Highlight
A mitochondrial protein–protein interaction network in wheat explains why microspores suffered from severe oxidative stress during pollen development, triggered premature tapetal PCD, and consequently resulted in pollen abortion.
Key words: Male sterility, mitochondrion proteomics, oxidative stress, protein network, tapetal programmed cell death, wheat.
Abstract
Plant male sterility has often been associated with mitochondrial dysfunction; however, the mechanism in wheat (Triticum aestivum L.) has not been elucidated. This study set out to probe the mechanism of physiological male sterility (PHYMS) induced by the chemical hybridizing agent (CHA)-SQ-1, and cytoplasmic male sterility (CMS) of wheat at the proteomic level. A total of 71 differentially expressed mitochondrial proteins were found to be involved in pollen abortion and further identified by MALDI-TOF/TOF MS (matrix-assisted laser desorption/ionization-time of fight/time of flight mass spectrometry). These proteins were implicated in different cellular responses and metabolic processes, with obvious functional tendencies toward the tricarboxylic acid cycle, the mitochondrial electron transport chain, protein synthesis and degradation, oxidation stress, the cell division cycle, and epigenetics. Interactions between identified proteins were demonstrated by bioinformatics analysis, enabling a more complete insight into biological pathways involved in anther abortion and pollen defects. Accordingly, a mitochondria-mediated male sterility protein network in wheat is proposed; this network was further confirmed by physiological data, RT-PCR (real-time PCR), and TUNEL (terminal deoxynucleotidyl transferase-mediated dUTP nick end labelling) assay. The results provide intriguing insights into the metabolic pathway of anther abortion induced by CHA-SQ-1 and also give useful clues to identify the crucial proteins of PHYMS and CMS in wheat.
Introduction
Plant male sterility has been observed in numerous species (Schnable and Wise, 1998; Aalto et al., 2013; Bachtrog et al., 2014) since it was first recorded by Kölreuter in 1763. It is characterized by the absence or non-function of plant pollen grains, or the inability of plants to produce or release functional pollen grains. Male-sterile mutants (including physiological male-sterility; PHYMS) are of tremendous value as genetic tools in male development research and the use of heterosis (Eckardt, 2006; Ba et al., 2014a). Despite it being suspected that it is closely related to mitochondrial function (Linke and Borner, 2005; J.W. Wang et al., 2013; Heng et al., 2014; Touzet and Meyer, 2014), the genetic and molecular mechanisms of male sterility in wheat remain to be further elucidated.
Mitochondria play a pivotal role in energy metabolism and the maintenance of cellular homeostasis, and their dysfunction is associated with male sterility in plants (Eckardt, 2006; Kumar et al., 2012; Chen and Liu, 2014; Horn et al., 2014; Hu et al., 2014; Touzet and Meyer, 2014). More recently, with the mitochondrial genome sequence completely elucidated in some higher plant species, there has been an increased application of genomic approaches to study anther development and pollen reproduction, for example in the monocotyledonous plants Oryza sativa (rice) (Notsu et al., 2002), Zea mays (maize) (Clifton et al., 2004), and Triticum species (wheat) (Ogihara et al., 2005), and dicotyledonous plants such as Arabidopsis thaliana (Unseld et al., 1997), Brassica napus (Handa, 2003), and Nicotiana tabacum (tobacco) (Sugiyama et al., 2005). Many studies suggest that the mitochondrial genome is the carrier of fertility-related genes (Hanson and Bentolila, 2004; Chen et al., 2010; Luo et al., 2013; Tang et al., 2014). Some of these genes and/or open reading frames (ORFs) encode pollen abortion-related proteins, such as ORF256 (7kDa) (Song and Hedgcoth, 1994), ORF13 (13kDa) (Klein et al., 2005), ORF138 (19kDa) (Yasumoto et al., 2009), ORF288 (32kDa) (Jing et al., 2012), ORF79 (8.9kDa) (Wang et al., 2006; Luan et al., 2013), PCF (25kDa) (Bentolila et al., 2002), and WA352 protein (Luo et al., 2013; Tang et al., 2014). However, how these proteins mediate metabolic-induced male sterility still needs to be elucidated, and the protein–protein interaction network remains largely unexplored, especially in wheat.
In the current study, physiological male-sterile Xinong 1376 (PHYMS-XN1376) plants (Zhang et al., 2013; Ba et al., 2014a, b; Liu et al., 2014), cytoplasmic male sterile Xinong 1376 (CMS-XN1376) plants, and their maintainer line (MF-XN1376) (Chen et al., 2009, 2010) were used. The isolation of floret mitochondria from wheat using Percoll density gradient methods is described. This is followed by the application of two-dimensional electrophoresis (2-DE) in combination with tandem mass spectrometry (MS/MS) analysis to compare the different expression patterns of floret mitochondrial proteins at the early uninucleate stage and the trinucleate stage. Seventy-one mitochondrial differentially expressed proteins (DEPs) were found to be involved in pollen abortion. Identification of these proteins combined with their changes in abundance as well as the impact of pollen development establishes a close linkage between mitochondria and male sterility. In the present study, a global view of the ubiquitous cellular changes associated with pollen abortion is revealed, and the existence of an intimate protein network elicited by mitochondrial mutation in male-sterile wheat is suggested. This network was further confirmed with other molecular biology techniques. The results provide insights into the molecular events of male sterility.
Materials and methods
Plant material and treatment
PHYMS-XN1376 plants were treated with the new type of chemical hybridizing agent (CHA)-SQ-1; it has broad-spectrum properties and can lead to complete male sterility, as has been shown through pharmacodynamic experimental field tests. It also has no side effects on agronomic traits. Currently, SQ-1 has been used to produce hybrid seeds on a large scale, as has been previously described (Ba et al., 2014a; Song et al., 2014). The PHYMS-XN1376, CMS-XN1376, and MF-XN1376 plants with the same nuclear background (T. aestivum nucleus) were grown conventionally in wheat fields at the experimental station of the Northwest Agriculture and Forestry University, Yangling, China (108°E, 34°15’N). The pollen developmental stages have been described previously (Song et al., 2014). Florets from which glumes and awns were removed (at the early uninucleate stage and the trinucleate stage, respectively) were harvested for isolation of mitochondria. Anthers at sequential stages (early uninucleate stage, binucleate stage, and trinucleate stage, respectively) were also collected.
Phenotypic characterization
Plant materials were photographed with a Nikon E995 digital camera (Nikon, Japan) mounted on a Motic K400 dissecting microscope (Preiser Scientific, Louisville, KY, USA). Fresh microspores were stained with 1% acetocarmine and 2% iodine–potassium iodide (2% I2–KI). For scanning electron microscopy, anthers were collected and processed essentially as described by Xu et al. (2014) and Zhang et al. (2010), and observed with a JSM-6360LV scanning electron microscope (JEOL).
Preparation of wheat mitochondria
Mitochondria were isolated from florets according to Chen et al. (2010) with minor modifications. Briefly, ~70g (fresh weight) of florets were homogenized with 700ml of ice-cold homogenization solution [0.3M mannitol, 50mM Na4P2O7, 2mM EGTA, 0.5% (w/v) polyvinylpyrrolidone (PVP)-40, 0.5% (w/v) bovine seum albumin (BSA), 20mM ascorbate, pH 8.0] using a pre-cooled Waring blender. After filtering through four layers of Miracloth (Calbiochem, San Diego, CA, USA), the filtrates were centrifuged at 3000 g for 15min, and then the supernatant was centrifuged at 18 000 g for 20min. The resultant organelle pellet was resuspended in 300ml of wash buffer [0.3M mannitol, 10mM TES-NaOH (pH 7.5), and 0.1% (w/v) BSA] with the aid of a clean soft-bristle paint brush, and the two centrifugation steps were repeated to produce a washed organelle pellet. The final organelle pellet was resuspended in 10ml of wash buffer and loaded onto a three-step Percoll gradient composed of, from bottom to top, 40% (v/v) Percoll, 24% (v/v) Percoll, and 20% (v/v) Percoll, each Percoll solution containing 0.3M mannitol, 0.1% (w/v) BSA, and 10mM TES-NaOH (pH 7.5), and centrifuged at 40 000 g for 50min. The light yellow-brown bands of mitochondria at the 40% and 24% interfaces were collected with an injection needle and washed through 5× dilutions in wash medium, and then centrifuged at 20 000 g for 10min. This was further purified by centrifugation at 40 000 g for 30min in a one-step Percoll gradient [28% Percoll in buffer containing 0.3M sucrose, 10mM TES-KOH (pH 7.5), and 0.1% (w/v) BSA]. The upper mitochondrial fraction was collected, washed twice by 1× dilutions in wash buffer (minus 0.1% BSA), and centrifuged at 20 000 g for 10min. The mitochondria-enriched pellet was resuspended in ~500 μl of wash buffer (minus 0.1% BSA), and used for the isolation of mitochondrial proteins. Mitochondrial activity was stained with 0.01M Janus green B as described previously (Chen et al., 2010), the integrity was assessed using a negative stain for transmission electron microscopy (TEM; Hitachi HT7700, Tokyo, Japan).
Two-dimensional gel electrophoresis
2-DE was performed following the method of Ye et al. (2013). Mitochondria were solubilized in 500 μl of lysis solution containing 7M urea, 2M thiourea, 4% (w/v) CHAPS, 65mM dithiothreitol (DTT), 0.5% (v/v) Bio-Lyte (pH 4–7), and 0.001% (w/v) bromphenol blue. After determining the protein concentration using the Bradford method as described by Zuo and Lundahl (2000), ~160 μg of mitochondrial protein was separated by loading the sample on a 17cm pH 4–7 linear immobilized pH gradient (IPG) strip (Bio-Rad) and actively rehydrated at 50V for 12h at 20 °C. Subsequently, focusing was performed using the IPGphor Isoelectric Focusing System apparatus under the following conditions: constant power (50 μA per IPG strip) at 250V for 1h, 500V for 1h, 1000V for 1h, 8000V for 4h, and 8000V for a total of 80 000 V-h (17cm, pH 4–7). The second electrophoretic dimension took place using 11% SDS–PAGE. Protein spots were visualized by silver staining.
Gel imaging and data analysis
Protein spots were analysed with PDQuest Software, version 8.0.1 (Bio-Rad) according to the manufacturer’s instructions. Briefly, the images were initially processed through transformation, filtering, automated spot detection, normalization, and matching. All protein spots detected in-gel were matched to the corresponding spots of the master gel (Euns-MF) and each spot density was normalized against the whole gel densities. The volume of the spot corresponded to the amount of protein expressed. Normalized spot volumes were used to compare different samples statistically and to determine fold change values. Only the spots altered >1.3-fold (P≤0.05) were considered as DEPs.
In-gel digestion and MALDI-TOF/MALDI-TOF-TOF MS analysis
DEPs detected in silver-stained gels were selected and excised manually for protein identification. In-gel digestion of protein spots was essentially performed according to the method of Ye et al. (2013). Matrix-assisted laser desorption/ionization (MALDI) samples were prepared according to a thin layer method as described by Gobom et al. (2001). Mass spectra were collected with a 5800 MALDI time of flight (TOF)/TOF™ analyser and analysed using TOF/TOF™ Series ExplorerTM Software V4.1.0 (AB Sciex, Foster City, CA, USA). Mass spectrometry (MS) spectra were recorded in a mass range from 700Da to 4000Da with a focus mass of 1700Da. For one main MS spectrum, 15 subspectra with 200 shots per subspectrum were accumulated, and for the MS/MS spectrum up to 25 subspectra with 250 shots per subspectrum were accumulated. Peptide mass fingerprinting (PMF) and MS/MS data were used to derive protein identity using the MASCOT search engine (Matrix Science) applied to the NCBInr or Swiss-Prot databases. The main parameters were set as follows: S/N ≥3.0; fixed modification, carbamidomethyl (Cys); variable modification, Gln→pyro-Glu (N-term Q), and oxidation (Met); maximum number of missing cleavages, 1; MS tolerance, ±100 ppm.; and MS/MS tolerance, ±0.3Da. Other search parameters are stated in the protein identification tables (Supplementary Table S1 available at JXB online).
Bioinformatic analysis of identified proteins
The function of the identified proteins was elucidated by using the gene index accompanied Uniprot accession number as input for the Uniprot database (http://www.uniprot.org). Hierarchical clustering was performed with the log-transformed data using Genesis 1.7.6 software. To better understand functions and interactions of identified proteins, a protein–protein interaction network was predicted. All identified proteins were blasted against the A. thaliana TAIR10 (http://www.arabidopsis.org/) and rice protein databases with the intention of obtaining annotated protein entries for protein–protein interaction tools (Jacoby et al., 2010). Interactions of the identified proteins were constructed with the online analysis tool STRING 9.1 (Szklarczyk et al., 2015). Cellular component and biological processes were predicted by Cytoscape plugin BiNGO 3.02 (Maere et al., 2005). The search parameters, TAIR and rice homologous proteins are listed in Supplementary Table S2 at JXB online.
Respiratory activity measurements
The activity of the cytochrome oxidase pathway (COP; V cyt) in anther was calculated by multiplying the total respiration rate (V t) and measured using the method of Vanlerberghe and McIntosh (1996).
Measurement of physiological indexes
O2 – and H2O2 content, and superoxide dismutase (SOD), catalase (CAT), and guaiacol peroxidase (POD) activity were established using the method of Gajewska and Sklodowska (2007). Lipid peroxidation was determined by estimating the malondialdehyde (MDA) content according to Hameed et al. (2012, 2014).
RT-PCR analysis
All primers are listed in Supplementary Table S3 at JXB online. Total RNA was isolated from anthers at different stages with the Trizol Reagent Kit (Invitrogen, Carlsbad, CA, USA) and subjected to first-strand cDNA synthesis using a PrimeScript™ RT reagent Kit (Takara Bio, Tokyo, Japan), according to the manufacturer’s protocol. To obtain reliable results, the Ct value was obtained from CFX96™ Real-Time PCR Detection System (Bio-Rad) using SYBR Premix EX Taq (Takara Bio). The geNorm and BestKeeper method algorithms (RefFinder, http://www.leonxie.com/referencegene.php?type=reference) were used to select the most suitable reference gene from the seven candidates. Both programs demonstrated that Actin was the best reference gene for quantifying gene expression (Supplementary Fig. S1). The RT-PCR was performed with TaKaRa Ex Taq DNA polymerase (Takara Bio) for 37 cycles of denaturation for 40 s at 94 °C, annealing for 40 s at 50–60 °C, and extension for 30 s at 72 °C, followed by a final extension for 10min at 72 °C. The PCR products were identified by 2% (w/v) agarose gels and visualized by ethidium bromide staining.
TUNEL assay
Anther sections (6 μm) were washed in phosphate-buffered saline (PBS) for 5min and fixed in 4% (w/v) paraformaldehyde in PBS for 15min, and then incubated in 20 μg ml–1 proteinase K (Promega) in PBS for 10min. Terminal deoxynucleotidyl transferase-mediated dUTP nick end labelling (TUNEL) was performed in a humid chamber for 60min in the dark at 37 °C using a Dead End Fluorometric TUNEL Kit (Promega, Madison, WI, USA) following the manufacturer’s instructions. All reactions were counterstained with 1 μg ml–1 propidium iodide (PI; Promega) in PBS for 15min to label all nuclei. Samples were analysed under a fluorescence confocal scanner microscope (A1R; Nikon, Tokyo, Japan). The green fluorescence of fluorescein (TUNEL signal) and red fluorescence of PI were analysed at 488nm (excitation) and 520nm (detection), and 488nm (excitation) and 620nm (detection), respectively.
Results
Validation of morphological defects in male-sterile mutants
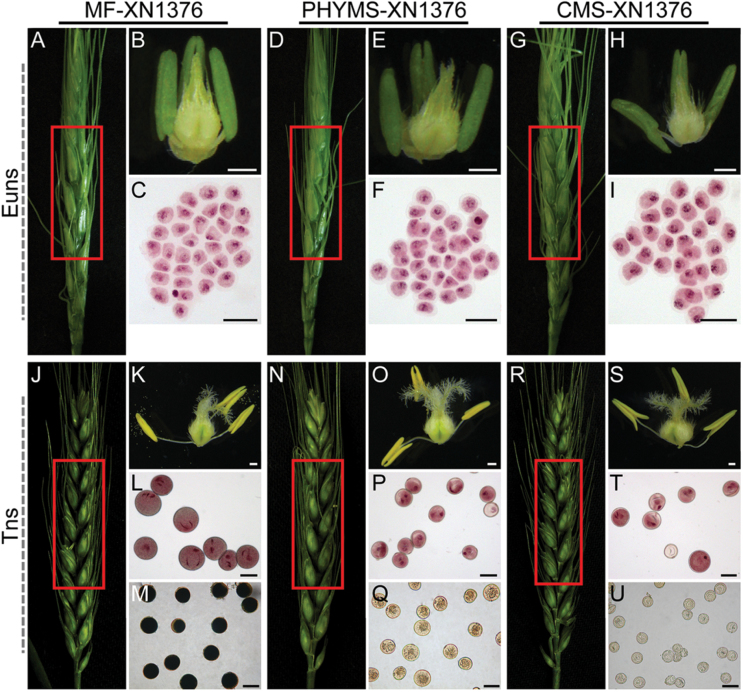
The morphological features of MF-XN1376, PHYMS- XN1376, and CMS-XN1376 plants were compared. All of these materials have the same nuclear background (T. aestivum nucleus). Compared with MF-XN1376, PHYMS-XN1376 and CMS-XN1376 showed normal spike and floral development but failed to produce any viable pollen at different stages of pollen development (Fig. 1A, D, G, J, N, R); however, the pistils of PHYMS-XN1376 and CMS-XN1376 were normal and able to produce normal seeds when they were backcrossed with fertile pollen (Fig. 1B, E, H, K, O, S). At the early uninucleate stage, there were no visible differences in pollen among the three samples after staining with 1% acetocarmine (Fig. 1C, F, I); however, the anthers of CMS-XN1376 were detectably different from the others (Fig. 1B, E, H), and there were no visible differences between MF-XN1376 and PHYMS-XN1376 (Fig. 1B, E). At the trinucleate stage, compared with MF-XN1376 anthers, the sterile anthers from both PHYMS-XN1376 and CMS-XN1376 plants were small and without mature pollen grains (Fig. 1K, O, S; Supplementary Fig. S2 at JXB online). Futhermore, fertile pollen grains of MF-XN1376 could be clearly distinguished from the sterile pollen of PHYMS-XN1376 and CMS-XN1376 by staining with 1% acetocarmine (Fig. 1L, P, T) and 2% I2–KI (Fig. 1M, Q, U). These results indicated that PHYMS-XN1376 and CMS-XN1376 plants were 100% pollen sterile.
Fig. 1.
Comparison of morphological features of PHYMS-XN1376, CMS-XN1376, and their maintainer line MF-XN1376. Comparison of spikes (A, D, G, J, N, R), and anthers and pistils (B, E, H, K, O, S) of CMS-XN1376, PHYMS-XN1376, and MF-XN1376 plants. The 1% acetocarmine (C, F, I, L, P, T) and 2% I2–KI (M, Q, U) staining of pollen grains. Euns, the early uninucleate stage; Tns, the trinucleate stage. Scale bars=1mm (B, E, H, K, O, S), 100 μm (C, F, I, L, M, P, Q, T, U).
Isolation of high-purity mitochondria from wheat florets
The isolation of pure organellar fractions from crude source material is the critical phase of any subproteome analysis. It was previously established that very pure mitochondria can be obtained from wheat (Chen et al., 2010). This established procedure was employed for the isolation of mitochondria with a recognized low contamination level. The function and integrity of the purified mitochondria were also assayed to ensure that proteins were not being lost through rupture during isolation and that key functions were maintained. Photomicrographs of mitochondria-enriched fractions generated with Percoll gradients are presented in Supplementary Fig. S3 at JXB online. The activity of the purified mitochondria was visualized using Janus green B staining under a light microscope (Supplementary Fig. S3A); they retained high activity after the 28% Percoll self-forming density centrifugation. The structural integrity of the purified mitochondria was confirmed by electron microscopy where most appeared morphologically intact (Supplementary Fig. S3B, C).
Establishment and analysis of mitochondrial proteome maps for male-sterile mutants and their maintainer line
Three independent samples of mitochondrial proteins isolated from wheat were separated by 2-DE (Supplementary Fig. S4 at JXB online). From a spot-to-spot comparison and statistical analysis, a total of 109, 105, and 128 reproducible protein spots were detected at the early uninucleate stage from MF-XN1376, PHYMS-XN1376, and CMS-XN1376 plants, respectively (Table 1; Supplementary Fig. S4A–C). The 2-DE gels also detected 129, 180, and 159 reproducible protein spots at the trinucleate stage in MF-XN1376, PHYMS-XN1376, and CMS-XN1376 plants, respectively (Table 1; Supplementary Fig. S4D–F). Table 1 presents the comparison of mitochondrial proteomics using different materials and an analysis of how these spots overlapped. Not surprisingly, there were large overlaps between these spots. In the same stages of these three materials, 96 spots overlapped at the early uninucleate stage in all three materials and 127 spots overlapped at the trinucleate stage. Additionally, in the same materials of these two stages, 101 spots in MF-XN1376, 94 spots in PHYMS-XN1376, and 119 spots in CMS-XN1376 overlapped between the two stages. Overall, 92 overlapping spots were seen in all of the 2-DE gels (Table 1; Supplementary Fig. S4).
Table 1.
Comparison of mitochondrial proteomics at the early uninucleate and trinucleate stages of the MF-XN1376, PHYMS-XN1376, and CMS-XN1376 varieties of wheat
| Pollen developmental stages | MF-XN1376 | PHYMS-XN1376 | CMS-XN1376 | Overlapped |
|---|---|---|---|---|
| Euns | 109 | 105 | 128 | 96 |
| Tns | 129 | 180 | 159 | 127 |
| Overlapped | 101 | 94 | 119 | 92 |
Overlapped; proteins showing the same expression patterns in both the early uninucleate stage (Euns) and trinucleate stage (Tns) of PHYMS-XN1376, CMS-XN1376, and MF-XN1376, respectively, or among MF-XN1376, PHYMS-XN1376, and CMS-XN1376 at the early uninucleate stage or the trinucleate stage.
Differentially expressed proteins among male-sterile mutants and their maintainer line
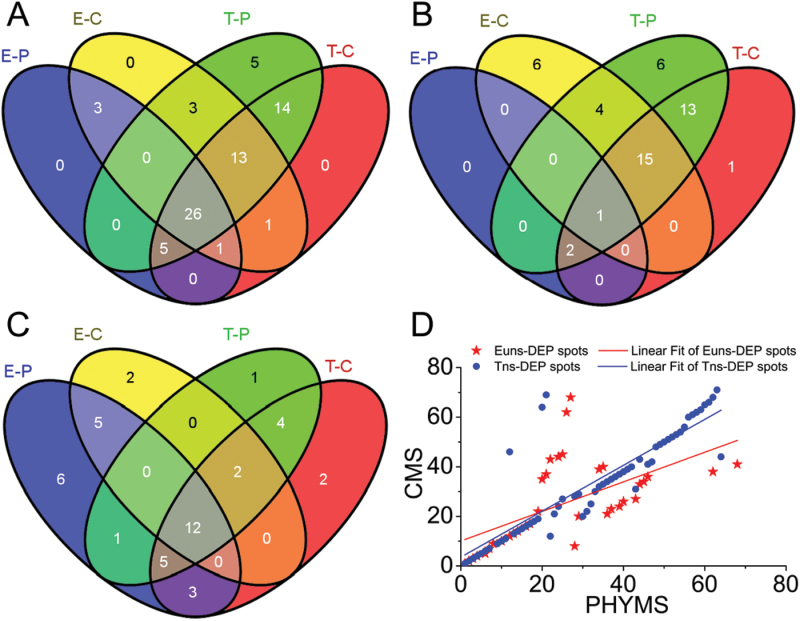
To determine the proteins whose abundance changed in male-sterile mutants, a threshold limit of 1.3-fold (P≤0.05) was set in this study as previously reported (Shen et al., 2009). Statistical analysis of 2-DE gels revealed that 71 protein spots (numbered from 1 to 71) were differentially expressed among MF-XN1376, PHYMS-XN1376, and CMS-XN1376 plants in both the early uninucleate stage and the trinucleate stage (Supplementary Figs S4, S5; Tables S4, S5 at JXB online). Of these, at the early uninucleate stage, there were 35 DEPs in PHYMS-XN1376 (three up- and 32 down-regulated) and 47 DEPs in CMS-XN1376 (21 up- and 26 down-regulated) (Fig. 2A–C; Supplementary Table S5). At the trinucleate stage, there were 66 DEPs in PHYMS-XN1376 (41 up- and 25 down-regulated) and 60 DEPs in CMS-XN1376 (32 up- and 28 down-regulated) (Fig. 2A–C; Supplementary Table S5). Additionally, overlapping DEPs were detected in all of the male-sterile plants, from 30 at the early uninucleate stage up to 58 at the trinucleate stage (Fig. 2A; Supplementary Table S5), and had a corresponding Pearson correlation coefficient from 0.33 up to 0.73 with a P-value <0.01 (Fig. 2D). Therefore, these observations document the changes in abundance of these DEPs along with the occurrence of pollen abortion. These changes show that there are different ways to induce male sterility under mitochondrial protein regulation in the initial stage of anther abortion, and ultimately this regulation makes these ways converge.
Fig. 2.
Venn diagram and scatter plot analysis of DEP spots in PHYMS-XN1376, CMS-XN1376, and MF-XN1376. The numbers of DEP spots which are up- or down-regulated are shown in the different segments; more details are presented in Supplementary Table S5 at JXB online. (A) All of the DEP spots; (B) the up-regulated protein spots; (C) the down-regulated protein spots. E/T-P, the early uninucleate stage/trinucleate stage of PHYMS-1376; E/T-C, the early uninucleate stage/trinucleate stage of CMS-XN1376. (D) The PHYMS-XN1376/CMS-XN1376 ratio of each DEP at the early uninucleate and trinucleate stages from PHYMS-XN1376 is plotted on the x-axis and correspondingly that from CMS-XN1376 is plotted on the y-axis.
Identification and functional classification of DEPs
In order to analyse further the physiological and biochemical properties of different proteins and their relationship to the abortive process, a total of 71 DEPs were analysed by MALDI-TOF/TOF MS (Supplementary Tables S1, S6 at JXB online). These proteins, associated with pollen development and tapetal activities in other plants, were found to be differentially expressed in PMS-XN1376 and CMS-XN1376 plants, as shown in Supplementary Tables S1 and S6. Among the 71 identified spots, 68 (~96%) have been functionally annotated in the current database, whereas three (spots 69–71) were annotated as unknown. To annotate these identities, their sequences were used as queries to search for homologues by BLASTP (NCBI). The corresponding homologues with the highest similarity are listed in Supplementary Table S7. All three proteins shared at least 80% positive identity with their homologues at the amino acid level, suggesting that they may have similar function. However, about one-third of the identified proteins were detected in multiple spots with different pIs or molecular masses (Supplementary Table S1), implying the existence of isoforms and post-translational modification.
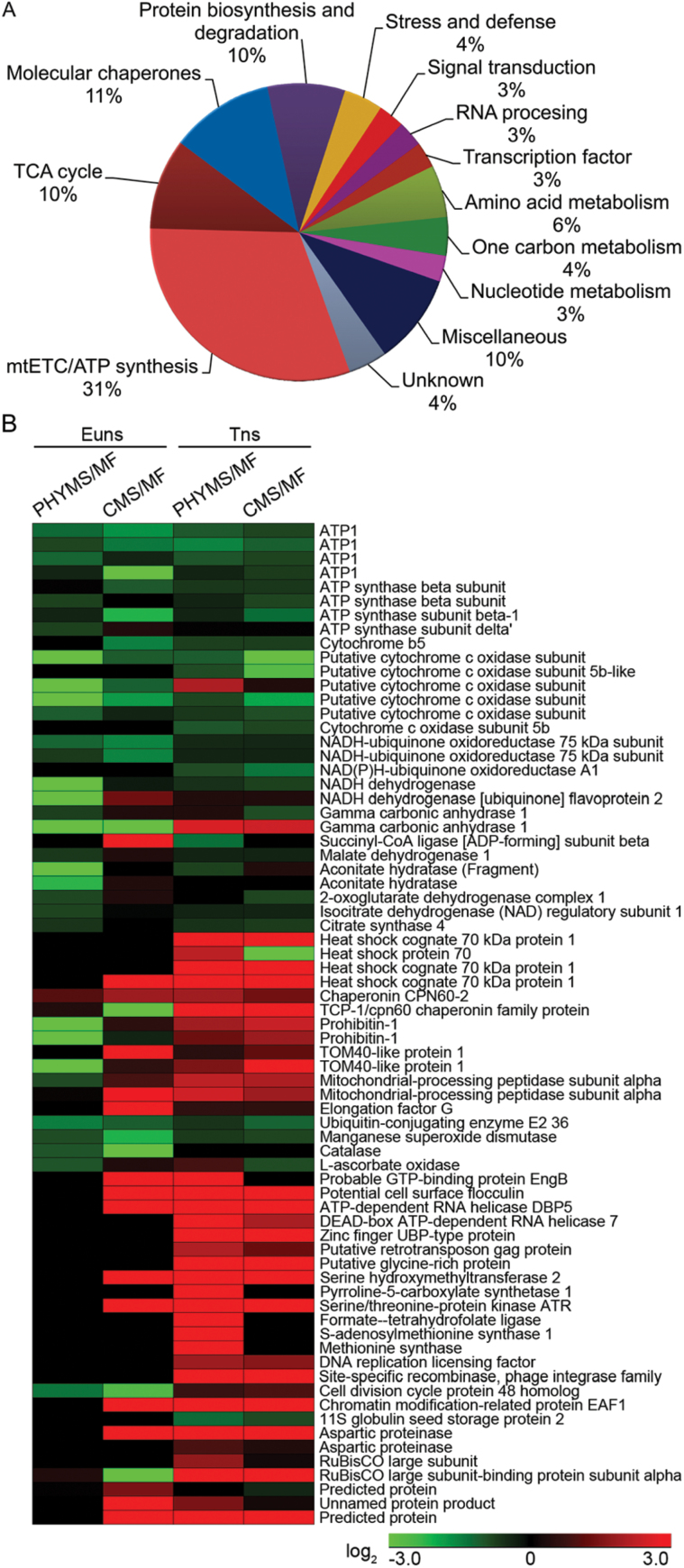
Furthermore, based on the biological process of each gene product according to annotations in these databases coupled with the metabolic and functional features of pollen, all of these identities were classified into 12 functional groups (Fig. 3A; Supplementary Table S1 at JXB online), which covered a wide range of pathways and functions, including mitochondrial electron transport (mtETC) and ATP synthesis, the tricarboxylic acid (TCA) cycle, molecular chaperones, protein biosynthesis and degradation, stress and defence, signal transduction, RNA processing, transcription factors, amino acid metabolism, one-carbon metabolism, and nucleotide metabolism. An impressive 62% of these identified proteins were implicated in the first four functional groups, whereas the largest functional group was proteins related to mtETC/ATP synthesis (31%). The other three predominant categories were proteins related to the TCA cycle (10%), molecular chaperones (11%), and protein biosynthesis and degradation (10%). Details of the function of the identified proteins and their abundance profiles in PHYMS-XN1376 and CMS-XN1376 (Fig. 3B) are discussed later.
Fig. 3.
Functional classification (A) and hierarchical clustering (B) of the identified proteins from florets of PHYMS-XN1376, CMS-XN1376, and MF-XN1376 at the early uninucleate and trinucleate stages. The hierarchical cluster analysis was conducted using the Genesis 1.7.6 procedure (Graz University of Technology, Austria, http://genome.tugraz.at/genesisclient/genesisclient_download.shtml) and the log2-transformed values of abundance change presented in Supplementary Fig. S5 and Table S5 at JXB online. Euns, the early uninucleate stage; Tns, the trinucleate stage; PHYMS, PHYMS-XN1376; CMS, CMS-XN1376; MF, MF-XN1376.
Protein–protein interaction analysis of identified proteins
Identified proteins were grouped into functional classes according to the biological processes in which they are involved. STRING and BiNGO, which offer an upgrade of the functional analysis, were used to visualize the protein–protein interaction, cellular component, and biological pathways. The STRING analysis revealed a protein association network (Supplementary Fig. S6 at JXB online), in which proteins associated with mtETC, ATP synthesis, the TCA cycle, protein metabolism, the S-adenosylmethionine (SAM) cycle, and antioxidant activity were highly clustered, and these clusters are highlighted with ellipses in Supplementary Fig. S6. Abbreviations of the specific protein names in the network are given in Supplementary Table S8. To obtain statistically over- or under-represented categories of cellular components and biological pathways related to male sterility, BiNGO was used to analyse the identified proteins (Supplementary Fig. S7). The results of cellular component analysis indicated that the identified proteins were more highly enriched in mitochondria (P=5.99e–24; Supplementary Fig. S7A; Table S9), but a number of non-mitochondrial proteins were identified in this study, which is a common problem in isolating mitochondria from plant tissue and also observed in many other mitochondrial proteomic studies (Taylor et al., 2005; Jacoby et al., 2010; Salvato et al., 2014). A complete list of the enriched Gene Ontology (GO) biological pathways for the proteins is presented in Supplementary Fig. S7B and Table S10. Of these, metabolic process (P=5.90e–6) and response to stimulus (P=7.28e–11) were significantly over-represented. Moreover, response to chemical stimulus (P=2.45e–11) was highly correlated with PHYMS-XN1376. Two other groups could be observed both in metabolic process and response to stimulus, namely response to oxidative stress (P=6.41e–7) and oxygen and reactive oxygen species metabolic process (P=3.90e–4), implying that male-sterile plants suffered from oxidative stress. The putative implications for the identified proteins in male sterility are discussed below. The network clusters obtained can provide a broader insight into the different roles of the identified proteins. These proteins are included in a wide range of biological pathways that are involved, either directly or indirectly, in male sterility of wheat.
Oxidative stress and tapetal programmed cell death (PCD) in anther
Mitochondria are not only the sites of oxygen consumption but also one of the sources of cellular reactive oxygen species (ROS) (Salvato et al., 2014). From the above results, the DEPs limited the electron transfer process and this was further confirmed by respiration assays (Supplementary Fig. S8 at JXB online); this might disrupt the balance of oxidative stress, and further trigger PCD in the anther. Therefore, to confirm these possibilities further, the ROS levels and DNA damage were analysed.
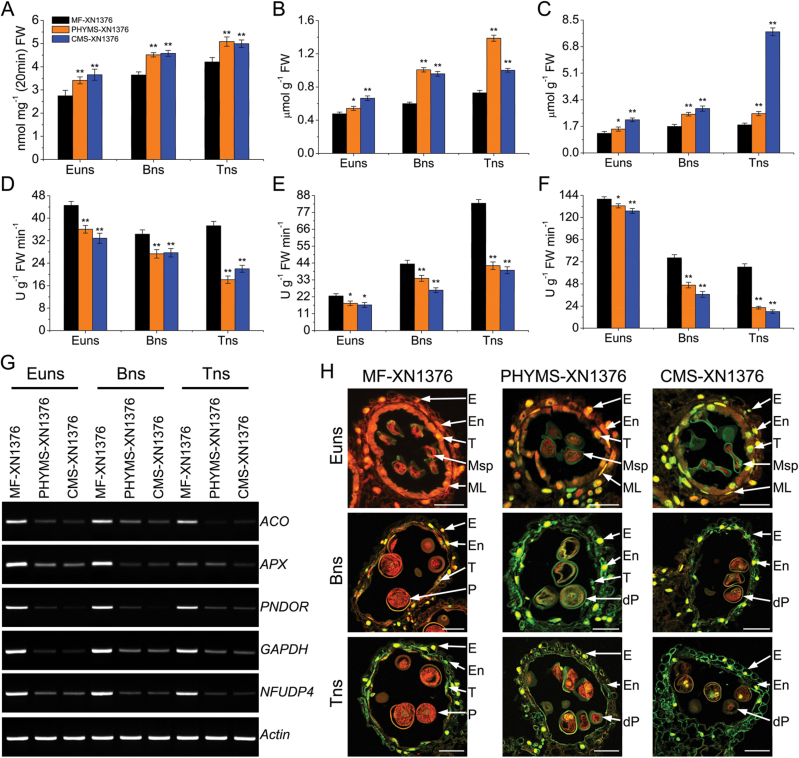
ROS contents and antioxidant enzyme activities were first measured (Fig. 4A–C). The levels of O2 – in PHYMS-XN1376 and CMS-XN1376 anthers were significantly higher than in MF-XN1376 from the early uninucleate stage to the trinucleate stage (Fig. 4A); the levels increased rapidly and remained 20% higher in PHYMS-XN1376, and reached a maximum value of 133% in CMS-XN1376. Excess O2 – was catalysed to form H2O2. The change of H2O2 content was similar to that of O2 –; it apparently increased with anther development in PHYMS-XN1376 and reached a maximum value of 190% at the trinucleate stage, and remained 35% higher in CMS-XN1376 (Fig. 4B). ROS scavenging depends on antioxidant enzymes, such as SOD, CAT, and POD. However, in this experiment, the activities of these enzymes were significantly decreased in PHYMS-XN1376 and CMS-XN1376 anthers when excess ROS were generated (Fig. 4D–F), and this further disrupted the oxidative/antioxidative balance. Subsequently, excess ROS degraded polyunsaturated lipids to form MDA, and its level was significantly increased in anthers of PHYMS-XN1376 (maximum value of 147%) and CMS-XN1376 (maximum value of 432%) (Fig. 4C). Moreover, RT-PCR analysis showed that two oxidation stress-related genes [ascorbate peroxidase (APX) and pyridine nucleotide-disulphide oxidoreductase (PNDOR)] were down-regulated in PHYMS-XN1376 and CMS-XN1376 (Fig. 4G). The above results indicate that the production of ROS overwhelmed the antioxidant capacity, which disrupted the balance between ROS production and clearance in male-sterile anthers; therefore oxidative stress occurred. Therefore, these results suggest that the oxidative stress of the anther in PHYMS-XN1376 was the same as that in CMS-XN1376.
Fig. 4.
ROS burst induces premature tapetal PCD. (A–F) The O2 – production rate (A), H2O2 (B), and MDA (C) contents, and the activities of SOD (D), POD (E), and CAT (F) in developing anthers. Data are means ±SD of three independent experiments. The significance of differences was assessed by Student’s t-test (*P<0.05, **P<0.01). Euns, the early uninucleate stage; Bns, the binucleate stage; Tns, the trinucleate stage. (G) Analysis of oxidation stress and energy supply genes by RT-PCR assay. Primers are listed in Supplementary Table S3 at JXB online. ACO, aconitase; APX, ascorbate peroxidase; PNDOR, pyridine nucleotide-disulphide oxidoreductase; GAPDH, glyceraldehydes-3-phosphate dehydrogenase; NFUDP4, nifU-like domain protein 4. (H) Detection of premature PCD by TUNEL assay in the anthers at different developmental stages. Red fluorescence represents staining with propidium iodide, and green fluorescence indicates TUNEL-positive nuclei of PCD cells. E, epidermis; En, endothecium; ML, middle layer; T, tapetum; Msp, microspore; P, pollen; dP, degenerated pollen. Scale bar=100 μm.
As described above, the ROS levels exhibited a time-dependent increase in male-sterile anthers. Excessive ROS in vivo may generate intermediate signals involved in PCD (Maxwell et al., 2002; Y. Wang et al., 2013). To investigate the abnormal PCD under oxidative stress in the anthers of PHYMS-XN1376 and CMS-XN1376 TUNEL, assays were then performed on anther sections (Fig. 4H; Supplementary Fig. S9 at JXB online). At the early uninucleate stage, the TUNEL fluorescence signal was detected in both PHYMS-XN1376 and CMS-XN1376 tapetal cells, suggesting that PCD occurred in the tapetum and was more strongly expressed in CMS-XN1376 than in PHYMS-XN1376. However, the TUNEL fluorescence signal was still not detected in microspores. Subsequently, from the binucleate stage to the trinucleate stage, TUNEL fluorescence showed stronger signals in the male-sterile tapetum and microspores. Then, the tapetal cells degraded completely and disappeared in PHYMS-XN1376 at the trinucleate stage and in CMS-XN1376 at the binucleate stage. These observations demonstrated that tapetum PCD commenced at the early uninucleate stage and it was degraded prematurely in PHYMS-XN1376 and CMS-XN1376. Furthermore, DNA fragmentation was apparently synchronized with excessive ROS levels.
These results indicated that down-regulation of DEPs inhibited the mtETC. This inhibition was correlated with excess production of ROS and reduced activity of antioxidant enzymes; the microspores suffered from severe oxidative stress during pollen development. Then, the chronic oxidative stress triggered PCD and consequently resulted in the abortion of microspores.
Discussion
DEPs preferentially disturbed the mtETC and ATP synthesis
Among the 12 functional groups, the largest of them was composed of proteins involved in mtETC/ATP synthesis, including 22 down-regulated proteins as listed in Supplementary Table S1 at JXB online (spots 1–22).
There were 12 proteins participating in mtETC (spots 9–20). In mitochondria, NADH dehydrogenase and NADH-ubiquinone oxidoreductase (also referred to as complex I) are the first and the largest enzyme of the mtETC. NADH initially binds to complex I and transfers high energy electrons to subsequent complexes (complex III. complex IV, and complex V). Remarkably, complex I is one of the main sites of premature electron leakage to oxygen (Moller, 2001) and has a role in triggering apoptosis (Petrussa et al., 2009; Chomova and Racay, 2010). In the present study, in both PHYMS-XN1376 and CMS-XN1376 plants, their down-regulation (spots 9–20; Fig. 3B; Supplementary Fig. S5 at JXB online) limited the electron transfer process, promoting ROS generation (Fig. 4A–C). If those ROS were not scavenged, superabundant ROS would then cause oxidative damage to cells, resulting in protein and DNA damage, lipid peroxidation, and even cell death (Foyer and Noctor, 2005). Here an inverse correlation was found between the level of intracellular ROS and the activities of anther SOD (Fig. 4D), CAT (Fig. 4E), and POD (Fig. 4F). Furthermore, applying the TUNEL assay, it was observed that anthers of PHYMS-XN1376 and CMS-XN1376 showed typical PCD characteristics (Fig. 4H; Supplementary Fig. S9). Additionlly, six COX subunits (spots 10–15) were down-regulated in two male sterility samples; this altered cytochrome c oxidase functionality or structure, which in turn inhibits electron transport from cytochrome c to oxygen. Therefore, the decreased abundance of the 12 proteins limited the mtETC and further affected pollen development.
Furthermore, representative subunits of ATP synthase were identified, including four α-subunits (spots 1–4), three β-subunits (spots 5–7), and one δ-subunit (spot 8). Mitochondrial membrane ATP synthase (F1F0 ATP synthase or complex V) plays an important role in energy metabolism by converting ADP into ATP in the presence of a transmembrane proton gradient (Sabar et al., 2003). The β-subunit is a catalytic site, which in combination with the α-subunit regulates the activity of ATP synthase. Recent studies of plant mitochondrial complexes indicate that several mitochondrial DNA regions encoding these ATP synthase subunits are associated with male sterility (Sabar et al., 2003; Eckardt, 2006; Ohta et al., 2010). In this study, similar patterns were also observed for these subunits (spots 1–8) that decreased in both PHYMS-XN1376 and CMS-XN1376 mitochondria (Fig. 3B; Supplementary Fig. S5 at JXB online). These might cause ATP synthase dysfunction and then impact on mitochondrial energy output and induce mitochondrial membrane potential changes, resulting in abnormal anther development with non-functional pollen.
Therefore, the present data suggest that the down-regulation of the 22 proteins not only slowed down the rate of electron transport in both PHYMS-XN1376 and CMS-XN1376 plants, but also caused excess electrons to combine with molecular oxygen to form ROS during pollen development, thus triggering PCD in the anther; these results are supported by the ROS measurement and TUNEL assay. More recently, research on plant male sterility has increase the focus of attention on ROS metabolism (Li et al., 2012; Luo et al., 2013; K. Wang et al., 2013). The aberrant expression of mitochondrial genes compromised the increased demand for respiratory function and cellular energy in the form of ATP during anther development. The data presented here also show that the same expression patterns are seen in different types of a male-sterile (PHYMS and CMS) wheat line; this provides insights into the molecular mechanisms of male sterility in wheat.
DEPs are largely implicated in enhanced protein biosynthesis and inhibited degradation of proteins
A total of 20 identified proteins (28% in total) in PHYMS-XN1376 and CMS-XN1376 plants were found to be associated with protein metabolism (Fig. 3B; Supplementary Table S1 at JXB online), including molecular chaperones (spots 30–37), protein biosynthesis and degradation (spots 38–43), RNA processing (spots 49 and 50), transcription factors (spots 51 and 52), and nucleotide metabolism (spots 60 and 61), and covered a wide range of DNA replication, RNA transcription, and protein synthesis/processing.
Most mitochondrial proteins are encoded in the nucleus. They are synthesized as precursor forms in the cytosol and must be imported into the mitochondria with the help of different protein translocases. Among these translocases, Tom40, the import channel of the mitochondrial outer membrane, plays an active role in sorting imported proteins. Mitochondrial Hsp70 and chaperonin CPN60 (Hsp60) are mitochondrial chaperones that are typically believed to be responsible for the transportation and refolding of pre-proteins from the cytoplasm into the mitochondrial matrix. Furthermore, the mitochondrial processing peptidase (MPP) removes the pre-sequences from the pre-proteins to form proteins (Becker et al., 2012). Ubiquitin-conjugating enzyme E2 (UBE2) belongs to the ubiquitin-conjugating enzyme family and performs the second step in the ubiquitination reaction that targets a protein for degradation via proteasomes (Miao et al., 2013). In the present study, the co-up-regulation of Tom40 (spots 38 and 39), mtHsp70 (spots 30–33), mtHsp60 (spots 34 and 35), and MPP (spots 40 and 41) was enhanced during novel protein biosynthesis. A variety of proteins (spots 36, 37, 42 49–52, 60, and 61) were directly or indirectly involved in initiation and elongation of the newly growing peptide chains during this synthesis. Conversely, protein degradation was inhibited by the down-regulation of UBE2 (spot 43). Ultimately, this resulted in an increase in the amount and abundance of proteins (Table 1).
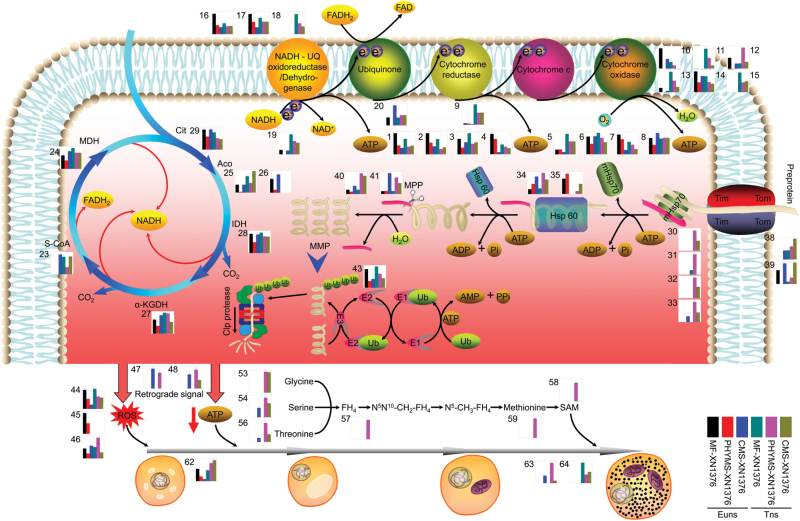
A possible mitochondria-mediated male sterility protein network in wheat
Plant male sterility plays an important role in developmental and molecular studies and hybrid breeding. In the present study, based on the putative functions and changes in the levels of the 68 identified proteins (Supplementary Fig. S5, S7B; Table S1 at JXB online), together with previously published results (Chen et al., 2009, 2010; Zhang et al., 2011, 2013; Ba et al., 2014a, b; Liu et al., 2014), an intriguing mitochondria-mediated male sterility protein network was proposed for wheat (Fig. 5). This network consists of several functional components, including inhibited mtETC and TCA cycle, accelerated biosynthesis and reduced degradation of proteins, generated ROS metabolism, disrupted cell division cycle, and epigenetics.
Fig. 5.
A protein network for the mechanism of mitochondria-mediated male sterility. Histograms and numbers represent protein identification and abundance listed in Supplementary Fig. S5 and Table S1 at JXB online. Cit, citrate synthase; Aco, aconitase; IDH, isocitrate dehydrogenase; α-KGDH, α-ketoglutarate dehydrogenase complex; S-CoA, succinyl-CoA synthetase; MDH, malate dehydrogenase; Tom, outer membrane translocases; Tim, import inner membrane translocases; mHsp70, matrix heat shock protein 70; Hsp60, heat shock protein 60; E1, ubiquitin-activating enzymes; E2, ubiquitin-conjugating enzymes; E3, ubiquitin-ligating enzymes; Ub, ubiquitin; FH4, tetrahydrofolate; SAM, S-adenosylmethionine; MPP, mitochondrial-processing peptidase; MMP, mature mitochondrial protein; Euns, the early uninucleate stage; Tns, the trinucleate stage.
In the present experiments, mtETC/ATP synthesis-related proteins were preferentially and greatly down-regulated in both PMS-XN1376 and CMS-XN1376 plants from the early uninucleate stage, including one NADH dehydrogenase (spot 19), three complex I proteins (spots 16–18), two complex II proteins (spots 9 and 20), six complex IV proteins (spots 10–15), and eight ATP synthases (spots 1–8). In addition, a range of antioxidant enzymes were down-regulated in this study (spots 44–46; Fig. 4D–F) and in previously published studies (Chen et al., 2009, 2010; Ye et al., 2009). These changes led to excess electrons directly combining with molecular oxygen to generate excessive ROS (Fig. 4A–C) while reducing ATP production in PHYMS-XN1376 and CMS-XN1376 plants. On the one hand, the excessive ROS caused peroxidation of membrane lipids (Fig. 4C) and resulted in increased permeability (Suzuki et al., 2012). In this case, a large number of pre-proteins were rapidly translocated through the outer and inner membranes into mitochondria; there they combined with the up-regulation of Tom40 (spots 38 and 39), mtHsp70 (spot 30–33), mtHsp60 (spots 34 and 35), and MPP (spots 40 and 41), and led to pre-proteins forming proteins; however, the down-regulation of UBE2 (spot 43) inhibited protein degradation. Eventually they increased the amount and abundance of mitochondrial proteins, including elongation factor G (spot 42), RNA helicase (spots 49 and 50), DNA replication licensing factor (spot 60), and site-specific recombinase (spot 61). Moreover, these extra proteins were involved in the biosynthesis of mitochondria-encoded proteins. Thus, the excessive ROS disturbed the balance between protein biosynthesis and degradation, which damaged the equilibrium of protein metabolism in mitochondria, and further affected pollen development. On the other hand, excessive ROS and reduced ATP as the primary signals, combined with the up-regulation of signal transduction proteins (spots 47 and 48), retrograde-regulated pollen development and ultimately led to pollen abortion. Furthermore, three proteins involved in the cell division cycle (spots 62 and 63) and nutrition storage (spot 64) displayed an abnormal change that hindered pollen development (Fig. 1P, T) and the biosynthesis of starch (Fig. 1Q, U); this further aggravated abortion rates. Thus, abnormal cell division and lack of nutrients are also important factors in pollen abortion. The present data also suggest that PHYMS-XN1376 and CMS-XN1376 shared the above pathway with respect to mitochondrial-mediated male sterility.
However, it should be noted that a number of enzymes (spots 23–29), belonging to the TCA cycle, are only down-regulated in PHYMS-XN1376 at the early uninucleate stage and/or at the trinucleate stage. It is possible that the inhibition of TCA cycle enzymes can limit NADH production, energy, and carbohydrate metabolism in PHYMS-XN1376, which indirectly inhibited the mtETC, which has been confirmed by RT-PCR analysis (three energy supply-related genes aconitase, GAPDH, and NFUDP4) and Zhang et al. (2011, 2013). This is unlike the abnormal mtETC in CMS-XN1376 plants which was directly inhibited by the related genes. Additionally, a variety of amino acid biosynthesis enzymes (spots 53, 54, and 56) were up-regulated in both PHYMS-XN1376 and CMS-XN1376 plants, and they generated excessive amino acids, including glycine, serine, and threonine. Tetrahydrofolate (spot 57), a one-carbon metabolism carrier only up-regulated in PHYMS-XN1376, converted these excessive amino acids into SAM (spots 58 and 59) which is involved in DNA methylation reactions. This is different from the process in CMS-XN1376 and has been documented by Ba et al. (2014b). Therefore, the two abnormal pathways only exist in PHYMS-XN1376 but not in CMS-XN1376.
2-DE analysis and putative functions revealed a highly interconnected protein network, which led to pollen abortion in wheat (Fig. 5). This network was further confirmed by physiological data, RT-PCR, and TUNEL assay (Fig. 4). The results showed that an increase in ROS levels was accompanied by inhibition of antioxidant enzyme activities in both PHYMS-XN1376 and CMS-XN1376 plants from the early uninucleate stage to the trinucleate stage (Fig. 4). Furthermore, RT-PCR analysis indicated that two oxidation stress-related genes (APX and PNDOR) and three energy supply-related genes (aconitase, GAPDH, and NFUDP4) were down-regulated in both PHYMS-XN1376 and CMS-XN1376 (Fig. 4G). These changes imply that the inhibition of mtETC causes excessive ROS generation and a decrease in ATP. Furthermore, the TUNEL assay showed that the PCD in anther tissues was initiated at the early uninucleate stage and extended to the binucleate and trinucleate stages (Fig. 4H; Supplementary Fig. S9 at JXB online). These results support the hypothesis that inhibition of mtETC causes excessive ROS generation and a decrease in ATP which induces premature tapetal degeneration and pollen apoptosis, ultimately leading to pollen abortion. Additionally, protein ubiquitination and DNA methylation have been documented by Liu et al. (2014) and Ba et al. (2014b), respectively.
Taken together, an intimate protein network of male sterility was proposed and confirmed based on proteomic screening and molecular biology. The results provide insight into the molecular mechanism of male sterility in wheat, which gives a useful guide for fertility restoration and the practical application of hybrid breeding.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Gene expression stability of the seven candidate reference genes for wheat anther using two approaches.
Figure S2. Scanning electron micrograph observations of anthers at the trinucleate stage.
Figure S3. Evaluation of mitochondria fraction.
Figure S4. 2-DE maps analysis of mitochondria proteomes from PHYMS-XN1376, CMS-XN1376, and MF-XN1376.
Figure S5. Abundance profiles of 71 differentially expressed proteins.
Figure S6. Analysis of protein interaction network by the STRING system.
Figure S7. Cellular component (A) and biological pathway (B) networks generated using the BiNGO plugin from the Cytoscape tool.
Figure S8. Analysis of respiratory activity.
Figure S9. Statistical analysis of apoptosis rate (%).
Table S1. Identification of differentially expressed proteins from isolated mitochondria of PHYMS-XN1376, CMS-XN1376, and MF-XN1376.
Table S2. Sixty-eight annotated proteins blasted against the TAIR and ORYSJ protein and TAMTG nucleotide databases.
Table S3. Specific primers used in this study.
Table S4. The abundance of differentially expressed proteins from isolated mitochondria of PHYMS-XN1376, CMS-XN1376, and MF-XN1376.
Table S5. The fold change and P-value of differentially expressed proteins in PHYMS-XN1376, and CMS-XN1376.
Table S6. Matched peptide sequences of identified proteins.
Table S7. Corresponding homologues of the three unknown proteins.
Table S8. Abbreviations of the specific protein names in the STRING network (Supplementary Fig. S6).
Table S9. Cellular component of differentially expressed proteins.
Table S10. Biological pathways of differentially expressed proteins.
Acknowledgements
We wish to thank members of the Z.M.H. lab from the Chinese Academy of Sciences for helpful discussion and critical reading of the manuscript. We would also like to thank the anonymous reviewers for their constructive comments on improving the manuscript. This work was supported by the National High Tech Research and Development Program of China (2011AA10A106), the National Support Program of China (2015BAD27B01), the National Natural Science Foundation of China (31171611 and 31371697), the Technological Innovation and Over Planning Projects of Shaanxi province (2014KTZB02-01-02), and Projects Opening up New Function of Precision Instrument of Northwest A&F University (dysb130210).
References
- Aalto EA, Koelewijn HP, Savolainen O. 2013. Cytoplasmic male sterility contributes to hybrid incompatibility between subspecies of Arabidopsis lyrata. G3-Genes Genomes Genetics 3, 1727–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ba QS, Zhang GS, Che HX, et al. 2014. a . Aliphatic metabolism during anther development interfered by chemical hybridizing agent in wheat. Crop Science 54, 1458–1467. [Google Scholar]
- Ba QS, Zhang GS, Wang JS, Niu N, Ma SC, Wang JW. 2014. b . Gene expression and DNA methylation alterations in chemically induced male sterility anthers in wheat (Triticum aestivum L.). Acta Physiologiae Plantarum 36, 503–512. [Google Scholar]
- Bachtrog D, Mank JE, Peichel CL, et al. 2014. Sex determination: why so many ways of doing it? PLoS Biology 12, e1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Bottinger L, Pfanner N. 2012. Mitochondrial protein import: from transport pathways to an integrated network. Trends in Biochemical Sciences 37, 85–91. [DOI] [PubMed] [Google Scholar]
- Bentolila S, Alfonso AA, Hanson MR. 2002. A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proceedings of the National Academy of Sciences, USA 99, 10887–10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Liu YG. 2014. Male sterility and fertility restoration in crops. Annual Review of Plant Biology 65, 579–606. [DOI] [PubMed] [Google Scholar]
- Chen RH, Liu W, Zhang GS, Ye JX. 2010. Mitochondrial proteomic analysis of cytoplasmic male sterility line and its maintainer in wheat (Triticum aestivum L.). Agricultural Sciences in China 9, 771–782. [Google Scholar]
- Chen RH, Ye JX, Zhang GS, Wang JS, Niu N, Ma SC, Zhao JX, Zhu JC. 2009. Differential proteomic analysis of anther proteins between cytoplasmic-nuclear male sterility line and its maintainer in wheat (Triticum aestivum L.). Progress in Biochemistry and Biophysics 36, 431–440. [Google Scholar]
- Chomova M, Racay P. 2010. Mitochondrial complex I in the network of known and unknown facts. General Physiology and Biophysics 29, 3–11. [DOI] [PubMed] [Google Scholar]
- Clifton SW, Minx P, Fauron CM, et al. 2004. Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiology 136, 3486–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckardt NA. 2006. Cytoplasmic male sterility and fertility restoration. The Plant Cell 18, 515–517. [Google Scholar]
- Foyer CH, Noctor G. 2005. Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. The Plant Cell 17, 1866–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajewska E, Sklodowska M. 2007. Effect of nickel on ROS content and antioxidative enzyme activities in wheat leaves. Biometals 20, 27–36. [DOI] [PubMed] [Google Scholar]
- Gobom J, Schuerenberg M, Mueller M, Theiss D, Lehrach H, Nordhoff E. 2001. Alpha-cyano-4-hydroxycinnamic acid affinity sample preparation. A protocol for MALDI-MS peptide analysis in proteomics. Analytical Chemistry 73, 434–438. [DOI] [PubMed] [Google Scholar]
- Hameed A, Goher M, Iqbal N. 2012. Heat stress-induced cell death, changes in antioxidants, lipid peroxidation, and protease activity in wheat leaves. Journal of Plant Growth Regulation 31, 283–291. [Google Scholar]
- Hameed A, Iqbal N, Malik SA. 2014. Effect ofd-mannose on antioxidant defense and oxidative processes in etiolated wheat coleoptiles. Acta Physiologiae Plantarum 36, 161–167. [Google Scholar]
- Handa H. 2003. The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Research 31, 5907–5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson MR, Bentolila S. 2004. Interactions of mitochondrial and nuclear genes that affect male gametophyte development. The Plant Cell 16 Suppl, S154–S169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng S, Wei C, Jing B, et al. 2014. Comparative analysis of mitochondrial genomes between the hau cytoplasmic male sterility (CMS) line and its iso-nuclear maintainer line in Brassica juncea to reveal the origin of the CMS-associated gene orf288. BMC Genomics 15, 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn R, Gupta KJ, Colombo N. 2014. Mitochondrion role in molecular basis of cytoplasmic male sterility. Mitochondrion 19, 198–205. [DOI] [PubMed] [Google Scholar]
- Hu J, Huang W, Huang Q, Qin X, Yu C, Wang L, Li S, Zhu R, Zhu Y. 2014. Mitochondria and cytoplasmic male sterility in plants. Mitochondrion 19, 282–288. [DOI] [PubMed] [Google Scholar]
- Jacoby RP, Millar AH, Taylor NL. 2010. Wheat mitochondrial proteomes provide new links between antioxidant defense and plant salinity tolerance. Journal of Proteome Research 9, 6595–6604. [DOI] [PubMed] [Google Scholar]
- Jing B, Heng SP, Tong D, et al. 2012. A male sterility-associated cytotoxic protein ORF288 in Brassica juncea causes aborted pollen development. Journal of Experimental Botany 63, 1285–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RR, Klein PE, Mullet JE, Minx P, Rooney WL, Schertz KF. 2005. Fertility restorer locus Rf1 of sorghum (Sorghum bicolor L.) encodes a pentatricopeptide repeat protein not present in the colinear region of rice chromosome 12. Theoretical and Applied Genetics 111, 994–1012. [DOI] [PubMed] [Google Scholar]
- Kumar P, Vasupalli N, Srinivasan R, Bhat SR. 2012. An evolutionarily conserved mitochondrial orf108 is associated with cytoplasmic male sterility in different alloplasmic lines of Brassica juncea and induces male sterility in transgenic Arabidopsis thaliana. Journal of Experimental Botany 63, 2921–2932. [DOI] [PubMed] [Google Scholar]
- Li SQ, Wan CX, Hu CF, Gao F, Huang Q, Wang K, Wang T, Zhu YG. 2012. Mitochondrial mutation impairs cytoplasmic male sterility rice in response to H2O2 stress. Plant Science 195, 143–150. [DOI] [PubMed] [Google Scholar]
- Linke B, Borner T. 2005. Mitochondrial effects on flower and pollen development. Mitochondrion 5, 389–402. [DOI] [PubMed] [Google Scholar]
- Liu HZ, Zhang GS, Zhu WW, et al. 2014. Differential proteomic analysis of polyubiquitin-related proteins in chemical hybridization agent-induced wheat (Triticum aestivum L.) male sterility. Acta Physiologiae Plantarum 36, 1473–1489. [Google Scholar]
- Luan J, Liu T, Luo W, Liu W, Peng M, Li W, Dai X, Liang M, Chen L. 2013. Mitochondrial DNA genetic polymorphism in thirteen rice cytoplasmic male sterile lines. Plant Cell Reports 32, 545–554. [DOI] [PubMed] [Google Scholar]
- Luo DP, Xu H, Liu ZL, et al. 2013. A detrimental mitochondrial–nuclear interaction causes cytoplasmic male sterility in rice. Nature Genetics 45, 573–577. [DOI] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. 2005. BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in biological networks. Bioinformatics 21, 3448–3449. [DOI] [PubMed] [Google Scholar]
- Maxwell DP, Nickels R, McIntosh L. 2002. Evidence of mitochondrial involvement in the transduction of signals required for the induction of genes associated with pathogen attack and senescence. The Plant Journal 29, 269–279. [DOI] [PubMed] [Google Scholar]
- Miao H, Ye Z, Silva JA, Qin Y, Hu G. 2013. Identifying differentially expressed genes in pollen from self-incompatible ‘wuzishatangju’ and self-compatible ‘shatangju’ mandarins. International Journal of Molecular Sciences 14, 8538–8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller IM. 2001. Plant mitochondria and oxidative stress: electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annual Review of Plant Physiologay and Plant Molecular Biology 52, 561–591. [DOI] [PubMed] [Google Scholar]
- Notsu Y, Masood S, Nishikawa T, Kubo N, Akiduki G, Nakazono M, Hirai A, Kadowaki K. 2002. The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Molecular Genetics and Genomics 268, 434–445. [DOI] [PubMed] [Google Scholar]
- Ogihara Y, Yamazaki Y, Murai K, et al. 2005. Structural dynamics of cereal mitochondrial genomes as revealed by complete nucleotide sequencing of the wheat mitochondrial genome. Nucleic Acids Research 33, 6235–6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta H, Ogino A, Kasai M, Sano Y, Kanazawa A. 2010. Fertility restoration by Ifr1 in rice with BT-type cytoplasmic male sterility is associated with a reduced level, but not processing, of atp6-orf79 co-transcribed RNA. Plant Cell Reports 29, 359–369. [DOI] [PubMed] [Google Scholar]
- Petrussa E, Bertolini A, Casolo V, Krajnakova J, Macri F, Vianello A. 2009. Mitochondrial bioenergetics linked to the manifestation of programmed cell death during somatic embryogenesis of Abies alba. Planta 231, 93–107. [DOI] [PubMed] [Google Scholar]
- Sabar M, Gagliardi D, Balk J, Leaver CJ. 2003. ORFB is a subunit of F1FO-ATP synthase: insight into the basis of cytoplasmic male sterility in sunflower. EMBO Reports 4, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvato F, Havelund JF, Chen M, Rao RS, Rogowska-Wrzesinska A, Jensen ON, Gang DR, Thelen JJ, Moller IM. 2014. The potato tuber mitochondrial proteome. Plant Physiology 164, 637–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Wise RP. 1998. The molecular basis of cytoplasmic male sterility and fertility restoration. Trends in Plant Science 3, 175–180. [Google Scholar]
- Shen Z, Li P, Ni RJ, et al. 2009. Label-free quantitative proteomics analysis of etiolated maize seedling leaves during greening. Molecular and Cellular Proteomics 8, 2443–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JS, Hedgcoth C. 1994. A chimeric gene (Orf256) is expressed as protein only in cytoplasmic male-sterile lines of wheat. Plant Molecular Biology 26, 535–539. [DOI] [PubMed] [Google Scholar]
- Song YL, Wang JW, Zhang PG, Zhang GS, Zhang LY, Zhao XL, Niu N, Ma SC. 2014. Cytochemical investigation at different microsporogenesis phases of male sterility in wheat, as induced by the chemical hybridising agent SQ-1. Crop and Pasture Science 65, 868–877. [Google Scholar]
- Sugiyama Y, Watase Y, Nagase M, Makita N, Yagura S, Hirai A, Sugiura M. 2005. The complete nucleotide sequence and multipartite organization of the tobacco mitochondrial genome: comparative analysis of mitochondrial genomes in higher plants. Molecular Genetics and Genomics 272, 603–615. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Koussevitzky S, Mittler R, Miller G. 2012. ROS and redox signalling in the response of plants to abiotic stress. Plant, Cell and Environment 35, 259–270. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, et al. 2015. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Research 43, D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang HW, Luo DP, Zhou DG, Zhang QY, Tian DS, Zheng XM, Chen LT, Liu YG. 2014. The rice Restorer rf4 for wild-abortive cytoplasmic male sterility encodes a mitochondrial-localized PPR protein that functions in reduction of WA352 transcripts. Molecular Plant 7, 1497–1500. [DOI] [PubMed] [Google Scholar]
- Taylor NL, Heazlewood JL, Day DA, Millar AH. 2005. Differential impact of environmental stresses on the pea mitochondrial proteome. Molecular and Cellular Proteomics 4, 1122–1133. [DOI] [PubMed] [Google Scholar]
- Touzet P, Meyer EH. 2014. Cytoplasmic male sterility and mitochondrial metabolism in plants. Mitochondrion 19, 166–171. [DOI] [PubMed] [Google Scholar]
- Unseld M, Marienfeld JR, Brandt P, Brennicke A. 1997. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nature Genetics 15, 57–61. [DOI] [PubMed] [Google Scholar]
- Vanlerberghe GC, McIntosh L. 1996. Signals regulating the expression of the nuclear gene encoding alternative oxidase of plant mitochondria. Plant Physiology 111, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW, Wang XL, Xu H, Tang HW, Zhang GS, Liu YG. 2013. Structural and expressional variation analyses of mitochondrial genomes reveal candidate transcripts for the S-V cytoplasmic male sterility in wheat (Triticum aestivum L.). Journal of Genetics and Genomics 40, 437–439. [DOI] [PubMed] [Google Scholar]
- Wang K, Gao F, Ji YX, Liu Y, Dan ZW, Yang PF, Zhu YG, Li SQ. 2013. ORFH79 impairs mitochondrial function via interaction with a subunit of electron transport chain complex III in Honglian cytoplasmic male sterile rice. New Phytologist 198, 408–418. [DOI] [PubMed] [Google Scholar]
- Wang Y, Loake GJ, Chu C. 2013. Cross-talk of nitric oxide and reactive oxygen species in plant programed cell death. Frontiers in Plant Science 4, 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZH, Zou YJ, Li XY, et al. 2006. Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. The Plant Cell 18, 676–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Ding ZW, Vizcay-Barrena G, Shi JX, Liang WQ, Yuan Z, Werck-Reichhart D, Schreiber L, Wilson ZA, Zhang DB. 2014. ABORTED MICROSPORES acts as a master regulator of pollen wall formation in Arabidopsis. The Plant Cell 26, 1544–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumoto K, Terachi T, Yamagishi H. 2009. A novel Rf gene controlling fertility restoration of Ogura male sterility by RNA processing of orf138 found in Japanese wild radish and its STS markers. Genome 52, 495–504. [DOI] [PubMed] [Google Scholar]
- Ye JX, Wang SP, Zhang FJ, Xie DQ, Yao YH. 2013. Proteomic analysis of leaves of different wheat genotypes subjected to PEG 6000 stress and rewatering. Plant Omics 6, 286–294. [Google Scholar]
- Ye JX, Zhang GS, Wang SP, Chen RH, Wang JS, Niu N, Ma SC, Li HX, Zhu JC. 2009. Differential proteomic studies on pollen grain proteins of wheat male sterile line induced by chemical hybridizing agent SQ-1. Chinese Journal of Biochemistry and Molecular Biology 25, 949–957. [Google Scholar]
- Zhang H, Liang WQ, Yang XJ, Luo X, Jiang N, Ma H, Zhang DB. 2010. Carbon starved anther encodes a MYB domain protein that regulates sugar partitioning required for rice pollen development. The Plant Cell 22, 672–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LY, Yuan L, Yang SL, Zhang GS, Wang JS, Song YL, Zhao ZJ, Niu N, Ma SC. 2011. Expression characteristic on TaPDC-E1α gene and its regulatory enzymes gene in male sterile line of wheat (Triticum aestivum). Acta Agronomica Sinica 37, 620–628. [Google Scholar]
- Zhang LY, Zhang GS, Zhao XL, Yang SL. 2013. Screening and analysis of proteins interacting with TaPDK from physiological male sterility induced by CHA in wheat. Journal of Integrative Agriculture 12, 941–950. [Google Scholar]
- Zuo SS, Lundahl P. 2000. A micro-Bradford membrane protein assay. Analytical Biochemistry 284, 162–164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.