Abstract
Perilipin 5 (PLIN5) is a lipid droplet (LD) protein highly expressed in oxidative tissues, including the fasted liver. However, its expression also increases in nonalcoholic fatty liver. To determine whether PLIN5 regulates metabolic phenotypes of hepatosteatosis under nutritional excess, liver targeted overexpression of PLIN5 was achieved using adenoviral vector (Ad-PLIN5) in male C57BL/6J mice fed high-fat diet. Mice treated with adenovirus expressing green fluorescent protein (GFP) (Ad-GFP) served as control. Ad-PLIN5 livers increased LD in the liver section, and liquid chromatography with tandem mass spectrometry revealed increases in lipid classes associated with LD, including triacylglycerol, cholesterol ester, and phospholipid classes, compared with Ad-GFP liver. Lipids commonly associated with hepatic lipotoxicity, diacylglycerol, and ceramides, were also increased in Ad-PLIN5 liver. The expression of genes in lipid metabolism regulated by peroxisome proliferator-activated receptor-α was reduced suggestive of slower mobilization of stored lipids in Ad-PLIN5 mice. However, the increase of hepatosteatosis by PLIN5 overexpression did not worsen glucose homeostasis. Rather, serum insulin levels were decreased, indicating better insulin sensitivity in Ad-PLIN5 mice. Moreover, genes associated with liver injury were unaltered in Ad-PLIN5 steatotic liver compared with Ad-GFP control. Phosphorylation of protein kinase B was increased in Ad-PLIN5-transduced AML12 hepatocyte despite of the promotion of fatty acid incorporation to triacylglycerol as well. Collectively, our data indicates that the increase in liver PLIN5 during hepatosteatosis drives further lipid accumulation but does not adversely affect hepatic health or insulin sensitivity.
Nonalcoholic fatty liver disease (NAFLD) is a metabolic disorder characterized by the accumulation of triacylglycerol (TG) in the liver in the absence of alcohol consumption. It affects up to 30% of the adult population in the United States and is currently the leading cause of chronic liver disease in the world (1). Health risks associated with NAFLD include the potential progression to liver cirrhosis and hepatocellular carcinoma, and the development of insulin resistance and type 2 diabetes (2–4). Although TG accumulation is the hallmark of NAFLD, its metabolites rather than TG itself are considered to cause dysregulation of insulin signaling and liver injury (5, 6). Free fatty acids (FAs), especially saturated FA such as palmitic and stearic acids are cytotoxic and provoke endoplasmic reticulum stress, oxidative stress, and inflammatory responses in the liver (5). The increase in diacylglycerol (DG) in steatotic liver is believed to activate protein kinase Cϵ and blunt insulin signaling (6). Therefore, factors that regulate storage and metabolism of TG in the liver are likely to be critically involved in the development of pathologies associated with NAFLD.
TG are primarily stored in lipid droplets (LDs) in hepatocytes. LDs are intracellular organelles composed of a neutral lipid core encased by a phospholipid monolayer that is studded with proteins to orchestrate the formation, movement, and use of stored lipids (7). Importantly, proteome analysis of hepatic LDs revealed a unique enrichment of multiple metabolic enzymes reflecting key roles of hepatic LDs in the regulation of hepatic metabolism (8). The perilipin (PLIN) family of proteins (PLIN1–PLIN5) is the prototypical LD proteins important for the formation of LDs (9). In mouse models of NAFLD such as diet-induced obese mice and ob/ob mice, PLIN2, PLIN3, and PLIN5 are increased (10–12). Both in vivo and in vitro studies imply that all 3 PLINs are involved in TG accumulation in hepatocytes. Reductions of PLIN2 prevent TG accumulation in hepatocytes in culture (13) and in the liver of diet-induced obese mice (14–17). Similarly, down-regulation of PLIN3 using antisense oligonucleotides improved diet-induced hepatosteatosis in mice (11). Lastly, TG contents were decreased in primary mouse hepatocytes after PLIN5 reduction (18) and in the liver from whole body PLIN5-deficient mice (12). The reduction of hepatosteatosis was associated with the improvement in insulin sensitivity when PLIN2 and PLIN3 were down-regulated in mice on high-fat diet (HFD) (11, 16, 17). The improvement in hepatic insulin sensitivity was also noted in whole body PLIN5-deficient mice but were fed regular rodent chow (NC) (19). Collectively, LD formation in the liver seems to precipitate insulin resistance.
However, there is also evidence indicating that the prevention of LD formation in the liver accelerates hepatic injury under excessive nutritional load. Microarray analysis of the liver from HFD-fed mice treated with antisense oligonucleotides against PLIN2 showed the increase in markers of hepatic proliferation (Afp and H19) and extracellular matrix remodeling (20). Markers of inflammation and oxidative stress were also increased in the liver of PLIN5-deficient mice on HFD (12). To add complexity, hepatic overexpression of PLIN2 in regular chow-fed mice caused hepatosteatosis with improvement in insulin sensitivity (21), indicating that the interplay between nutritional environment and PLINs may determine how TG accumulation modifies insulin signaling and hepatic injuries in NAFLD. To understand the nature of interactions between PLINs and nutritional environment, knowledge regarding how different PLINs regulate accumulation of TG in the liver and hepatocyte health in vivo will be important. Although all 3 PLINs (PLIN2, PLIN3, and PLIN5) seem to participate in TG accumulation in the liver, each PLIN potentially has distinct roles due to unique structural characteristics. PLIN5 interacts with key lipases adipose triglyceride lipase (ATGL) and its colipase comparative gene identification-58 (CGI-58) (22–24) and may be linked to mitochondria to better facilitate FA oxidation (25). Notably, PLIN5 knockdown in primary mouse hepatocytes increased lipolysis and FA oxidation, whereas the knockdown of PLIN2 and PLIN3 in AML12 hepatocyte cell line did not affect lipolysis (18, 26).
Here, PLIN5 was overexpressed in the liver of mice on HFD via adenovirus to determine how PLIN5 will affect markers of glucose homeostasis and hepatic injuries along with hepatic lipid profiles. We used high-resolution accurate mass spectrometry to identify lipid species differentially regulated by PLIN5 overexpression and may modulate insulin sensitivity and hepatic health in hepatosteatosis.
Materials and Methods
Animal studies
Experiments were performed in accordance with the Institutional Animal Care and Use Committee guidelines of Eastern Virginia Medical School. Three-month-old male C57Bl/6J (Bl6) (The Jackson Laboratory) were housed in 12-hour light, 12-hour dark cycle at 22°C and fed HFD containing 45 kcal% fat (D124551; Research Diets) or NC (Harlan 2018) ad libitum for 6 weeks (regular chow and HFD) or 16 weeks (HFD only). Thereafter, type 5 adenoviruses expressing green fluorescent protein (GFP) (Ad-GFP) or mouse PLIN5 (Ad-PLIN5) under cytomegalovirus promoter, all from Vector Biolabs, were delivered through tail vein injection at 1 × 109 plaque forming units per mouse. Mice continued to consume HFD throughout the study up until harvest. Ten to fourteen days after adenovirus injection, mice fed ad libitum were euthanized by carbon dioxide asphyxiation in late morning. Cardiac blood was collected for serum analyses. Left lateral lobe of the liver was fixed in 10% formalin for histological analyses. The rest of the liver were snap frozen in liquid nitrogen upon harvest and stored at −80°C until analyses.
Glucose homeostasis
For glucose tolerance test (GTT), mice fasted overnight received glucose ip at 1-mg/g body weight (HFD) or 1.5-mg/g body weight (NC). For insulin tolerance test (ITT), mice fasted for 4 hours in the morning received insulin lispro (Eli Lilly) ip at 1.0-mIU/g body weight (HFD) or 0.75-mIU/g body weight (NC). Tail blood glucose was measured at indicated times using a One Touch Ultra hand-held glucometer (LifeScan, Inc).
Colorimetric assays
Cardiac blood collected at harvest was centrifuged at 7000 revolutions per minute for 10 minutes at room temperature to obtain serum. TG (Stanbio Laboratories), β-hydroxybutyrate (Stanbio), nonesterified FA (Wako Chemicals), and mouse insulin (Mercodia) were measured in the serum according to manufacturers' instructions. Liver TG were measured using an aforementioned TG colorimetric assay after Folch extraction as we published and corrected for protein contents (27).
Cell culture and adenovirus transduction
AML12 hepatocyte cells obtained from American Type Culture Collection and grown in 50/50 DMEM/Ham's F12 media supplemented with 10% fetal bovine serum; insulin, transferrin, and selenium (Gibco); and 40-ng/mL dexamethasone (Sigma) were transduced in suspension at 80 plaque forming units per cell of Ad-PLIN5 or Ad-GFP for 1 hour in serum-free growth medium, and then cultured overnight in 10% fetal bovine serum growth medium at 37°C in 5% CO2. For labeling studies, transduced cells were incubated with 1.2-Ci/mL [3H]oleic acid (OA) (specific activity 10 mCi/mol; NEN) and the 16- to 18-hour incorporation of [3H]OA to the TG fraction was determined as in Ref. 28. Lipolysis in response to cAMP was determined as the reduction of [3H]OA-labeled TG in AML12 cells treated with or without 1 mmol/L 8-bromoadenosine 3′,5′-cyclic monophosphate (Sigma) in the presence of 9.5 μmol/L triacsin C (Enzo Life Sciences) to prevent reincorporation of [3H]OA to TG. Data were normalized to cellular protein contents.
mRNA and quantitative PCR
Total RNA from liver tissues (50–100 mg) were prepared using QIAGEN RNeasy kit (QIAGEN) according to the manufacture's protocol and reverse transcribed with iScript (Bio-Rad). Gene expressions were assessed using ABI TaqMan commercial primers (Applied Biosystems) and results were expressed taking β-actin as an internal standard.
Western blotting
Tissues were homogenized in lysis buffer with the addition of protease and phosphatase inhibitor cocktails (both from Sigma) as published (15). Western blottings were performed using 20–40 μg/lane of protein measured by Bio-Rad DC Protein Assay (Bio-Rad), and signals were visualized by chemiluminescence. Densitometric analyses were performed with ImageJ using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as an internal control. Primary antibodies were guinea pig anti-PLIN5 (Progen Biotechnik) at 1:1000 and guinea pig anti-PLIN2 (Fitzgerald) at 1:1000. Rabbit anti-GAPDH, rabbit antiphosphorylated Akt S473, and mouse anti-Akt (pan) were all diluted at 1:1000 and all from Cell Signaling Technology. Secondary antibodies were horseradish peroxidase-conjugated goat antiguinea pig-IgG and horseradish peroxidase-conjugated goat antirabbit-IgG (Santa Cruz Biotechnology, Inc) both at 1:5000.
Histological analyses of the liver
Paraffin-embedded liver sections were incubated with primary antibodies followed by visualization using fluorescent secondary antibodies as previously published (15) except that slides were incubated with Sudan Black B (MP Biomedicals) at 0.1% for PLIN5 and 0.4% for PLIN2 for 25 minutes after the incubation with secondary antibodies to reduce autofluorescence (29). Primary antibodies were guinea pig anti-PLIN5 (Progen) at 1:200 and guinea pig anti-PLIN2 at 1:800 (Fitzgerald). Secondary antibodies were Cyanine3 antiguinea pig antibodies at 1:1000 from Jackson ImmunoResearch. Nuclei were visualized with 1-μg/mL 4′,6-diamidino-2-phenylindole (Life Technologies). Images of liver sections were recorded with AxioObserver Z.1 fluorescent microscope (Carl Zeiss) or a confocal microscope (LSM510; Carl Zeiss).
Determination of liver lipids by liquid chromatography with tandem mass spectrometry (LC-MS/MS)
Lipids were extracted from 400 mg of the same area of the liver across the samples as described (28). A total of 10μM lipid standards to include a monoacylglycerol, DG, and TG mix (Supelco), palmitic acid, and cholesterol ester (CholE) (both from Sigma) was spiked into each tissue lysate before lipid extraction. Samples were analyzed on the Q-Exactive Orbitrap platform (Thermo Fisher Scientific) via LC-MS/MS (30, 31) between 70K and 140K resolutions for the Full MS and up to 35K resolution for product ion scan data (MS2). Product ion spectrum at 35K was obtained using higher collision dissociation fragmentation for the 5 most abundant ions observed from the Full MS spectrum. The overfill ratio for the Orbitrap was setup at 2e3 ions that is also considered as the threshold for the dynamic range of the spectrum below which no product ion MS2 ions were collected for lipid identification. Instrument settings are described in detail within the Supplemental Methods.
Peak analysis was performed by Lipid Search algorithm (Mitsubishi Knowledge Industry) that performs peak detection with raw files, MSn, and precursor ion accurate masses (28). Lipid Search algorithm identifies lipids by comparing the raw file of MS2 with a comprehensive database of over 106 lipid species. The identification of each product ion is ranked by mass tolerance, matched to a fragmentation library and predicted retention time to determine the molecular structure of the precursor ion. For the dataset, the mass tolerance was set to 5 parts per million for the precursor mass. Relative quantitation for the analytes is performed using the precursor ion data based on the high-resolution accurate mass information. MS/MS spectra were manually inspected and species assigned based on known lipid fragmentation. Spectra corresponding to the peak apex of each extracted ion chromatogram were examined for relationships between adducts, isotopes and dimers using accurate mass measurements. Related ions with similar retention times were grouped together into a component table represented by the largest adduct peak.
For lipid quantification, identified species are aligned across Ad-PLIN5 and Ad-GFP liver groups and the integrated precursor extracted ion chromatograms are calculated to compare area differences for the individual lipid species. LC-MS data were corrected for the mean background obtained from the average of 2 blank solvent injections. Livers from 3 mice per group (biological triplicates) were analyzed with technical duplicate for Ad-PLIN5 group yielding 3 values for Ad-GFP and 6 values for Ad-PLIN5 groups. For the 6 values in Ad-PLIN5, outliers defined as outside of median-(3× median absolute deviation) and median+(3× median absolute deviation) were removed. The statistical comparison of individual lipid species between Ad-GFP and Ad-PLIN5 was performed using Wilcoxon rank test. The abundance of the lipid class was determined as the sum of peak area of lipid species that belong to a lipid class. For Ad-PLIN5 group, the average of technical duplicates was used to represent the liver from each mouse.
Statistics
Data are presented as mean ± SEM. Differences of numeric parameters between 2 groups were assessed with Student's t tests or Mann-Whitney test, and those for multiple group comparisons were assessed with one-way ANOVA (Tukey post hoc test). P < .05 was considered significant.
Results
Adenovirus-mediated overexpression of PLIN5 promotes hepatic TG and LD accumulations in Bl6 mice on HFD
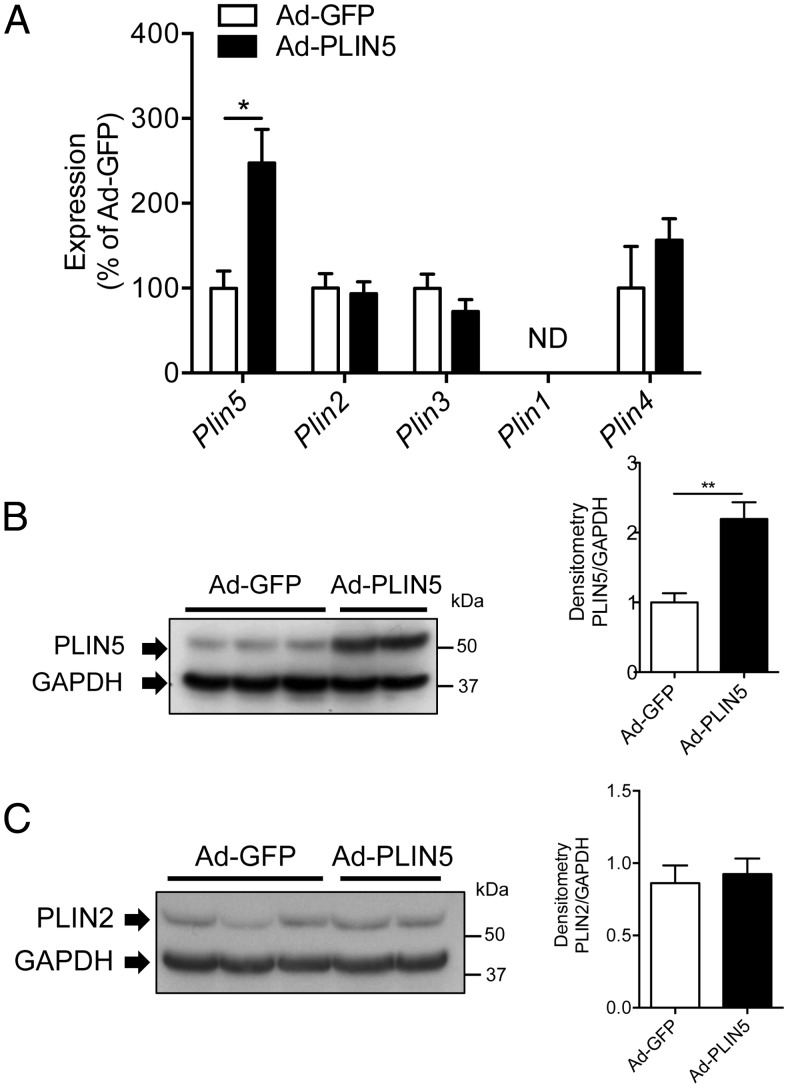
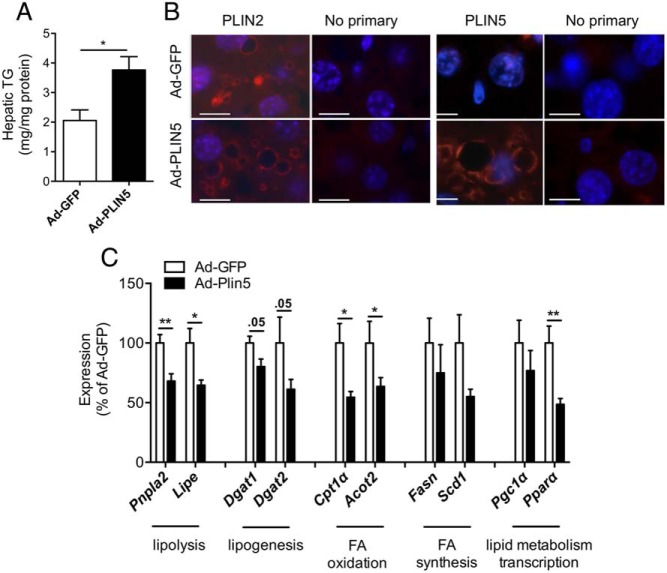
In mouse models of NAFLD such as diet-induced obese mice and ob/ob mice, PLIN5 is reported to be increased (Supplemental Figure 1A) (12, 26). To determine the consequence of PLIN5 overexpression in the liver in the setting of HFD, adenoviruses expressing Plin5 (Ad-PLIN5) or control (Ad-GFP) were injected into the tail vein of male Bl6 mice fed HFD for 6 weeks before injection. Two weeks after adenovirus administration, livers from Ad-PLIN5 mice on the HFD showed significant increases in Plin5 mRNA levels but did not show changes in the hepatic expressions of other members of the Plin family, including Plin2 (Figure 1A). Western blotting of the liver showed PLIN5 protein expression was successfully increased to 2-fold in mice treated with Ad-PLIN5 over Ad-GFP (Figure 1B). In contrast to the heart (32) and pancreatic islets (28) where the induction of PLIN5 promoted an increase in PLIN2 protein without changes in Plin2 mRNA expression, hepatic PLIN2 protein did not increase after the overexpression of hepatic PLIN5 in mice with diet-induced hepatosteatosis (Figure 1C). In agreement with in vitro data in which PLIN5 expression increased cellular TG contents in AML12 cells (18), in vivo overexpression of PLIN5 in the liver significantly increased hepatic TG contents in Ad-PLIN5 mice (Figure 2A). Hematoxylin eosin-stained liver sections revealed that 6 weeks of HFD resulted in steatosis in both Ad-GFP and Ad-PLIN5 mice but with more prominent and diffuse appearance of large LD in Ad-PLIN5 mice (Supplemental Figure 1B). In agreement with hematoxylin eosin, PLIN2 staining showed large PLIN2-coated LDs were quite abundant in the livers of Ad-PLIN5 mice compared with Ad-GFP mice (Figure 2B). Immunofluorescence using PLIN5 antibody showed ring like staining typical of LDs in Ad-PLIN5 liver, whereas such staining was rarely detected in Ad-GFP liver (Figure 2B) within the sensitivity of antibody currently available for immunofluorescence. PLIN5-coated LD was not detected in hepatic stellate cells nor vimentin positive cells, including Kupffer cells, leukocytes, and endothelia cells (Supplemental Figure 1, C and D). Taken together, these data implies that PLIN5 overexpression in the liver exacerbates hepatosteatosis biochemically and histologically in mice on HFD.
Figure 1.
Adenovirus-mediated expression of PLIN5 in liver of mice on HFD. A, Quantitative PCR of Plin1-Plin5 levels in livers of Ad-GFP- and Ad-PLIN5-treated mice on 6 week of HFD presented taking the average levels from Ad-GFP mice as 100% and normalized using β-actin as an internal control. ND, not detected. Data are mean ± SEM; n = 5–6 per group; *, P < .05. B and C, Western blotting of PLIN5 (B) and PLIN2 (C) in Ad-GFP and Ad-PLIN5 livers using GAPDH as an internal control. Representative pictures on the left and band density of PLIN5 (B) and PLIN2 (C) taking the average expression levels from Ad-GFP-treated mice as 1 on the right. Data are mean ± SEM; n = 5–6 per group; **, P < .01.
Figure 2.
Change in lipids and LD in HFD mice treated with Ad-PLIN5. A, Hepatic TG quantitated in livers of Ad-GFP- and Ad-PLIN5-treated mice after 6 week of HFD. Data are mean ± SEM; n = 5–6 per group; *, P < .05. B, Liver sections from Ad-GFP (top row)- and Ad-PLIN5 (bottom row)-treated mice immunostained with PLIN2 and immunostained with PLIN5 along with negative control (no primary antibodies). Scale bar, 10 μm. C, Quantitative PCR determined the expression of genes involved in major lipid metabolic pathways in livers of Ad-PLIN5 male mice on HFD for 6 weeks. Levels are presented taking the levels of Ad-GFP on the corresponding duration of HFD as 100%. Expression levels were normalized using β-actin as an internal control. Data are mean ± SEM; n = 5–6; *, P < .05; **, P < .01. Pnpla2, ATGL; Lipe, hormone-sensitive lipase (aka HSL); Dgat1 and Dgat2, DG acyltransferases 1 and 2; Cpt1a, carnitine palmitoyltransferase 1α; Fasn, FA synthase; Scd1, stearoyl-CoA desaturase-1; Acot2, acyl-CoA thioesterase 2; Ppargc1a, PPAR-γ coactivator 1α.
Despite its location on the surface of LD, the down-regulation of PLIN5 in the liver and cultured cells affects the expressions of a wide range of genes in lipid metabolism (12, 18). In the current study, the overexpression of PLIN5 in HFD mice showed significant reductions of genes involved in lipolysis (Pnpla2 and Lipe) and FA oxidation (Cpt1α and Acot2, P < .05 vs Ad-GFP) (Figure 2C). There were also trends for the reduction in genes for lipogenesis (Dgat1 and Dgat2) in Ad-PLIN5 mice (P = .05 vs Ad-GFP) (Figure 2C). No significant differences in FA synthesis-related genes (Fasn and Scd1) were measured with PLIN5 overexpression compared with Ad-GFP control. The common transcriptional regulator of these genes for lipid metabolism, peroxisome proliferator-activated receptor (PPAR)-α (Ppara) was previously shown to be down-regulated when PLIN5 was overexpressed in the cardiac muscle (32, 33). Comparably, we observed a significant reduction in hepatic Ppara expression in Ad-PLIN5 mice that may explain the decreased expressions of other hepatic lipid enzymes (Figure 2C). Overall hepatic PLIN5 overexpression results in the reduction in PPAR-α and PPAR-α-regulated genes, the opposite of changes observed in PLIN5-deficient liver (12).
LC-MS/MS reveals changes in the lipidome of liver overexpressing PLIN5
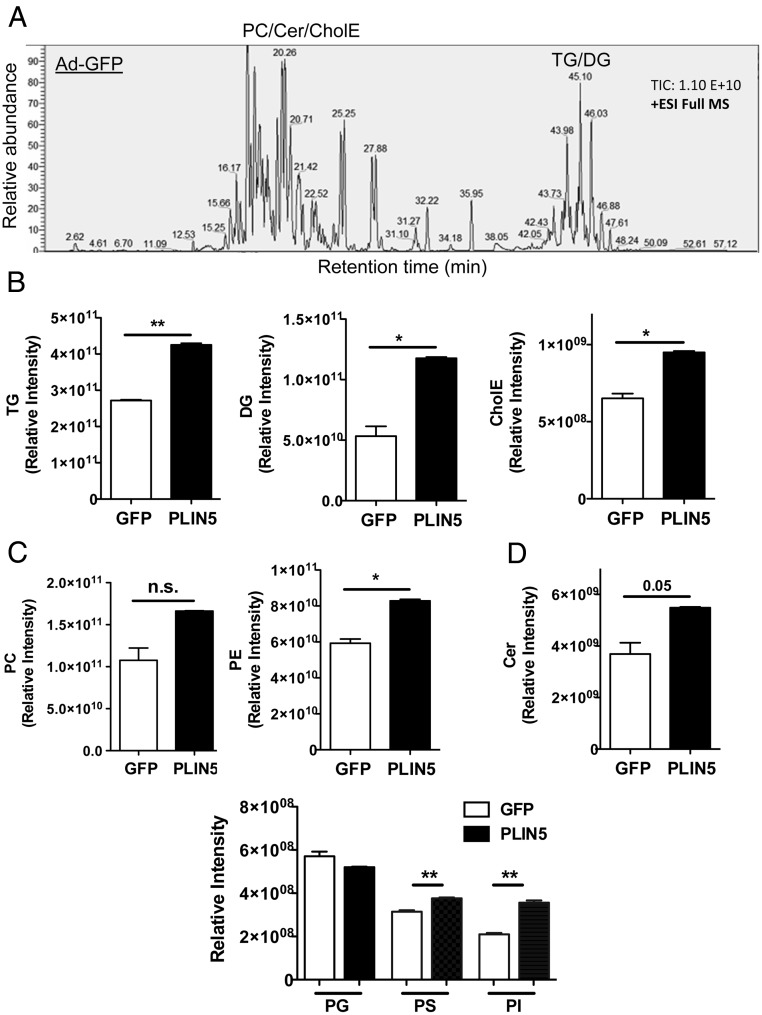
To gain a more detailed characterization of lipidomes in Ad-PLIN5 liver, we performed unbiased, shotgun comparison of lipids between the livers of Ad-GFP- and Ad-PLIN5-treated Bl6 mice on HFD using high resolution Q-Exactive LC-MS/MS. Using the condition previously described (30, 31), we identified phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylserine, phosphatidylglycerol, phosphatidylinositol, ceramides (Cers), CholE, TG, and DG in lipids extracted from the livers. In Figure 3A, the representative total ion chromatogram depicts mass spectral peaks across all m/z in positive ion mode. Neutral lipids commonly enriched in LDs, including TG, DG, and CholE (34), were accurately detected with the Q-Exactive. The abundance of these 3 lipid classes was significantly increased in the liver of Ad-PLIN5 mice with the most prominent rise in DG at the fold increase of 2.6-fold compared with Ad-GFP control (Figure 3B). PC and PE, phospholipids abundantly found at the surface of LDs (35), were also detected in our assays with the significant increase for PE in PLIN5 overexpressing liver (Figure 3C). Among other less abundant phospholipids, phosphatidylserine and phosphatidylinositol were substantially increased in the Ad-PLIN5 liver (Figure 3C). In addition, Cer showed a trend of increase in the Ad-PLIN5 liver (Figure 3D), indicating that PLIN5 overexpression drives the increase in a wide range of lipid classes.
Figure 3.
Changes in the hepatic lipidome of HFD mice treated with Ad-PLIN5. Lipids extracted from the livers of 6 weeks HFD mice treated with Ad-GFP or Ad-PLIN5 and harvested at fed ad libitum in late morning as in Materials and Methods were analyzed by Q-Exactive LC-MS/MS as in Materials and Methods. A, The representative total ion chromatogram of lipids in Ad-GFP liver in full MS. B–D, Relative intensity of neutral lipids (B), phospholipids (C), and Cers (D) determined as in Materials and Methods was compared between liver from Ad-GFP and Ad-PLIN5 mice. PG, phosphatidylglycerol; PS, phosphatidylserine; PI, phosphatidylinositol. Data are mean ± SEM; n = 3; *, P < .05; **, P < .01; n.s., not significant.
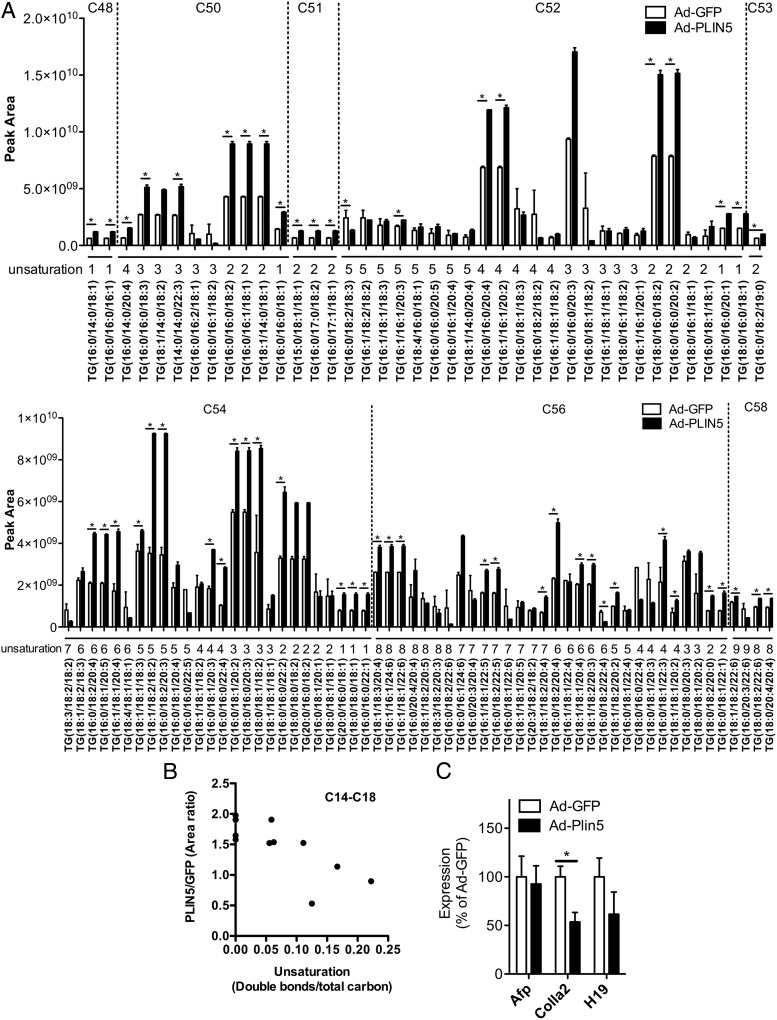
We identified 609 TG species with the sum of 3 FA chains from C34 to C74 in both Ad-GFP and Ad-PLIN5 livers, of which 50 species were increased more than 2-fold and 8 species were reduced to less than 0.5-fold in Ad-PLIN5 liver (P < .05) (Supplemental Tables 1 and 2). The 100 TG species most abundant in Ad-GFP liver (Figure 4A) represented 76% of total TG in abundance for both Ad-GFP and Ad-PLIN5 livers. These TG species mostly consisted of C50 to C56, which is in agreement with total carbon distribution previously reported in the mouse livers (34). Interestingly, Ad-PLIN5 liver contained more saturated FA compared with Ad-GFP liver when the extent of unsaturation was compared in individual FA chain length between C14 and C18 (Spearman r = −0.80, P < .01) (Figure 4B).
Figure 4.
Profiles of the TG lipid class in the livers of HFD mice overexpressing PLIN5. A, Relative abundance (peak area) of the 100 most abundant TG species identified in the liver of Ad-GFP (white)- and Ad-PLIN5 (black)-treated mice (6-week HFD). TGs are grouped based on their total number of carbons (c) and unsaturation (number of double bonds). Data are mean ± SEM; n = 3; *, P < .05. B, Correlation between the unsaturation of C14 to C18 fatty acyl chains expressed as the number of double bonds/total carbon and abundance of fatty acyl chains expressed as an area ratio in Ad-PLIN5 over Ad-GFP liver of the TG species as appeared in A. C, Quantitative PCR determined the expression of genes involved in liver injury in livers of Ad-PLIN5 male mice fed HFD (6-week HFD). Levels are presented taking the levels of Ad-GFP on the corresponding duration of HFD as 100%. Expression levels were normalized using β-actin as an internal control. Data are mean ± SEM; n = 5–6; *, P < .05; **, P < .01. Afp, α-fetoprotein (AFP); Col1a2, collagen α1(II chain); H19, imprinted maternally expressed transcript (nonprotein coding).
We identified 154 DG species that ranged from C24 to C56, of which 92 species were increased more than 2-fold and 2 DG species were reduced to less than 0.5-fold in Ad-PLIN5 liver reflecting the significant rise in total DG in Ad-PLIN5 liver compared with Ad-GFP liver (P < .05) (Supplemental Tables 3 and 4). Most DG belonged to C34 and C38 for both groups followed by C36 and C40 (Supplemental Figure 2). Five CholE species were identified in the liver, of which CholE(20:4) and CholE(22:6) contributed to the increase in total CholE in Ad-PLIN5-treated liver compared with Ad-GFP control liver (Supplemental Figure 3A). Our lipidomics also identified 31 Cer species present in both Ad-PLIN5 and Ad-GFP liver with many of them significantly increased in Ad-PLIN5 liver compared with Ad-GFP control. Cers with d18:01 backbone was most abundant. However, we also identified those with d18:2 and d18:0 (Supplemental Figure 3B). Cer(d18:1/16:0) has been implicated as a contributor to insulin resistance in the liver (36, 37) and was also the most prominent Cer present in both livers in our study. At the same time, very long chain Cers (C22 to C24) that may have the opposite effect of C16 Cers (37) were highly abundant and significantly increased in Ad-PLIN5 liver (Supplemental Figure 3B).
We identified 120 PC species and 130 PE species (Supplemental Figures 4 and 5A). Although total PC did not show statistically significant alterations, many PC species were increased in Ad-PLIN5 liver compared with Ad-GFP liver leaving only 5 species of PC to be significantly reduced to less than 0.5-fold in Ad-PLIN5 liver (Supplemental Table 5). PC (16:0/18:1) previously identified as a ligand for PPAR-α (38), was the most prominent PC species and significantly increased in Ad-PLIN5 liver (Supplemental Figure 4). Collectively, Ad-PLIN5 expression in the liver resulted in the increase of a broad spectrum of lipids both in terms of type of lipids and chain compositions within each lipid classes.
However, the increase of lipids mediated by PLIN5 overexpression had little impact on markers associated with liver injury that were previously noted to be increased in the liver of mice on HFD compared with regular chow-fed mice (20). In Figure 4C, quantitative PCR revealed the Colla2 was significantly reduced in the livers of Ad-PLIN5 mice on 6-week HFD (P < .05 vs Ad-GFP), and there were no major changes in genes associated with hepatic proliferation in response to hepatic injury (Afp and H19). In Supplemental Figure 5B, adenovirus treatment itself did not seem to increase these genes in the liver. In comparison, the reduction of PLIN2 or PLIN5 in the livers of HFD mice was reported to up-regulate genes of liver injuries (12, 20). Thus, PLIN5 seems to aid in the expansion of the hepatic neutral lipid pool without activating genes associated with steatohepatitis.
Up-regulation of hepatic PLIN5 does not accelerate insulin resistance in mice with steatotic liver
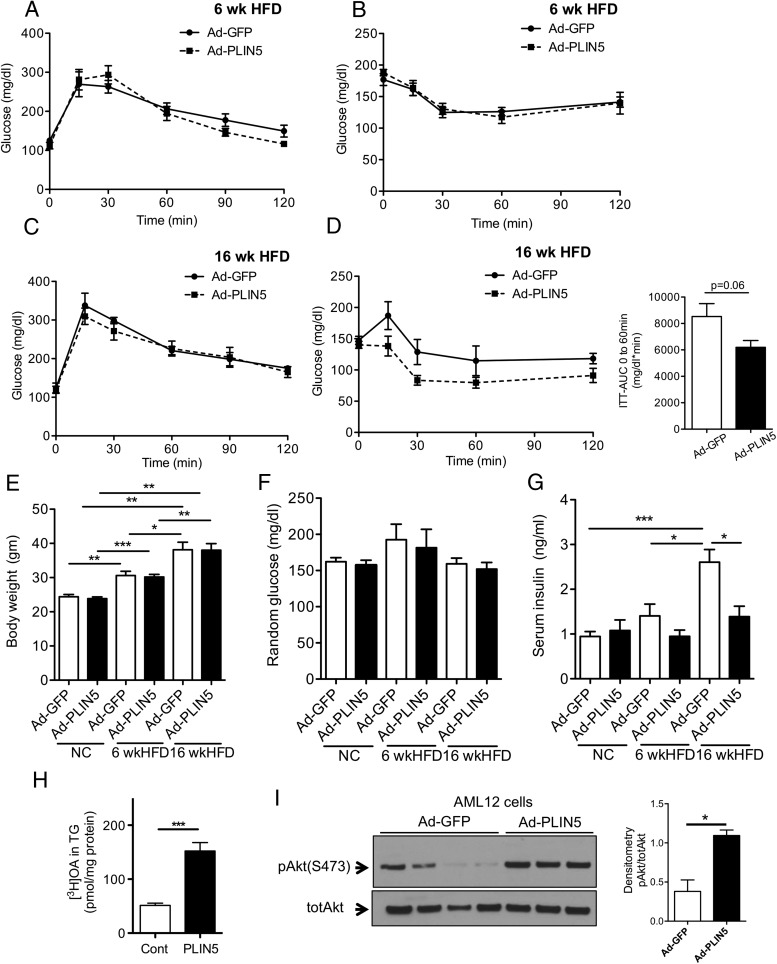
We then evaluated the effect of hepatic PLIN5 overexpression on whole body glucose homeostasis. Despite the increase in liver TG, GTT (Figure 5A) and ITT (Figure 5B) revealed no impairment in glucose or insulin tolerance in AD-PLIN5 mice compared with Ad-GFP mice on 6-week HFD. In addition, we tested glucose and insulin tolerance when PLIN5 were overexpressed in the liver of mice on a longer term of HFD (16 weeks) and those maintained on NC for 6 weeks (Supplemental Figure 6, A and B). Glucose and insulin tolerance were similar between Ad-GFP and Ad-PLIN5 nice on NC (Supplemental Figure 6, C and D). In Figure 5C, the overexpression of PLIN5 in the liver did not lead to significant changes in glucose tolerance in 16-week HFD mice. However, there was a trend for the improvement in the insulin tolerance of 16-week HFD Ad-PLIN5 mice compared with Ad-GFP control (P = .06 for area under the curve, time 0–60 minutes) (Figure 5D). Characterization of metabolic parameters, including body weight, blood glucose, serum TG, serum β-hydroxybutyrate, and serum nonesterified FA levels, showed no differences between Ad-PLIN5 and Ad-GFP mice on either NC, short or long-term HFD, again indicating relatively quiescent expansion of hepatic lipid pools driven by PLIN5 expression in the liver (Figure 5, E and F, and Supplemental Figure 6, E and F). Notably, the incremental rise in weight and serum insulin levels without significant changes in blood glucose in Ad-GFP mice on NC, short- and long-term HFD confirm that adenovirus administration preserves the widely accepted progression of insulin resistance in Bl6 on HFD (39). Histology (Supplemental Figure 1A) and liver TG contents (Supplemental Figure 6G) also indicate that hepatosteatosis is present in Ad-GFP mice on HFD. Interestingly, the liver targeted overexpression of PLIN5 prevented the progressive rise of serum insulin seen in AD-GFP mice. Serum insulin levels in 16-week HFD mice were clearly lower compared with Ad-GFP control mice (Figure 5G), which agrees with the trend of the improvement in ITT (Figure 5D). Collectively, the overexpression of hepatic PLIN5 in the setting of diet-induced hepatosteatosis did not promote insulin resistance in spite of increased liver TG and LD (Figure 2); rather overexpression of PLIN5 was associated with reduced serum insulin levels implicating an improvement in insulin resistance.
Figure 5.
Glucose homeostasis of short- and long-term HFD mice treated with Ad-PLIN5. A–D, Mice were placed on a 6- or 16-week HFD, then treated with Ad-GFP (closed line) and Ad-PLIN5 (dashed line). Intraperitoneal glucose tolerance (A and C) and insulin tolerance (B and D) were evaluated in mice on the separate HFD time plans. Area under the curve (AUC) between time 0 and 60 minutes. E–G, Body weight (E), glucose in late morning fed ad libitum (F), and serum insulin (G) measured in Ad-GFP- and Ad-PLIN5-treated male C56BL/6J mice on NC, and 6- or 16-week HFD taken at harvest. Data are mean ± SEM; n = 3–6; *, P < .05; **, P < .01. H, Overnight incorporation of [3H]OAs into triglycerides (TG) in AML12 cells transduced with Ad-PLIN5 (PLIN5) and nontransduced cells (Cont). Data are mean ± SEM; n = 4; ***, P < .001. I, Representative Western blotting of phosphorylated protein kinase B (pAkt S473) and total protein kinase B (totAkt) in AML12 cells transduced with Ad-PLIN5 and Ad-GFP. Band density expressed taking the average expression levels of from Ad-GFP as 1 relative to totAkt.
To address the potential implication of PLIN5 expression in hepatic insulin sensitivity, murine hepatocyte cell line AML12 was transduced with Ad-PLIN5. [3H]OA labeling confirmed that Ad-PLIN5 promotes incorporation of FA into TG (Figure 5H) without a change in cellular uptake of [3H]OA (Supplemental Figure 6H). In this setting, Ad-PLIN5-transduced AML12 cells grown in culture medium as in Materials and Methods showed higher phosphorylation of protein kinase B (pAkt) compared with control cells (Figure 5I). Taken together, the overexpression of PLIN5 in a hepatocyte cell line increased TG formation but enhanced a parameter of insulin signaling.
Discussion
PLIN5 is increased along with PLIN2 in fat cakes of the liver from mice on HFD (26), indicating that PLIN5 is an important constituent of LDs in diet-induced hepatosteatosis. Here, we demonstrated that adenovirus-mediated overexpression of PLIN5 in the liver was sufficient to drive further LD formation and alter lipid profiles in the liver of Bl6 on HFD. Ad-PLIN5 mice on HFD showed an up-regulation in both the size and number of hepatic LD and the increase in hepatic TG, CholE, and DG determined by LC-MS/MS. However, the increase in hepatic LD driven by PLIN5 overexpression by adenovirus did not impair glucose homeostasis at least within the time frame we observed. Rather, serum insulin levels were reduced in Ad-PLIN5 mice, indicating improvement in insulin sensitivity. Also, there were no increases in hepatocyte proliferation markers, or collagen. Thus, the expression of PLIN5 in the setting of HFD in the liver expands hepatic lipid pools without compromising health and insulin sensitivity of the liver.
One potential mechanism responsible for the increase in the TG in Ad-PLIN5 liver is the prevention of lipolysis by PLIN5. Interestingly, the increase of TG in Ad-PLIN5 liver was not associated with the rise in PLIN2 protein indicating that PLIN5 being overexpressed might replace PLIN2 in the coating of LD and support the increase in TG. PLIN5 but not PLIN2 has been shown to interact with ATGL and colipase CGI-58, 2 proteins that play regulatory roles in lipolysis in wide ranges of tissue (40). The reduction of CGI-58 in the liver by antisense oligonucleotides and liver specific deletion of ATGL both resulted in hepatosteatosis, indicating that they are key regulators of lipolysis in the liver (41, 42). The interaction of PLIN5 with ATGL and CGI-58 is believed to hamper lipolysis in the liver environment at least in a basal state, as the down-regulation of PLIN5 in AML12 cells and hepatocytes increased lipolysis (12, 18). Conversely, the overexpression of PLIN5 in AML12 cells reduced basal lipolysis (24). However, PLIN5 increases lipolysis in a cAMP dependent manner in AML12 cells, COS 7cells, and MIN6 β-cell lines (Supplemental Figure 6I) (24, 28, 43). To determine the contribution of lipolysis in the exacerbation of hepatosteatosis by Ad-PLIN5, a future study should address how HFD affects phosphorylation status of PLIN5 in vivo and its interaction with ATGL and CGI-58 in the liver.
In addition to the direct interaction with ATGL and CGI-58, PLIN5 may reduce lipolysis through the regulation of gene expression. The reduction in both pnpla2 (ATGL) and lipe expression was noted in Ad-PLIN5 liver. In contrast, genes for FA synthesis (Fasn) and TG synthesis (Dgat1 and Dgat2) were unchanged or reduced implying that slowing in TG use rather than increased synthesis plays the major role in LD accumulation by PLIN5. The change in genes involved in lipid metabolism we observed in Ad-PLIN5 liver is similar to those seen when PLIN5 was overexpressed in skeletal muscle and heart (32, 33, 44), and is summarized as the reduction in PPAR-α target genes. Of note, the data from ATGL knockout mice indicates that lipolysis by ATGL produces signals that regulate PPAR-α activity (45, 46). Considering that PLIN5 affects the rate of lipolysis in cells, overexpression of PLIN5 in the liver may regulate PPAR-α activity through the alteration in lipolytic signals. Interestingly, unbiased lipidomics of the liver, we performed in the current study showed PC (16:0/18:1) that is shown to serve as a ligand for PPAR-α (38) is increased with Ad-PLIN5 overexpression in the liver leaving the nature of signals modulating PPAR-α target genes for future determination.
Dissociation of hepatosteatosis and insulin resistance was previously seen in several models. Deficiency of histone deacetylase 3 in the liver improved insulin sensitivity despite marked hepatosteatosis (21). This improvement in insulin sensitivity did not require the change in insulin signaling but was mediated by shifting metabolic substrates from gluconeogenesis to lipid storage in PLIN2-coated LD. Actually, adenovirus-mediated overexpression of PLIN2 in the liver was sufficient to improve insulin sensitivity (21). Another potential mechanism is the reduction of lipotoxic metabolites. The CGI-58 down-regulation in the liver by antisense oligonucleotides prevented glucose intolerance in mice on HFD despite the increase in hepatosteatosis and higher DG contents. However, the DG accumulation under CGI-58 reduction was limited to LD/endoplasmic reticulum fraction, whereas DG in the plasma membrane was reduced resulting in lower activation of protein kinase Cϵ, one of the molecules considered to be responsible for the impairment in insulin signaling in response to hepatosteatosis (47). In addition to favorable effects on insulin sensitivity implied in the current study, PLIN5 may play an important cytoprotective role in cells under nutritional stress. The liver and heart from PLIN5-deficient mice show increases in oxidative stress (12, 48). Thus, PLIN5 may aid in the spatial distribution of FA through its incorporation to LDs while reducing release of lipid metabolites through the down-regulation of PPAR-α target genes. The relatively benign nature of lipid accumulation driven by PLIN5 up-regulation was also noted in heart and skeletal muscle (32, 44).
Q-Exactive LC-MS/MS revealed that up-regulation of lipids by PLIN5 overexpression in the liver is not limited to TG but includes DG, CholE, and phospholipids as well. Indeed, the fold increase in DG was higher than that of TG, implying that LDs in Ad-PLIN5 liver is enriched with DG. Considering that the expression of the final enzymes responsible for TG synthesis (Dgat1 and Dgat2) was lower in Ad-PLIN5 liver, this increase may reflect the reduced conversion of DG to TG, which was also seen in aforementioned hepatosteatosis due to CGI-58 down-regulation (47). The increase in phospholipids is likely critical to support expansion of LD as it serves as a surfactant to prevent coalescence of LD (49). DG may also support LD biogenesis through its interaction with proteins and phospholipids (50). It is of interest to note that the increase in DG and PC was also noted in LDs isolated from hepatocytes of mice after fasting and HFD, 2 conditions that are known to increase PLIN5 expression in the liver (34).
Although PLIN5 seems to drive proportional increase in many of lipid species we detected in the liver (Figure 4 and Supplemental Figures 1–4), we observed a couple of unique alterations in FA chain characteristics with PLIN5 overexpression. For TG, saturation tends to be higher in C14 to C18 FA chains in Ad-PLIN5 liver compared with control (Figure 4B). The increase in CholE was mostly driven by those with FA with long chains. It requires further studies to define whether difference in chain length and saturation of these neutral lipids affects formation and mobilization of LD, generation of lipid derived metabolites, and hepatocyte health.
Adenovirus-mediated gene transfer is widely used to modulate the expression of genes in the liver and known to primarily transduce hepatocytes (51–55). However, the method has limitations that adenovirus will be cleared by host immune cells and will not sustain the transduction. Thus, our observation is limited to short term (2 weeks) effects of PLIN5 overexpression in the liver. This might have reduced sensitivity to detect some of changes such as GTT and ITT. Future studies will require approaches such as transgenic mice or transduction by adeno-associated virus to achieve long-term expression of PLIN5 by gene transfer. Also, the increase in baseline inflammation due to host immune cells could modify phenotypes observed using adenovirus. Thus, the interpretation of parameters such as liver injury and inflammation should take this limitation into account. Despite these limitations, we demonstrated that Ad-GFP-treated mice progressively increase weight, serum insulin, and liver steatosis supporting that metabolic hallmarks associated with HFD are maintained after adenovirus transduction and that these mice serve as appropriate controls for the current study.
In summary, adenovirus-mediated overexpression of PLIN5 in the liver resulted in expansion of liver lipids that is not limited to TG but include DG, CholE, phospholipids, and part of Cer species, without apparent impairment in insulin sensitivity or activation of markers associated with steatohepatitis.
Acknowledgments
We thank the Histology Core of Eastern Virginia Medical School for technical assistance.
Author contributions: Y.I. conceived the study; M.B.T., D.M.-H., Y.M., T.K., and Y.I. (all aspects), J.N. (quantitative PCR and immunofluorescence), and E.V.G. (in vivo expression of adenoviral vectors), A.P. and S.D. (LC-MS/MS) were responsible for the acquisition and analysis of the data; M.B.T. and Y.I. contributed to research design, interpreted the data, drafted the manuscript, and critically revised the manuscript for important intellectual content. All authors revised and approved the final version of the manuscript.
This work was supported by National Institutes of Health Grants R01-DK090490 and R01-DK090490-02W1 (to Y.I.).
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- Ad-GFP
- adenoviruses expressing GFP
- ATGL
- adipose triglyceride lipase
- Bl6
- C57Bl/6J
- Cer
- ceramide
- CGI-58
- comparative gene identification-58
- CholE
- cholesterol ester
- DG
- diacylglycerol
- FA
- fatty acid
- GAPDH
- glyceraldehyde 3-phosphate dehydrogenase
- GFP
- green fluorescent protein
- GTT
- glucose tolerance test
- HFD
- high-fat diet
- ITT
- insulin tolerance test
- LC-MS/MS
- liquid chromatography with tandem mass spectrometry
- LD
- lipid droplet
- NAFLD
- nonalcoholic fatty liver disease
- NC
- regular rodent chow
- OA
- oleic acid
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PLIN
- perilipin
- PPAR
- peroxisome proliferator-activated receptor
- TG
- triacylglycerol.
References
- 1. Loomba R, Sanyal AJ. The global NAFLD epidemic. Nat Rev Gastroenterol Hepatol. 2013;10:686–690. [DOI] [PubMed] [Google Scholar]
- 2. Lazo M, Hernaez R, Bonekamp S, et al. Non-alcoholic fatty liver disease and mortality among US adults: prospective cohort study. BMJ. 2011;343:d6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Argo CK, Caldwell SH. Epidemiology and natural history of non-alcoholic steatohepatitis. Clin Liver Dis. 2009;13:511–531. [DOI] [PubMed] [Google Scholar]
- 4. Starley BQ, Calcagno CJ, Harrison SA. Nonalcoholic fatty liver disease and hepatocellular carcinoma: a weighty connection. Hepatology. 2010;51:1820–1832. [DOI] [PubMed] [Google Scholar]
- 5. Neuschwander-Tetri BA. Hepatic lipotoxicity and the pathogenesis of nonalcoholic steatohepatitis: the central role of nontriglyceride fatty acid metabolites. Hepatology. 2010;52:774–788. [DOI] [PubMed] [Google Scholar]
- 6. Perry RJ, Samuel VT, Petersen KF, Shulman GI. The role of hepatic lipids in hepatic insulin resistance and type 2 diabetes. Nature. 2014;510:84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Olofsson SO, Boström P, Andersson L, Rutberg M, Perman J, Borén J. Lipid droplets as dynamic organelles connecting storage and efflux of lipids. Biochim Biophys Acta. 2009;1791:448–458. [DOI] [PubMed] [Google Scholar]
- 8. Crunk AE, Monks J, Murakami A, et al. Dynamic regulation of hepatic lipid droplet properties by diet. PLoS One. 2013;8:e67631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bickel PE, Tansey JT, Welte MA. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim Biophys Acta. 2009;1791:419–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Motomura W, Inoue M, Ohtake T, Takahashi N, et al. Up-regulation of ADRP in fatty liver in human and liver steatosis in mice fed with high fat diet. Biochem Biophys Res Commun. 2006;340:1111–1118. [DOI] [PubMed] [Google Scholar]
- 11. Carr RM, Patel RT, Rao V, et al. Reduction of TIP47 improves hepatic steatosis and glucose homeostasis in mice. Am J Physiol Regul Integr Comp Physiol. 2012;302:R996–R1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang C, Zhao Y, Gao X, et al. Perilipin 5 improves hepatic lipotoxicity by inhibiting lipolysis. Hepatology. 2015;61:870–882. [DOI] [PubMed] [Google Scholar]
- 13. Magnusson B, Asp L, Boström P, et al. Adipocyte differentiation-related protein promotes fatty acid storage in cytosolic triglycerides and inhibits secretion of very low-density lipoproteins. Arterioscler Thromb Vasc Biol. 2006;26:1566–1571. [DOI] [PubMed] [Google Scholar]
- 14. Chang BH, Li L, Paul A, et al. Protection against fatty liver but normal adipogenesis in mice lacking adipose differentiation-related protein. Mol Cell Biol. 2006;26:1063–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Imai Y, Varela GM, Jackson MB, Graham MJ, Crooke RM, Ahima RS. Reduction of hepatosteatosis and lipid levels by an adipose differentiation-related protein antisense oligonucleotide. Gastroenterology. 2007;132:1947–1954. [DOI] [PubMed] [Google Scholar]
- 16. McManaman JL, Bales ES, Orlicky DJ, et al. Perilipin-2-null mice are protected against diet-induced obesity, adipose inflammation, and fatty liver disease. J Lipid Res. 2013;54:1346–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Varela GM, Antwi DA, Dhir R, et al. Inhibition of ADRP prevents diet-induced insulin resistance. Am J Physiol Gastrointest Liver Physiol. 2008;295:G621–G628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li H, Song Y, Zhang LJ, et al. LSDP5 enhances triglyceride storage in hepatocytes by influencing lipolysis and fatty acid β-oxidation of lipid droplets. PLoS One. 2012;7:e36712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mason RR, Mokhtar R, Matzaris M, et al. PLIN5 deletion remodels intracellular lipid composition and causes insulin resistance in muscle. Mol Metab. 2014;3:652–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Imai Y, Boyle S, Varela GM, et al. Effects of perilipin 2 antisense oligonucleotide treatment on hepatic lipid metabolism and gene expression. Physiol Genomics. 2012;44:1125–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sun Z, Miller RA, Patel RT, et al. Hepatic Hdac3 promotes gluconeogenesis by repressing lipid synthesis and sequestration. Nat Med. 2012;18:934–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Granneman JG, Moore HP, Mottillo EP, Zhu Z. Functional interactions between Mldp (LSDP5) and Abhd5 in the control of intracellular lipid accumulation. J Biol Chem. 2009;284:3049–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Granneman JG, Moore HP, Mottillo EP, Zhu Z, Zhou L. Interactions of perilipin-5 (Plin5) with adipose triglyceride lipase. J Biol Chem. 2011;286:5126–5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang H, Bell M, Sreenivasan U, et al. Unique regulation of adipose triglyceride lipase (ATGL) by perilipin 5, a lipid droplet-associated protein. J Biol Chem. 2011;286:15707–15715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang H, Sreenivasan U, Sreenevasan U, et al. Perilipin 5, a lipid droplet-associated protein, provides physical and metabolic linkage to mitochondria. J Lipid Res. 2011;52:2159–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bell M, Wang H, Chen H, et al. Consequences of lipid droplet coat protein downregulation in liver cells: abnormal lipid droplet metabolism and induction of insulin resistance. Diabetes. 2008;57:2037–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Faleck DM, Ali K, Roat R, et al. Adipose differentiation-related protein regulates lipids and insulin in pancreatic islets. Am J Physiol Endocrinol Metab. 2010;299:E249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Trevino MB, Machida Y, Hallinger DR, et al. Perilipin 5 regulates islet lipid metabolism and insulin secretion in a cAMP-dependent manner: implication of its role in the postprandial insulin secretion. Diabetes. 2015;64:1299–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Viegas MS, Martins TC, Seco F, do Carmo A. An improved and cost-effective methodology for the reduction of autofluorescence in direct immunofluorescence studies on formalin-fixed paraffin-embedded tissues. Eur J Histochem. 2007;51:59–66. [PubMed] [Google Scholar]
- 30. Bird SS, Marur VR, Sniatynski MJ, Greenberg HK, Kristal BS. Lipidomics profiling by high-resolution LC-MS and high-energy collisional dissociation fragmentation: focus on characterization of mitochondrial cardiolipins and monolysocardiolipins. Anal Chem. 2011;83:940–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bird SS, Marur VR, Sniatynski MJ, Greenberg HK, Kristal BS. Serum lipidomics profiling using LC-MS and high-energy collisional dissociation fragmentation: focus on triglyceride detection and characterization. Anal Chem. 2011;83:6648–6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang H, Sreenivasan U, Gong DW, et al. Cardiomyocyte-specific perilipin 5 overexpression leads to myocardial steatosis and modest cardiac dysfunction. J Lipid Res. 2013;54:953–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pollak NM, Schweiger M, Jaeger D, et al. Cardiac-specific overexpression of perilipin 5 provokes severe cardiac steatosis via the formation of a lipolytic barrier. J Lipid Res. 2013;54:1092–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chitraju C, Trötzmüller M, Hartler J, et al. Lipidomic analysis of lipid droplets from murine hepatocytes reveals distinct signatures for nutritional stress. J Lipid Res. 2012;53:2141–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bartz R, Li WH, Venables B, et al. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J Lipid Res. 2007;48:837–847. [DOI] [PubMed] [Google Scholar]
- 36. Raichur S, Wang ST, Chan PW, et al. CerS2 haploinsufficiency inhibits β-oxidation and confers susceptibility to diet-induced steatohepatitis and insulin resistance. Cell Metab. 2014;20:687–695. [DOI] [PubMed] [Google Scholar]
- 37. Turpin SM, Nicholls HT, Willmes DM, et al. Obesity-induced CerS6-dependent C16:0 ceramide production promotes weight gain and glucose intolerance. Cell Metab. 2014;20:678–686. [DOI] [PubMed] [Google Scholar]
- 38. Chakravarthy MV, Lodhi IJ, Yin L, et al. Identification of a physiologically relevant endogenous ligand for PPARα in liver. Cell. 2009;138:476–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roat R, Rao V, Doliba NM, et al. Alterations of pancreatic islet structure, metabolism and gene expression in diet-induced obese C57BL/6J mice. PLoS One. 2014;9:e86815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kimmel AR, Sztalryd C. Perilipin 5, a lipid droplet protein adapted to mitochondrial energy utilization. Curr Opin Lipidol. 2014;25:110–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brown JM, Betters JL, Lord C, et al. CGI-58 knockdown in mice causes hepatic steatosis but prevents diet-induced obesity and glucose intolerance. J Lipid Res. 2010;51:3306–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu JW, Wang SP, Alvarez F, et al. Deficiency of liver adipose triglyceride lipase in mice causes progressive hepatic steatosis. Hepatology. 2011;54:122–132. [DOI] [PubMed] [Google Scholar]
- 43. Pollak NM, Jaeger D, Kolleritsch S, et al. The interplay of protein kinase a and perilipin 5 regulates cardiac lipolysis. J Biol Chem. 2015;290:1295–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bosma M, Sparks LM, Hooiveld GJ, et al. Overexpression of PLIN5 in skeletal muscle promotes oxidative gene expression and intramyocellular lipid content without compromising insulin sensitivity. Biochim Biophys Acta. 2013;1831:844–852. [DOI] [PubMed] [Google Scholar]
- 45. Zechner R, Zimmermann R, Eichmann TO, et al. FAT SIGNALS–lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 2012;15:279–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ong KT, Mashek MT, Davidson NO, Mashek DG. Hepatic ATGL mediates PPAR-α signaling and fatty acid channeling through an L-FABP independent mechanism. J Lipid Res. 2014;55:808–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cantley JL, Yoshimura T, Camporez JP, et al. CGI-58 knockdown sequesters diacylglycerols in lipid droplets/ER-preventing diacylglycerol-mediated hepatic insulin resistance. Proc Natl Acad Sci USA. 2013;110:1869–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kuramoto K, Okamura T, Yamaguchi T, et al. Perilipin 5, a lipid droplet-binding protein, protects heart from oxidative burden by sequestering fatty acid from excessive oxidation. J Biol Chem. 2012;287:23852–23863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Krahmer N, Guo Y, Wilfling F, et al. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab. 2011;14:504–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pol A, Gross SP, Parton RG. Review: biogenesis of the multifunctional lipid droplet: lipids, proteins, and sites. J Cell Biol. 2014;204:635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Waddington SN, McVey JH, Bhella D, et al. Adenovirus serotype 5 hexon mediates liver gene transfer. Cell. 2008;132:397–409. [DOI] [PubMed] [Google Scholar]
- 52. Duffy MR, Bradshaw AC, Parker AL, McVey JH, Baker AH. A cluster of basic amino acids in the factor X serine protease mediates surface attachment of adenovirus/FX complexes. J Virol. 2011;85:10914–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li F, Xiang Y, Potter J, Dinavahi R, Dang CV, Lee LA. Conditional deletion of c-myc does not impair liver regeneration. Cancer Res. 2006;66:5608–5612. [DOI] [PubMed] [Google Scholar]
- 54. Saini C, Liani A, Curie T, et al. Real-time recording of circadian liver gene expression in freely moving mice reveals the phase-setting behavior of hepatocyte clocks. Genes Dev. 2013;27:1526–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Deng Y, Wang ZV, Tao C, et al. The Xbp1s/GalE axis links ER stress to postprandial hepatic metabolism. J Clin Invest. 2013;123:455–468. [DOI] [PMC free article] [PubMed] [Google Scholar]