Abstract
Background
Even though altered metabolism representing a hallmark of cancer was proposed nearly a century ago, recent technological advances have allowed investigators to continue uncovering a previously unrecognized complexity of metabolic programs that drive tumorigenesis beyond that of aerobic glycolysis.
Methods
The bioenergetic state of a diverse panel of glioblastoma models, including isogenic lines derived from a genetically engineered adult astrocytic mouse model and patient-derived glioblastoma stem cells, was determined at baseline and in stressed conditions. Mechanisms contributing to the discovered metabolic phenotypes were determined through molecular and chemical perturbation, and their biological consequences were evaluated in vivo and in patient samples.
Results
Attenuated mitochondrial reserve capacity was identified as a common metabolic phenotype in glioblastoma lines. This phenotype was linked mechanistically with the capacity of Ras-mediated signaling to inhibit pyruvate dehydrogenase (PDH) activity through downregulation of PDH phosphatase (PDP) expression. PDP1 repression was validated clinically in patient-derived samples, suggesting that aberrant cellular signaling typical of glioblastoma actively modulates PDH activity. This phenotype was reversed through both chemical and molecular perturbation. Restoration of PDH activity through stable expression of PDP1-impaired tumorigenic potential.
Conclusions
These findings support the central role that PDH regulation plays as a downstream consequence of aberrant signaling associated with gliomagenesis and the scientific rationale to continue to develop and test clinical strategies designed to activate PDH as a form of anticancer therapy in glioblastoma.
Keywords: glioblastoma, pyruvate dehydrogenase, Ras, spare respiratory capacity, tumor metabolism
The current concept that altered metabolism represents a hallmark of cancer was initially proposed by Otto Warburg nearly a century ago.1,2 His seminal observation involved the unique capacity of cancer to perform aerobic glycolysis, or high fermentative metabolism of glucose, resulting in production and release of lactic acid even in the presence of adequate oxygen. It has been inferred that these observations were a direct and passive consequence of damaged mitochondria in cancer cells; however, subsequent studies have not supported this notion, demonstrating oxidative phosphorylation to continue to play an active role in tumor metabolism.3,4 Further, continued investigations are uncovering a previously unrecognized complexity of metabolic programs that actively drive tumorigenesis beyond that of aerobic glycolysis. Through dynamic metabolic remodeling, which includes anabolic metabolism, anapleurosis, lipid remodeling, and altered redox balance, specific cells are able to acquire a selective advantage within a specific microenvironment.5–8
The World Health Organization (WHO) classifies glioma into grades I–IV, based on histological features that play a central role in prognosis and guiding clinical management. For example, patients with grade I tumors are typically cured following surgical resection, while patients diagnosed with grade IV tumors (ie, glioblastoma [GBM]) have a median survival of approximately 1 year despite aggressive multimodality treatment consisting of surgery, radiation therapy, and chemotherapy.9 Beyond histological features, considerable progress has been made in understanding the underlying biology of glioblastoma. For example, whole genome sequencing performed by The Cancer Genome Atlas (TCGA) Research Network has identified 3 clear nodes of genetic alterations, which include alterations in receptor tyrosine kinase signaling, p53, and Rb pathways.10 The phosphatidylinositol 3′ kinase (PI3K) signaling pathway represents one of the common signaling pathways that is aberrantly activated in GBM. Stimulation initiates a signaling cascade that ultimately results in activation of the prosurvival signal Akt, which is associated with increasing tumor grade, decreased levels of apoptosis, and adverse clinical outcome in glioma.11 A parallel pathway commonly activated in GBM is the ERK/MAPK pathway, which leads to cellular proliferation and tumor progression.12,13
Despite these advancements in our understanding of upstream events signaling tumorigenesis, their functional consequence or resultant metabolic alterations contributing to their aggressive phenotype, remain unclear. Since tumors have access to a wide variety of genetic and/or epigenetic modifications, this level of understanding may be particularly relevant because there are a limited number of metabolic strategies that cancer cells can employ to drive an aggressive phenotype, suggesting the potential for aberrant metabolic programs to serve as cancer-specific therapeutic targets. Accordingly, recent technological advancements have provided investigators with the opportunity to gain further insight into the underlying metabolism of tumors. One approach is through metabolomic profiling, which is the global quantitative assessment of endogenous metabolites within a biological system. This line of investigation has identified targetable metabolic phenotypes that are consistent with anabolic metabolism in GBM corresponding with clinical outcomes.5,14 Another approach is through understanding the metabolic phenotype of GBM by studying its bioenergetic state, which provides insight into relative reliance on glycolysis and/or oxidative phosphorylation.15,16 Although knowledge of the metabolic state of cells at baseline may be informative, since the typical tumor microenvironment presents diverse metabolic challenges, we hypothesized that unique metabolic responses elicited during stressed conditions, including glycolytic and/or mitochondrial reserve capacities, could provide more relevant insight into metabolic phenotypes associated with tumorigenesis. In this report using a diverse panel of GBM models, we identified attenuated mitochondrial reserve capacity as a consistent metabolic phenotype. We then went on to demonstrate that this phenotype was driven by Ras-mediated signaling, which actively regulated pyruvate dehydrogenase (PDH) activity through PDH phosphatase (PDP) expression. Further, we demonstrated that this phenotype could be reversed through both chemical and molecular perturbation, and more importantly, restoring PDH activity impaired tumorigenesis, supporting the therapeutic potential for targeting this metabolic program in GBM.
Materials and Methods
Cell Culture
Human GBM cell lines U251 and T98G, G179, G144, and Human Astrocyte-SV40 (NHA) were obtained and grown in conditions described previously.17,18 T, TP, TR, and TRP lines were generated and grown as previously described.19 Mesenchymal (MES83, MES326) and proneural (PN19 and PN84) glioma stem cells (GSCs) were generated and grown as previously described.20 AZD6244 (1 µM) and MK2206 (100 nM) were purchased from Selleckchem.
Cellular Bioenergetics
Extracellular acidification rate (ECAR), oxygen consumption rate (OCR), and spare respiratory capacity were measured using the Seahorse XF24 (Seahorse Bioscience).14 For spare respiratory capacity measurements, the mitochondrial stress kit (Seahorse Bioscience) was used, consisting of oligomycin (1 µM), carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP) (300 nM), rotenone (1 µM), and antimycin (1 µM). Mitochondrial isolation was performed using methods previously described14 with equimolar concentrations of glucose and pyruvate (1 mM). Floating cells were analyzed by placing 100 000 cells in a volume of 10 µL into the central well of the microplate and allowing them to stabilize for 10 minutes. A screen specific for the microplate was then firmly laid over the well without overflow of the media; 500 µL of assay media were then added to the cells and left in a non-CO2 incubator for 45 minutes prior to obtaining readings. Readings were normalized to total protein, and data were analyzed using Seahorse software, with the unpaired t test being applied to test the significance of change. Results were representative of at least 3 independent experiments.
Pyruvate Dehydrogenase Activity
A PDH enzyme activity microplate assay kit (Abcam) was used to determine PDH activity. Results are representative of at least 3 independent experiments.
Western Blot and Reverse Transcriptase Polymerase Chain Reaction
Western blot, reverse transcriptase (RT-)PCR, and image quantification were performed using methods previously described.18 Antibodies used for Western blot included PDH 459400, (Invitrogen), phospho-PDH ab177461 (Abcam), PDP1 AF720 (R&D Systems), and actin A5316 (Sigma Aldrich). RT-PCR primers included:PDK1(5′-CCAAGACCTCGTGTTGAGACC-3′;5′-AATACAGCTTCAGGTCTCCTTGG-3′), PDK2 (5′GAGCCTCCTGGACATCATGG-3′; 5′-TACTCAAGCACGCCTTGTGC-3′), PDK3(5′-ACTGTATTCCATGGAAGGAGTGG-3′;5′-CTCCAATCATCGGCTTCAGG-3′), PDK4 (5′-AACTGTGATGTGGTAGCAGTGG-3′;5′-GATGTGAATTGGTTGGTCTGG-3′), PDP1(5′-GGACTTACTGGCAAGAGCTTAT-3′;5′-GCCTCCAAGGAGATGTCATTAT-3′), PDP2 (5′-ACCACCTCCGTGTCTATTGG-3′;5′-CCAGCGAGATGTCAGAATCC-3′), and β-actin(5′-AGAGCTACGAGCTGCCTGAC-3′; 5′-AGCACTGTGTTGGCGTACAG-3′).
Establishment of Stable Pyruvate Dehydrogenase Phosphatase 1-overexpressing Cells
Human PDP1 catalytic subunit 1 cDNA/ORF (ccsbBroad304_03447) was cloned into vector pLX304-Blast-V5 in DH5α obtained as glycerol stock. Plasmids were isolated from the culture, quantitated, and transfected into TR cells using methods previously described.14
Animal Handling
All in vivo experiments were performed according to institutional guidelines and approved by the Institutional Animal Care and Use Committee. Flank-xenografts were established in athymic nu/nu mice (Charles River Laboratories) using methods previously described.14
Statistical Analysis
Statistical analysis was done using the Student t test unless otherwise indicated.
Results
Baseline Bioenergetics of Glioblastoma Cell Lines
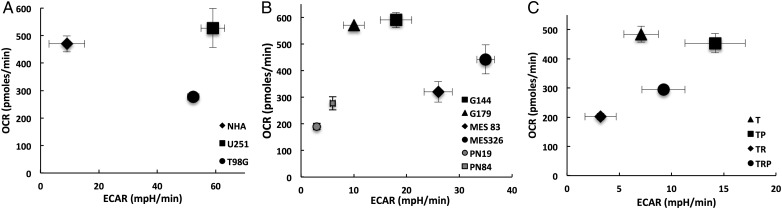
We utilized a diverse panel of glioma cell models to provide insight into relative reliance on glycolysis and/or oxidative phosphorylation in GBM. These included established GBM cell lines (U251 and T98G), normal human astrocytes (NHAs), glioma neural stem (GNS) lines (G144 and G179),17 patient-derived proneural (PN84, PN19), and mesenchymal (M83, M326) GSCs, which recapitulate the aggressive, invasive phenotype of GBM.20 In addition, we applied a novel, genetically engineered mouse (GEM) cell line model to provide insight into specific signaling pathways that may be driving observed differences in metabolism. Specifically, primary lines of astrocytes were generated from a series of conditional GEM models in which 1 or 2 of the 3 core GBM pathways were genetically targeted, as previously described.19,21 After Cre-mediated recombination, these mice express (i) N-terminal 121-amino acid truncation mutant of SV40 large T antigen (T) from the human glial fibrillary acidic protein promoter, which inactivates all 3 Rb family proteins; (ii) constitutively active KrasG12D mutant (R); and/or (iii) homozygous Pten deletion (P). Although Ras is an uncommon stochastic mutation in GBM, its downstream signaling intermediates are frequently aberrant in GBM, and causal roles of Ras network aberration in GEM high-grade astrocytoma have been validated in several engineered mouse studies.22 Systematic characterization of primary astrocytes generated from this model have demonstrated that both Rb inactivation and Ras activation (TR) are required for high-grade glioma tumorigenesis, and the incidence of both tumor and high-grade glioma development was increased when combined with Pten loss (TRP). As an initial investigation, we evaluated baseline metabolism of the described lines using the Seahorse Extracellular Flux Analyzer (XF24). The capacity of this platform to measure OCR and proton production (ECAR), indicative of oxidative phosphorylation and glycolysis, respectively, can thereby provide an overall assessment of the metabolic state of each individual line.14,16,23 Specifically, by plotting OCR as a function of ECAR, relative utilization of glycolysis and oxidative phosphorylation can be defined along with the energetic state of an individual cell line;23 (Supplementary material, Fig. S1A).
Clear metabolic heterogeneity was observed in cell lines grown in culture (Fig. 1). As expected with the established cell lines, normal astrocytes had a high OCR (470 ± 29 pmoles/min) and low ECAR (9 ± 6 mpH/min), suggesting a high reliance on oxidative phosphorylation in these nonmalignant cells (Fig. 1A). Further, both GBM cell lines (U251 and T98G) demonstrated expected high levels of ECAR (59 ± 4 and 52 ± 2 mpH/min, respectively), which is consistent with aerobic glycolysis. However, these lines also demonstrated relatively high levels of baseline oxidative phosphorylation, with U251 cells utilizing mitochondrial respiration at rates similar to those of NHA (527 ± 70 pmoles/min). The GNS lines also demonstrated high levels of mitochondrial respiration (G144: 590 ± 29 and G179: 571 ± 2 pmoles/min) but relatively low ECAR (G144: 18 ± 3 and G179: 10 ± 1 mpH/min) when compared with established GBM lines (Fig. 1B). Interestingly, the aggressive mesenchymal GSCs had significantly higher levels of ECAR (M83: 26 ± 3 and M326: 35 ± 2 mpH/min) and modestly increased OCR (M83: 320 ± 38and M326: 442 ± 55 pmoles/min) when compared with the proneural GSCs (PN19: 3 ± 1 mpH/min and 190 ± 12 pmoles/min; PN84: 6 ± 1 mpH/min and 277 ± 25 pmoles/min), suggesting a lower energetic state in these less aggressive cells. This supports the concept of metabolic heterogeneity between GBM subtypes and the role metabolism may play in their underlying aggressiveness. However, this metabolic pattern was not recapitulated in the T/R/P model system (Fig. 1C). When evaluating OCR, the T and TP lines, which have a low incidence of tumorigenicity and/or are more consistent with low-grade glioma when grown in vivo,19,21 demonstrated relatively high levels of OCR (T: 484 ± 27 and TP 454 ± 33 pmoles/min) when compared with the more aggressive TR and TRP lines (TR: 202 ± 12 and; TRP 294 ± 11 pmoles/min), however, the TR and TRP lines were not associated with an expected increase in ECAR (T: 7 ± 2; TP 14 ± 3; TR: 3 ± 2; TRP 9 ± 2 mpH/min). Collectively, based on the heterogeneity observed in these studies, baseline metabolism was inconsistent in both differentiating the metabolic phenotype of cancer cells from nontransformed cells and predicting the underlying aggressiveness of an individual cancer line.
Fig. 1.
Baseline metabolic profiles of glioblastoma cells. The oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were plotted against one another to define the bioenergetic state of the described cell lines.
Glioblastoma Cell Lines Have an Attenuated Spare Respiratory Capacity
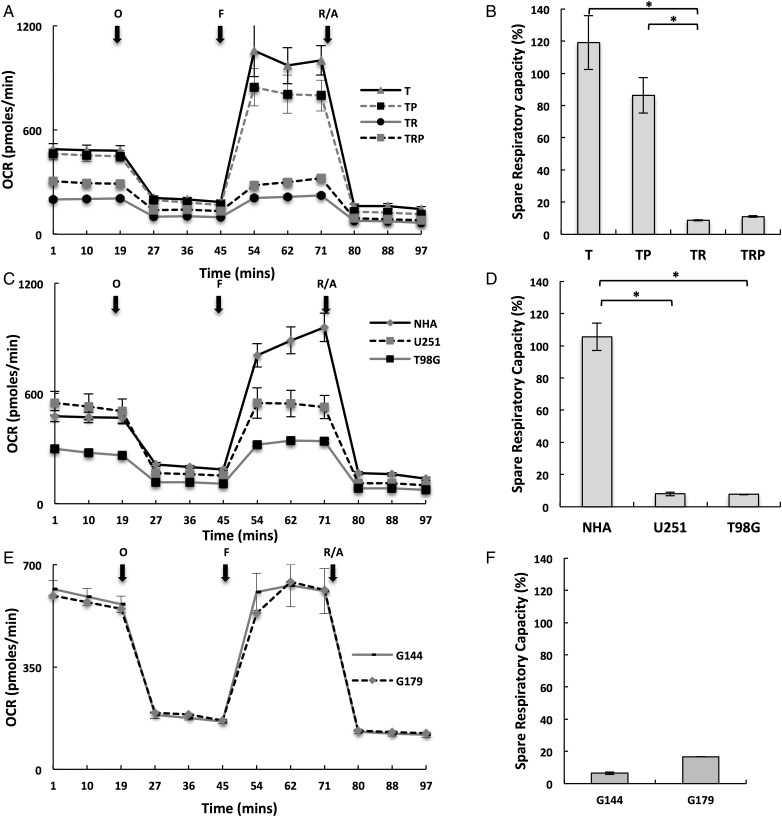
Since baseline cellular metabolism demonstrated a limited capacity in providing insight into tumor-specific metabolism, we extended our investigation to further characterize the respiratory phenotype of these lines by defining their mitochondrial reserve capacities. As the above studies suggested, Ras signaling (TR and TRP) attenuated baseline OCR, and these specific lines were associated with the aggressive phenotype of GBM; initial investigations were performed using the isogenic T/R/P model. These studies were performed by metabolically perturbing individual lines by adding specific compounds in succession and thereby shifting the bioenergetic profile of the cells, which represents a methodology derived from established approaches for studying mitochondria.15 In these studies, cells were first treated with the ATP synthase inhibitor oligomycin, thereby decreasing OCR associated with ATP synthesis. Next, cells were exposed to FCCP, which is an uncoupling agent that dissipates the mitochondrial membrane potential and leads to rapid and maximal OCR without generating ATP. The spare respiratory capacity (SRC) of a cell line is defined as the quantitative difference between the maximal uncontrolled OCR and initial basal OCR, as described in Supplementary material, Fig. S1B. Cells were subsequently exposed to the mitochondrial inhibitors rotenone and antimycin A, thereby, shutting down mitochondrial respiration and allowing calculation of the mitochondrial and nonmitochondrial fractions contributing to respiration. Both the Rb mutated T and TP lines, which again typically do not develop high-grade gliomas in vivo,19,21 demonstrated similar respiratory phenotypes with significant increases in OCR following decoupling that resulted in SRCs of 120% ± 17% and 86% ± 11%, respectively. However, the addition of a mutation in Ras, demonstrated in both TR and TRP lines, exhibited attenuated maximal OCR following decoupling that only returned back to baseline levels and resulted in diminished SRC (TR = 8% ± 0.4% and TRP = 11% ± 0.4%; Fig. 2A and B). This pattern was consistent when tested in our other model systems. For example, the respiratory phenotype of normal astrocytes was similar to the T and TP lines and resulted in a significant SRC of 106% ± 8% (Fig. 2C and D). However, both the established GBM lines and invasive GNS lines demonstrated an attenuated SRC similar to the TRP and TR lines (Fig. 2E and F). This concept was further supported when we compared the respiratory phenotype between GSC subtypes. The proneural GSCs, which demonstrated slow growth in vivo with minimal angiogenesis and necrosis,20 demonstrated a mitochondrial phenotype consistent with normal astrocytes (ie, SRC of 112% ± 3% and 114% ± 8% in PN19 and PN84 cells, respectively). The rapidly growing, invasive mesenchymal GSC lines, however, demonstrated an attenuated SRC (Supplementary material, Fig. S2A and B). Collectively, these findings suggest that an attenuated SRC may represent a metabolic phenotype of GBM and contribute towards its aggressive biology. We extended our investigations to determine if mitochondrial potential could serve as another tool for determining cell-autonomous metabolic heterogeneity. Despite clear differences in metabolism between T and TR cells, no appreciable differences were observed in how this translated to steady-state mitochondrial function (as measured by membrane potential) (Supplementary material, Fig. S3). This suggests that many factors may influence a cell's energy requirements and that baseline evaluation of metabolism in vitro may be limited for elucidating how they translate into its metabolic phenotype.
Fig. 2.
Glioblastoma cells demonstrate an attenuated spare respiratory capacity. (A, C and E) The respiratory phenotype was defined in the described cell lines using established methods to study mitochondrial function that involved sequentially exposing cells to oligomycin (O), carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP) (F), and rotenone/antimycin (R/A) in real-time on the Seahorse platform. (B, D and F) The relatively spare respiratory capacity (%) was calculated as (oxygen consumption rate [OCR] following FCCP – baseline OCR)/baseline OCR. * P < .01.
Ras-mediated Signaling Actively Modulates Spare Respiratory Capacity
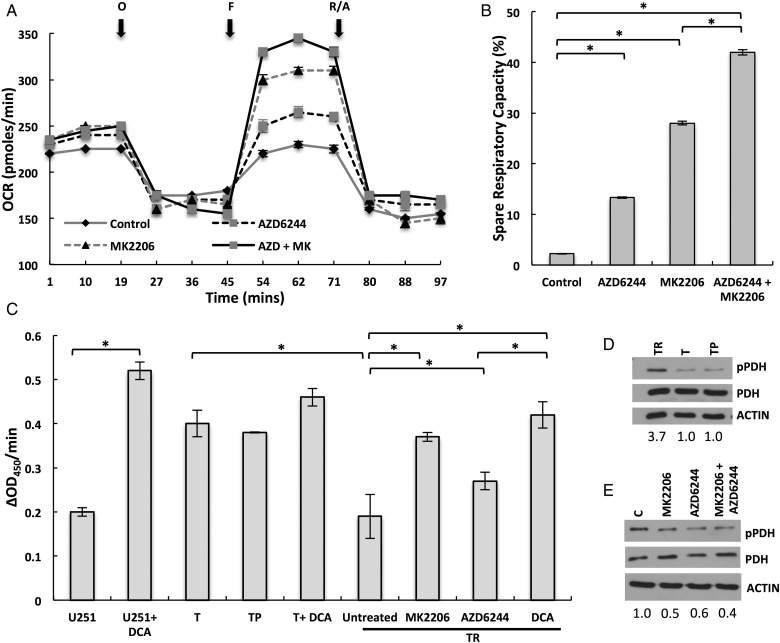
Next, we investigated the potential mechanism underlying the observed changes in respiratory phenotype. As in the initial investigation, we determined if observed changes in reserve capacity were hardwired in the mitochondria or if they could be modulated through cell signaling. Mitochondria were isolated from both U251, which demonstrated an attenuated SRC, and normal astrocytes by using methods previously described.14 Interestingly, SRC was identical in isolated mitochondria in these lines, suggesting that the observed changes in mitochondrial phenotype may be actively modulated through cellular signaling (Supplementary material, Fig. S2C). Since our GEM lines demonstrated that Ras-pathway activation contributed to the observed attenuated SRC, we next sought to determine if this respiratory phenotype could be reversed through signaling inhibition. To test this, we exposed TR cells to the MEK inhibitor AZD6244 and the Akt inhibitor MK2206, which represent downstream mediators of 2 of the most extensively studied effectors of Ras signaling (ie, Raf and PI3K, respectively24) with initial studies confirming target engagement (Supplementary material, Fig. S2D). In the TR line, baseline OCR and OCR remained unchanged following exposure to the ATP synthase inhibitor oligomycin when compared with vehicle control following 24 hour pretreatment with each agent (Fig. 3A). However, both Akt and MEK inhibition independently demonstrated the capacity to reverse the previously described attenuated respiratory capacity following decoupling to determine maximal OCR and SRC, resulting in 13% ± 0.1% and 28% ± 0.4% increases in reserve capacities, respectively (Fig. 3A and B). Interestingly, combining Akt and MEK inhibition led to a further increase in SRC (38% ± 0.5%), suggesting redundancy in the ability of signaling pathways to influence the mitochondrial phenotype. Similarly, MEK and/or Akt inhibition reverted the attenuated oxidative reserve capacity in U251 and G179 cells, although their relative reliance on each pathway appeared cell-line dependent (Supplementary material, Fig. S4A–D). Collectively, these findings suggest that Ras-mediated signaling actively modulates SRC. We went on to determine if baseline levels of Akt or MEK activation could be used to determine a cell’s SRC and/or differential response to pathway inhibition. Unfortunately, this line of investigation failed to provide further insight into our findings (Supplementray material, Fig. S5). For example, in the isogenic TRP model, Ras mutation increased Akt activation, while ERK remained relatively unchanged, which supported this line's increased metabolic response to Akt inhibition. U251 cells demonstrated relatively low Akt activation, supporting its minimal response to Akt inhibition when compared with MEK; however, these MEK responsive cells had less ERK activity when compared with the other models. Further, both PN lines, which are less aggressive and have increased SRC when compared with mesenchymal lines, demonstrated relatively high levels of both Akt and MEK activation. Collectively, these findings suggest that the downstream sequelae of pathway activation likely involves a complex signaling network that eventually translates to their metabolic consequences and reiterates the difficulty in identifying functional” signaling pathways in a given tumor.
Fig. 3.
Ras-mediated signaling modulates oxygen consumption rate (OCR) and spare respiratory capacity through inhibition of pyruvate dehydrogenase (PDH) activity. (A and B) The respiratory phenotype was defined in TR cells exposed to either vehicle control or the described agents 24 hours prior to analysis. To study mitochondrial function, cells were sequentially exposed to oligomycin (O), carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP) (F), and rotenone/antimycin (R/A) in real time. The relative spare respiratory capacity (%) was calculated as (OCR following FCCP – baseline OCR)/baseline OCR. (C) PDH enzyme activity in the described cell lines and treatment conditions using a microplate assay kit. (D) Western blot was performed to determine levels of phosphorylated PDH (pPDH) in the described cell lines. Levels were quantified by determining the pPDH/PDH ratio for each line and then normalized to the ratio of T cells. (E) Western blot was performed to determine levels of pPDH in TR cells exposed to vehicle control or the stated agents. Levels were quantified by determining the pPDH/PDH ratio for each treatment condition and then normalizing to the ratio of vehicle control cells. *P < .05.
Ras-mediated Signaling Modulates Oxygen Consumption Rate Through Inhibition of Pyruvate Dehydrogenase Activity
Next, we investigated potential mechanistic underpinnings of the observed Ras signaling-mediated regulation of OCR. Since PDH represents a key gatekeeping enzyme that regulates glycolytic flux into the mitochondria, we hypothesized that oncogenic signaling associated with Ras pathway activation inhibited enzyme activity, thereby actively attenuating mitochondrial flux and maximal OCR. Although the role of PDH in modulating glycolytic flux and baseline oxygen consumption has been established, its capacity to regulate maximal OCR, and thereby a cell’s mitochondrial SRC, has yet to be explored. As an initial investigation to determine if changes in PDH activity could modulate SRC, we evaluated OCR in cells treated with the metabolic modulator dichloroacetate (DCA). DCA has been previously shown to inhibit PDH kinase (PDK), which is an endogenous inhibitor of PDH. By inhibiting PDK, DCA in turn activates PDH, allowing for increased glycolytic flux into the mitochondria and oxidative phosphorylation.25 TR cells exposed to DCA (1mM) demonstrated an expected increase in baseline OCR. In addition to baseline increases in OCR, DCA was also shown to increase SRC at levels consistent with MEK and/or Akt inhibition (Supplementary material, Fig. S4E and F). We next evaluated the potential of these signaling pathways to modulate PDH activity. As demonstrated in Fig. 3C, the addition of DCA to U251 cells led to an expected increase in PDH activity, increasing activity 2.6-fold. Interestingly, baseline PDH activity of both T and TP lines was 2-fold higher than U251 cells, and the addition of DCA only led to modest increases in enzyme activity in these less aggressive lines. Conversely, the aggressive TR lines demonstrated a similar level of baseline PDH activity as U251 cells. Twenty-four hour exposure of TR lines to MK2206 and AZD6244 led to increases in PDH activity (2.0 and 1.4-fold, respectively), which was consistent with their differential capacity for modulating mitochondrial reserve capacity (Fig. 3B). Interestingly, the levels of PDH activation following Akt inhibition in the TR line were equivalent to DCA. Similar findings were observed in U251 cells (Supplementary material, Fig. S4G); although consistent with the above data (Supplementary material, Fig. S4A and B), this line was more responsive to MEK inhibition. PDH activity is regulated by reversible phosphorylation. Phosphorylation of PDH by PDH kinases (PDK1-4) inhibits its action and attenuates pyruvate use in the mitochondria for oxidative phosphorylation, whereas dephosphorylation by PDH phosphatases (PDP1-2) stimulates enzyme activity. We evaluated the phosphorylation state of PDH in the T/R/P lines. Consistent with enzyme activity studies, the TR line demonstrated a high baseline level of PDH phosphorylation, which was increased 3.7-fold when compared with both T and TP lines (Fig. 3D). Further, these levels were decreased by ∼50% following both MEK and Akt inhibition (Fig. 3E). Similar findings of increased SRC and decreased PDH phosphorylation were observed in TR cells following inhibition of AKT expression (Supplementary material, Fig. S4H). Collectively, these findings further support the role PDH activity plays in regulating maximal OCR and SRC and how aberrant signaling pathways typical to GBM modulate this phenotype.
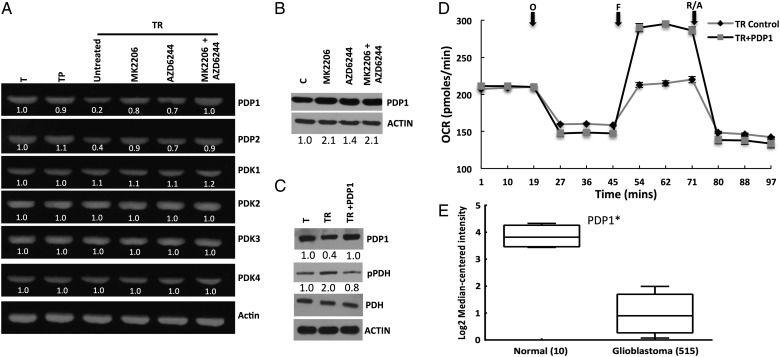
Attenuated Pyruvate Dehydrogenase Activity and Spare Respiratory Capacity Are Mediated Through Pyruvate Dehydrogenase Phosphatase Downregulation
Since we had established that Ras-mediated signaling resulted in deceased PDH activity, we next sought to determine its mechanistic underpinnings. Since PDKs and PDPs represent proteins regulating PDH activity, we determined if these enzymes were aberrantly expressed in our model. We identified PDP1(and PDP2 to a lesser extent) to be downregulated in the TR line, with an 80% and 60% decrease in messenger RNA levels, respectively (Fig. 4A). Interestingly, their levels were restored back to the levels in both T and TP following Akt and MEK inhibition. Similar findings were observed when we evaluated the protein expression of PDP1, with an approximately 2-fold increase in expression in TR following MEK and/or Akt inhibition (Fig. 4B). We next sought to demonstrate a causal link between PDP1 expression and OCR/SRC since we had already established that Ras-mediated signaling attenuates maximal OCR and SRC and had demonstrated its potential to inhibit PDH activity through downregulation of PDP1. We transiently overexpressed PDP1 in TR, resulting in an expected decrease in PDH phosphorylation (Fig. 4C). Next, we directly evaluated the role of PDP1 expression on the cells’ mitochondrial phenotype. Similar to studies in TR cells following MEK and Akt inhibition, transient overexpression of PDP1 did not influence baseline levels of OCR; it did, however, result in restoration of SRC by increasing it 36% ± 0.6% (Fig. 4D). Since we had already established the capacity of aberrant signaling pathways typical in GBM to downregulate PDP1 expression, we went on to evaluate for PDP1 expression in human samples using the TCGA database,10 which demonstrated significantly decreased levels of PDP1 in GBM when compared with normal brain (Fig. 4E) and further supported the interplay between Ras-mediated oncogenic signaling in GBM and PDH regulation.
Fig. 4.
Attenuated pyruvate dehydrogenase (PDH) activity and mitochondrial reserve capacity in glioblastoma are mediated through downregulation of pyruvate dehydrogenase phosphatase (PDP) expression. (A) The PDH regulatory proteins PDP1-2 and PDK1-4 were evaluated by reverse transcriptase PCR in the described cell lines and treatment conditions. Levels were quantified relative to actin and then normalized to the T cell line. (B) Western blot was performed to determine levels of PDP1 expression in TR cells treated in the described conditions. (C) Western blot was performed to determine levels of the stated proteins in T, TR, and the TR line overexpressing PDP1 (TR + PDP1). (D) The respiratory phenotype of the described cell lines was determined using the Seahorse platform. (E) PDP1 expression in normal brain and GBM was evaluated on the The Cancer Genome Atlas platform. *P < .01.
PDP1-mediated Regulation of Pyruvate Dehydrogenase Activity Contributes To Glioblastoma Tumorigenesis
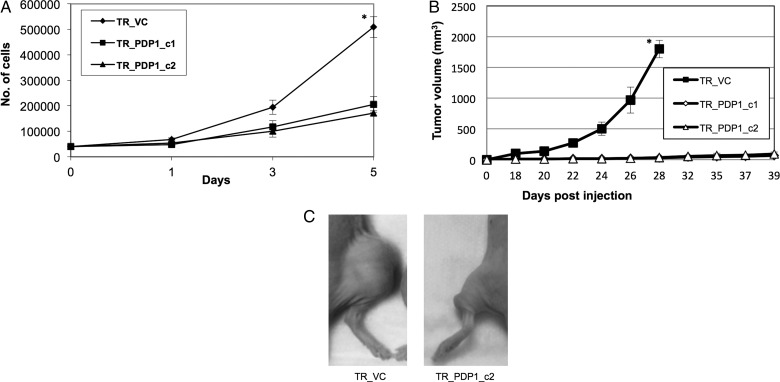
To determine if observed changes in PDH activity and SRC influenced tumorigenesis, we generated stable TR lines that overexpressed PDP1. We confirmed PDP1 overexpression in these lines, along with their expected decrease in PDH phosphorylation and increase in SRC (Supplementary material, Fig. S6A–C). We then evaluated the growth kinetics of these lines in vitro. Although PDP1-overexpressing clones maintained viability, they grew at a decreased rate when compared with vector controls (Fig. 5A). Next, we evaluated in vivo growth rate using an s.c. mouse xenograft model. Although a tumor was palpable in TR clones overexpressing PDP1, they failed to grow larger than 30 mm3 when compared with vector controls (Fig. 5B and C).
Fig. 5.
Modulation of pyruvate dehydrogenase (PDH) activity through PDP1 expression influences glioblastoma growth. (A) TR vector control cells (TR_VC) and clones overexpressing PDP1 (TR_PDP1_c1/2) were evaluated growth kinetics were evaluated in vitro. (B) Tumor growth curves were obtained in the described cell lines at stated times in an s.c. mouse xenograft model (n = 10/cell line) using perpendicular diameter measurements of each tumor with digital calipers; volumes were calculated using the formula (LxWxW)/2. (C) A representative image of described cell lines grown in an s.c. mouse xenograft model. *P < .01.
Discussion
In this report, we have presented several novel findings that involve unique metabolic states associated with gliomagenesis and how these processes are modulated through oncogenic signaling pathways. Initial studies focused on cellular metabolism at baseline to define relative metabolism through oxidative phosphorylation and aerobic glycolysis, which collectively suggested that baseline metabolism has a limited capacity to determine the tumorigenicity and/or aggressiveness of a given cell line when grown in vitro. These results are not completely unexpected because the metabolic programs evolved during gliomagenesis are likely a direct consequence of their need to adapt to the tumor microenvironment, which is likely not recapitulated in in-vitro conditions devoid of oxygen and nutrient restrictions. Furthermore, it is likely that several factors play a role in maintaining an individual cell's baseline oxidative phosphorylation state including reliance on glycolysis or alternate forms of energy metabolism (eg, glutamine metabolism and/or fatty acid oxidation, and baseline levels of oxidative stress). These factors likely play a role in the observed metabolic heterogeneity in these lines.
Based on these potential limitations associated with studying the metabolic state of cell lines in baseline conditions, we extended our investigations to determine if differential response to bioenergetic stress were associated with and/or contributed towards gliomagenesis. We uncovered an actively regulated oxidative state involving attenuated mitochondrial SRC associated with gliomagenesis. Our understanding about the potential relevance of SRC, which is defined as the difference between the maximal unregulated respiration and the initial basal respiration of an individual cell line15 in both normal and cancer cells, is continuing to evolve. It has been suggested that reserve capacity is utilized to provide for increased energy demands during stressed conditions. This was demonstrated to be particularly relevant in cardiac tissue, which is rich in mitochondria and capable of dynamically responding to increased bioenergetic demands during stressed conditions.23 In our studies, this metabolic adaptation appeared to be maintained in the nontumorigenic normal lines tested but was consistently lost in GBM lines. Interestingly, recent studies support this as an emerging tumor-specific metabolic phenotype. Sandulache et al identified that loss of mitochondrial reserve capacity was attributed to p53 mutation in squamous cell carcinoma, conferring therapeutic resistance.26 Anso et al identified a loss of SRC following myc-induced malignant transformation of osteocytes to osteogenic sarcoma.27 Collectively, these findings, coupled with our results performed on a diverse panel of GBM lines, suggest that an attenuated SRC represents a metabolic shift associated with tumorigenesis. However, the specific biological advantage(s) offered to tumor cells by this metabolic shift remain unclear. Again, our current understanding of the SRC is that it allows normal cells to adapt to increased bioenergetic demands during periods of short-term stress. However, the GBM microenvironment poses a constant array of stresses to an individual cell. Although vascularized tumors, newly formed vessels in GBM are tortuous with compromised vascular integrity. This limits tissue perfusion, resulting in a microenvironment with variable levels of hypoxia and nutrient deprivation.28,29 Our findings, which demonstrate the capacity of signaling pathways to attenuate maximal OCR and SRC, may be particularly relevant in the context of intermittent hypoxia,30 as we hypothesize that it may be disadvantageous for cancer cells to rely on compensatory increases in OCR long-term; this could potentially lead to increased generation of reactive oxygen species and cellular damage associated with increased oxidative phosphorylation.31,32
Next, we demonstrated that the observed changes in SRC were a direct consequence of PDH activity. Although the central regulatory role PDH plays in baseline oxidative phosphorylation through modulation of glycolytic flux into the mitochondria is well established, the secondary role of this gatekeeping enzyme has yet to be described. Further, its tight regulation by oncogenic signaling reinforces its potential implications in tumorigenesis, contributing to tumorigenesis. We proceeded to mechanistically link changes in PDH activity and SRC with Ras pathway activation and demonstrated the ability to restore SRC through targeted inhibition of its downstream effectors. These findings add to the growing body of data demonstrating the capacity of Ras signaling for modulating metabolic programs at multiple levels in cancer. Chiaradonna et al demonstrated the role of oncogenic Ras in enhancing utilization of the glycolytic pathway,33 and Ying et al identified the potential of Ras to control glycolysis at several rate-limiting steps, which led to shunting of glucose metabolism towards anabolic metabolism.8 Our data provide further insight into the dynamic role this oncogene plays in tumor metabolism. Interestingly, we demonstrated that multiple signaling pathways have the capacity to modulate PDP1 expression. Redundant signaling pathways are commonly observed in cancer and allow cells to maintain activity of key downstream circuits in the presence of altered upstream ligand-dependent signaling and/or to dynamically acquire resistance to targeted inhibition of a single pathway. Further, the redundancy of AKT and MEK/ERK pathways has been described previously.34,35 Therefore, the observed redundancy of these pathways further underscores the potential biological importance of PDH regulation in contributing to the aggressive phenotype of GBM and the potential therapeutic limitations of targeting these upstream pathways individually.
Lastly, we demonstrated that Ras-mediated regulation of PDH activity and mitochondrial function was mediated through the capacity of this pathway to attenuate expression of PDH regulatory proteins PDPs. The potential for oncogenic signaling typical of GBM to downmodulate expression of PDPs was validated in patient samples, with PDP1 expression being significantly repressed in GBM compared with normal brain tissue when evaluated from the TCGA dataset. More importantly, restoring expression of PDP1 in our model diminished tumorigenicity. Targeting the PDH/PDP1 axis may also have application in GSCs, which studies have shown to have a unique metabolic state when compared with differentiated cells.16 Although the multiple roles Ras signaling plays in tumorigenesis are well established, our findings demonstrated that targeting this single downstream consequence of pathway activation can completely revert tumorigenic potential of this oncoprotein. These findings are consistent with emerging data reinforcing the central role played by alterations of PDH activity in the oncogenic transformation following genetic mutation. Recent discoveries involving the mechanistic underpinnings of oncogene-induced senescence represent one of the most notable examples describing this important interplay between a driver mutation and its functional, metabolic consequence.36 In these studies, it was shown that PDH was a crucial mediator of senescence induced by BRAFV600E in normal melanocytes, leading to activation of PDH through suppression of PDK1 and induction of PDP2. This resulted in increased cellular respiration and subsequent senescence secondary to redox stress. Abrogating this mutation-induced metabolic program led to oncogenic transformation and, similar to our findings, restoring PDH activity diminished tumorigenic potential. Our model demonstrated that Ras signaling can modulate PDH activity solely through regulation of PDPs without requiring alterations in PDK1. These finding are consistent with TCGA data, which demonstrated only aberrant expression of PDP1 in clinical samples. This suggests a complex interplay between a specific mutation and an individual cell's capacity to regulate PDH activity through a balance between PDH activators and inhibitors that require further investigation. In addition, further work is still required to better understand the mechanisms contributing to the differential metabolic response of normal cells and premalignant and/or tumor cells to a specific genetic mutation. These studies support the functional consequence of alterations in PDH activity playing a central event driving tumorigenesis. The therapeutic potential of these findings shows strong promise and is being actively investigated, including recent work demonstrating antitumor activity of the PDH activator DCA in GBM.25
In summary, using a diverse panel of GBM and normal lines, we demonstrated that an attenuated SRC was a common metabolic phenotype of GBM. We established that this phenotype was driven by signaling pathways typical to GBM and was a consequence of their capacity to inhibit PDH activity by attenuating expression PDP1 and normalizing this metabolic program's diminished tumorigenic potential. These findings provide further support for the central role that PDH regulation plays in tumorigenesis and the scientific rationale to continue to develop and test clinical strategies designed to activate PDH as a form of anticancer therapy in GBM.
Supplementary Material
Funding
US Army Medical Research and Materiel Command, National Functional Genomics Center project, under award number W81XWH-08-2-0101 (P.C.), The American Cancer Society (RSG-11-029-01-CSM; P.C.), Florida Department of Health, Bankhead-Coley Cancer Research Program (4BB03; P.C.), and the Southeastern Brain Tumor Foundation (P.C.).
Conflict of interest statement. None declared.
Supplementary Material
References
- 1.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124(3215):269–270. [PubMed] [Google Scholar]
- 2.Warburg O, Posener K, Negelein E. Uber den Stoffwechsel der Carcinomzelle. Biochem Zeitschr. 1924;152:309–344. [Google Scholar]
- 3.Marin-Valencia I, Yang C, Mashimo T, et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. 2012;15(6):827–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moreno-Sanchez R, Rodriguez-Enriquez S, Marin-Hernandez A, Saavedra E. Energy metabolism in tumor cells. FEBS J. 2007;274(6):1393–1418. [DOI] [PubMed] [Google Scholar]
- 5.Chinnaiyan P, Kensicki E, Bloom G, et al. The metabolomic signature of malignant glioma reflects accelerated anabolic metabolism. Cancer Res. 2012;72(22):5878–5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Locasale JW, Grassian AR, Melman T, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43(9):869–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Possemato R, Marks KM, Shaul YD, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476(7360):346–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ying H, Kimmelman AC, Lyssiotis CA, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149(3):656–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 10.TCGA. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chakravarti A, Zhai G, Suzuki Y, et al. The prognostic significance of phosphatidylinositol 3-kinase pathway activation in human gliomas. J Clin Oncol. 2004;22(10):1926–1933. [DOI] [PubMed] [Google Scholar]
- 12.Lopez-Gines C, Gil-Benso R, Benito R, et al. The activation of ERK1/2 MAP kinases in glioblastoma pathobiology and its relationship with EGFR amplification. Neuropathology. 2008;28(5):507–515. [DOI] [PubMed] [Google Scholar]
- 13.Nicoletti NF, Erig TC, Zanin RF, et al. Mechanisms involved in kinin-induced glioma cells proliferation: the role of ERK1/2 and PI3K/Akt pathways. J Neurooncol. 2014;120(2):235–244. [DOI] [PubMed] [Google Scholar]
- 14.Prabhu A, Sarcar B, Kahali S, et al. Cysteine catabolism: a novel metabolic pathway contributing to glioblastoma growth. Cancer Res. 2014;74(3):787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi SW, Gerencser AA, Nicholls DG. Bioenergetic analysis of isolated cerebrocortical nerve terminals on a microgram scale: spare respiratory capacity and stochastic mitochondrial failure. J Neurochem. 2009;109(4):1179–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vlashi E, Lagadec C, Vergnes L, et al. Metabolic state of glioma stem cells and nontumorigenic cells. Proc Natl Acad Sci USA. 2011;108(38):16062–16067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pollard SM, Yoshikawa K, Clarke ID, et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell. 2009;4(6):568–580. [DOI] [PubMed] [Google Scholar]
- 18.Prabhu A, Sarcar B, Kahali S, et al. Targeting the unfolded protein response in glioblastoma cells with the fusion protein EGF-SubA. PLoS One. 2012;7(12):e52265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vitucci M, Karpinich NO, Bash RE, et al. Cooperativity between MAPK and PI3K signaling activation is required for glioblastoma pathogenesis. Neuro Oncol. 2013;1510:1317–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mao P, Joshi K, Li J, et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci USA. 2013;110(21):8644–8649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song Y, Zhang Q, Kutlu B, et al. Evolutionary etiology of high-grade astrocytomas. Proc Natl Acad Sci USA. 2013;110(44):17933–17938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guha A, Feldkamp MM, Lau N, et al. Proliferation of human malignant astrocytomas is dependent on Ras activation. Oncogene. 1997;15(23):2755–2765. [DOI] [PubMed] [Google Scholar]
- 23.Hill BG, Dranka BP, Zou L, et al. Importance of the bioenergetic reserve capacity in response to cardiomyocyte stress induced by 4-hydroxynonenal. Biochem J. 2009;424(1):99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3(1):11–22. [DOI] [PubMed] [Google Scholar]
- 25.Michelakis ED, Sutendra G, Dromparis P, et al. Metabolic modulation of glioblastoma with dichloroacetate. Sci Transl Med. 2010;2(31):31ra34. [DOI] [PubMed] [Google Scholar]
- 26.Sandulache VC, Skinner HD, Ow TJ, et al. Individualizing antimetabolic treatment strategies for head and neck squamous cell carcinoma based on TP53 mutational status. Cancer. 2012;118(3):711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anso E, Mullen A, Felsher D, et al. Metabolic changes in cancer cells upon suppression of MYC. Cancer Metab. 2013;1(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jain RK. Molecular regulation of vessel maturation. Nat Med. 2003;9(6):685–693. [DOI] [PubMed] [Google Scholar]
- 29.Vartanian A, Singh SK, Agnihotri S, et al. GBM's multifaceted landscape: highlighting regional and microenvironmental heterogeneity. Neuro Oncol. 2014;16(9):1167–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dewhirst MW. Intermittent hypoxia furthers the rationale for hypoxia-inducible factor-1 targeting. Cancer Res. 2007;67(3):854–855. [DOI] [PubMed] [Google Scholar]
- 31.Brand KA, Hermfisse U. Aerobic glycolysis by proliferating cells: a protective strategy against reactive oxygen species. FASEB J. 1997;11(5):388–395. [DOI] [PubMed] [Google Scholar]
- 32.Sohal RS, Weindruch R. Oxidative stress, caloric restriction, and aging. Science. 1996;273(5271):59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiaradonna F, Sacco E, Manzoni R, et al. Ras-dependent carbon metabolism and transformation in mouse fibroblasts. Oncogene. 2006;25(39):5391–5404. [DOI] [PubMed] [Google Scholar]
- 34.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11(2):85–95. [DOI] [PubMed] [Google Scholar]
- 35.Logue JS, Morrison DK. Complexity in the signaling network: insights from the use of targeted inhibitors in cancer therapy. Genes Dev. 2012;26(7):641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaplon J, Zheng L, Meissl K, et al. A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature. 2013;498(7452):109–112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.