53 knockdown disintegrates early S-phase replication foci preferentially.
DNA loops are dissociated from the nuclear scaffold when BAF53 is depleted
According to the multi-loop subcompartment model, a chromosomal subdomain is a cluster of DNA loops (Munkel et al., 1999). Because the dissociation of DNA loops from the nuclear scaffold would result in the disintegration of the chromosomal subdomain, we investigated whether BAF53 knockdown affects the attachment of the DNA loop to the nuclear scaffold. We measured the chromatin loop sizes using the maximum fluorescent halo technique (Vogelstein et al., 1980). Nuclei were extracted with 2 M NaCl, and then were exposed for 1 min to short-wave ultraviolet (UV) light in the presence of ethidium bromide to observe the DNA halo. The nuclear scaffold was distinguished from the halo by immunostaining with anti-lamin A/C antibody (Fig. 4).
Fig. 4.
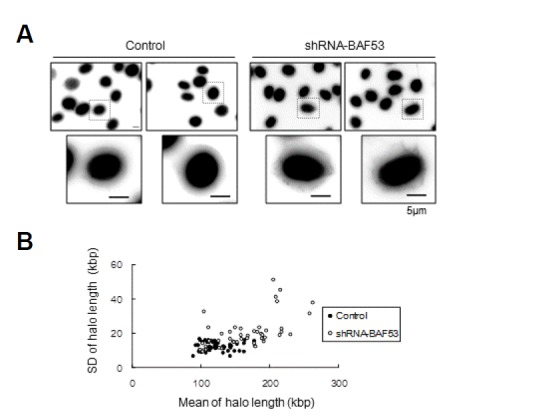
The dissociation of DNA loops from the nuclear scaffold in BAF53 knockdown cells. (A) Nonfixed nuclei on coverslips were submitted to the maximum fluorescent halo technique (MFHT) for DNA loop-size measurements. Immunostaining with an anti-lamin antibody delimitated the matrix from DNA loops. Scale bar, 5 μm. (B) The mean of the halo length of each cell was plotted versus the standard deviation of the halo length of the cell. t-test; P < 0.001.
The mean loop size estimated from NIH3T3 cell nuclei was 124 ± 12.4 kbp, which was comparable to the values reported for other somatic cell nuclei (Buongiorno-Nardelli et al., 1982; Vogelstein et al., 1980). Two changes in DNA loop sizes were recognized after BAF53 knockdown. First, mean loop size was increased to 165 ± 20 kbp. Second, the size of loops in a single nucleus became largely irregular. The standard deviation of the loop size was less than 16.6 kbp in the control cell nuclei, indicating that the size of DNA loops is rather constant in the control cells. In the BAF53-knockdown cell nuclei, however, the standard deviation of the loop size was largely increased up to 51.2 kbp. Stochastic dissociation of DNA loops from the nuclear scaffold could account for these increases in mean loop size and the standard deviation of the loop size. We conclude that BAF53 is required for the association of DNA loops with the nuclear scaffold.
DISCUSSION
We previously reported that CTs expanded after BAF53 knockdown. Loss of the internal meshwork such as interactions between adjacent chromatin fibers or the anchorage of the chromatin fiber onto a protein-rich scaffold could account for the expansion of CTs. Because the internal meshwork is likely to act as a primary force that constitutes chromosomal subdomains, loss of the internal meshwork is expected to result in the disintegration of chromosomal subdomains.
In both of the approaches examined here, the data support that chromosomal subdomains become disintegrated when BAF53 is not available. Visualization of higher-order chromatin structure by the expression of H2B-GFP provides images closest to its natural state. Although the beaded features of chromosomal subdomains were blurred by out-of-focus light from highly overlapped structures, we found the beaded features became consistently less evident after BAF53 knockdown. In addition, we noticed that the loss of beaded features selectively occurred at the nucleoplasm, while bead images at the nuclear periphery and perinucleolar regions were less affected.
It is generally accepted that replication focus is another manifestation of chromosomal subdomains (Cayrou et al., 2010; Muck and Zink, 2009). Replication foci are formed by the association of several active replicons that fire DNA replication more or less simultaneously. If replicons adjacent to each other in a chromosome build a stable structure, the synchronous initiation of replicons within a given structure would result in the formation of replication foci. A study in which Chinese hamster ovary cells were incubated in [3H] thymidine for 30 min before fiber autoradiography demonstrated that adjacently located replicons fire DNA replication at approximately the same time (Lark et al., 1971). The grouping of replicons located adjacently in a chromosome and quasi-synchronous firing of all the replicons in each group have been further supported by DNA combing (Michalet et al., 1997). In this study, we demonstrated that BAF53 is essential for maintaining the integrity of BrdU-prelabeled replication foci. Furthermore, early S-phase replication foci were preferentially affected by BAF53 depletion. Previously, we examined the effect of BAF53 knockdown on BrdU-labeling during the progress of DNA replication. We found that BAF53 was required for BrdU-labeling at the early stage of DNA replication (Kwon and Kwon, 2012). On the other hand, BrdU-labeling at the mid and late stages of DNA replication was not decreased by BAF53 knockdown. It is well consistent with the finding of this study that early S-phase replication foci were preferentially affected by BAF53 depletion. Because early S-phase replication foci are known to be located in the nucleoplasm, and mid and late S-phase replication foci are located at nuclear periphery and perinucleolar regions (Kennedy et al., 2000; O’Keefe et al., 1992; Zink et al., 1999), this finding is well consistent with the observation that beaded features of H2B-GFP images were lost selectively in the nucleoplasm after BAF53 depletion.
It has been proposed that early S-phase replication foci are mostly composed of euchromatin abundant with active genes while late S-phase replication foci comprise heterochromatin (Kennedy et al., 2000; Leonhardt et al., 2000; Muck and Zink, 2009; O’Keefe et al., 1992). Therefore, our observations suggest that BAF53 is essential for the formation of chromosomal subdomains of euchromatin but not that of heterochromatin. Because BAF53 knockdown left a residual amount of BAF53 in the cells, we cannot rule out the possibility that chromosomal subdomains of heterochromatin still require BAF53 but are resistant to the disintegration in the presence of low concentration of BAF53. Taken together, our data support the idea that the same structural change in a higher-order chromatin structure is responsible for the disruption of replication foci and disintegration of chromosomal subdomains. All of this evidence further supports the idea that the stable architecture of replication foci is a fundamental unit of interphase chromatin and that BAF53 plays an essential role in the formation of fundamental units of chromosomal architecture, namely chromosomal subdomains.
The detailed structure of chromosomal subdomains remains obscure. Nevertheless, DNA loops anchored to a protein scaffold have been a predominant model of the higher-order chromatin architecture of interphase chromatin. DNA loop structures have been observed using electron microscopy in the Miller spreading of the 75S RNA genes located at Balbiani rings of the fly Chironomus (Andersson et al., 1980). The lampbrush chromosomes in amphibian oocytes were seen to be organized into a series of DNA loops, emanating from a chromosome axis (Spring and Franke, 1981). The DNA loop structure of cultured cells in general can be visualized using the maximum fluorescent halo technique in which nuclei permeabilized with NP-40 were exposed to high salt concentrations to extract histones and non-histone chromatin proteins from DNA, preserving the attachment sites of DNA loops to the nuclear scaffold. Under this treatment, the DNA halo representing DNA loops appears as a smooth contour of a concentric circle, implying remarkable size regularity of DNA loops. In this study, we showed that BAF53 depletion led to the increase in the loop size from 124 ± 12.4 kbp to 165 ± 20 kbp, providing unequivocal evidence to support the idea that DNA loops dissociate from the nuclear scaffold when BAF53 is depleted. The loop size increases erratically, as demonstrated by the increase in the standard deviation of the loop size from 16.6 kbp to 51.2 kbp. This suggests that the dissociation of DNA loops from the scaffold does not proceed in unison but may occur in a stochastic manner. These results identify BAF53 as a candidate for a regulator protein for a chromatin-anchoring protein scaffold. It must be noted, however, that the dissociation of DNA loops from the nuclear scaffold is incomplete and stochastic after BAF53 knockdown. A low level of BAF53 remaining after BAF53 knockdown may explain this incomplete and stochastic dissociation. On the other hand, this result can be equally well explained based on the fractal globule model. Attractive interactions in chromatin are prerequisite for the formation of the fractal globule. It has been demonstrated that the removal of attractive interactions allows the region of the fractal globule to open up due to the chain entropy (Mirny, 2011). Because several different interactions could contribute to attractive interactions for the fractal globule, BAF53 may be involved in one of the attractive interactions in the fractal globule. If that is the case, BAF53 depletion would result in an incomplete and stochastic dissociation of DNA loops from the nuclear scaffold as shown in the DNA halo assay.
Our studies indicate that BAF53 plays a role in linking DNA to the nuclear scaffold, which is essential for the formation of chromosomal subdomains. Certainly, the elucidation of the molecular mechanism by which BAF53 links DNA to the nuclear scaffold would be the next key question. BAF53 is an actin-related protein, and has been shown to mediate the formation of branching networks of actin filaments in vitro (Rando et al., 2002). An actin branching network is the most versatile and dynamic scaffold present in the cells. Accordingly, branching networks of actin filaments are an ideal candidate for the internal meshwork, which is responsible for the formation of chromosomal subdomains. While raising many fundamental questions regarding the architectural basis for chromosomal subdomains, our data, which have revealed the requirement of BAF53 for the formation of chromosomal subdomains, would provide a framework toward further understanding of the higher order architecture of chromatin in interphase.
Acknowledgments
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF-2010-0022664). We are grateful to Dr. Kanda for providing the H2B-GFP expression vector.
REFERENCES
- Albiez H., Cremer M., Tiberi C., Vecchio L., Schermelleh L., Dittrich S., Küpper K., Joffe B., Thormeyer T., von Hase J., et al. Chromatin domains and the interchromatin compartment form structurally defined and functionally interacting nuclear networks. Chromosome Res. 2006;14:707–733. doi: 10.1007/s10577-006-1086-x. [DOI] [PubMed] [Google Scholar]
- Andersson K., Bjorkroth B., Daneholt B. The in situ structure of the active 75 S RNA genes in Balbiani rings of Chironomus tentans. Exp. Cell Res. 1980;130:313–326. doi: 10.1016/0014-4827(80)90008-7. [DOI] [PubMed] [Google Scholar]
- Belmont A.S., Bruce K. Visualization of G1 chromosomes: a folded, twisted, supercoiled chromonema model of interphase chromatid structure. J. Cell Biol. 1994;127:287–302. doi: 10.1083/jcb.127.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buongiorno-Nardelli M., Micheli G., Carri M.T., Marilley M. A relationship between replicon size and supercoiled loop domains in the eukaryotic genome. Nature. 1982;298:100–102. doi: 10.1038/298100a0. [DOI] [PubMed] [Google Scholar]
- Cayrou C., Coulombe P., Mechali M. Programming DNA replication origins and chromosome organization. Chromosome Res. 2010;18:137–145. doi: 10.1007/s10577-009-9105-3. [DOI] [PubMed] [Google Scholar]
- Cremer T., Cremer M. Chromosome territories. Cold Spring Harb. Perspect. Biol. 2010;2:a003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T., Kreth G., Koester H., Fink R.H., Heintzmann R., Cremer M., Solovei I., Zink D., Cremer C. Chromosome territories, interchromatin domain compartment, and nuclear matrix: an integrated view of the functional nuclear architecture. Crit. Rev. Eukaryot. Gene Expr. 2000;10:179–212. [PubMed] [Google Scholar]
- Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D.A., Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji L., Xu R., Lu L., Zhang J., Yang G., Huang J., Wu C., Zheng C. TM6, a novel nuclear matrix attachment region, enhances its flanking gene expression through influencing their chromatin structure. Mol. Cells. 2013;36:127–137. doi: 10.1007/s10059-013-0092-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda T., Sullivan K.F., Wahl G.M. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol. 1998;8:377–385. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- Kennedy B.K., Barbie D.A., Classon M., Dyson N., Harlow E. Nuclear organization of DNA replication in primary mammalian cells. Genes Dev. 2000;14:2855–2868. doi: 10.1101/gad.842600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S.J., Kwon H. Actin-related protein BAF53 is essential for the formation of replication foci. Anim. Cells Syst. 2012;16:183–189. [Google Scholar]
- Lark C.G., Consigli R., Toliver A. DNA replication in Chinese hamster cells: evidence for a single replication fork per replicon. J. Mol. Biol. 1971;58:873–875. doi: 10.1016/0022-2836(71)90046-5. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Chang S.H., Shim J.H., Lee J.Y., Yoshida M., Kwon H. Cytoplasmic localization and nucleocytoplasmic shuttling of BAF53, a component of chromatin-modifying complexes. Mol. Cells. 2003;16:78–83. [PubMed] [Google Scholar]
- Lee K., Kang M.J., Kwon S.J., Kwon Y.K., Kim K.W., Lim J.H., Kwon H. Expansion of chromosome territories with chromatin decompaction in BAF53-depleted interphase cells. Mol. Biol. Cell. 2007;18:4013–4023. doi: 10.1091/mbc.E07-05-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt H., Rahn H.P., Weinzierl P., Sporbert A., Cremer T., Zink D., Cardoso M.C. Dynamics of DNA replication factories in living cells. J. Cell Biol. 2000;149:271–280. doi: 10.1083/jcb.149.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O., et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalet X., Ekong R., Fougerousse F., Rousseaux S., Schurra C., Hornigold N., van Slegtenhorst M., Wolfe J., Povey S., Beckmann J.S., Bensimon A., et al. Dynamic molecular combing: stretching the whole human genome for high-resolution studies. Science. 1997;277:1518–1523. doi: 10.1126/science.277.5331.1518. [DOI] [PubMed] [Google Scholar]
- Mirny L.A. The fractal globule as a model of chromatin architecture in the cell. Chromosome Res. 2011;19:37–51. doi: 10.1007/s10577-010-9177-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muck J., Zink D. Nuclear organization and dynamics of DNA replication in eukaryotes. Front. Biosci. 2009;14:5361–5371. doi: 10.2741/3600. [DOI] [PubMed] [Google Scholar]
- Muller W.G., Walker D., Hager G.L., McNally J.G. Large-scale chromatin decondensation and recondensation regulated by transcription from a natural promoter. J. Cell Biol. 2001;154:33–48. doi: 10.1083/jcb.200011069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller W.G., Rieder D., Kreth G., Cremer C., Trajanoski Z., McNally J.G. Generic features of tertiary chromatin structure as detected in natural chromosomes. Mol. Cell Biol. 2004;24:9359–9370. doi: 10.1128/MCB.24.21.9359-9370.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkel C., Eils R., Dietzel S., Zink D., Mehring C., Wedemann G., Cremer T., Langowski J. Compartmentalization of interphase chromosomes observed in simulation and experiment. J. Mol. Biol. 1999;285:1053–1065. doi: 10.1006/jmbi.1998.2361. [DOI] [PubMed] [Google Scholar]
- O’Keefe R.T., Henderson S.C., Spector D.L. Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific alpha-satellite DNA sequences. J. Cell Biol. 1992;116:1095–1110. doi: 10.1083/jcb.116.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S., Aiden E.L. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando O.J., Zhao K., Janmey P., Crabtree G.R. Phosphatidylinositol-dependent actin filament binding by the SWI/SNF-like BAF chromatin remodeling complex. Proc. Natl. Acad. Sci. USA. 2002;99:2824–2829. doi: 10.1073/pnas.032662899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spring H., Franke W.W. Transcriptionally active chromatin in loops of lampbrush chromosomes at physiological salt concentrations as revealed by electron microscopy of sections. Eur. J. Cell Biol. 1981;24:298–308. [PubMed] [Google Scholar]
- Sung Y.H., Choi E.Y., Kwon H. Identification of a nuclear protein ArpN as a component of human SWI/SNF complex and its selective association with a subset of active genes. Mol. Cells. 2001;11:75–81. [PubMed] [Google Scholar]
- Tumbar T., Sudlow G., Belmont A.S. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J. Cell Biol. 1999;145:1341–1354. doi: 10.1083/jcb.145.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., Pardoll D.M., Coffey D.S. Supercoiled loops and eucaryotic DNA replicaton. Cell. 1980;22:79–85. doi: 10.1016/0092-8674(80)90156-7. [DOI] [PubMed] [Google Scholar]
- Zhao K., Wang W., Rando O.J., Xue Y., Swiderek K., Kuo A., Crabtree G.R. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
- Zink D., Bornfleth H., Visser A., Cremer C., Cremer T. Organization of early and late replicating DNA in human chromosome territories. Exp. Cell Res. 1999;247:176–188. doi: 10.1006/excr.1998.4311. [DOI] [PubMed] [Google Scholar]


