Abstract
Background: The manipulation of pregnancy diets in animals can lead to changes in DNA methylation with phenotypic consequences in the offspring. Human studies have concentrated on the effects of nutrition during early gestation. Lacking in humans is an epigenome-wide association study of DNA methylation in relation to perturbations in nutrition across all gestation periods.
Methods: We used the quasi-experimental setting of the Dutch famine of 1944–45 to evaluate the impact of famine exposure during specific 10-week gestation periods, or during any time in gestation, on genome-wide DNA methylation levels at age ∼ 59 years. In addition, we evaluated the impact of exposure during a shorter pre- and post-conception period. DNA methylation was assessed using the Illumina 450k array in whole blood among 422 individuals with prenatal famine exposure and 463 time- or sibling-controls without prenatal famine exposure.
Results: Famine exposure during gestation weeks 1–10, but not weeks 11–20, 21–30 or 31-delivery, was associated with an increase in DNA methylation of CpG dinucleotides cg20823026 (FAM150B), cg10354880 (SLC38A2) and cg27370573 (PPAP2C) and a decrease of cg11496778 (OSBPL5/MRGPRG) (P < 5.9 × 10−7, PFDR < 0.031). There was an increase in methylation of TACC1 and ZNF385A after exposure during any time in gestation (P < 2.0 × 10−7, PFDR = 0.034) and a decrease of cg23989336 (TMEM105) after exposure around conception. These changes represent a shift of 0.3–0.6 standard deviations and are linked to genes involved in growth, development and metabolism.
Conclusion: Early gestation, and not mid or late gestation, is identified as a critical time-period for adult DNA methylation changes in whole blood after prenatal exposure to famine.
Keywords: DNA methylation, prenatal, Dutch Famine, Hunger Winter, exposure, nutrition
Introduction
Measures of prenatal adversity have been associated with adult disease,1 but identifying the causal factors has been challenging.2 Quasi-experimental studies using the setting of the of the Dutch Hunger Winter of 1944–45,3 a 6-month famine, show that exposure to famine during pregnancy may increase the risk of obesity, type 2 diabetes, dyslipidaemia and schizophrenia in the offspring, and have highlighted early gestation as a sensitive period.4
In animal experiments, perturbation of prenatal nutrition has been shown to alter epigenetic marks such as DNA methylation which control stable changes in gene expression potential5 and are associated with adult phenotype.6 Experiments perturbing early maternal nutrition have shown that blastocyst development and the period around implantation is a critical developmental window during which DNA methylation differences may arise.7,8 This may be related to the dynamic nature of DNA methylation during this period, as the genome is demethylated after fertilization and remethylated in the period after implantation.9 Most animal studies, however, evaluate nutritional perturbations across gestation in relation to DNA methylation differences.10,11
In humans, we have shown that prenatal famine exposure is associated with persistent differences in DNA methylation in adulthood and that the relation can depend on of the gestational timing of the exposure.12,13 We previously only examined famine exposure during the first or last 10 weeks of gestation in a candidate gene approach12,13 and the first 10 weeks in a genome-scale study.14 Studies by us and others on prenatal famine,12–15 seasonal food shortage16 and micro-nutrient supplementation17,18 have focused on early gestation as being one period that is critically sensitive in this respect. DNA methylation differences may also arise later in gestation, for example from smoking during pregnancy.19
It is unknown during which other specific periods of gestation the human methylome may be sensitive to prenatal perturbations in nutrition, as a systematic genome-wide investigation is still missing. Here we report on an epigenome-wide association study (EWAS) for famine exposure during specific gestation periods and for exposure to famine in any period during gestation.
Methods
Study setting
The Dutch Hunger Winter was a 6-month famine at the end of World War II that resulted from punitive measures imposed by occupying German forces after a national railway strike, winter conditions and fuel shortages. Food rations were distributed centrally and, during the famine, rations were below 900 kcal/day between 26 November 1944 and 15 May 1945. The percentage of calories from proteins, fat and carbohydrates in the diet was constant as the food rations diminished.20,21
Study subjects
The Dutch Hunger Winter Families study is described in detail elsewhere.22 We identified live-born infants in three institutions in famine-exposed cities in the western Netherlands and selected the 2417 singleton births between 1 February 1945 and 31 March 1946 of infants whose mothers were exposed to the famine during or immediately preceding that pregnancy. The study population is characterized by low socioeconomic status (SES) pregnancies and does not differ on SES of origin. We sampled 890 births from 1943 and 1947, to include infants whose mothers were not exposed to famine during this pregnancy as time-controls. A current address was obtained for 2300 individuals (70% of the birth cohort). They were invited by mail to participate in a telephone interview and in a clinical examination, together with a same-sex sibling not exposed to the famine to serve as a family-control. This means that any of the famine-exposed individuals or time-controls from the birth series could have a matched unexposed sibling control. We conducted 1075 interviews and 971 clinical examinations (of 345 clinic births without a matched sibling and 313 with a matched sibling) between 2003 and 2005.
The study was approved by the Institutional Review Board (IRB) of Columbia University Medical Center and by the Medical Ethics Committee (MEC) of Leiden University Medical Center. Study participants provided verbal consent at the start of the telephone interview and written informed consent at the start of the clinical examination.
Famine exposure
We used the date of the last menstrual period (LMP) from the hospital records to define the start of gestation unless it was missing or implausible (12%). Otherwise, we estimated LMP from annotations on the birth record and from birthweight and date of birth.22 We characterized exposure to famine during gestation by determining the gestational ages (in weeks after LMP) during which the mother was exposed to an official ration of less than 900 kcal/day. As in prior studies of the population starting in 2007,23 we considered the mother exposed in gestational weeks 1–10, 11–20, 21–30, or 31 to delivery if these gestational time-windows were entirely contained within this period. Pregnancies with LMP between 26 November 1944 and 4 March 1945 were thus considered exposed in weeks 1–10; between 18 September 1944 and 24 December 1944 in weeks 11–20; between 10 July 1944 and 15 October 1944 in weeks 21–30; and between 2 May 1944 and 24 August 1944 in week 31 to delivery. Some participants therefore could be considered exposed to famine during two adjacent 10-week periods. Individuals exposed in at least one of the 10-week gestation periods were considered to have had ‘any’ prenatal famine exposure. No individuals were exposed throughout gestation. The exposure definition outlined above represents an average exposure to famine post conception of < 900 kcal/day during an entire gestation period of 10 weeks. This definition does not cover individuals with LMPs between 5 March 1945 and 15 May 1945, who were exposed to extreme famine but for shorter periods and do not meet this definition. To evaluate the impact of shorter periods of famine on these births, we evaluated changes in DNA methylation outcomes in this group. We also explored the potential influence of the duration of famine exposure during gestation and DNA methylation at age ∼ 58 years.
Characteristics in 2003–2005
At interview and examination, we collected data on several adult characteristics that have been associated with differential DNA methylation in whole blood, including smoking,24 dietary intake of macronutrients25 or micronutrients26 and SES.27 These were evaluated as potential confounders in our analyses. Smoking was classified as never, current or past. Dietary intake in the past 12 months was ascertained from a 140-item food frequency questionnaire developed to assess dietary habits in an elderly Dutch population. This questionnaire provides estimates of macronutrient (total energy and fat, protein and carbohydrate) and micronutrient intake (folic acid, vitamin B12, and vitamin B6).28 We classified study participants on a five-level education scale, identifying individuals with primary (ages 6–12 years), lower or middle level vocational, secondary, higher vocational and university education.
DNA methylation data generation and processing
Genome-wide DNA methylation data were generated using the Illumina Infinium Human Methylation 450k BeadChip (450k array). Briefly, 500 ng of genomic DNA from whole blood isolated using the salting-out technique was bisulfite-treated using the EZ-96 DNA methylation kit (Zymo Research, Orange County, USA). DNA of study participants was treated together with that of their control sibling on 96-well plates with even distributions of exposure periods, sex ratios and mean ages per 96-well plate and 450k array. Sibling pairs were placed on the same row of the 450k array with random allocation to the left or right column. Participants without a control sibling were randomly placed on a separate set of 96-well plates with even distributions in exposure periods, sex ratios and mean ages.
The 450k arrays were measured at the Erasmus Medical Center Human Genotyping Facility in Rotterdam, The Netherlands. The quality of the generated 450k array data was assessed using both sample dependent and sample independent quality metrics using the Bioconductor package MethylAid (29) with default settings. Bisulfite conversion efficiency was assessed using the dedicated probes on the 450k array and by Sanger Sequencing four random samples per 96-well plate. To prevent loss of power due to sample swaps, a subset (N = 21) of the genotypes of single nucleotide polymorphisms (SNPs) yielded by the 450k array were compared by those measured with MASSARRAY and sample sexes were checked using X-chromosomal CpG dinucleotides (Supplementary Methods, available as Supplementary data at IJE online).
Normalization of the dataset was performed by Functional Normalization30 implemented in the minfi package31 using six principal components. All measurements with a detection P-value > 0.01 or zero intensity value were set as missing. The measurement success rate per sample was > 99%. Probes that did not map to unique genomic locations32 or with a < 95% measurement success rate were then removed.
Because of background signal and measurement characteristics, few of the CpG dinucleotides have a methylation estimate (approximated by the array β value) equal to zero or one. It is known from bisulfite sequencing data that a significant proportion of CpG dinucleotides are fully methylated or unmethylated and show no interindividual variation.14 After consultation of 17 whole genome bisulfite sequencing datasets covering the major blood cell types, namely lymphocytes, neutrophils and monocytes,33,34 such uninformative CpG dinucleotides were identified and removed (Supplementary Methods, available as Supplementary data at IJE online).
After further removal of the X and Y chromosomal probes, outlier detection was performed using principal component analysis (PCA) on all remaining probes. High-quality DNA methylation data were obtained for 944 of the 971 individuals who completed the clinical examination for 342 596 autosomal CpG dinucleotides. The first three principal components of the variation in the data account for 16.5% of the total variation (Supplementary Figures 3 and 4, available as Supplementary data at IJE online) and reflect the cellular heterogeneity of whole-blood specimens, as indicated by a high correlation between these components and imputed cell mixture proportions.35 The next 12 principal components each explained less than 1.6% of the variation and mainly reflect technical variation. Of these 944 individuals, 348 individuals were exposed during any 10-week period and 74 individuals were conceived during the famine between 5 March 1945 and 15 May 1945. As controls we used for each EWAS 160 individuals born in the same institutions in 1943 or 1947 (time-controls) and 303 same-sex siblings recruited from the individuals in the 1943, 1945–46 and 1947 births series.
Statistical analysis
We used generalized estimation equations (GEE) with a Gaussian link function to evaluate the association between DNA methylation percentage (as reflected by the 450k array β-value) and famine exposure. Six separate EWAS analyses were run, one each for exposures to a specific 10-week period of gestation, one for ‘any’ exposure during gestation and one for conceptions from March to early May 1945. Thus for each EWAS, the individuals meeting an exposure definition were compared with all 463 unexposed time- and family-controls. GEE offer a flexible statistical framework to handle within-family correlations, unaffected from confounding by unmeasured family-level factors.36 Using GEE, we controlled for correlation within sibships and we additionally adjusted for age, sex, row on the 450k array and batch type (bisulfite conversion plate and scan batch), and adjusted for cell heterogeneity by incorporating the first three principal variance components as their proxy measure. We additionally adjusted for potential confounders including smoking status, current macronutrient and micronutrient intake and socioeconomic status where noted. In view of sex-specific differences previously reported by us,13 we evaluated possible interactions between famine exposure and sex for the outcomes of interest by incorporating both interaction and main effects in the model. In the absence of statistical evidence for sex interactions, we present sex-adjusted results combining men and women. All P-values reported for the associations between DNA methylation and famine exposure are two-sided. P-values were corrected for genome-wide inflation by multiplying the standardized robust error with the square root of the inflation factor λ (Supplementary Methods, available as Supplementary data at IJE online)37 and were adjusted for multiple testing using false discovery rate correction (FDR).38 Methylation percentages in the figures and tables reflect β-value estimates. We used a 10kb-window around CpG dinucleotides associated with prenatal famine to investigate the existence of additional (nominal) significant associations. In a Dutch Reference panel39 (GoNL version 5) we found no overlap between single nucleotide polymorphisms (SNPs) and the probes measuring the CpG dinucleotides associated with prenatal famine exposure. Annotation of the genomic regions to regulatory features such as enhancers was done using ENCODE annotations.40
Results
A total of 348 individuals met our definition of being exposed during any of the four 10-week gestation periods. Of these, 73 individuals were classified as exposed during weeks 1–10 of gestation, 123 during weeks 11–20, 143 during weeks 21–30 and 128 during weeks 31 through to delivery. Some individuals meet the definition for exposure in two adjacent gestation periods. Of individuals born in 1945 or 1947 in the same institutions as the famine-exposed individuals, 160 are time-controls. The famine-exposed individuals and the time-controls had 303 unexposed same-sex siblings serving as family-controls (Table 1).
Table 1.
Individual characteristics by famine exposure in selected gestation periods
| Characteristics | Any exposure | Weeks 1–10 of gestation | Weeks 11–20 of gestationd | Weeks 21–30 of gestatione | Weeks 31-deliveryf | Time-controls: 1943 & 1947 | Family-controls: siblings |
|---|---|---|---|---|---|---|---|
| N | 348 | 73 | 123 | 143 | 128 | 160 | 303 |
| Age (y) [SD]* | 58.9 [0.5] | 58.6 [0.4] | 58.8 [0.5] | 59.0 [0.5] | 59.1 [0.5] | 59.2 (2.0) | 57.3 [6.4] |
| % male | 46 | 46.6 | 46.3 | 49.7 | 48.4 | 45.0 | 41.9 |
| Paternal SESa [SD] | 1.4 [0.8] | 1.4 [0.7] | 1.4 [0.8] | 1.4 [0.8] | 1.5 [0.9] | 1.5 [0.8] | 1.4 [0.8] |
| SESa [SD] | 2.2 [1.3] | 1.9 [1.2] | 2.2 [1.3] | 2.2 [1.2] | 2.5 [1.3] | 2.2[1.1] | 2.2 [1.2] |
| % smokersb | 25.3 | 24.7 | 27.6 | 23.8 | 24.2 | 20.2 | 22.4 |
| Current dietary intakec | |||||||
| Kcal [SD] | 2249 [628] | 2334 [612] | 2293 [719] | 2265 [684] | 2233 [574] | 2167[637] | 2168 [631] |
| Protein (g) [SD] | 96 [28] | 100 [29] | 100 [33] | 98 [29] | 94 [24] | 95[26] | 91 [26] |
| Fat (g) [SD] | 91 [33] | 94 [33] | 94 [38] | 92 [36] | 89 [29] | 86[33] | 86 [32] |
| Carbohydrates (g) [SD] | 234 [74] | 243 [73] | 234 [81] | 235 [77] | 235 [71] | 226[74] | 232 [74] |
| Folic acid (μg) [SD] | 228 [131] | 236 [73] | 230 [80] | 233 [188] | 215 [65] | 216[71] | 209 [63] |
| Vitamin B12 (μg) [SD] | 7.1 [6.2] | 7.3 [4.5] | 7.4 [5.0] | 7.4 [8.0] | 6.4 [4.1] | 6.7[4.0] | 5.9 [3.1] |
| Vitamin B6 (μg) [SD] | 1.9 [0.5] | 1.9 [0.5] | 1.9 [0.6] | 1.9 [0.5] | 1.8 [0.5] | 1.9[0.5] | 1.8 [0.5] |
aSocioeconomic status as assessed by attained educational level (1–5 scale with 1 as lowest category).
bThe percentage of current smokers.
cEstimated from food frequency questionnaire. Entries include: total energy intake (kcal); macronutrient intake of protein, fat and carbohydrates (in grams); and micronutrients intake of folic acid, vitamin B12 and vitamin B6 (in micrograms).
d24 individuals were classified as exposed in both weeks 1–10 and weeks 11–20.
e52 individuals were classified as exposed in both weeks 11–20 and 21–30.
f43 individuals were classified as exposed in both weeks 21–30 and 31-delivery.
*P < 0.01, comparing age at examination of family-controls with time-controls and famine exposed individuals. Other characteristics show no differences between exposure categories with omnibus tests across exposure categories.
Comparing the individuals with prenatal famine exposure, time-controls and family-controls, we found a difference in the mean age (P < 0.001), with sibling-controls being slightly younger than the prenatal famine-exposed individuals (Table 1). Omnibus tests showed no other differences between exposure groups for any of the remaining characteristics. There were no differences in the proportion of complete sibships for exposure and time-control groups (Supplementary Table 1, available as Supplementary data at IJE online).
DNA methylation after famine exposure in specific 10-week gestation periods
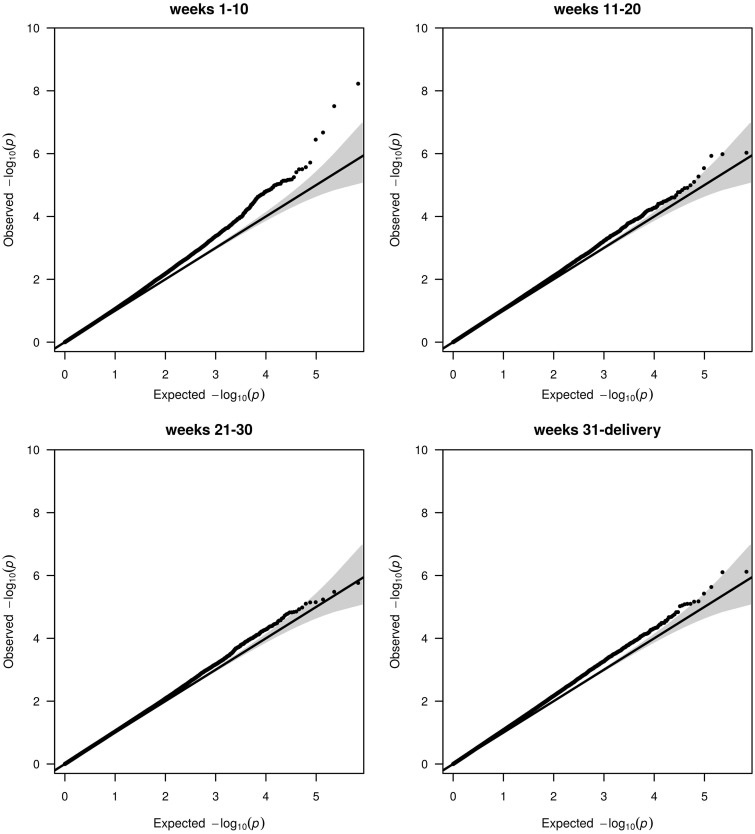
We performed a separate EWAS of famine exposure during each of the four 10-week gestation periods. Quantile-Quantile plots (QQ-plots) representing the observed vs the expected test statistic for each evaluated CpG nucleotide given the number of performed tests are shown in Figure 1. For several CpGs, nominal P-values were lower than the expected distribution of P-values for famine exposure in weeks 1–10 of gestation. In contrast, no evidence for an association between DNA methylation and famine exposure in weeks 11–20, 21–30 or 31 to delivery was observed.
Figure 1.
QQ-plots of prenatal famine exposure during specific 10-week periods. Plots depicting the Observed statistic (y-axis) with the statistic as expected by chance given the number of tests (x-axis). The 95% confidence interval of this relationship is given by the grey area around the expected line (black) for the instance that the observed statistic exactly follows the expected statistic. Each dot is the test statistic for one CpG dinucleotide. Enrichments for associations that go beyond that expected by chance can be seen as deviations upward from the expected line and 95% CI area. The P-values were corrected for the inflation factor [weeks 1–10 (λ = 1.12), weeks 11–20 (λ = 1.12), weeks 21–30 (λ = 1.12), weeks 31-delivery (λ = 1.20)] and uncorrected QQ-plots are shown in Supplementary Figure 8, available as Supplementary data at IJE online.
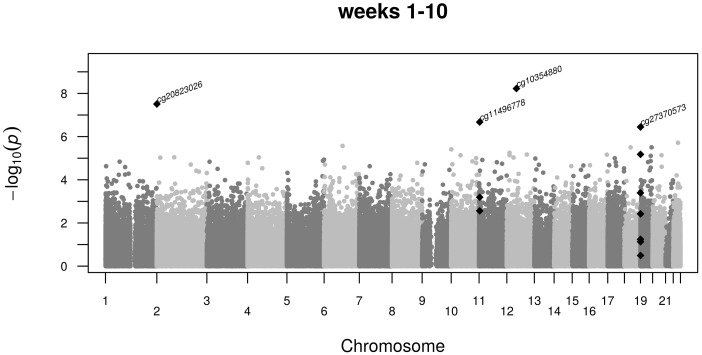
Four CpG dinucleotides (cg20823026, cg11496778, cg10354880 and cg27370573) were associated with famine exposure in weeks 1–10 of gestation after multiple testing correction (PFDR < 0.031, Figure 2). No association was found for these CpG dinucleotides in any of the other exposure periods (Table 2). The effect sizes did not change after further adjustment for variables that could potentially mediate the effects of prenatal nutrition on DNA methylation, including current lifestyle factors such as smoking and dietary characteristics and SES.
Figure 2.
Manhattan plot of specific CpG associations with prenatal famine exposure during weeks 1–10 of gestation. Shown are the −log10 P-values (y-axis) of the association between DNA methylation at single CpG dinucleotides and famine exposure along the autosomal chromosomes (x-axis). Marked by the CpG dinucleotide identifier are the CpG dinucleotides significant after multiple testing. These and adjacent nominally significant CpG dinucleotides are depicted as black diamonds.
Table 2.
CpG dinucleotides associated with famine exposure during weeks 1–10 of gestation
| CpG dinucleotide | Any exposure | Weeks 1–10 | Weeks 11–20 | Weeks 21–30 | Weeks 31-delivery | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CpG | Location (hg19) | Mean Beta-value, % (SD) controls | Estimate (95% CI)a | PFDRb | Estimate, (95% CI) | PFDR | Estimate (95% CI) | PFDR | Estimate (95% CI) | PFDR | Estimate (95% CI) | PFDR |
| cg20823026 | chr2:366,113 | 84.8 (4.8) | 0.5 (−0.1–1.1) | 0.89 | 2.3 (1.5–3.1) | 5.27x10−3 | 0.6 (−0.3–1.4) | 0.94 | 0.8 (0.0–1.5) | 0.90 | 0.4 (−0.5–1.3) | 0.95 |
| cg11496778 | chr11:3,225,076 | 26.4 (4.7) | −0.5 (−1.0–0.0) | 0.86 | −2.3 ( −3.1–−1.5) | 0.024 | −1.1 (−1.8–−0.5)b | 0.69 | −0.3 (−1.0–0.4) | 0.96 | 0.1 (−0.6–0.8) | 0.98 |
| cg10354880 | chr12:46,737,123 | 87.6 (1.3) | 0.1 (−0.1–0.2) | 0.95 | 0.7 (0.5–0.9) | 2.03x10−3 | 0.1 (−0.1–0.3) | 0.96 | 0.1 (−0.1–0.3) | 0.98 | 0.1 (−0.1–0.3) | 0.93 |
| cg27370573 | chr19:292,167 | 84.2 (4.1) | 0.7 (0.1–1.2) | 0.81 | 2.7 (1.7–3.7) | 0.031 | 0.9 (0.1 −1.7) | 0.86 | 0.3 (−0.5–1.1) | 0.97 | 0.8 (0.0–1.6) | 0.84 |
Estimates, confidence intervals and P-values are given for an analysis for famine exposure using a GEE within the dependent βeta value and. as covariates. technical batch, row on the 450k array, age, gender and principal components 1, 2 and 3.
a95% confidence interval (CI) is related to the nominal P-values.
bP-value corrected for multiple testing.
CpG cg20823026 shows a 2.3% increase in methylation [95% confidence interval (CI): 1.5 to 3.1, P = 3.1 × 10−8] among individuals exposed in weeks 1–10 compared with time- and family-controls, representing a shift of 0.50 standard deviations (SD), and is located in an intergenic region between FAM150B (genomic distance 77 kb) and TMEM18 (302 kb). CpG cg11496778 shows a 2.3% decrease in methylation (95% CI: −3.1 to −1.5, P = 2.1 × 10−7), a shift of 0.50 SD, and is located in a poised promoter distal from the OSBPL5 (distance 37 kb) and MRGPRG (distance 14kb) genes. Two adjacent CpGs show a nominally significant difference in methylation between the individuals prenatally exposed to famine and the time- and sibling controls (Supplementary Table 2, available as Supplementary data at IJE online). CpG cg10354880 shows an increase of 0.7% (95% CI: 0.5 to 0.9, P = 5.9 × 10−7), representing a shift of 0.56 SD in methylation. This CpG is located in an enhancer mapping to the downstream SLC38A2 gene (15 kb distance). CpG cg27370573 shows an increase of 2.7% (95% CI: 1.7 to 3.7, P = 3.6 × 10−7), representing a 0.68-SD higher methylation. This CpG is located in the proximal promoter of PPAP2C (<1 kb) together with adjacent CpG dinucleotides that show a nominally significant difference in methylation (Supplementary Table 3, available as Supplementary data at IJE online).
There was no interaction of famine exposure and sex, and the cell composition of blood did not affect the observed patterns of famine exposure and methylation (Supplementary Table 4, available as Supplementary data at IJE online), except for cg11496778 (OSBPL5/ MRGPRG) for which the methylation change after famine exposure became more pronounced after cellular adjustment (from −1.5% to −2.3%).
DNA methylation and any exposure to famine
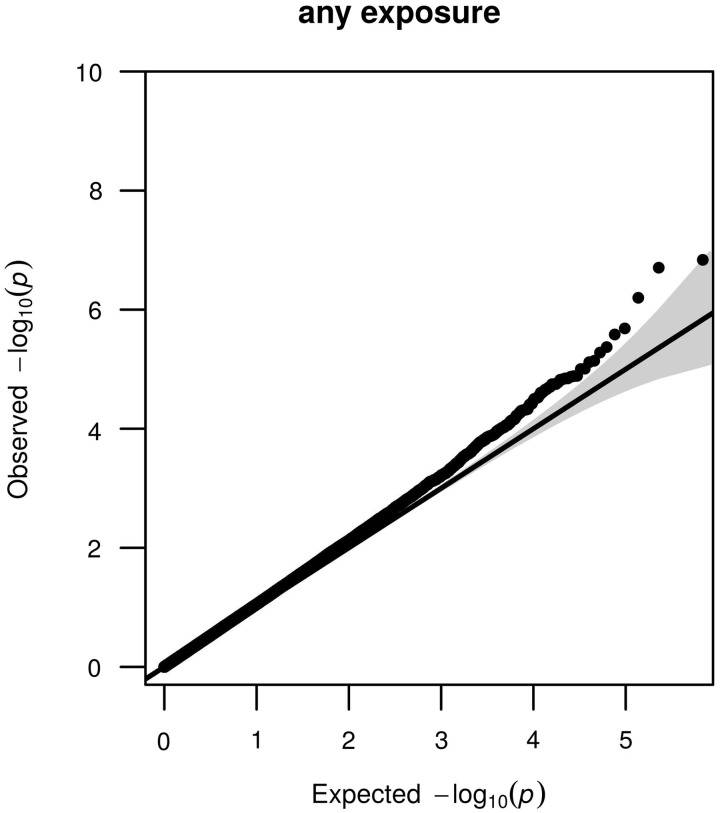
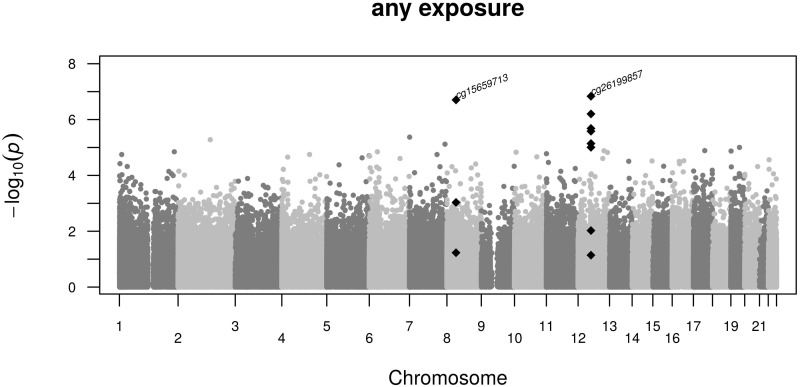
Next we performed an EWAS of famine exposure in any of the gestation periods. A QQ-plot representing the observed vs the expected test statistic given the number of performed tests for each evaluated CpG nucleotide is shown in Figure 3. Methylation differences were seen for the CpG dinucleotides cg15659713 and cg26199857 (PFDR = 0.034, Figure 4). The estimates did not change after additional adjustment for smoking, current dietary characteristics or SES. There was no interaction between famine exposure and sex. The estimates were not affected by cell composition (Supplementary Table 4, available as Supplementary data at IJE online). Neither of these two CpG dinucleotides was associated with famine exposure limited to weeks 1–10, 11–20, 21–30 or weeks 31 to delivery, and the direction and the magnitude in each of these four exposure periods were similar to the estimate for the aggregate (Table 3). We further considered whether DNA methylation of these two CpG dinucleotides was dependent on the number of weeks of famine exposure. No relation between the number of weeks of famine exposure and DNA methylation of cg15659713 and cg26199857 was found in the individuals with any exposure to famine.
Figure 3.
QQ-plots of any prenatal famine exposure. Plots depicting the Observed statistic (y-axis) with the statistic as expected by chance given the number of tests (x-axis). The 95% confidence interval of this relationship is given by the grey area around the expected line (black) for the instance that the observed statistic exactly follows the expected statistic. Each dot is the test statistic for one CpG dinucleotide. Enrichments for associations that go beyond that expected by chance can be seen as deviations upward from the expected line and 95% CI area. The P-values were corrected for the inflation factor (λ = 1.14).
Figure 4.
Manhattan plot of specific CpG associations with any prenatal famine exposure. Shown are the –log10 P-values (y-axis) of the association between DNA methylation at single CpG dinucleotides and famine exposure along the autosomal chromosomes (x-axis). Marked by the CpG dinucleotide identifier are the CpG dinucleotides significant after multiple testing. These and adjacent nominally significant CpG dinucleotides are depicted as black diamonds.
Table 3.
CpG dinucleotides associated with any prenatal famine exposure
| CpG dinucleotide | Any exposure | Weeks 1–10 | Weeks 11–20 | Weeks 21–30 | Weeks 31-delivery | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CpG | Location (hg19) | Mean Beta-value, % (SD) controls | Estimate, (95% CI)a | PFDRb | Estimate (95% CI) | PFDR | Estimate (95% CI) | PFDR | Estimate (95% CI) | PFDR | Estimate (95% CI) | PFDR |
| cg15659713 | chr8:38,586,183 | 24.2 (3.9) | 1.2 (0.8–1.7) | 0.034 | 1.1 (0.2–1.9) | 0.74 | 1.5 (0.8–2.1) | 0.47 | 1.2 (0.7–1.8) | 0.47 | 1.7 (1.1–2.3) | 0.09 |
| cg26199857 | chr12:54,764,265 | 68.1 (5.8) | 2.0 (1.3–2.7) | 0.034 | 1.9 (0.6–3.2) | 0.68 | 1.7 (0.7–2.7) | 0.68 | 1.9 (1.0–2.9) | 0.53 | 2.3 (1.3–3.4) | 0.41 |
Estimates, confidence intervals and P-values are given for an analysis for famine exposure using a GEE within the dependent βeta-value and, as covariates, technical batch, row on the 450k array, age, gender and principal components 1, 2 and 3.
a95% confidence interval (CI) is related to the nominal P-values.
bP-value corrected for multiple testing.
CpG cg15659713 shows a 1.2% increase (95% CI: 0.8 to 1.7, P = 2.0 × 10−7), representing a 0.32-SD shift in methylation which extends to two neighbouring CpG dinucleotides (Supplementary Table 5, available as Supplementary data at IJE online). These three CpG dinucleotides are located in the first exon of the TACC1 gene. CpG cg26199857 shows a 2.0% methylation increase (95% CI: 1.3 to 2.7, P = 1.5 × 10−7), representing an effect size of 0.35 SD. Six neighbouring CpG dinucleotides show a nominal significant difference in methylation (Supplementary Table 6, available as Supplementary data at IJE online). These six CpG dinucleotides are located in the last exons of ZNF385A and in the promoter region of GPR84 (6kb, which is located telomeric from ZNF385A).
Conceptions in March, April and May 1945
Our focus on famine exposures defined by 10-week gestation periods with rations averaging less than 900 kcal/day excludes individuals conceived during the famine in March, April and early May 1945. These individuals (N = 74) were conceived during extreme famine, but exposed for a shorter period in gestation and also before conception. These individuals had both pre- and post-conception famine exposure and we performed an EWAS on this group. The QQ-plot showed several CpGs with a nominal P-value lower than the expected distribution of P-values (Supplementary Figure 1, available as Supplementary data at IJE online). One CpG dinucleotide, cg23989336, was associated with pre- and post-conception famine exposure (PFDR < 0.037). No association was found however for cg23989336 in any of the 10-week gestation periods (Supplementary Table 7 available as Supplementary data at IJE online). The effect size did not change after adjustment for potentially confounding variables, including current lifestyle factors such as smoking and dietary characteristics or for SES.
CpG cg23989336 was associated with a −3.5% decrease in methylation (95% CI: −4.6 to −2.3, P = 1.0 × 10−7), representing a shift of 0.54 standard deviations (SD), and is located in an enhancer region between TMEM105 (1 kb) and SLC38A10 (15 kb). The association extended to four adjacent CpG dinucleotides (Supplementary Table 8, Supplementary Figure 2, available as Supplementary data at IJE online).
Discussion
We found that DNA methylation in four CpG dinucleotides measured at age ∼ 58 years in whole blood was associated with prenatal famine exposure during the first 10 weeks of gestation. No associations were found however with famine exposure in other 10-week periods (weeks 11–20, 21–30 or 31 to delivery). We found an association between any famine exposure and DNA methylation of two CpG dinucleotides. Individuals conceived in March, April and early May 1945 with pre- and post-conception famine exposure showed an association with DNA methylation of one CpG dinucleotide. The CpG dinucleotides identified are linked to genes with roles in growth, differentiation and metabolism.
Of the four CpG dinucleotides, CpG cg20823026 is located between FAM150B, involved in cell growth and differentiation,41 and TMEM18, a known obesity gene.42 CpG cg10354880 lies within a strong enhancer43 mapping to the SLC38A2 gene, whose function includes matching nutrient supply to fetal demand, together with IGF2,44 for which we previously reported differential methylation in a subset of this cohort.12,15 CpG dinucleotide cg11496778 is located distal from OSBPL5 which is implicated with intracellular cholesterol transport;45 and CpG dinucleotide cg27370573 lies in a CpG island in the proximal promoter of PPAP2C involved in cell cycle regulation.46
With regard to the two CpG dinucleotides associated with any famine exposure, the region around CpG dinucleotide cg26199857 overlaps several exons of ZNF385A. Moreover, ENCODE data47 indicate that the region overlaps an enhancer that interacts with the promoter of ZNF385A under the influence of CTCF protein binding.48 ZNF385A is implicated in cell division and survival upon genotoxic stress49 and early adipogenesis.50 CpG cg15659713 is located in an exon of TACC1, which is dynamically expressed during development51 and has a role in cell division52 and thyroid hormone and retinoic acid signalling cascades.53
We also found evidence for an association in the group with pre- and post-conception famine exposure. CpG dinucleotide cg23989336 overlaps an enhancer and a CpG island downstream of TMEM105, a gene of unknown function, and SLC38A10, shown to influence body size in mouse knock-out studies.54 This association was not found after prenatal famine exposure throughout any of the 10-week gestation periods, including weeks 1–10. More detailed studies on famine exposure before and around the time of conception should therefore be undertaken.
The CpG dinucleotides identified can be linked to genes in pathways involved in growth, development and lipid metabolism and are mostly located in exons and regulatory regions such as enhancers, matching findings from a previous genome-scale study of DNA methylation and early gestation famine exposure in a small subset of this population.14 In that study we measured more CpG dinucleotides (1.2 M) and more of these important regulatory regions55,56 which are sparsely covered by the 450k array. In that study we found 181 of these regions associated with famine exposure during early gestation that were not covered in our current study; overall there was an overlap of just 9000 CpG dinucleotides between studies. The preferential coverage of CpG islands and promoters by the 450k array, assessing ∼ 1.5% of all CpG dinucleotides in the genome, may explain why we only see a limited number of CpG dinucleotides associated with prenatal famine. In both studies we covered only a fraction of CpG dinucleotides in regulatory regions33 and of the 28 M CpG dinucleotides in the genome; this highlights the need for new cost-effective high-throughput platforms for assessing DNA methylation genome-wide.57 We previously studied DNA methylation of several candidate regions in a subset of the cohort with EpiTYPER.13 The LEP, INSIGF and ABCA1 promoter regions were represented by one or more CpG dinucleotides in the 450k array, and although these assays do not cover the same CpG dinucleotides, methylation of the LEP and INSIGF promoters was also associated with famine exposure during weeks 1–10 in the 450k data (data not shown).
In our current and previous studies,12–15 we found differences in DNA methylation percentages that are small in absolute terms, but that are of medium effect size (∼ 0.5 standard deviations) from an epidemiological perspective.58 Such small methylation differences are often found in epigenetic epidemiology57 and are thought to exert an effect through the modulation of gene networks59,60 or to mark other larger molecular differences in a regulatory region, such as in histone modifications.57 Animal experiments suggest that small absolute differences in methylation of medium effect size may still exert an effect.10
The mechanisms underlying DNA methylation differences from prenatal exposures await elucidation, but the observed timing-specific associations in blood may hint at an intrinsic sensitivity of newly developing tissues.61 We measured DNA methylation in whole blood, and the haematopoietic system is formed during prenatal weeks 1–10. Animal8,62 and human studies16 suggest that similar methylation changes are likely to be found in other tissues. Consultation of an external dataset suggests a broad agreement in methylation levels across six other tissues for the seven CpG dinucleotides in humans.63 We do not have tissue specimens available other than whole blood in this population and are unable to address tissue-specificity of the differential methylation we observed here. Associations with any prenatal famine exposure may have arisen by other mechanisms than those associated with early gestation, as also suggested by animal experiments,7 and may be independent of the dynamics of DNA methylation during early development. A recent study on prenatal smoking also described select methylation changes after any smoking during pregnancy.19 In contrast to our study, some of these associations showed a relation between the duration of the prenatal exposure and DNA methylation. Further studies that are able to adequately account for potential confounding by postnatal and life-course factors will be required to unravel the mechanisms involved.61
In view of previous sex-specific methylation outcomes,13 we tested for exposure by sex interactions for the loci of interest, but none were detected. We therefore combined men and women in all analyses and added sex as a covariate. Because of the recognized low power of formal interaction tests in this setting, sex-specific effects cannot be excluded but will need further evaluation in larger study samples.
Our study has other limitations. DNA methylation was measured six decades after the exposure; the differences could have been larger at birth, could have changed during life or could represent sub-clinical health differences.64 The associations persisted after adjustment for variables that could potentially mediate the effect of prenatal nutrition on DNA methylation, including current lifestyle factors such as smoking and dietary characteristics and SES. It will be interesting to study the relation between famine exposure, health outcomes and the methylation of single CpG dinucleotides or along entire pathways, as both approaches appear to be promising in relation to complex phenotypes.65 Confounding by postnatal and life-course factors remains possible, but this requires that selected confounding factors for individuals born after exposure in different gestational windows and DMPs only apply to births in the early gestation window. After prenatal smoking, DNA methylation differences present in blood at birth may persist for up to two decades in the same individuals.19 Likewise, the current DNA differences associated with prenatal famine exposure could reflect the situation at birth. More longitudinal studies, both in animal experiments and in human cohorts, will be required to gain additional insights into this important issue in epigenetic epidemiology.57 Finally, the small number of individual CpG dinucleotides associated with prenatal famine exposure may not only be related to the sparse coverage of the 450k array, but also to the limited number of famine-exposed individuals.
Although our study findings await replication, the results of candidate locus studies previously reported for this population12,13 were also sensitive to gestational diabetes,66 prenatal smoking67 and folic acid supplementation.18 Except for prenatal smoke exposure and gestational diabetes, no genome-wide studies on other prenatal exposures using the 450k array have been reported. With the exception of prenatal smoking,19 these studies have been small in terms of sample size. There was no overlap in the CpG dinucleotides differentially methylated after prenatal famine exposure and 450k array studies on prenatal smoking and gestational diabetes.19,68–71
Our study builds on a strong design. It exploits the quasi-experimental conditions of the Dutch famine that imposed severe nutrition restrictions upon previously well-fed populations, avoiding confounding by social characteristics related to nutrition in pregnancy. The famine was well defined in place and time, so that exposed and unexposed populations can be distinguished from their place and date of birth. Moreover, we enrolled same-sex siblings to serve as family-controls. This avoids confounding due to unmeasured family factors common to siblings.
In summary, we show in a genome-wide DNA methylation study that prenatal famine exposure in early, but not in mid or late gestation, affects DNA methylation at specific CpG dinucleotides. These findings provide additional evidence that the early gestation period is a critical time-window during which the prenatal environment may affect the human blood methylome. The functional implications of these findings need further exploration.
Supplementary Data
Supplementary data are available at IJE online.
Funding
This study was supported by the U.S. National Institutes of Health [AG042190 to LHL and BTH, HL067914 to LHL], and the European Union's Seventh Framework Program IDEAL [259679 to P.E.S.]. Funding to pay the Open Access publication charges for this article was provided by NIH, grant AG042190. The funders had no role in study design, data collection, analysis, decision to publish or preparation of the manuscript.
Supplementary Material
Acknowledgements
We express our gratitude to the participants of the Dutch Hunger Winter Families study and the staff of TNO Quality of Life for contact tracing. We wish to acknowledge the staff of the department of Gerontology and Geriatrics Study Center at the Leiden University Medical Center for the physical examinations and the Central Clinical Chemical Laboratory for extracting DNA.
Conflict of interest: None declared.
References
- 1.Barker DJ. The origins of the developmental origins theory. J Intern Med 2007;261:412–17. [DOI] [PubMed] [Google Scholar]
- 2.Joseph KS, Kramer MS. Review of the evidence on fetal and early childhood antecedents of adult chronic disease. Epidemiol Rev 1996;18:158–74. [DOI] [PubMed] [Google Scholar]
- 3.Stein ZA, Susser M, Saenger G, Marolla F. Famine and Human Development: The Dutch Hunger Winter of 1944-1945. New York, NY: Oxford University Press, 1975. [Google Scholar]
- 4.Lumey LH, Stein AD, Susser E. Prenatal famine and adult health. Annu Rev Public Health 2011;32:237–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 2003;33(Suppl):245–54. [DOI] [PubMed] [Google Scholar]
- 6.Waterland RA, Michels KB. Epigenetic epidemiology of the developmental origins hypothesis. Annu Rev Nutr 2007;27:363–88. [DOI] [PubMed] [Google Scholar]
- 7.Bogdarina I, Haase A, Langley-Evans S, Clark AJ. Glucocorticoid effects on the programming of AT1b angiotensin receptor gene methylation and expression in the rat. PLoS One 2010;5:e9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morgan HD, Jin XL, Li A, Whitelaw E, O'Neill C. The culture of zygotes to the blastocyst stage changes the postnatal expression of an epigentically labile allele, agouti viable yellow, in mice. Biol Reprod 2008. ;79:618–23. [DOI] [PubMed] [Google Scholar]
- 9.Guo H, Zhu P, Yan L, et al. The DNA methylation landscape of human early embryos. Nature 2014;511:606–10. [DOI] [PubMed] [Google Scholar]
- 10.Lillycrop KA, Phillips ES, Torrens C, Hanson MA, Jackson AA, Burdge GC. Feeding pregnant rats a protein-restricted diet persistently alters the methylation of specific cytosines in the hepatic PPAR alpha promoter of the offspring. Br J Nutr 2008;100:278–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKay JA, Xie L, Harris S, Wong YK, Ford D, Mathers JC. Blood as a surrogate marker for tissue-specific DNA methylation and changes due to folate depletion in post-partum female mice. Mol Nutr Food Res 2011;55:1026–35. [DOI] [PubMed] [Google Scholar]
- 12.Heijmans BT, Tobi EW, Stein AD, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A 2008;105:17046–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tobi EW, Lumey LH, Talens RP, et al. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet 2009;18:4046–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tobi EW, Goeman JJ, Monajemi R, et al. DNA methylation signatures link prenatal famine exposure to growth and metabolism. Nat Commun 2014;5:5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tobi EW, Slagboom PE, van Dongen J, et al. Prenatal famine and genetic variation are independently and additively associated with DNA methylation at regulatory loci within IGF2/H19. PLoS One 2012;7:e37933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dominguez-Salas P, Moore SE, Baker MS, et al. Maternal nutrition at conception modulates DNA methylation of human metastable epialleles. Nat Commun 2014;5:3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoyo C, Murtha AP, Schildkraut JM, et al. Methylation variation at IGF2 differentially methylated regions and maternal folic acid use before and during pregnancy. Epigenetics 2011;6:928–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steegers-Theunissen RP, Obermann-Borst SA, Kremer D, et al. Periconceptional maternal folic acid use of 400 microg per day is related to increased methylation of the IGF2 gene in the very young child. PLoS One 2009;4:e7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richmond RC, Simpkin AJ, Woodward G, et al. Prenatal exposure to maternal smoking and offspring DNA methylation across the lifecourse: Findings from the Avon Longitudinal Study of Parents and Children (ALSPAC). Hum Mol Genet 2015;24:2201–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Burger GCE, Drummond JC, Sandstead HR. Malnutrition and starvation in Western Netherlands, September 1944-July 1945.The Hague, The Netherlands: General State Printing Office,1948.
- 21.Trienekens G. The food supply in the Netherlands during the Second World War. In: Smith DF Phillips J Food, Science, Policy and Regulation in the Twentieth Century. International and Comparative Perspectives. London: Routledge, 2000. [Google Scholar]
- 22.Lumey LH, Stein AD, Kahn HS, et al. Cohort profile: the Dutch Hunger Winter families study. Int J Epidemiol 2007;36:1196–204. [DOI] [PubMed] [Google Scholar]
- 23.Stein AD, Kahn HS, Rundle A, Zybert PA, van der Pal-de Bruin, Lumey LH. Anthropometric measures in middle age after exposure to famine during gestation: evidence from the Dutch famine. Am J Clin Nutr 2007;85:869–76. [DOI] [PubMed] [Google Scholar]
- 24.Zeilinger S, Kuhnel B, Klopp N, et al. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS One 2013;8:e63812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bouchard L, Rabasa-Lhoret R, Faraj M, et al. Differential epigenomic and transcriptomic responses in subcutaneous adipose tissue between low and high responders to caloric restriction. Am J Clin Nutr 2010;91:309–20. [DOI] [PubMed] [Google Scholar]
- 26.Fiorito G, Guarrera S, Valle C, et al. B-vitamins intake, DNA-methylation of one carbon metabolism and homocysteine pathway genes and myocardial infarction risk: the EPICOR study. Nutr Metab Cardiovasc Dis 2014;24:483–88. [DOI] [PubMed] [Google Scholar]
- 27.Borghol N, Suderman M, McArdle W, et al. Associations with early-life socio-economic position in adult DNA methylation. Int J Epidemiol 2012;41:62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stein AD, Rundle A, Wada N, Goldbohm RA, Lumey LH. Associations of gestational exposure to famine with energy balance and macronutrient density of the diet at age 58 years differ according to the reference population used. J Nutr 2009;139:1555–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Iterson M, Tobi EW, Slieker RC, et al. MethylAid: visual and interactive quality control of large Illumina 450k datasets. Bioinformatics 2014;30:3435–37. [DOI] [PubMed] [Google Scholar]
- 30.Fortin JP, Labbe A, Lemire M, et al. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol 2014;15:503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014;30:1363–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen YA, Lemire M, Choufani S, et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 2013;8:203–09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ziller MJ, Gu H, Muller F, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature 2013;500:477–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams D, Altucci L, Antonarakis SE, et al. BLUEPRINT to decode the epigenetic signature written in blood. Nat Biotechnol 2012;30:224–26. [DOI] [PubMed] [Google Scholar]
- 35.Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 2012;13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frisell T, Oberg S, Kuja-Halkola R, Sjolander A. Sibling comparison designs: bias from non-shared confounders and measurement error. Epidemiology 2012;23:713–20. [DOI] [PubMed] [Google Scholar]
- 37.Devlin B, Roeder K. Genomic control for association studies. Biometrics 1999;55:997–1004. [DOI] [PubMed] [Google Scholar]
- 38.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 1995;57:289–300. [Google Scholar]
- 39.Genome of The Netherlands Consortium. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet 2014;46:818–25. [DOI] [PubMed] [Google Scholar]
- 40.Ernst J, Kheradpour P, Mikkelsen TS, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 2011;473:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang H, Pao LI, Zhou A, et al. Deorphanization of the human leukocyte tyrosine kinase (LTK) receptor by a signaling screen of the extracellular proteome. Proc Natl Acad Sci U S A 2014;111:15741–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pei YF, Zhang L, Liu Y, et al. Meta-analysis of genome-wide association data identifies novel susceptibility loci for obesity. Hum Mol Genet 2014;23:820–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chadwick LH. The NIH Roadmap Epigenomics Program data resource. Epigenomics 2012;4:317–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Constancia M, Angiolini E, Sandovici I, et al. Adaptation of nutrient supply to fetal demand in the mouse involves interaction between the Igf2 gene and placental transporter systems. Proc Natl Acad Sci U S A 2005;102:19219–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du X, Kumar J, Ferguson C, et al. A role for oxysterol-binding protein-related protein 5 in endosomal cholesterol trafficking. J Cell Biol 2011;192:121–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morris KE, Schang LM, Brindley DN. Lipid phosphate phosphatase-2 activity regulates S-phase entry of the cell cycle in Rat2 fibroblasts. J Biol Chem 2006;281:9297–306. [DOI] [PubMed] [Google Scholar]
- 47.Heidari N, Phanstiel DH, He C, et al. Genome-wide map of regulatory interactions in the human genome. Genome Res 2014;24:1905–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell 2009;137:1194–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Das S, Raj L, Zhao B, Kimura Y, et al. Hzf determines cell survival upon genotoxic stress by modulating p53 transactivation. Cell 2007;130:624–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kawagishi H, Wakoh T, Uno H, et al. Hzf regulates adipogenesis through translational control of C/EBPalpha. EMBO J 2008;27:1481–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lauffart B, Dimatteo A, Vaughan MM, Cincotta MA, Black JD, Still IH. Temporal and spatial expression of TACC1 in the mouse and human. Dev Dyn 2006;235:1638–47. [DOI] [PubMed] [Google Scholar]
- 52.Lauffart B, Howell SJ, Tasch JE, Cowell JK, Still IH. Interaction of the transforming acidic coiled-coil 1 (TACC1) protein with ch-TOG and GAS41/NuBI1 suggests multiple TACC1-containing protein complexes in human cells. Biochem J 2002;363(Pt 1):195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guyot R, Vincent S, Bertin J, Samarut J, Ravel-Chapuis P. The transforming acidic coiled coil (TACC1) protein modulates the transcriptional activity of the nuclear receptors TR and RAR. BMC Mol Biol 2010;11:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skarnes WC, Rosen B, West AP, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 2011;474:337–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siggens L, Ekwall K. Epigenetics, chromatin and genome organization: recent advances from the ENCODE project. J Intern Med 2014;276:201–14. [DOI] [PubMed] [Google Scholar]
- 56.Thurman RE, Rynes E, Humbert R, et al. The accessible chromatin landscape of the human genome. Nature 2012;489:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mill J, Heijmans BT. From promises to practical strategies in epigenetic epidemiology. Nat Rev Genet 201314:585–94. [DOI] [PubMed] [Google Scholar]
- 58.Cohen J. Statistical Power Analysis for the Behavioral Sciences. 2nd edn. New York, NY: Hillsdale, 1988. [Google Scholar]
- 59.Jiao Y, Widschwendter M, Teschendorff AE. A systems-level integrative framework for genome-wide DNA methylation and gene expression data identifies differential gene expression modules under epigenetic control. Bioinformatics 2014;30:2360–66. [DOI] [PubMed] [Google Scholar]
- 60.Stoger R. The thrifty epigenotype: an acquired and heritable predisposition for obesity and diabetes? Bioessays 2008;30:156–66. [DOI] [PubMed] [Google Scholar]
- 61.Heijmans BT, Tobi EW, Lumey LH, Slagboom PE. The epigenome: archive of the prenatal environment. Epigenetics 2009;4:526–31. [DOI] [PubMed] [Google Scholar]
- 62.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol 2003;23:5293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Slieker RC, Bos SD, Goeman JJ, et al. Identification and systematic annotation of tissue-specific differentially methylated regions using the Illumina 450k array. Epigenetics Chromatin 2013;6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heijmans BT, Mill J. Commentary: The seven plagues of epigenetic epidemiology. Int J Epidemiol 2012;41:74–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pidsley R, Viana J, Hannon E, et al. Methylomic profiling of human brain tissue supports a neurodevelopmental origin for schizophrenia. Genome Biol 2014;15:483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bouchard L, Thibault S, Guay SP, et al. Leptin gene epigenetic adaptation to impaired glucose metabolism during pregnancy. Diabetes Care 2010;33:2436–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Murphy SK, Adigun A, Huang Z, et al. Gender-specific methylation differences in relation to prenatal exposure to cigarette smoke. Gene 2012;494:36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Finer S, Mathews C, Lowe R, et al. Maternal gestational diabetes is associated with genome-wide DNA methylation variation in placenta and cord blood of exposed offspring. Hum Mol Genet 2015 Jan 29. 10.1093/hmg/ddv013. [DOI] [PubMed] [Google Scholar]
- 69.Joubert BR, Haberg SE, Bell DA, et al. Maternal smoking and DNA methylation in newborns: in utero effect or epigenetic inheritance? Cancer Epidemiol Biomarkers Prev 2014;23:1007–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Quilter CR, Cooper WN, Cliffe KM, et al. Impact on offspring methylation patterns of maternal gestational diabetes mellitus and intrauterine growth restraint suggest common genes and pathways linked to subsequent type 2 diabetes risk. FASEB J 2014;28:4868–79. [DOI] [PubMed] [Google Scholar]
- 71.Ruchat SM, Houde AA, Voisin G, et al. Gestational diabetes mellitus epigenetically affects genes predominantly involved in metabolic diseases. Epigenetics 2013;8:935–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.