Highlight
The characterization of three BrAOP2 paralogues, crucial candidates for engineering beneficial glucosinolates, revealed that functional divergence occurred during the evolution of triplicated AOP2 genes in Brassica rapa.
Key words: Brassica rapa, BrAOP2, expression pattern, functional divergence, glucosinolates, paralogues.
Abstract
The glucosinolate biosynthetic gene AOP2 encodes an enzyme that plays a crucial role in catalysing the conversion of beneficial glucosinolates into anti-nutritional ones. In Brassica rapa, three copies of BrAOP2 have been identified, but their function in establishing the glucosinolate content of B. rapa is poorly understood. Here, we used phylogenetic and gene structure analyses to show that BrAOP2 proteins have evolved via a duplication process retaining two highly conserved domains at the N-terminal and C-terminal regions, while the middle part has experienced structural divergence. Heterologous expression and in vitro enzyme assays and Arabidopsis mutant complementation studies showed that all three BrAOP2 genes encode functional BrAOP2 proteins that convert the precursor methylsulfinyl alkyl glucosinolate to the alkenyl form. Site-directed mutagenesis showed that His356, Asp310, and Arg376 residues are required for the catalytic activity of one of the BrAOP2 proteins (BrAOP2.1). Promoter–β-glucuronidase lines revealed that the BrAOP2.3 gene displayed an overlapping but distinct tissue- and cell-specific expression profile compared with that of the BrAOP2.1 and BrAOP2.2 genes. Quantitative real-time reverse transcription-PCR assays demonstrated that BrAOP2.1 showed a slightly different pattern of expression in below-ground tissue at the seedling stage and in the silique at the reproductive stage compared with BrAOP2.2 and BrAOP2.3 genes in B. rapa. Taken together, our results revealed that all three BrAOP2 paralogues are active in B. rapa but have functionally diverged.
Introduction
Glucosinolates are a class of nitrogen- and sulfur-rich secondary metabolites common in the family Brassicaceae (or Cruciferae), which includes the model plant Arabidopsis thaliana and economically important Brassica crops such as turnip (Brassica rapa ssp. rapa), broccoli (Brassica oleracea var. italica), and cauliflower (B. oleracea var. botrytis) (Halkier and Gershenzon, 2006). Glucosinolates and their breakdown products have been the subject of extensive studies because of their roles in plant defence against microbial pathogens and herbivorous insects (Kroymann et al., 2003; Clay et al., 2009).
Some glucosinolates and their breakdown products act as anti-carcinogenic compounds. For example, Met-derived 4-methylsulfinylbutyl glucosinolate [glucoraphanin (GRA)] has been the focus of considerable attention because its degradation product, sulforaphane, which was first isolated from broccoli, exhibits pronounced anti-carcinogenic activity (Zhang et al., 1994; Fahey et al., 1997; Gamet-Payrastre et al., 2000; Talalay and Fahey, 2001; Fahey et al., 2002). However, sulforaphane can undergo further reactions to generate 2-hydroxy-3-butenyl glucosinolate (progoitrin), which is goitrogenic and lacks notable anti-cancer activity (Faulkner et al., 1998). Thus, it will be advantageous to manipulate glucosinolate structures to alter the nutritional and economic value of Brassica vegetables.
Glucosinolates are derived from glucose and an amino acid, and can be classified as aliphatic, aromatic, or indole glucosinolates based on the precursor amino acid (Gershenzon, 2006; Sonderby et al., 2010). Their biosynthesis occurs in three independent stages: (i) side-chain elongation of the precursor amino acid; (ii) formation of the core structure; and (iii) modification of the side chain. Elongation and modification of the side chain (R-group) generates diverse glucosinolate compounds, of which more than 200 structures have been identified (Clarke, 2010).
In Arabidopsis thaliana, a small gene family of 2-oxoglutarate-dependent dioxygenases (AtAOP1, AtAOP2, and AtAOP3) that resulted from gene duplication was identified by fine-scale mapping (Hall et al., 2001; Kliebenstein et al., 2001b ). A heterologous expression and in vitro enzyme assay showed that AOP2 catalysed the conversion of 3-methylsulfinylpropyl- and 4-methylsulfinylbutyl glucosinolates to the corresponding alkenyl glucosinolates 2-propenyl and 3-butenyl, respectively, while AOP3 displayed only weak catalytic activity in the conversion of 3-methylsulfinylpropyl glucosinolate to 3-hydroxypropyl glucosinolate (Kliebenstein et al., 2001b ). AtAOP1 is believed to be the ancestral gene that gave rise to AtAOP2 and AtAOP3 through a series of gene duplication events; however, its function is unclear (Kliebenstein et al., 2001b ). Arabidopsis thaliana has been reported to display differential AOP expression, whereby particular accessions expressed either AtAOP2 or AtAOP3 or neither, but not both. Moreover, in some Arabidopsis accessions the absence of both functional enzymes has led to the accumulation of the precursor methylsulfinylalkyl glucosinolate (Kliebenstein et al., 2001a ). The mechanism by which a particular accession transcribes AtAOP2 or AtAOP3 but not both was found to involve the complete inversion of the two structural genes resulting in AtAOP3 being expressed from the AtAOP2 promoter (Chan et al., 2010; Kliebenstein et al., 2001b ). Arabidopsis accessions have also been shown to have three different AtAOP2 alleles with variations in exon 2; for example, AtAOP2 from the Columbia (Col-0) ecotype contains a 5bp frame-shift deletion that led to the accumulation of methylsulfinylalkyl glucosinolates (Kliebenstein et al., 2001b ; Neal et al., 2010).
Brassica crops are of great economical and nutritional importance to humans. In B. oleracea, BoAOP1 and BoAOP2 were found to be duplicated, while BoAOP3 was absent (Gao et al., 2004), and the BoAOP2 homologue (BoGSL-ALK) from collard (B. oleracea var. viridis) was reported to catalyse the conversion of methylsulfinylalkyl glucosinolate to the alkenyl form in plants (Li and Quiros, 2003). Broccoli (B. oleracea var. italica), on the other hand, contains a non-functional allele of BoGSL-ALK because of a 2bp deletion in exon 2, which causes the accumulation of GRA (Li and Quiros, 2003). More recently, another non-functional BoAOP2 gene contributing to the accumulation of GRA because of the presence of a premature stop codon has been found in B. oleracea (Liu et al., 2014).
In B. rapa, which underwent an additional whole-genome triplication after its divergence from a common ancestor of Arabidopsis thaliana, three orthologues of the BrAOP loci were found, each containing the tandem duplicated genes BrAOP1 and BrAOP2 but not BrAOP3 (Wang et al., 2011). These duplicated BrAOP2 genes may enhance the potential resources for quantitative variation of a particular trait (Li et al., 2008). To the best of our knowledge, however, no studies have investigated whether all three BrAOP2 genes are functional.
Here, we investigated the consequence of polyploidy on BrAOP2 structure, phylogeny, gene expression, and function both in vivo and in vitro. Our results highlight the importance of BrAOP2 paralogues in controlling the conversion of beneficial glucosinolates to harmful ones and the expression divergence of duplicated BrAOP2 genes. These findings will help in enriching beneficial aliphatic glucosinolates (e.g. GRA) and in reducing anti-nutritional aliphatic glucosinolates (e.g. progoitrin and gluconapin) in B. rapa for the benefit of humans.
Materials and methods
Plant material
B. rapa accessions yellow sarson L143 and Chiifu-401/42 were germinated and grown in greenhouses at the Chinese Academy of Agricultural Sciences (Beijing, China) during the spring of 2011. Leaves were collected from Chiifu-401/42 for cloning of BrAOP2 genes. Accession L143 plants were used to collect different organs (root, stem, leaves, inflorescence, and siliques) to analyse BrAOP2 expression patterns. We collected samples of three biological replicates under normal growth conditions 2 and 10 weeks after sowing. Two to three healthy, undamaged fresh leaves were collected, snap frozen in liquid nitrogen, and kept at –80°C until use.
Arabidopsis thaliana ecotype Col-0 was used for functional complementation studies in vivo. Col-0 seeds were plated on soil and cold treated at 4 °C for 2 d in the dark. After stratification, the seeds were transferred into a temperature-controlled growth chamber at 22 °C, with a 16/8h photoperiod, light intensity of 120 mmol m−2 s−1, and 40% relative humidity.
The seeds of plants grown on Petri dishes were briefly surface sterilized with 75% (v/v) ethanol for 8min and then washed three times with sterile water. Seeds were sown on Murashige and Skoog (MS) agar medium (half-strength MS salt, pH 5.8) and cold treated at 4 °C for 2 d in the dark, and then placed in the growth chamber. Transgenic plants were selected by germination on half-strength MS medium containing 60 µg ml−1 of kanamycin and 30 µg ml−1 of hygromycin antibiotics and were treated subsequently as wild-type plants.
Sources of genome data
B. rapa gene sequences for synteny analyses were obtained from BRAD (v.1.5; http://brassicadb.org) (Cheng et al., 2011). AOP1 and AOP2 sequences and genome datasets for Arabidopsis thaliana were downloaded from The Arabidopsis Information Resource (TAIR9; http://www.arabidopsis.org/index.jsp). Arabidopsis AOP3 sequences were from GenBank (http://www.ncbi.nlm.nih.gov/). The genomic dataset for Arabidopsis lyrata was downloaded from the Joint Genome Initiative database (Gene model 6; http://genome.jgi-psf.org/Araly1/Araly1.home.html) (Hu et al., 2011), gene and genome data for Thellungiella halophila were from Yang et al. (2013), Schrenkiella parvula and Thellungiella salsuginea datasets were from Dassanayake et al. (2011) and Wu et al. (2012), Leavenworthia alabamica, Sisymbrium irio, and Aethionema arabicum genomic data sets were from Haudry et al. (2013), Camelina sativa and B. oleracea genomic datasets were from Kagale et al. (2014) and Liu et al. (2014), and the Raphanus sativus genomic dataset was obtained from Kitashiba et al. (2014).
Phylogenetic analysis and motif identification
AOP genes from other sequenced Brassicaceae species such as Arabidopsis lyrata, S. parvula, T. salsuginea, T. halophila, L. alabamica, S. irio, Aethionema arabicum, Camelina sativa, B. oleracea, and R. sativus were identified based on BRAD multi-syntenic orthologues with Arabidopsis thaliana (http://brassicadb.org/brad/searchSyntenytPCK.php). We aligned full-length sequences of AOP proteins from the sequenced Brassicaceae genomes using CLUSTAL W with default parameters (Larkin et al., 2007). The phylogenetic tree was reconstructed based on the conserved sequences in the N-terminal and C-terminal parts of AOP proteins using the neighbour-joining method with MEGA v.6.0 software (Tamura et al., 2011), and bootstrap values with 1000 replicates were calculated. MEME version 4.9.1 (Bailey et al., 2009) was used to identify the conserved motifs of syntenic AOP proteins in the sequenced Brassicaceae species. The parameters for the analysis were as follows: number of repetitions, 0 or 1; maximum number of motifs, 14; and optimum motif width, 6–100. The MAST program (Bailey and Gribskov, 1998) was used to search for each of the motifs in the AOP sequences.
Heterologous expression and enzyme assays
Total leaf RNA was isolated using a Total RNA Extraction kit according to the manufacturer’s instructions (Sangon, Shanghai, China). First-strand cDNA was synthesized from approximately 2 µg of total RNA using a TransScript First Strand cDNA Synthesis SuperMix kit (TransGen Biotech, Beijing, China) with oligo(dt) as a primer in a 20 µl reaction. Full-length cDNAs of BrAOP2.1, BrAOP2.2, and BrAOP2.3 were amplified using gene-specific primers (Supplementary Table S3, available at JXB online) and cloned into pET-32a expression vectors (Novagen, Madison, WI, USA). This procedure placed the cDNAs into a fusion protein with thioredoxin under the control of a T7 promoter. After cloning, the inserted cDNA was sequenced to verify the junctions and to ensure that no mutations were introduced by PCR. The construct was expressed in Escherichia coli strain BL21(DE3) grown in Luria–Bertani medium to an OD of 0.6 at 600nm. Induction of recombinant protein synthesis was initiated by the addition of 0.5mM isopropyl β-d-thiogalactopyranoside overnight at 16 °C. Cells were harvested by centrifugation at 8000g. After resuspension in 1M Tris/HCl (pH 7.5), 200mg of lysozyme was added and incubated on ice for 30min. The cells were then sonicated twice with a microprobe at 55% of full power for a 5min 20% cycle. Cell debris was precipitated by centrifugation at 12 000g and the supernatant was used for enzyme purification by the BugBuster® Ni-NTA His Bind Purification kit following the manufacturer’s instructions (Novagen). The purified lysate was tested by SDS-PAGE and then used for the enzyme assays.
Enzyme assays were conducted as described by Kliebenstein et al. (2001b ) with some modifications. The assay mixture contained 200mM sucrose, 10mM oxoglutarate, 10mM ascorbate, 100 µl of purified intact GRA, 200 µM FeSO4, and 400 µl of enzyme preparation in a final volume of 4ml. The reactions were allowed to proceed for 4h at 28 °C, and the glucosinolates were then purified and analysed by high-performance liquid chromatography (HPLC).
Subcellular localization of BrAOP2 proteins
To identify the subcellular localization of the three BrAOP2 proteins, ProCAMV35S:BrAOP2:GFP (green fluorescent protein) constructs were made. BrAOP2 coding sequences without stop codons were isolated and cloned into the C-terminal GFP fusion vector pSPYCE-35S/pUC-SPYCE. The resulting constructs were verified by DNA sequencing. The subcellular localization of BrAOP2 proteins was detected by monitoring the transient expression of GFP in B. rapa mesophyll protoplast cells (Yoo et al., 2007) under a confocal laser-scanning microscope (Nikon, Tokyo, Japan). GFP fluorescence was imaged at an excitation wavelength of 488nm, and the emission signal was detected at 495–530nm for GFP and at 643–730nm for chlorophyll autofluorescence.
Site-directed mutagenesis of BrAOP2
To identify the key active-site residues of BrAOP2, site-directed mutagenesis was performed according to the procedure of Ho et al. (1989) by overlap extension using PCR. The mutagenic primers used to randomly change two histidines to leucine (H308L and H356L), aspartic acid to alanine (D310A), and arginine to tryptophan (R376W) are shown in Supplementary Table S4, available at JXB online. The mutated sequences were analysed by sequencing after being cloned into the pEASY-T1 vector (TransGen Biotech). The heterologous expression and enzyme assays of the mutant proteins were also analysed as described above.
Generation of transgenic plants overexpressing BrAOP2
The coding sequences of BrAOP2 genes were isolated and amplified using Chiifu-401/42 genomic cDNA as a template with gene-specific primers including restriction sites (XbaI/SmaI) and ligated to the pEASY-T1 vector. Sequencing analysis was performed and the pEASY-T1:BrAOP2 constructs were digested with XbaI/SmaI and inserted into a pBI121 vector driven by the cauliflower mosaic virus (CaMV) 35S promoter. The resulting construct was verified by DNA sequencing and subsequently transformed into Agrobacterium tumefaciens (strain GV3101). The binary vector pBI121 contains a kanamycin resistance gene to aid in the selection of transformed Arabidopsis lines. The floral infiltration method (Clough and Bent, 1998) was used to transform Col-0 plants. The T1 generation was first screened on kanamycin selection medium (half-strength MS salt, 60mg l−1 of kanamycin) and then transferred to soil. The T2 generation derived from selected plants was used to identify homozygous transformed lines. T3 generation homozygous plants were subsequently used in HPLC analysis.
Histochemical analysis of transgenic plants expressing ProBrAOP2:GUS fusion constructs
The BrAOP2 promoter regions (~1.2kb) were amplified from the genomic DNA of Chiifu-401/42 plants and cloned into the pEASY-1 vector. To drive β-glucuronidase gene (GUS) expression under the control of BrAOP2 promoters, the plant transformation vectors pBI121 and pCambia 1300 were recombined with HindIII/XbaI (BrAOP2.1 Pro) and PstI/XbaI (BrAOP2.2 Pro and BrAOP2.3 Pro) reactions (Supplementary Table S5, available at JXB online). The identified ProBrAOP2:GUS clones were transformed into Agrobacterium tumefaciens strain GV3101 and Arabidopsis thaliana Col-0 plants. The histochemical localization of GUS in several independent transgenic lines harbouring the ProBrAOP2:GUS construct was performed as described by Jefferson et al. (1987) with some modifications. Sample tissues were infiltrated with reaction buffer (50mM Na2HPO4/NaH2PO4, pH 7.0, 0.5mM K3Fe(CN)6, 0.5mM K4Fe(CN)6, containing 2mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid as substrate), and incubated at 37°C overnight. Plant pigments were destained with 75% ethanol, and GUS staining patterns were recorded under a binocular microscope (Stemi 2000-C; Zeiss).
Glucosinolate extraction and HPLC analysis
Glucosinolates were extracted and measured as described previously (Hongju and Schnitzler, 2002). Lyophilized samples (0.20g) were weighed accurately in 15ml plastic tubes and immersed in boiling methyl alcohol (5ml) containing 100 µl of benzyl glucosinolate as the internal standard. After 20min of additional shaking, samples were cooled at 4 °C and centrifuged at 3000g for 10min. The supernatant fraction (extract) was cleaned twice with 70% methyl alcohol, loaded on DEAE Sephadex A-25 columns and desulfated overnight at room temperature using purified sulfatase (Sigma, E.C. 3.1.6.) prior to HPLC. The column was then washed three times with 0.5ml of deionized water, and the eluent filtrated through a 0.45 µm membrane was used for HPLC analysis. Glucosinolates were identified by comparing retention times and UV absorption spectra with purified standards. The concentration of individual glucosinolates was calculated in nmol mg−1 of dry weight relative to the area of the internal standard peak using the respective response factors reported previously (Brown et al., 2003).
Reverse transcriptase (RT)-mediated first-strand synthesis and real-time quantitative (q)RT-PCR analysis
Total RNA was isolated from different organs using a Total RNA Extraction kit according to the manufacturer’s instructions (Sangon) and then treated with DNase I (Sigma-Aldrich, MO, USA) to eliminate DNA. The RNA purity was determined spectrophotometrically, and the quality was determined by examining rRNA bands on 1% agarose gels. cDNA was synthesized from approximately 2 µg of total RNA using TransScript First-Strand cDNA Synthesis SuperMix (TransGen Biotech) with oligo(dT) as a primer in a 20 µl reaction.
Primer specificity for the three BrAOP2 genes and BrGAPDH was verified by DNA sequencing after cloning the PCR products into the pEASY-T1 vector. The efficiency of gene-specific BrAOP2 and BrGAPDH primers was ascertained initially using a 4-fold serial dilution of L143 cDNA. A linear correlation coefficient (r 2) of 0.95 and above was observed over a 64-fold dilution range, indicating the high efficiency of each primer pair. qRT-PCR was performed in a total volume of 15 µl, including 2 µl of diluted cDNA, 0.5 µl of each primer (10 pM), and 7.5 µl 2×SYBR Green Master Mixes (Thermo Fisher, MA, USA) on an Eppendorf Real-Time PCR System (Eppendorf, Hamburg, Germany) according to the manufacturer’s instructions. The qRT-PCR program was conducted at 95 °C for 2min, followed by 40 cycles of 95 °C for 30 s and 60°C for 60 s. The expression level of BrGAPDH (glyceraldehyde 3-phosphate dehydrogenase) was used as an internal control and the expression of other genes was computed using the 2–ΔΔCT method (Livak and Schmittgen, 2001). Data were analysed from three independent sets of biological replicates with three technical replicates for each. The primers used in this work are listed in Supplementary Table S6, available at JXB online.
Statistical analysis
Data from different experimental sets were analysed for statistical significance using one-way analysis of variance (ANOVA) with a Duncan post-hoc test with SPSS software. A P value of <0.05 was considered significant.
Results
Paralogous relationship of the three BrAOP2 genes
In B. rapa, three AOP2 paralogues produced by genome replication and distributed in different chromosomes have been annotated in the Brassica Database (BRAD, http://brassicadb.org); namely, BrAOP2.1 (Bra03418, A09), BrAOP2.2 (Bra000848, A03), and BrAOP2.3 (Bra018521, A02). We isolated and sequenced the BrAOP2 genes from B. rapa accession Chiifu-401/42 based on their sequences in BRAD (Cheng et al., 2011). The three BrAOP2 genes contain two conserved (exon 1 and exon 3) and one variable (exon 2) exons compared with AtAOP2 (Fig. 1). As a result, the open reading frames varied from 1287 to 1323bp, encoding proteins of 439, 440, and 428 aa, respectively (Table 1). The deduced amino acid sequences from the three BrAOP2 genes were 74–81% identical (Supplementary Table S1, available at JXB online), and shared high similarity (74–96%) with the functional BoAOP2 protein, and lower similarity (55–60%) with AtAOP2 ecotype Cvi (Supplementary Table S1).
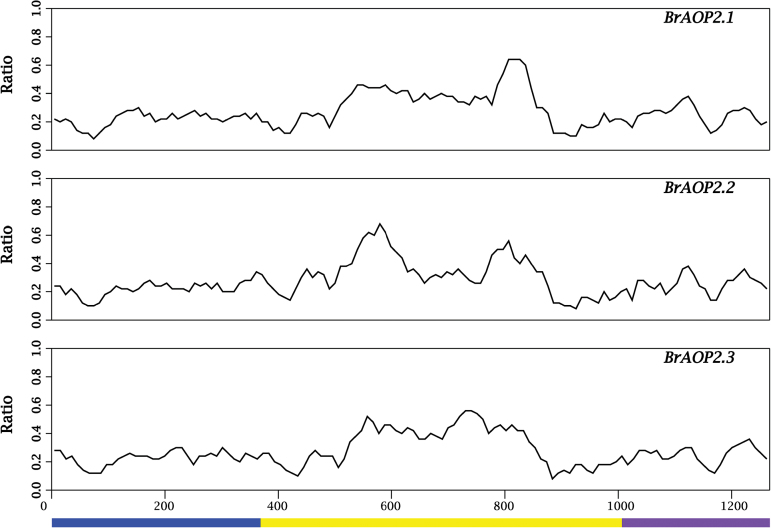
Fig. 1.
Nucleotide polymorphisms in the three BrAOP2 and AtAOP2 sequences. Each BrAOP2 sequence was compared with the AtAOP2 sequence, and the relative frequency of nucleotide substitution between them was tested. The vertical axis reflects the relative frequency of nucleotide substitution between BrAOP2 and AtAOP2 sequences per sliding window. A sliding window of 50bp with a step width of 10bp was used. Alignment gaps were included in scaling the horizontal axis. The three exons of AtAOP2 are depicted under the horizontal axis. (This figure is available in colour at JXB online.)
Table 1.
Summary of the BrAOP2 sequences used in the study
| Gene ID | Coding sequence (bp) | Protein (aa) | No. of exons (size in bp) | No. of introns (size in bp) |
|---|---|---|---|---|
| BrAOP2.1 (Bra034180) | 1320 | 439 | 3 (368 691 261) | 2 (293 167) |
| BrAOP2.2 (Bra000848) | 1323 | 440 | 3 (371 691 261) | 2 (16 881 358) |
| BrAOP2.3 (Bra018521) | 1287 | 428 | 3 (371 655 261) | 2 (398 853) |
Alignment of the amino acids confirmed the presence of two conserved domains, DIOX-N and 2OG-FeII_Oxy, at the N-terminal and C-terminal regions of the BrAOP2s, respectively (Fig. 2). These two conserved domains are known to be responsible for 2-oxoglutarate/Fe(II)-dependent dioxygenase activity, which is associated with an important class of enzymes that mediate a variety of oxidative reactions (Prescott and Lloyd, 2000). In contrast to the two highly conserved domains, the middle part of BrAOP2 proteins only showed patches of similarity (Fig. 2). In this variable region, the deduced BrAOP2 proteins were all composed of three motifs (e.g. motif 12, 13, 14), with one additional motif 14 involved in BrAOP2.2 and BrAOP2.3, respectively (Supplementary Fig. S1 and Table S7, available at JXB online). The structural divergence of BrAOP2 proteins might have consequences for their functional diversification.
Fig. 2.
Multiple alignments of the BrAOP2, BoAOP2, and AtAOP2 protein sequences. Multiple alignments were performed using MEGA v.6.0. AtAOP2 is a known functional protein from Arabidopsis thaliana (ecotype Cvi), and BoAOP2 is from B. oleracea (collard). The solid lines above the alignment indicate the consensus sequences for the DIOX-N and 2OG-FeII_Oxy domains identified using the Conserved Domain Database (Marchler-Bauer et al., 2011)). The black shading indicates that at least four proteins share the same amino acid site. Asterisks show four active-site residues in the 2OG-FeII_Oxy domain.
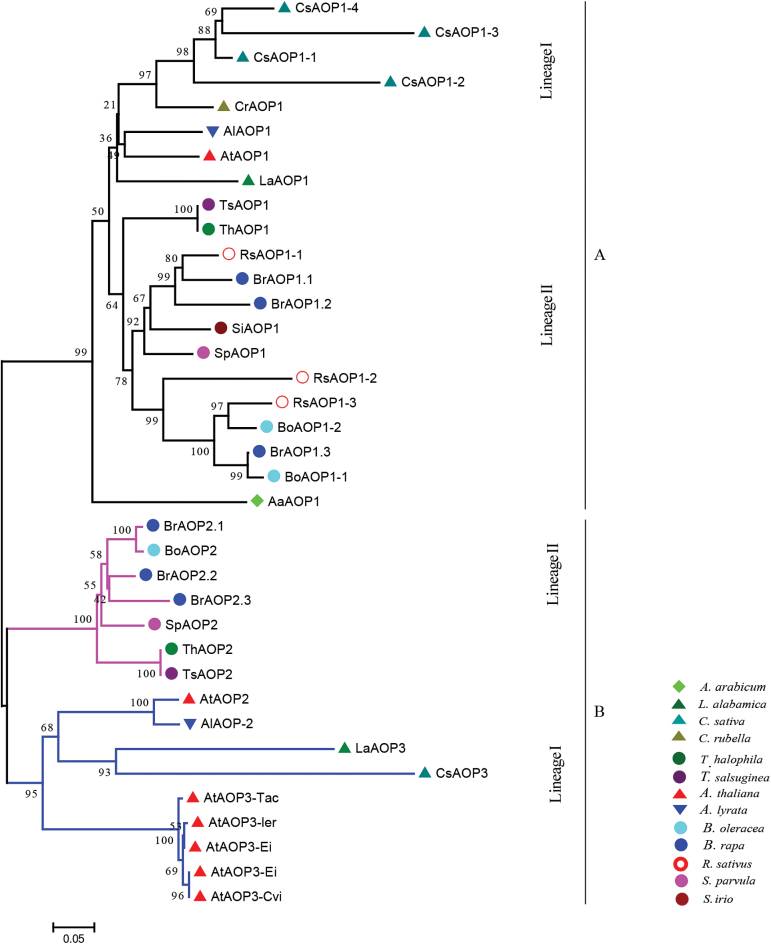
The Brassicaceae is a medium-sized family that can be split into two major groups: the Aethionema group and the core group (Franzke et al., 2011). Three major lineages (I, II, and III) of the core group have been proposed based on the sequences of the chloroplast NADH dehydrogenase gene ndhF (Beilstein et al., 2006) and supported by subsequent studies (Koch et al., 2007; Beilstein et al., 2008; Couvreur et al., 2010). To investigate further the evolutionary origin of the three BrAOP2 genes, we first identified all AOP genes in the 13 sequenced Brassicaceae species in BRAD (Cheng et al., 2011) according to their genome annotation information and their syntenic relationship with Arabidopsis thaliana AOP genes (http://brassicadb.org/brad/searchSyntenytPCK.php) (Supplementary Table S2, available at JXB online). We next reconstructed a phylogenetic tree of all deduced AOP amino acid sequences using the maximum-likelihood method (Tamura et al., 2011) (Fig. 3).
Fig. 3.
Evolutionary relationships and motif structures of Brassicaceae AOP proteins. Phylogenetic analysis was performed based on the conserved sequences at the N and C terminal parts of AOP proteins using the neighbour-joining method (bootstrap test with 1000 replicates) (Tamura et al., 2011). The sequenced Brassicaceae species in BRAD included: lineage I species Arabidopsis thaliana, Arabidopsis lyrata, Capsella rubella, Camelina sativa, and L. alabamica; lineage II species T. halophila, T. salsuginea, S. parvula (synonym of T. parvula), S. irio, B. rapa, B. oleracea, and R. sativus; and an early branching sister of the core Brassicaceae, Aethionema arabicum. Black branches indicate AOP1 proteins, coloured branches indicate AOP2 and AOP3 proteins. The tree is drawn to scale; bar, number of substitutions per site.
Phylogenetic analyses revealed that all sequenced Brassicaceae species in core Brassicaceae retained AOP1 genes derived from AaAOP1 of Aethionema arabicum (Fig. 3, group A). Most of the lineage II species (excluding S. irio and R. sativus) retained AOP2 genes. Most of the lineage I species possessed AOP3 genes, which demonstrated further structural divergence in the middle part from AOP2 (Fig. 3, group B; Supplementary Fig. S1 and Supplementary Table S1). BrAOP2.1 grouped with BoAOP2 with high bootstrap support. Although AtAOP2 and AtAOP3 clustered together in Arabidopsis thaliana accessions, they are not co-expressed because of their inverted gene structure (Chan, et al., 2010). All AOP2 genes of the genus Brassica were more similar to those of the genus Thellungiella than the genus Arabidopsis.
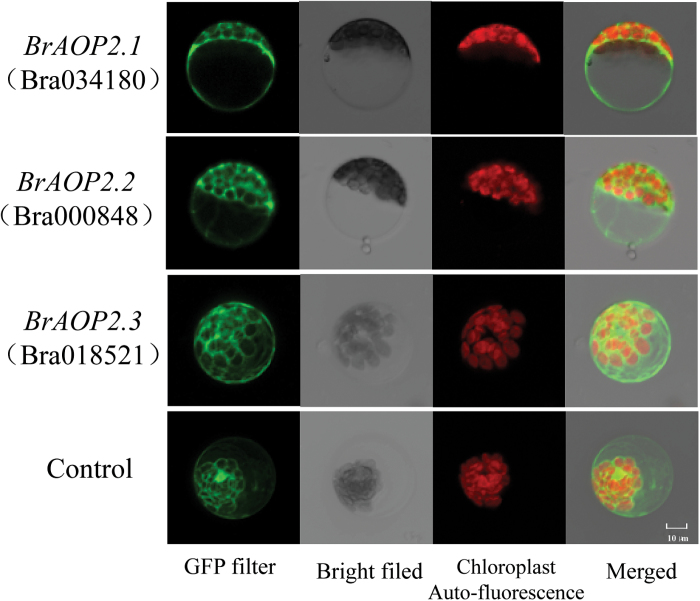
The three BrAOP2 proteins are all located in the cytoplasm
To investigate the subcellular localization of the three BrAOP2 proteins, three ProCAMV35S:BrAOP2:GFP vectors were constructed and their localization was detected by monitoring the transient expression of GFP in B. rapa mesophyll protoplast cells (Fig. 4). A strong green fluorescent signal was observed in the cytoplasm of transiently transformed cells, demonstrating that the three BrAOP2 proteins were predominantly cytoplasmic, consistent with the subcellular localization for the secondary modification of aliphatic glucosinolates in Arabidopsis (Sonderby et al., 2010).
Fig. 4.
Subcellular localization of BrAOP2 proteins in B. rapa protoplasts. Images were taken in a dark field for green fluorescence and chloroplast autofluorescence (red), while the outlook of cells was photographed in a bright field. Bar, 10 µm.
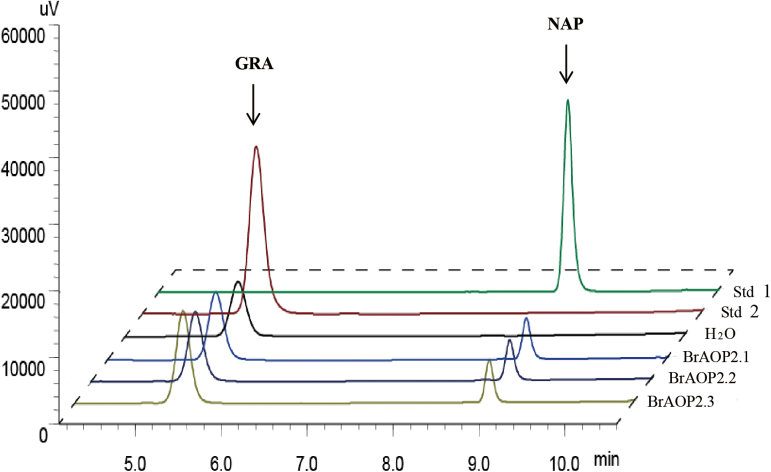
The three BrAOP2 paralogues are all active in B. rapa
The in vitro catalytic activity of the three BrAOP2 proteins can be monitored readily when they are expressed heterologously by measuring the conversion of GRA to 3-butenyl glucosinolate (NAP). We therefore heterologously expressed and purified thioredoxin fusion proteins for the three BrAOP2 genes in E. coli. As shown in Fig. 5, all three BrAOP2 proteins successfully catalysed the conversion of GRA to NAP, showing that they all had the capacity to convert methylsulfinylalkyl glucosinolates to the alkenyl form (GSL-ALKs).
Fig. 5.
Enzymatic activity of BrAOP2 heterologously expressed in E. coli. HPLC results (monitored at 229nm) of purified desulfoglucosinolates from bacterial extracts containing heterologously expressed BrAOP2 fusion proteins is shown. The compound identities were confirmed by comparing retention times and UV light absorption profiles with those of authentic standards. Std 2 indicates desulfated GRA standard, Std 1 indicates desulfated NAP standard, BrAOP2.1, BrAOP2.2, and BrAOP2.3 indicate GRA treated with these three BrAOP2 enzymes, and H2O shows GRA treated with ddH2O as the negative control.
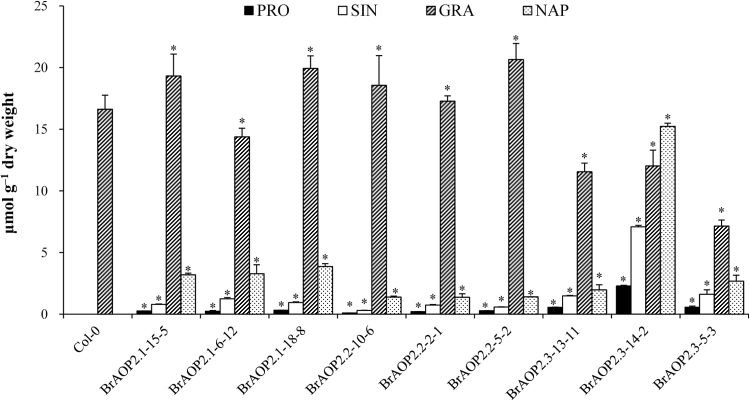
The in vivo functional contribution of each BrAOP2 gene to the conversion of methylsulfinylalkyl glucosinolate to alkenyl glucosinolates can be assessed in AOP2 mutant Arabidopsis by the complementation test. Arabidopsis Col-0 contains a non-functional AtAOP2 allele with natural variations that lead to the accumulation of GRA (Kliebenstein et al., 2001b ), which is regarded as an AOP2 mutant in the complementation test. All three BrAOP2 genes (under the control of the constitutive CaMV 35S promoter) were introduced into Arabidopsis Col-0, and at least three independent homozygous lines of each BrAOP2 gene were analysed for total and individual glucosinolate fractions in 6-week-old rosette leaves.
The functional complementation among the BrAOP2 genes catalysed the conversion of the C3 and C4 methylsulfinylalkyl glucosinolate to alkenyl glucosinolates (e.g. sinigrin and NAP); this conversion was not observed in the Col-0 control (Fig. 6). Line BrAOP2.3-14-2 showed a high level of conversion of the precursor methylsulfinylalkyl glucosinolate to sinigrin and NAP as well as to 2-hydroxy-3-butenyl-glucosinolate (PRO). PRO was further hydroxylated by the action of the GS-OH locus product (Mithen et al., 1995). The two lines BrAOP2.1-15-5 and BrAOP2.1-18-8 both showed a slightly lower conversion of the precursor methylsulfinylalkyl glucosinolate to sinigrin and NAP. All three BrAOP2.2 lines showed relatively low conversions of the precursor methylsulfinylalkyl glucosinolate to sinigrin and NAP, producing trace amounts of PRO. Thus, the mutant complementation analysis in Arabidopsis thaliana clearly suggested that all three BrAOP2 genes had biological activities in vivo.
Fig. 6.
Functional complementation analyses of BrAOP2 genes in Arabidopsis thaliana Col-0. The glucosinolate content and profile were determined in 6-week-old rosette leaves. Three independent mutant-complemented lines for each BrAOP2 gene were analysed, and average foliar glucosinolates from 30 individual plants are represented along with their standard errors. Asterisks indicate significant differences in glucosinolate content compared with Col-0 (P<0.05, one-way ANOVA analysis with a Duncan post-hoc test). PRO, 2-hydroxy-3-butenyl-glucosinolate; SIN, sinigrin.
His356, Asp310, and Arg376 residues are required for the catalytic activity of BrAOP2.1
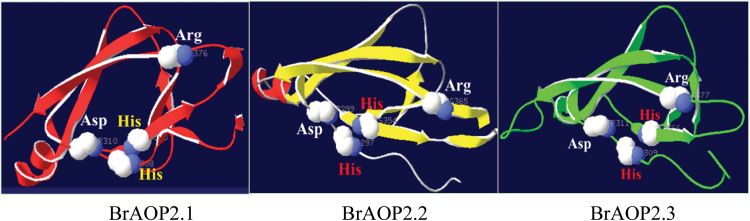
The two conserved domains DIOX-N and 2OG-FeII_Oxy at the N- and C-terminal regions of BrAOP2 (Fig. 2) are essential for 2-oxoglutarate/Fe(II)-dependent dioxygenase activity (Pfam accession number PF03171), which can act on a variety of substrates (Schofield and Zhang, 1999; Costas et al., 2004). The catalytically active site is formed by a mononuclear non-haem Fe centre co-ordinated by two histidine residues and one carboxylate moiety (Hegg and Que, 1997; Zhou et al., 1998; Neidig et al., 2004; Purpero and Moran, 2007). We identified four active-site residues, His308, His356, Asp310, and Arg376, in the conserved 2OG-FeII_Oxy domain of BrAOP2.1 using the PROSITE database (http://prosite.expasy.org/) (Fig. 7).
Fig. 7.
Four active-site residues identified in three BrAOP2 2OG-FeII_Oxy domains. The tertiary structure of each BrAOP2 protein was predicted with the program SWISS-MODEL, and the representation was made with Swiss Pdb-Viewer 4.01 (Arnold et al, 2006). (This figure is available in colour at JXB online.)
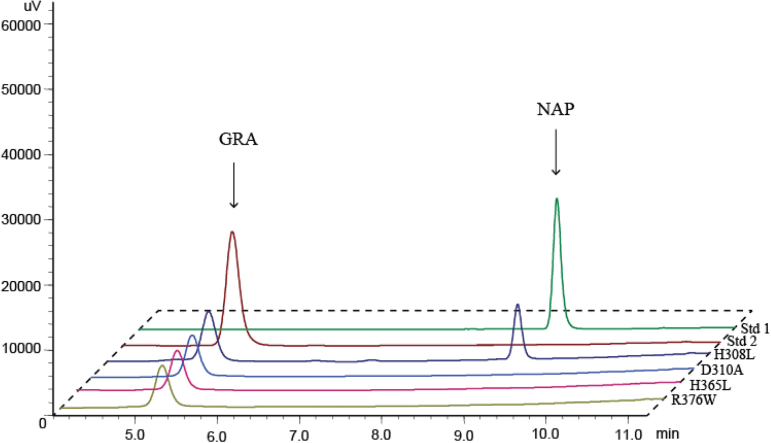
To test the hypothesis that strictly conserved amino acid residues are required for AOP2 activity, we mutated the nucleotides that encode the conserved amino acids of BrAOP2.1 by site-directed mutagenesis. The Asp310 codon was changed to encode alanine (D310A), the histidine codons at positions 308 and 356 were changed to encode leucine (H308L and H356L), and the codons for arginine residue 376 were changed to encode tryptophan (R376W). Heterologous expression and enzyme assays were conducted for the mutant proteins, and the purified H308L mutant enzyme was shown to still catalyse the conversion of GRA to NAP, suggesting that His308 may be not essential for the catalytic mechanism (Fig. 8). By contrast, when His356, Asp310, or Arg376 were changed to leucine, alanine, and tryptophan, respectively, no catalytic activity was detected in any of the mutant BrAOP2.1 proteins (Fig. 8). These results suggested that these three highly conserved amino acid residues shown to be necessary for iron binding in the 2OG-FeII_Oxy domain are crucial for enzymatic activity in BrAOP2.
Fig. 8.
Enzymatic activity of heterologously expressed site-directed mutant BrAOP2.1 proteins. The purified mutant proteins were assayed with GRA and the products were extracted and analysed by HPLC as described in Materials and methods. Site-directed mutagenesis of BrAOP2.1 was conducted four times to test the four active-site residues. (This figure is available in colour at JXB online.)
BrAOP2 genes exhibit distinct expression levels in B. rapa
Genome polyploidy events are often associated with variable expression of homologous gene pairs within a genome. Expression divergence between duplicate genes has long been of interest to geneticists and evolutionary biologists (Ohno, 1970; Ferris and Whitt, 1979) because divergence is considered the first step in functional divergence between duplicate genes, which increases the chance of retention of duplicate genes in a genome (Ohno, 1970). Indeed, the divergence of gene expression between duplicate genes has been reported in many studies (Blanc and Wolfe, 2004; Casneuf et al., 2006; Duarte et al., 2006; Tuskan et al., 2006).
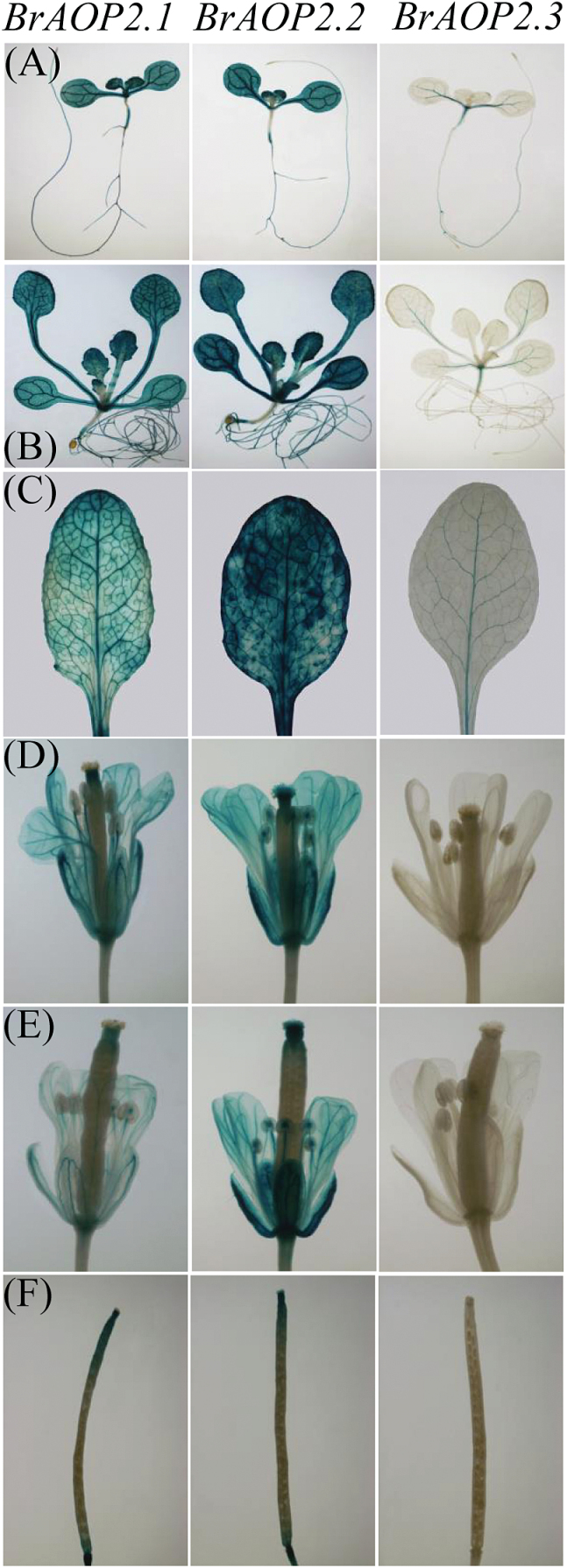
To better understand the expression of BrAOP2 genes, we generated transgenic plants carrying a GUS reporter driven by each BrAOP2 promoter (ProBrAOP2:GUS). Reporter gene expression in transgenic Arabidopsis lines revealed that BrAOP2.1 and BrAOP2.2 paralogues showed similar and stable expression patterns, while ProBrAOP2.3:GUS lines showed distinct expression patterns during the developing stages of Arabidopsis thaliana (Fig. 9).
Fig. 9.
Histochemical GUS staining of ProBrAOP2:GUS transgenic Arabidopsis lines at different developmental stages. (A) One-week-old seedlings, (B) 3-week-old seedlings, (C) 5-week-old rosette leaves, (D) flowers, (E) immature silique, and (F) mature silique. Plants from stages (A) and (B) were grown on MS medium, and plants from stage (C–F) were grown on soil. A GUS histochemical assay was carried out on three independent single-copy transgenic lines of each BrAOP2 paralogue in the T3 generation and showed the same result.
Histochemical analysis revealed higher GUS activity in the seedlings, mature rosette leaves, flowers, and siliques of ProBrAOP2.1:GUS transgenic lines compared with ProBrAOP2.2:GUS lines. In ProBrAOP2.3 transgenic lines, prominent GUS staining was detected in the cotyledon of seedlings and mature rosette leaves (Fig. 9A–C), with undetectable GUS expression in the roots of 1-week-old seedlings, flowers, and siliques. In the rosette leaves of Arabidopsis, the BrAOP2 promoter showed cell-specific GUS activity. This promoter showed maximal activity in the whole leaves, including the vasculature and mesophyll cells, whereas the predominant activity of the BrAOP2.1 and BrAOP2.2 promoters was observed in the mid-vein, primary, and secondary veins (Fig. 9B), and that of the BrAOP2.3 promoter was mainly in the mid-vein and primary veins. The non-uniform and cell-specific expression patterns of the BrAOP2 genes within leaves, roots, and reproductive tissues might have important implications for the accumulation of NAP across different regions of the plant. Thus, the GUS histochemical data obtained using ProBrAOP2:GUS Arabidopsis lines confirmed that the BrAOP2.3 promoter has overlapping but distinct cell and tissue expression patterns.
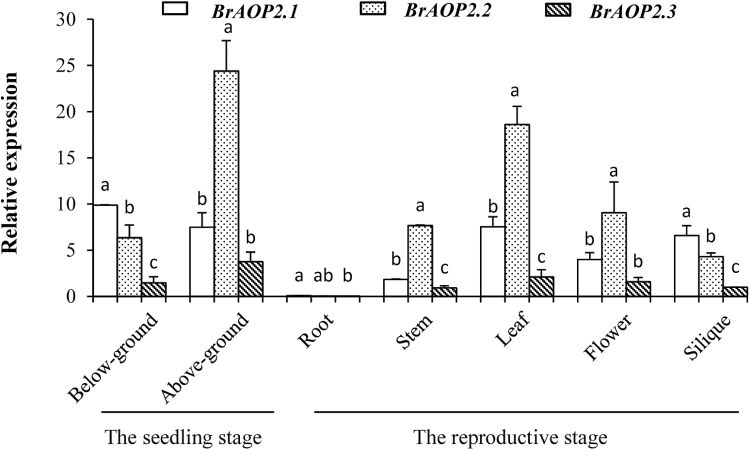
We further measured BrAOP2 expression at the transcript level during different developmental stages and in different tissue types in accession L143 (a yellow sarson accession) using qRT-PCR analysis. In general, BrAOP2.1 showed a slightly different pattern of expression in below-ground tissue at seedling stage and silique at reproductive stage compared to BrAOP2.2 and BrAOP2.3 genes (Fig. 10). In the seedling stage, the expression of BrAOP2.1 was higher than that of BrAOP2.2 and BrAOP2.3 in below-ground tissue, while in above-ground tissues the expression of BrAOP2.2 was higher than that of BrAOP2.1 and BrAOP2.3. In the reproductive stage, the three BrAOP2 genes were expressed abundantly in glucosinolate-synthesizing tissues such as leaves, siliques, and flowers, as well as in transporting tissue stems; only a trace accumulation of these transcripts was detected in the roots. In above-ground tissues, BrAOP2.2 was highly expressed in the leaves, stems, and inflorescence, but was less expressed in the siliques. BrAOP2.1 was abundant in the siliques and leaves, while the expression of BrAOP2.3 was reduced in all above-ground tissues tested.
Fig. 10.
Relative expression profiles of BrAOP2 genes in L143 organs at the seedling stage (2 weeks) and reproductive stage (10 weeks) (horizontal axis). The vertical axis indicates gene expression levels relative to BrAOP2.3 (Bra000848) expression in siliques. Error bars represent the standard deviation from three biological repeats. Different lowercase letters indicate significant differences at P<0.05 using ANOVA analysis followed by a Duncan post-hoc test.
Discussion
Nutritionally and economically important Brassica crops such as cabbage, broccoli, and cauliflower are rich sources of glucosinolates, which are present in all organs, especially aliphatic glucosinolates. Here, we have reported the characterization, functional analysis, and expression of BrAOP2 paralogues in B. rapa. The three BrAOP2 genes exhibited overlapping but distinct tissue- and cell-specific expression patterns, suggesting that they play a co-ordinated role in catalysing the conversion of beneficial GRA to the harmful gluconapin. This information will be helpful to breeders aiming to modify glucosinolate levels in B. rapa.
All three BrAOP2 genes catalysed the conversion of GRA to NAP
Genome polyploidization is widespread in the plant kingdom where it plays a major role in generating diversity and providing abundant genetic material for the evolution or expansion of gene families and the formation of new genes (Hittinger and Carroll, 2007; Spillane et al., 2007). The evolutionary consequences of duplicated genes after polyploidy include loss or silencing (non-functionalization), retention of the original function, or functional divergence through subfunctionalization or neofunctionalization (Lynch, 2000; Zhang, 2003).
In our study, overexpression of the BrAOP2 paralogues in Arabidopsis thaliana Col-0 and E. coli demonstrated that the three encoded BrAOP2 proteins are involved in the conversion of methylsulfinylbutyl glucosinolates to corresponding alkenyl glucosinolates. The enzyme activity suggested that the three BrAOP2 genes might be involved in a similar reaction in B. rapa. Multiple alignments of the amino acid sequences of the three BrAOP2 proteins showed significant structural conservation in the C- and N-terminal regions. However, a frame-shift mutation in exon 2 results in an absent or incomplete 2OG-FeII_Oxy domain (at the N terminus), giving rise to a non-functional AOP2 allele in other Brassicaceae (e.g. BoAOP2 from broccoli and AtAOP2 from Col-0; Li and Quiros, 2003; Neal et al., 2010). Site-directed mutagenesis analyses showed that all three BrAOP2 proteins shared three key active-site residues in their 2OG-FeII_Oxy domains that are crucial for enzymatic activity. Therefore, the 2OG-FeII_Oxy domain and its key active-site residues may be responsible for BrAOP2s retaining their dioxygenase activity across plant development stages and tissue/cell types, and under variable environmental conditions. Thus, polyploidization of the Brassica genomes does not seem to have altered the basic BrAOP2 gene function, and all AOP2 homologues seem to have retained the subdivision of gene function in most polyploidy Brassica crops.
AOP variation and speciation in the Brassicaceae
The Brassicaceae contains 338 genera and 3709 species, including many economically important crops (Warwick et al., 2006). The core group has undergone three ancient whole-genome duplication events (Franzke et al., 2011), that have played a crucial role in the genetic diversification and species radiation of lineages in the Brassicaceae. Furthermore, whole-genome triplication events have occurred in Brassica (Br-α), L. alabamica (La-α), and Camelina sativa (Cs-α), as determined by analyses of their recently sequenced genomes (Haudry et al., 2013; Slotte et al., 2013; Cheng et al., 2014). Additionally, the genomes of 13 crucifer species have been completely or partially sequenced, and this has provided the opportunity to clarify the evolution of AOP genes.
Our phylogenetic analyses showed that the core Brassicaceae species have retained AOP1, while AOP2 is retained by most of the lineage II species (excluding S. irio and R. sativus), and AOP3 by lineage I species. The variation in AOP2/AOP3 has led to different aliphatic glucosinolate profiles in each lineage (Al-Shehbaz and Al-Shammary, 1987). Our phylogenetic tree is consistent with the finding that two different gene duplication events occurred in the Brassicaceae AOP locus (Kliebenstein et al., 2001b ); the first event led to the separation of AOP1 and the AOP2/AOP3 progenitor, while the second gave rise to the formation of AOP2 and AOP3. Following gene duplications, the separate copies evolved new enzymatic functions. Thus, it is likely that some copies were lost when the genetic backgrounds of species altered, or as a result of environmental adaptation. The variations in lineage specificity of AOP2 versus AOP3 in Brassicaceae species agrees with previous data showing that GS-ALK (AOP2)/GS-OHP (AOP3) variation occurred in the distantly related Arabidopsis and in the genera Thlaspi and Malcolmia (Daxenbichler et al., 1991). In the current study, the lineage-specific evolution of AOP in the core Brassicaceae group was assumed to proceed alongside the split of the three major lineages, and may have been driven by the whole-genome duplication and maintained by environmental pressure.
Expression divergence of BrAOP2 genes in B. rapa
Studies have shown that gene duplication enables duplicates to become specialized in various organs of the plant or in response to different environmental stimuli (Ferris and Whitt, 1979; Hughes, 1994; Adams, 2007; Ha et al., 2007; Kliebenstein, 2008). Thus, duplicate copy genes would be expected to have more diversified expression profiles than single-copy genes in a group of closely related organisms (Li et al., 2005). Polyploidy has considerable effects on duplicate gene expression, including silencing and up- or downregulation of one of the duplicated genes through increased variation in dosage-regulated gene expression, altered regulatory interactions, and rapid genetic and epigenetic changes (Osborn et al., 2003; Adams and Wendel, 2005).
In the current study, the expression profiles of three BrAOP2 paralogues were shown to vary in different organs of B. rapa (Fig. 10), indicating that they are differentially regulated during plant development. For example, in the L143 reproductive stage, only trace expression of BrAOP2 was detected in the roots, but expression was abundant in other organs, similar to the expression profiles reported previously for AtAOP2 (Neal et al., 2010). The histochemical analysis of GUS activity revealed that the three BrAOP2 genes showed non-uniform and cell-specific expression patterns during the development stages of Arabidopsis (Fig. 9). When an approximately 1.2kb region of the sequence upstream of the three BrAOP2 genes was scanned using the Plant Cis-Acting Regulatory Elements database (Lescot et al., 2002), several specific cis-regulatory elements related to tissue-dependent expression and elements responsive to auxin, glucose signalling, and abiotic and biotic responses were observed (see Supplementary Table S8, available at JXB online). The disparity of cis-regulatory elements observed among the three BrAOP2 promoters could contribute to the differential expression patterns of BrAOP2 (Chen et al., 2010). Moreover, the distinct developmental and spatial expression patterns of BrAOP2 within Arabidopsis also suggest a role in distributing glucosinolate content in B. rapa, which in turn may have important consequences for plant defence against environmental stresses.
BrAOP2: a crucial candidate for engineering beneficial glucosinolates in B. rapa
The hydrolytic breakdown products of GRA, especially the isothiocyanates, are beneficial bioactive constituents that have cancer-preventative properties in humans. However, GRA can further react with the goitrogenic compound progoitrin, which has other detrimental effects on animal health. Therefore, the enrichment of beneficial glucosinolates and the reduction of detrimental glucosinolates has been the focus of much attention in the breeding programmes of Brassica crops.
In the current study, we found that NAP was predominant in the total glucosinolate contents of B. rapa, which is consistent with the results of other studies (Padilla et al., 2007; Lou et al., 2008; Kim et al., 2010). Three functional BrAOP2 genes, each encoding an AOP2 enzyme, were identified as giving rise to the abundant NAP found in B. rapa. In B. oleracea, GRA accumulation was found to be caused by non-functional GSL-ALK genes (Li and Quiros, 2003), which are homologous to the AOP genes in Arabidopsis thaliana (Neal et al., 2010). In B. oleracea, a 2bp GSL-ALK deletion caused the gene to become non-functional, which abolished the conversion of methylsulfinyl glucosinolates into alkenyl glucosinolates, resulting in an enrichment of GRA (Li and Quiros, 2003). In B. napus, the application of a new germplasm with reduced detrimental glucosinolates and increased beneficial glucosinolates was achieved by using RNA interference to downregulate the expression of GSL-ALK (Liu et al., 2012). Thus, it is possible to block the side-chain modification to produce GRA-enriched B. rapa vegetables.
The work described in this study has helped increase our understanding of BrAOP2 genes in glucosinolate side-chain modifications of polyploidy B. rapa. Our findings provide evidence of expression partitioning and the evolution of AOP2 gene family homologues in determining the aliphatic glucosinolate profile of B. rapa. The information reported here should facilitate the improvement of aliphatic glucosinolate traits in Brassica crops.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Motif structures of AOP genes in Brassicaceae.
Supplementary Table S1. Amino acid sequence identity (%) of AOP2 among Arabidopsis thaliana, B. rapa, and B. oleracea.
Supplementary Table S2. AOP genes identified in 13 Brassicaceae species in the Brassica Database.
Supplementary Table S3. BrAOP2 primers used to clone the coding sequences for heterologous expression vector construction.
Supplementary Table S4. Mutagenic primers used to construct mutant BrAOP2 genes.
Supplementary Table S5. BrAOP2 primers used to clone the promoter sequences for vector construction.
Supplementary Table S6. Primer sequences for BrAOP2 and BrGAPDH used in qRT-PCR analysis.
Supplementary Table S7. The sequence of motifs in the middle part of AOP2 and AOP3 identified by MEME.
Supplementary Table S8. Summary of cis-regulatory elements present within a 1.2kb upstream region of BrAOP2 homologues.
Acknowledgements
We thank Chao Sun for his helpful suggestions. This work was supported by the National Program Fund for Key Basic Research Projects, the 973 Program of China (2013CB127000 and 2012CB113900), and the National High Technology R&D Program of China (2012AA100101). This research work was carried out in the Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, China. The authors declare no competing financial interests.
Glossary
Abbreviations:
- ANOVA
analysis of variance
- CaMV
cauliflower mosaic virus
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GFP
green fluorescent protein
- GRA
glucoraphanin
- GSL-ALK
alkenyl form of methylsulfinylalkyl glucosinolate
- GUS
β-glucuronidase
- HPLC
high-performance liquid chromatography
- MS
Murashige and Skoog
- NAP
3-butenyl glucosinolate
- PRO
2-hydroxy-3-butenyl-glucosinolate
- qRT-PCR
quantitative real-time reverse transcription-PCR.
References
- Adams KL. 2007. Evolution of duplicate gene expression in polyploid and hybrid plants. Journal of Heredity 98, 136–141. [DOI] [PubMed] [Google Scholar]
- Adams KL, Wendel JF. 2005. Novel patterns of gene expression in polyploid plants. Trends in Genetics 21, 539–543. [DOI] [PubMed] [Google Scholar]
- Al-Shehbaz IA, Al-Shammary KI. 1987. Distribution and chemotaxonomic significance of glucosinolates in certain Middle-Eastern cruciferae. Biochemical Systematics and Ecology 15, 559–569. [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Research 37, W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Gribskov M.1998. Combining evidence using p-values: application to sequence homology searches. Bioinformatics 12, 48–54. [DOI] [PubMed] [Google Scholar]
- Beilstein MA, Al-Shehbaz IA, Kellogg EA. 2006. Brassicaceae phylogeny and trichome evolution. American Journal of Botany 93, 607–619. [DOI] [PubMed] [Google Scholar]
- Beilstein MA, Al-Shehbaz IA, Mathews S, Kellogg EA. 2008. Brassicaceae phylogeny inferred from phytochrome A and Ndhf sequence data: tribes and trichomes revisited. American Journal of Botany 95, 1307–1327. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. 2004. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. The Plant Cell 16, 1679–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PD, Tokuhisa JG, Reichelt M, Gershenzon J. 2003. Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana . Phytochemistry 62, 471–481. [DOI] [PubMed] [Google Scholar]
- Casneuf T, De Bodt S, Raes J, Maere S, Van de Peer Y. 2006. Nonrandom divergence of gene expression following gene and genome duplications in the flowering plant Arabidopsis thaliana . Genome Biology 7, R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EK, Rowe HC, Kliebenstein DJ. 2010. Understanding the evolution of defense metabolites in Arabidopsis thaliana using genome-wide association mapping. Genetics 185, 991–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KN, Zhang YB, Tang TA, Shi SH. 2010. Cis-regulatory change and expression divergence between duplicate genes formed by genome duplication of Arabidopsis thaliana . Chinese Science Bulletin 55, 2359–2365. [Google Scholar]
- Cheng F, Liu S, Wu J, Fang L, Sun S, Liu B, Li P, Hua W, Wang X. 2011. BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biology 11, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Wu J, Wang X. 2014. Genome triplication drove the diversification of Brassica plants. Horticulture Research 1, 14024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke DB. 2010. Glucosinolates, structures and analysis in food. Analytical Methods 2, 310–325. [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. 2009. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323, 95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costas M, Mehn MP, Jensen MP, Que L. 2004. Dioxygen activation at mononuclear nonheme iron active sites: Enzymes, models, and intermediates. Chemical Reviews 104, 939–986. [DOI] [PubMed] [Google Scholar]
- Couvreur TL, Franzke A, Al-Shehbaz IA, Bakker FT, Koch MA, Mummenhoff K. 2010. Molecular phylogenetics, temporal diversification, and principles of evolution in the mustard family (Brassicaceae). Molecular Biology and Evolution 27, 55–71. [DOI] [PubMed] [Google Scholar]
- Dassanayake M, Oh DH, Haas JS, et al. 2011. The genome of the extremophile crucifer Thellungiella parvula . Nature Genetics 43, 913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daxenbichler ME, Spencer GF, Carlson DG, Rose GB, Brinker AM, Powell RG. 1991. Glucosinolate composition of seeds from 297 species of wild plants. Phytochemistry 30, 2623–2638. [Google Scholar]
- Duarte JM, Cui L, Wall PK, Zhang Q, Zhang X, Leebens-Mack J, Ma H, Altman N, dePamphilis CW. 2006. Expression pattern shifts following duplication indicative of subfunctionalization and neofunctionalization in regulatory genes of Arabidopsis . Molecular Biology and Evolution 23, 469–478. [DOI] [PubMed] [Google Scholar]
- Fahey JW, Zhang Y, Talalay P. 1997. Broccoli sprouts: an exceptionally rich source of inducers of enzymes that protect againstchemicalcarcinogens. Proceedings of the National Academy of Sciences, USA 94, 10367–10372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey JW, Haristoy X, Dolan PM, Kensler TW, Scholtus I, Stephenson KK, Talalay P, Lozniewski A. 2002. Sulforaphane inhibits extracellular, intracellular, and antibiotic-resistant strains of Helicobacter pylori and prevents benzo[a]pyrene-induced stomach tumors. Proceedings of the National Academy of Sciences, USA 99, 7610–7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner K, Mithen R, Williamson G. 1998. Selective increase of the potential anticarcinogen 4-methylsulphinylbutyl glucosinolate in broccoli. Carcinogenesis 19, 605–609. [DOI] [PubMed] [Google Scholar]
- Ferris SD, Whitt GS. 1979. Evolution of the differential regulation of duplicate genes after polyploidization. Journal of Molecular Evolution 12, 267–317. [DOI] [PubMed] [Google Scholar]
- Franzke A, Lysak MA, Al-Shehbaz IA, Koch MA, Mummenhoff K. 2011. Cabbage family affairs: the evolutionary history of Brassicaceae. Trends in Plant Science 16, 108–116. [DOI] [PubMed] [Google Scholar]
- Gamet-Payrastre L, Li PF, Lumeau S, Cassar G, Dupont MA, Chevolleau S, Gasc N, Tulliez J, Terce F. 2000. Sulforaphane, a naturally occurring isothiocyanate, induces cell cycle arrest and apoptosis in HT29 human colon cancer cells. Cancer Research 60, 1426–1433. [PubMed] [Google Scholar]
- Gao M, Li G, Yang B, McCombie WR, Quiros CF. 2004. Comparative analysis of a Brassica BAC clone containing several major aliphatic glucosinolate genes with its corresponding Arabidopsis sequence. Genome 47, 666–679. [DOI] [PubMed] [Google Scholar]
- Gershenzon BAHaJ. 2006. Biology and biochemistry of glucosinolates. Annual Review of Plant Biology 57, 303–333. [DOI] [PubMed] [Google Scholar]
- Ha M, Li WH, Chen ZJ. 2007. External factors accelerate expression divergence between duplicate genes. Trends in Genetics 23, 162–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkier BA, Gershenzon J. 2006. Biology and biochemistry of glucosinolates. Annual Review of Plant Biology 57, 303–333. [DOI] [PubMed] [Google Scholar]
- Hall C, McCallum D, Prescott A, Mithen R. 2001. Biochemical genetics of glucosinolate modification in Arabidopsis and Brassica. Theoretical and Applied Genetics 102, 369–374. [Google Scholar]
- Haudry A, Platts AE, Vello E, Hoen DR, et al. 2013. An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nature Genetics 45, 891–898. [DOI] [PubMed] [Google Scholar]
- Hegg EL, Que L., Jr. 1997. The 2-His-1-carboxylate facial triad—an emerging structural motif in mononuclear non-heme iron(II) enzymes. European Journal of Biochemistry 250, 625–629. [DOI] [PubMed] [Google Scholar]
- Hittinger CT, Carroll SB. 2007. Gene duplication and the adaptive evolution of a classic genetic switch. Nature 449, 677–681. [DOI] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59. [DOI] [PubMed] [Google Scholar]
- Hongju H HC, Schnitzler W H. 2002. Glucosinolate composition and contents in Brassica vegetables. Scientia Agricultural Sinica (China) 35, 192–197. [Google Scholar]
- Hu TT, Pattyn P, Bakker EG, et al. 2011. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nature Genetics 43, 476–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL. 1994. The evolution of functionally novel proteins after gene duplication. Proceedings of the Royal Society B: Biological Sciences 256, 119–124. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO Journal 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S, Koh C, Nixon J, et al. 2014. The emerging biofuel crop Camelina sativa retains a highly undifferentiated hexaploid genome structure. Nature Communications 5, 3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Chu SM, Kim SJ, Lee DJ, Lee SY, Lim SH, Ha S-H, Kweon SJ, Cho HS. 2010. Variation of glucosinolates in vegetable crops of Brassica rapa L. ssp. pekinensis. Food Chemistry 119, 423–428. [Google Scholar]
- Kitashiba H, Li F, Hirakawa H, et al. 2014. Draft sequences of the radish (Raphanus sativus L.) genome. DNA Research 21, 481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Kroymann J, Brown P, Figuth A, Pedersen D, Gershenzon J, Mitchell-Olds T. 2001. a Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiology 126, 811–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Lambrix VM, Reichelt M, Gershenzon J, Mitchell-Olds T. 2001. b Gene duplication in the diversification of secondary metabolism: tandem 2-oxoglutarate-dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis . The Plant Cell 13, 681–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ. 2008. A role for gene duplication and natural variation of gene expression in the evolution of metabolism. PLoS One 3, e 1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch MA, Dobes C, Kiefer C, Schmickl R, Klimes L, Lysak MA. 2007. Supernetwork identifies multiple events of plastid trnF(GAA) pseudogene evolution in the Brassicaceae. Molecular Biology and Evolution 24, 63–73. [DOI] [PubMed] [Google Scholar]
- Kroymann J, Donnerhacke S, Schnabelrauch D, Mitchell-Olds T. 2003. Evolutionary dynamics of an Arabidopsis insect resistance quantitative trait locus. Proceedings of the National Academy of Sciences, USA 100 (Suppl. 2 ), 14587–14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S. 2002. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research 30, 325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Quiros CF. 2003. In planta side-chain glucosinolate modification in Arabidopsis by introduction of dioxygenase Brassica homolog BoGSL-ALK. Theoretical and Applied Genetics 106, 1116–1121. [DOI] [PubMed] [Google Scholar]
- Li J, Hansen BG, Ober JA, Kliebenstein DJ, Halkier BA. 2008. Subclade of Flavin-Monooxygenases Involved in Aliphatic Glucosinolate Biosynthesis. Plant Physiology 148, 1721–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WH, Yang J, Gu X. 2005. Expression divergence between duplicate genes. Trends in Genetics 21, 602–607. [DOI] [PubMed] [Google Scholar]
- Liu S, Liu Y, Yang X, et al. 2014. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nature Communications 5, 3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Hirani AH, McVetty PB, Daayf F, Quiros CF, Li G. 2012. Reducing progoitrin and enriching glucoraphanin in Brassica napus seeds through silencing of the GSL-ALK gene family. Plant Molecular Biology 79, 179–189. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lou P, Zhao J, He H, Hanhart C, Del Carpio DP, Verkerk R, Custers J, Koornneef M, Bonnema G. 2008. Quantitative trait loci for glucosinolate accumulation in Brassica rapa leaves. New Phytologist 179, 1017–1032. [DOI] [PubMed] [Google Scholar]
- Lynch M. 2000. The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, et al. 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Research 39, D225–D229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithen R, Clarke J, Lister C, Dean C. 1995. Genetics of aliphatic glucosinolates.3. side-chain structure of aliphatic glucosinolates in Arabidopsis thaliana . Heredity 74, 210–215. [Google Scholar]
- Neal CS, Fredericks DP, Griffiths CA, Neale AD. 2010. The characterisation of AOP2: a gene associated with the biosynthesis of aliphatic alkenyl glucosinolates in Arabidopsis thaliana . BMC Plant Biology 10, 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidig ML, Kavana M, Moran GR, Solomon EI. 2004. CD and MCD studies of the non-heme ferrous active site in (4-hydroxyphenyl)pyruvate dioxygenase: Correlation between oxygen activation in the extradiol and alpha-KG-dependent dioxygenases. Journal of the American Chemical Society 126, 4486–4487. [DOI] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication . Berlin, New York: Springer-Verlag. [Google Scholar]
- Osborn TC, Pires JC, Birchler JA, et al. 2003. Understanding mechanisms of novel gene expression in polyploids. Trends in Genetics 19, 141–147. [DOI] [PubMed] [Google Scholar]
- Padilla G, Cartea ME, Velasco P, de Haro A, Ordas A. 2007. Variation of glucosinolates in vegetable crops of Brassica rapa . Phytochemistry 68, 536–545. [DOI] [PubMed] [Google Scholar]
- Prescott AG, Lloyd MD. 2000. The iron(II) and 2-oxoacid-dependent dioxygenases and their role in metabolism (1967 to 1999). Natural Product Reports 17, 367–383. [DOI] [PubMed] [Google Scholar]
- Purpero V, Moran GR. 2007. The diverse and pervasive chemistries of the alpha-keto acid dependent enzymes. Journal of Biological Inorganic Chemistry 12, 587–601. [DOI] [PubMed] [Google Scholar]
- Schofield CJ, Zhang ZH. 1999. Structural and mechanistic studies on 2-oxoglutarate-dependent oxygenases and related enzymes. Current Opinion in Structural Biology 9, 722–731. [DOI] [PubMed] [Google Scholar]
- Slotte T, Hazzouri KM, Agren JA, et al. 2013. The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nature Genetics 45, 831–835. [DOI] [PubMed] [Google Scholar]
- Sonderby IE, Geu-Flores F, Halkier BA. 2010. Biosynthesis of glucosinolates—gene discovery and beyond. Trends in Plant Science 15, 283–290. [DOI] [PubMed] [Google Scholar]
- Spillane C, Schmid KJ, Laoueille-Duprat S, Pien S, Escobar-Restrepo JM, Baroux C, Gagliardini V, Page DR, Wolfe KH, Grossniklaus U. 2007. Positive Darwinian selection at the imprinted MEDEA locus in plants. Nature 448, 349–352. [DOI] [PubMed] [Google Scholar]
- Talalay P, Fahey JW. 2001. Phytochemicals from cruciferous plants protect against cancer by modulating carcinogen metabolism. Journal of Nutrition 131, 3027S–3033S. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, et al. 2006. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604. [DOI] [PubMed] [Google Scholar]
- Wang H, Wu J, Sun S, Liu B, Cheng F, Sun R, Wang X. 2011. Glucosinolate biosynthetic genes in Brassica rapa . Gene 487, 135–142. [DOI] [PubMed] [Google Scholar]
- Warwick SI, Francis A, Al-Shehbaz IA. 2006. Brassicaceae: Species checklist and database on CD-Rom. Plant Systematics and Evolution 259, 249–258. [Google Scholar]
- Wu HJ, Zhang Z, Wang JY, et al. 2012. Insights into salt tolerance from the genome of Thellungiella salsuginea . Proceedings of the National Academy of Sciences, USA 109, 12219–12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang RL, Jarvis DE, Chen H, et al. 2013. The reference genome of the halophytic plant Eutrema salsugineum . Frontiers in Plant Science 4, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. 2007. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhang J. 2003. Evolution by gene duplication: an update. Trends in Ecology & Evolution 18, 292–298. [Google Scholar]
- Zhang Y, Kensler TW, Cho CG, Posner GH, Talalay P. 1994. Anticarcinogenic activities of sulforaphane and structurally related synthetic norbornyl isothiocyanates. Proceedings of the National Academy of Sciences, USA 91, 3147–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Gunsior M, Bachmann BO, Townsend CA, Solomon EI. 1998. Substrate binding to the α-ketoglutarate-dependent non-heme iron enzyme clavaminate synthase 2: coupling mechanism of oxidative decarboxylation and hydroxylation. Journal of the American Chemical Society 120, 13539–13540. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.