Highlight
A tobacco (Nicotiana tobaccum) SVP family MADS-box gene is identified as a new regulator working in pedicel elongation for optimal inflorescence architecture by directly regulating an Arabidopsis BP homologue NtBPL
Key words: BP, gibberellin, MADS-box transcription factor, pedicel, SVP, tobacco.
Abstract
Optimal inflorescence architecture is important for plant reproductive success by affecting the ultimate number of flowers that set fruits and for plant competitiveness when interacting with biotic or abiotic conditions. The pedicel is one of the key contributors to inflorescence architecture diversity. To date, knowledge about the molecular mechanisms of pedicel development is derived from Arabidopsis. Not much is known regarding other plants. Here, an SVP family MADS-box gene, NtSVP, in tobacco (Nicotiana tabacum) that is required for pedicel elongation was identified. It is shown that knockdown of NtSVP by RNA interference (RNAi) caused elongated pedicels, while overexpression resulted in compact inflorescences with much shortened pedicels. Moreover, an Arabidopsis BREVIPEDECELLUS/KNAT1 homologue NtBP-Like (NtBPL) was significantly up-regulated in NtSVP-RNAi plants. Disruption of NtBPL decreased pedicel lengths and shortened cortex cells. Consistent with the presence of a CArG-box at the NtBPL promoter, the direct binding of NtSVP to the NtBPL promoter was demonstrated by yeast one-hybrid assay, electrophoretic mobility shift assay, and dual-luciferase assay, in which NtSVP may act as a repressor of NtBPL. Microarray analysis showed that down-regulation of NtBPL resulted in differential expression of genes associated with a number of hormone biogenesis and signalling genes such as those for auxin and gibberellin. These findings together suggest the function of a MADS-box transcription factor in plant pedicel development, probably via negative regulation of a BP-like class I KNOX gene. The present work thus postulates the conservation and divergence of the molecular regulatory pathways underlying the development of plant inflorescence architecture.
Introduction
Inflorescence architecture plays critical roles in plant reproductive success by affecting the ultimate number of flowers that set fruits. Optimal inflorescence structure increases the competitiveness of plants when interacting with adverse environments (Evers et al., 2011; Iwata et al., 2012). Pedicel length is one of the key contributors to the structural diverstiy of plant inflorescences. In Arabidopsis, the class I KNOX gene BREVIPEDICELLUS (BP) was among the first to be found to affect pedicel development and inflorescence architecture (Venglat et al., 2002). BP encodes the homeodomain protein KNAT1, a member of the KNOX (KNOTTED1-LIKE HOMEODOMAIN) family including SHOOT MERISTEMLESS (STM), KNAT2, and KNAT6 that are involved in promoting stem cell division and delaying differentiation in the shoot apical meristem (SAM) (Hamant and Pautot, 2010; Hay and Tsiantis, 2010). Mutation of BP causes shortened pedicels and internodes because of fewer cell divisions. More profound defects were found for cell differentiation, elongation, and growth on the abaxial side than on the adaxial side of the pedicel, causing downward-pointing flowers and a compact inflorescence architecture, in addition to its pleitropic effect on the inflorescence stem and style.
BP is regulated by a number of genes. The phenotypes of the BP mutation become more severe in the mutants of ERECTA (ER) (van Zanten et al., 2009), which may play a role redundant to BP in plant architecture regulation. ASYMMETRIC LEAVES 1 (AS1), a MYB transcription factor, acts in conjunction with AS2, a LATERAL ORGAN BOUNDARIES domain (LBD) transcription factor, to repress BP expression (Ori et al., 2000; Guo et al., 2008; Xu et al., 2008) by recruiting components of the Polycomb-repressive complex2 to its promoter to generate an inactive chromatin state (Phelps-Durr et al., 2005; Luo et al., 2012; Lodha et al., 2013). Recently, CINCINNATA-like TEOSINTE BRANCHED 1-CYCLOIDEA-PCF (TCP) transcription factors have been reported to interact with AS2 in the repression of class I KNOX genes (Li et al., 2012). These studies suggest that KNOX repression may require different types of transcription factors in the development of different organs. Similar to their role in other plant organs, phytohormones such as auxins have been shown to be implicated in pedicel development. Mutation of BIG/CRM1, an auxin transport-related gene, causes shortened pedicels and internodes (Venglat et al., 2002; Xu et al., 2003; Douglas and Riggs, 2005; Yamaguchi et al., 2007; Ragni et al., 2008; Yamaguchi and Komeda, 2013). The severity of the corymb-like inflorescence in crm1/big mutants correlated with increased levels of PIN1, indicaitng that CRM1/BIG controls the elongation of the pedicels and stem internodes through auxin action (Yamaguchi and Komeda, 2013). Gain of function of the meristem identity regulator LEAFY (LFY) also produces downward-bending pedicels (Yamaguchi et al., 2012). LFY can bind to the AS2 promoter to enhance its activity directly, and subsequently suppresses BP expression (Xu et al., 2003; Yamaguchi et al., 2012).
MADS-box genes play important roles during plant development, especially floral organ development. The SHORT VEGETATIVE PHASE (SVP) MADS-box genes are of divergent functions in different plants. In Arabidopsis, the two SVP-clade genes SVP and AGL24 have opposite functions in controlling the floral transition, with AGL24 functioning as a promoter and SVP as a repressor (Hartmann et al., 2000; Michaels et al., 2003). In addition to this, SVP and AGL24 act redundantly to control the identity of the floral meristem and to repress expression of class B, C, and E genes for regulating flower development (Gregis et al., 2006, 2008, 2009). JONTLESS (J) is the tomato SVP homologue that is required for abscission zone (AZ) development (Mao et al., 2000) and works with other MADS-box proteins SlMBP21 and MACROCALYX (MC) in the form of ternary protein complexes (Nakano et al., 2012; Liu et al., 2014). Mutation of J also causes an indeterminate inflorescence, suggesting its additional function to suppress the sympodial growth of tomato inflorescences (Szymkowiak and Irish, 2006). In snapdragon (Antirrhinum majus), the SVP homologue INCOMPOSITA (INCO) is responsible for prophyll development as well as floral meristem identity (Masiero et al., 2004). Importantly, the involvement of SVP-like genes in determining inflorescence architecture was recently found to be conserved in Arabidopsis and rice (Liu et al., 2013). Thus, study of the functions of SVP genes in additional species such as common tobacco (Nicotiana tabacum L.) would be interesting.
In this work, an SVP-like MADS-box gene NtSVP was identified in tobacco, another member of the Solanaceae family (Reinhardt and Kuhlemeier, 2002) in addition to tomato. It was found that, instead of working in flower AZ development and inflorescence determinacy like the tomato SVP-like gene J, NtSVP may play a major role in pedicel elongation. Moreover, NtSVP may directly regulate a downstream BP-like class I KNOX gene NtBPL as a transcription repressor. These findings should expand our understanding of the molecular mechanism that underlies plant inflorescence development.
Materials and methods
Plant material and growth conditions
Seeds of tobacco cv. W38 were obtained from the Tobacco Research Institute, Chinese Academy of Agricultural Sciences. All plants were grown at 24 °C in a greenhouse under long-day conditions (16h light/8h dark) with auxiliary light from sodium lamps.
RACE and TAIL-PCR
THe NtSVP full-length cDNA sequence was amplified by the rapid amplification of cDNA ends (RACE) method. For 5’ RACE, primers NtSVPgstWR and Gene Racer 5’ primer were used. For 3’ RACE, primers AP, AUAP, NtSVP-F1, and NtSVP-F2 were used. The promoter sequence of NtBPL was amplified by thermal asymmetric interlaced PCR (TAIL-PCR) as previously described (Liu and Whittier, 1995) by AD primers (AD1–AD13) and three NtBPL-specific primers, NtBPL-SP1, NtBPL-SP2, and NtBPL-SP3, which were designed using expressed sequence tag (EST) sequences. All PCR constructs were subcloned into the pGEM-T Easy vector (Promega, USA), transformed into Escherichia coil DH5a cells, and sequenced. The primers used are shown in Supplementary Table S2 available at JXB online.
Binary vector construction and tobacco transformation
RNA interference (RNAi) constructs were generated using pKANNIBAL (Wesley et al., 2001) and pART27 vectors (Gleave, 1992). A 296bp region from the C-terminus of NtSVP and a 367bp region from the C-terminus of NtBPL were inserted into the pKANNIBAL vector twice in opposite directions with an intron between them to create a hairpin. The hairpin structure was then inserted into the binary vector pKART27. For construction of gene overexpression vectors, full-length open reading frames (ORFs) of NtSVP and NtBPL were inserted into pCHF3 binary vectors (Hajdukiewicz et al., 1994) with gene-specific primers (Supplementary Table S2 at JXB online). After the sequences of the resulting constructs were confirmed, all vectors were transformed into the tobacco cv. W38 by Agrobacterium tumefaciens- (strain C58C1) mediated transformation using the leaf disc procedure (Horsch et al., 1985) with timentin used as the bacteriostatic agent. Transgenic seeds were screened on half-strength Murashige and Skoog (MS) medium containing 100mg l–1 kanamycin.
Quantitative RT-PCR (qRT-PCR)
Total RNA was extracted using an RNAprep pure plant kit (Tiangen, China) according to the manufacturer’s instructions. A 2 μg aliquot of total RNA was used for cDNA synthesis using TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (Transgen, China). qRT-PCR was performed on a ABI7300 qRT-PCR system (Applied Biosystems, USA) according to the manufacturer’s instructions using SYBR Premix Ex Taq™ II (Takara, Japan). The relative expression of each gene was calculated according to the 2–ΔΔCt method (Livak and Schmittgen, 2001). All reactions were performed with three technical and three biological replicates. The data were normalized to the housekeeping gene NtACTIN9. The primers used are shown in Supplementary Table S2 at JXB online.
Yeast one-hybrid assays
Bait plasmids were simultaneously heat transformed into yeast strain Golden yeast and selected on SD/–Ura agar medium. SMART technology synthesizes NtSVP-containing ends that are homologous to the end of the linearized pGADT7-Rec2 prey plasmid (Clontech). The linearized pGADT7-Rec vector and NtSVP in the bait-specific reporter strain were co-transformed into yeast cells and plated on aureobasidin A-containing selective medium according to the manufacturer’s instructions (Clontech, USA). The primers used are listed in Supplementary Table S2 at JXB online.
Electrophoretic mobility shift assays (EMSAs)
The NtSVP full-length ORF was cloned into the pMAL-C2 vector (NEB, USA) using primer pair BamHI-NtSVP Fw and XhoII-NtSVP Rv (Supplementary Table S2 at JXB online), and transformed into E. coli BL21 (DE3). Proteins were extracted from bacterial cells by ultrasonic crushing, and the cell lysate was purified by amylose resin affinity chromatography (NEB, USA) according to the manufacturer’s protocol. Probes containing the CArG-box and several flanking bases were labelled with biotin at the 3’ end. Unlabelled oligonucleotides of the same sequence were used as competitors. Oligonucleotides with a mutant CArG-box (AAATTATAAT) were used as negative control. A 1 μg aliquot of purified maltose-binding protein (MBP)–NtSVP protein and 50fmol biotin-labelled probes were used for the binding reaction for each sample. Florescence of biotin-labelled DNA was detected using the LightShift Chemiluminescent EMSA Kit (Pierce, USA). The oligonucleotides used are listed in Supplementary Table S2 at JXB online.
Transient expression assay using a dual-luciferase system
The dual-luciferase assay was performed following a previously described method (Hellens et al., 2005; Meng et al., 2013). The effector plasmid 35S:NtSVP was constructed with the pCAMBIA2300 vector using the primer pair BamHI-NtSVP Fw and XhoII-NtSVP Rv (Supplementary Table S2 at JXB online). The recombinant promoter containing the ~1kb native promoter of NtBPL amplified by the primer pair SalI-proNtBPL-Fw and BamHI-proNtBPL-RV and fused to the minimum 35S promoter was cloned into the vector pGreen-0800-LUC. Besides the wild-type CArG-box, two other reporter plasmids containing a mutant CArG-box in the NtBPL promoter were generated as controls using primers m1CArG-F, m1CArG-R, m2CArG-F, and m2CArG-R.
The reporter strain was incubated either alone or as a mixture with the effector strain (at the reporter:effector ratio of 1:1). The agrobacterial suspension in a 10ml syringe (without the metal needle) was carefully press-infiltrated onto healthy leaves of 1-month-old N. benthamiana. Plants were left in the dark for 1 d, and then moved to a greenhouse with long-day conditions (16h light/8h dark) for 2 d. Infiltrated leaf samples were sprayed with luciferin (1mM luciferin and 0.01% Triton X-100), and photographed. The luciferase activity was measured using the dual-luciferase reporter assay system (Promega, USA) according to the manufacturer’s instructions. The relative luciferase activity was calculated as the ratio between the firefly luciferase and the control Renilla luciferase activity. Five biological repeats were measured for each sample.
Morphological analyses
To determine the mean pedicel lengths, pedicels were photographed at the anthesis stage, and measured using IMAGE J software. Values are means ±SE, based on 50 (n=10×5) pedicels from five individual plants per genotype. To determine the angles between pedicels and the inflorescence stem, 50 (n=10×5) pedicels from five plants were measured as a replication, and three replications were conducted.
For histological analysis, longitudinal sectioning was performed on pedicels fixed in FAA (3.7% formaldehyde, 5% acetic acid, 50% absolute ethanol). For cortex cell length and cortex cell number, 15 (n=3×5) pedicels at anthesis from five individual plants for each construct were sectioned longitudinally and photographed. To determine the mean pedicel cortex cell length, 30 cortex cells in a middle longitudinal cortex row were measured using IMAGE J software in each photograph. To determine the mean pedicel cortex cell number, all cortex cells at the middle sections were counted. This number was used to represent the total number of cortex cells in each pedicel. The number of cortex cells was counted in 15 sectioned pedicels for each genotype and averaged.
Microarray analysis
Microarray assay was conducted by Shanghai Bio Corporation with two independent biological replicates, using an Agilent Whole Tobacco Genome Oligo Microarray (4×44K) (Agilent Technologies, Palo Alto, CA, USA). The chip hybridization results were scanned by the Agilent Microarray Scanner System and were normalized and analysed with Agilent Feature Extraction software. Differentially expressed genes with a significance threshold of P<0.05 and a >2-fold change between the NtBPL-RNAi transgenic plants and the wild type were selected. The description of each gene was annotated with Arabidopsis genes, which were obtained by a tblastx search against TAIR10 transcripts. Gene Ontology (GO) enrichment analysis was carried out by using proteins of Arabidopsis as templates at the AGRIGO website (http://bioinfo.cau.edu.cn/agriGO/). The Arabidopsis gene information was used for MapMan annotation (Thimm et al., 2004)
Results
Identification of the NtSVP gene in tobacco
In light of the diverse functions of the SVP family genes, it was planned to explore the roles of SVP-like genes in tobacco. One homologue was cloned from N. tabacum cv. W38 using primers designed from the tomato J sequence and was named NtSVP. NtSVP is 1062bp in length with eight exons and seven introns, and encodes a 238 amino acid protein that shares ~77% similarity with the tomato J protein. It encodes a protein with four typical domains: a MADS-box domain, a variable I domain, a conserved K domain, and a C-terminal region (Supplementary Fig. S1A at JXB online). Phylogenetic analysis showed that NtSVP indeed belongs to the SVP-like lineage, with J and CaSVP as its closest homologues (Supplementary Fig. S1B available at JXB online). Interestingly, in contrast to most other members of the SVP-clade, NtSVP was highly expressed in pedicels and carpels in addition to vegetative organs such as leaf, stem, and root (Supplementary Fig. S1C available at JXB online).
NtSVP regulates pedicel elongation
To characterize the function of NtSVP, RNAi transgenic lines were produced using a construct containing the less conserved C-terminal portion of NtSVP (Supplementary Fig. S2A at JXB online). Meanwhile, overexpression lines were generated using the DNA fragment containing the complete ORF. A total of 17 independent NtSVP RNAi lines (NtSVP-RNAi) and 11 independent NtSVP overexpression lines (NtSVP-OE) were obtained with evident inflorescence alteration when compared with that of the wild type (Fig. 1A–D). Unexpectedly, no obvious defect was observed in the pedicel AZ, suggesting functional divergence of NtSVP when compared with the tomato SVP homologue J (Supplementary Fig. S2B at JXB online).
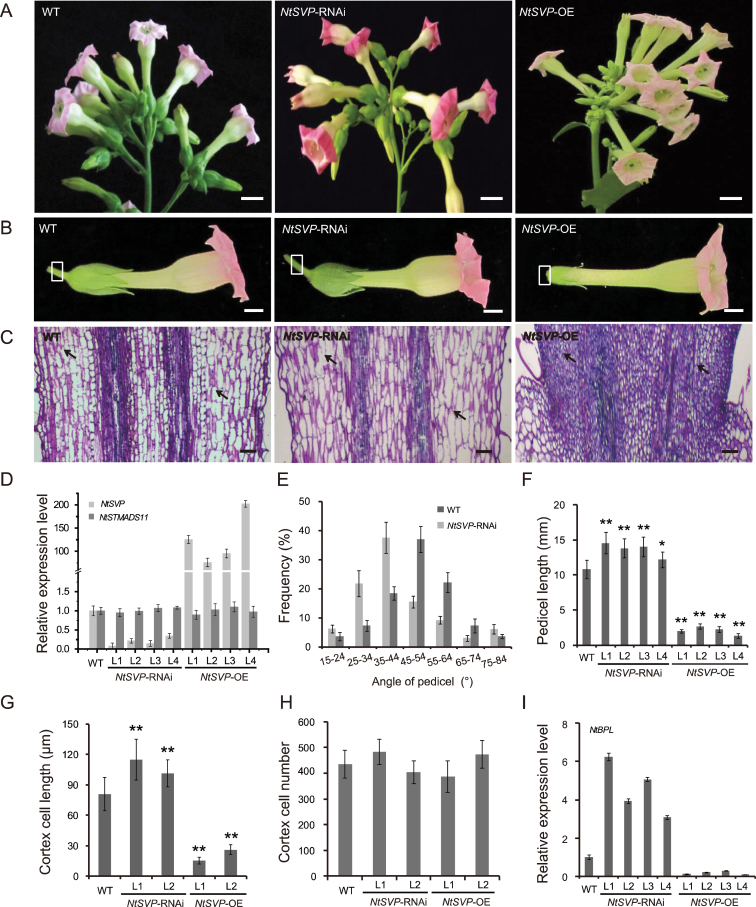
Fig. 1.
Characterization of NtSVP transgenic plants. (A) Inflorescence morphology of wild-type (WT), NtSVP-RNAi, and NtSVP-OE plants. Scale bars=20mm. (B) Representative flowers from WT, NtSVP-RNAi, and NtSVP-OE plants at anthesis. The portion of pedicel used for histological observation is indicated by a white box. Scale bars=10mm. (C) Longitudinal sections of the pedicels at anthesis of WT and NtSVP transgenic plants. Arrows point to cortex cell areas that are used for measurement. Scale bars=100 μm. (D) Detection of NtSVP and NtSTMADS11 expression in NtSVP transgenic lines by qRT-PCR. The tobacco NtACTIN9 gene was used as an internal control. (E) Distribution of the angles between the pedicels and the inflorescences in WT and NtSVP-RNAi plants. In total, 50 pedicels from five plants were measured as a replicate (n=10×5), and three replications were performed. Values are means ±SE. (F) Lengths of pedicels in WT, NtSVP-RNAi, and NtSVP-OE plants at anthesis (n=10×5, 50 pedicels from five individual plants per genotype). Asterisks indicate significant differences compared with the WT (*P<0.05, **P<0.01, Student’s t-test). (G) Quantification of pedicel cortex cell lengths of WT and NtSVP transgenic plants at anthesis (n=30×3×5, 15 pedicel sections from five individual plants per genotype were used and 30 cortex cells were measured for each section). Values are means ±SE. Asterisks indicate significant differences compared with the WT (**P<0.01, Student’s t-test). (H) Cell numbers in the longitudinal cortex file of pedicels in WT and NtSVP transgenic plants at anthesis (n=3×5, 15 sections from five individual plants per genotype were used). Values are means ±SE. (I) Detection of NtBPL expression in NtSVP transgenic lines by qRT-PCR. NtACTIN9 was used as an internal control.
Interestingly, RNAi lines had markedly stretched inflorescences when compared with those of wild-type plants (Fig. 1A). A closer observation revealed increased angles between the pedicel and the inflorescence. As shown in Fig. 1E, the range of pedicel angles in the wild-type plants was between 25 ° and 54 °, with a mean value of 42.8 °. In contrast, the average pedicel angle in RNAi lines was increased to 56.7 °, a 32.5% increase compared with the wild-type plants. Moreover, pedicels of RNAi lines were longer than those of the wild-type plants (Fig. 1B, F). The average length of pedicels in the RNAi transgenic lines was up to 13.7mm as measured from 50 flowers per phenotype, an increase of 26.2% when compared with the mean length of wild-type pedicels. Such elongation of pedicels persisted during flower development (Supplementary Fig. S2C, D at JXB online).
On the other hand, overexpression of NtSVP caused much shorter stature of the plants compared with the wild type (Supplementary Fig. S2E, F at JXB online). Remarkably, inflorescences became compacted due to significantly shortened pedicels (Fig. 1A, B). It was almost impossible to measure their lengths due to extremely small sizes. The average pedicel length for NtSVP-OE transgenic plants was ~2mm at anthesis (Fig. 1F), much shorter than those of the wild type (10.8mm). A longitudinal section of the pedicels showed altered elongated cortex cells at the pedicels of NtSVP-RNAi transgenic plants, while cortex cells of the overexpression lines were small and densely organized (Fig. 1C). To distinguish whether the change of pedicel length is caused by increased cell elongation or promoted cell division, cell lengths were measured at the cortical regions. The results showed that cortex cells were significantly elongated in RNAi lines. As shown in Fig. 1G, the average cortex cell length increased to >100 μm on average, when compared with 80 μm in wild-type pedicel cortex cells. On the other hand, cortex cells in NtSVP overexpression plants were extremely short, only 18–28% of the wild-type mean. Despite this, no significant difference was found for cell numbers in the transgenic plants and the wild-type control (Fig. 1H). These data suggest that NtSVP regulates pedicel development by affecting cell elongation, rather than cell proliferation. To test the possible off-target effect of RNAi, the correlation between expression levels of transgenes and pedicel lengths was studied. As shown in Fig. 1D, qRT-PCR assay showed that NtSVP transcript levels were indeed proportional to pedicel lengths in the transgenic plants (Fig. 1F), while another homologous MADS-box gene NtSTMADS11 was not affected. These results demonstrate that the effect on pedicel elongation was indeed caused by altered expression of NtSVP.
NtSVP suppresses the expression of NtBPL, a tobacco BP/KNAT1 homologue
In Arabidopsis, BP has been known as the major player in pedicel development (Venglat et al., 2002; Douglas and Riggs, 2005). To study the functions of BP homologues in tobacco, the tobacco EST sequences were searched using Arabidopsis BP as a query. One EST (AF544053) showed high similarity to Arabidopsis BP and was named NtBPL (Nicotiana tabacum BP-Like). Full-length cDNA was then obtained using 5’ and 3’ RACE experiments. NtBPL protein contained four domains typically found in class I KNOX proteins, such as KNOX I and KNOX II domains (Supplementary Fig. S3A at JXB online). It shares 55.7% sequence similarity to the Arabidopsis BP and is closely related to thetomato class I KNOX gene TKN1 (Supplementary Fig. S3B at JXB online). Similar to Arabidopsis BP, NtBPL was found to have the highest expression in tobacco pedicels (Supplementary Fig. S3C at JXB online). The NtBPL expression level in pedicels of NtSVP transgenic plants was determined and it was found that NtBPL was up-regulated in NtSVP-RNAi lines, and significantly suppressed when NtSVP was overexpressed (Fig. 1I). Furthermore, the expression level of NtBPL increased along with pedicel development stages, when the expression of NtSVP decreased from stage 1 to stage 4, indicating possible co-ordinated expression between NtSVP and NtBPL during tobacco pedicel development (Supplementary Fig. S4 at JXB online).
NtBPL promotes pedicel elongation in tobacco
To identify whether NtBPL plays a role in tobacco pedicel development, transgenic plants were generated comprising 18 NtBPL-RNAi lines and 15 NtBPL-OE lines. Four lines for each genotype were further studied; these exhibited clear morphological changes in their inflorescences compared with wild-type plant (Fig. 2A, C). As shown in Fig. 2C, the expression of NtBPL was down-regulated in RNAi lines and increased in OE lines, while two closely related genes NTH20 and NTH22 were not affected. Unlike NtSVP transgenic plants, transgenic manipulation of NtBPL did not affect tobacco plant height (Supplementary Fig. S5A at JXB online), nor did this have a clear effect on pedicel angles (Supplementary Fig. S5B at JXB online). Despite this, knockdown of NtBPL reduced pedicel lengths an average of 1.8mm, or 16% of that of the wild type, while overexpression of NtBPL caused a significant increase in pedicel length, up to 13.8mm, or a 27.8% increase, as measured from 50 flowers per phenotype (Fig. 2B, D). These data indicate that NtBPL is closely correlated with pedicel elongation and indeed required for pedicel development. Accordingly, cortex cells were found to be reduced ~10% in NtBPL-RNAi plants and increased >15% when NtBPL was overexpressed (Fig. 2E; Supplementary Fig. S5C at JXB online). Because the total number of cortex cells remained statistically unchanged, the variation of pedicel lengths can be attributed to the elongation of cortex cells (Supplementary Fig. S5D at JXB online).
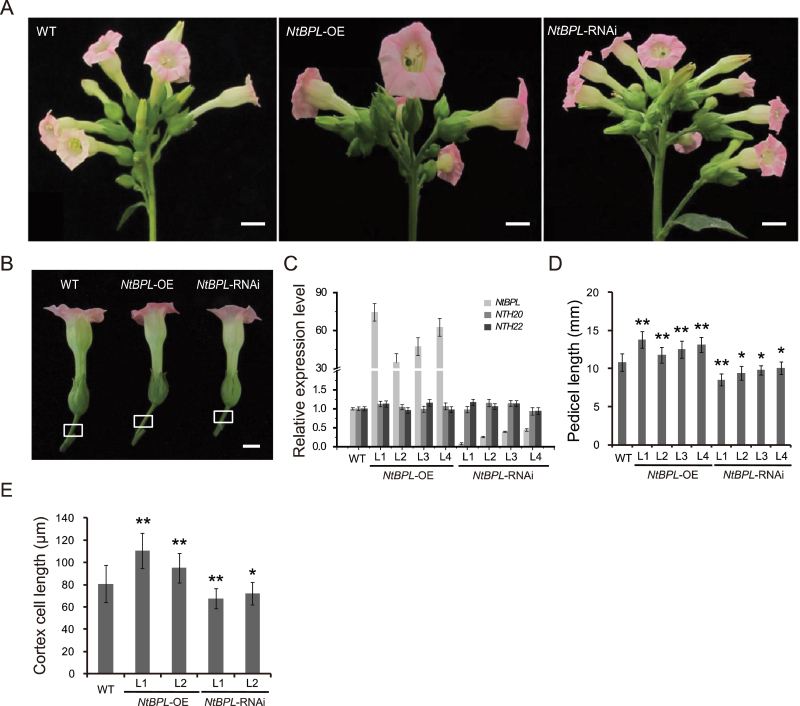
Fig. 2.
Characterization of NtBPL transgenic plants. (A) Inflorescence morphology of wild-type (WT), NtBPL-OE, and NtBPL-RNAi plants. Scale bar=20mm. (B) The flowers of WT, NtBPL-OE, and NtBPL-RNAi, with pedicels of different lengths as indicated by white boxes. Scale bars=10mm. (C) Detection of the expression levels of NtBPL and two closely related homologues NTH20 and NTH22 in NtBPL transgenic lines by qRT-PCR. NtACTIN9 was used as an internal control. (D) Lengths of pedicels in WT, NtBPL-OE, and NtBPL-RNAi plants at anthesis (n=8×5, 40 pedicels from five individual plants per genotype). Values are means ±SE. Asterisks indicate significant differences compared with the WT (*P<0.05, **P<0.01, Student’s t-test). (E) Quantification of pedicel cortex cell lengths of WT and NtBPL transgenic plants at anthesis (n=30×3×5, 15 pedicel photographs were prepared from five individual plants, and 30 cortex cells were measured for each photograph). Values are means ±SE. Asterisks indicate significant differences compared with the WT (*P<0.05, **P<0.01, Student’s t test).
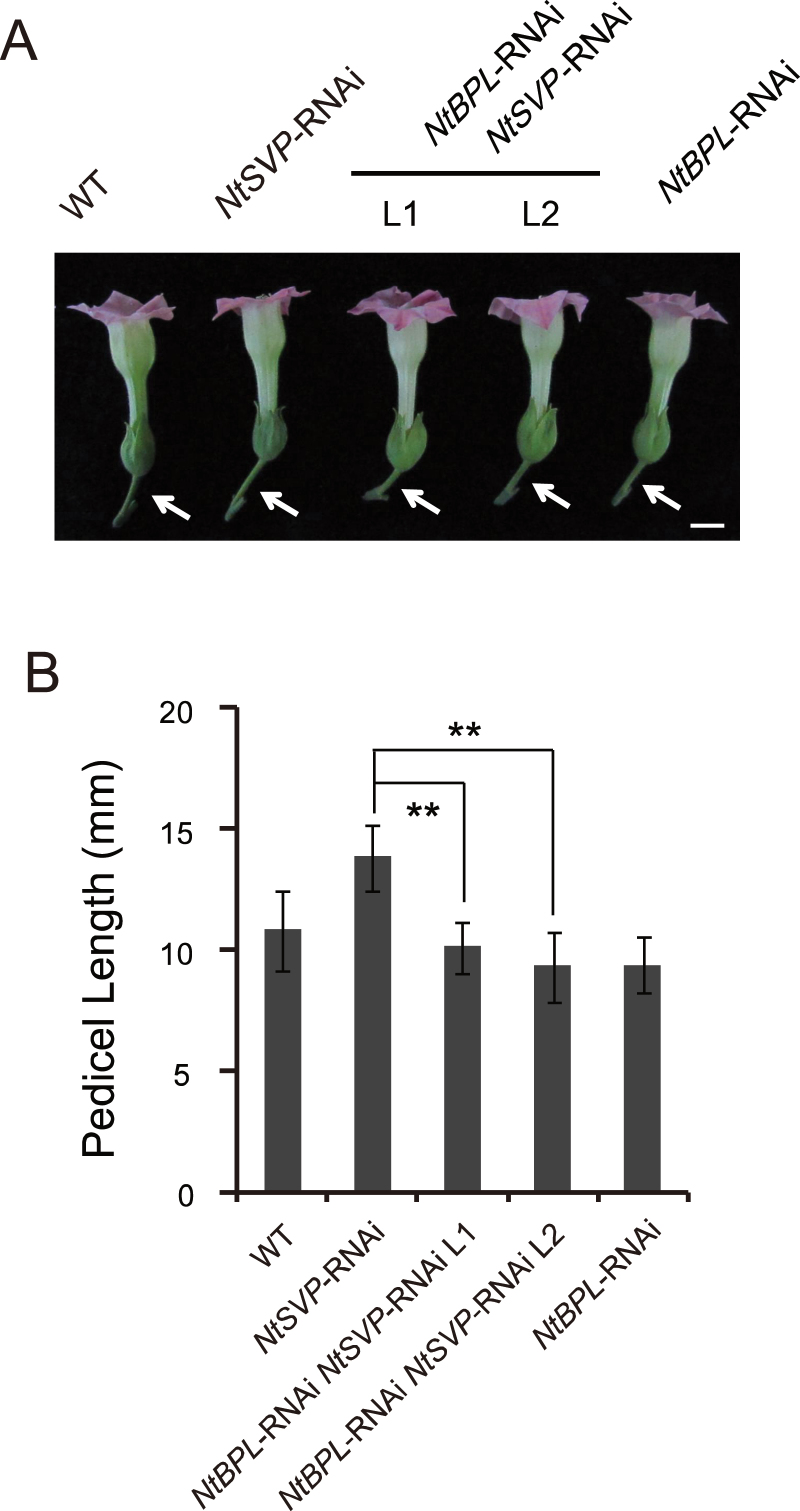
The expression level of NtSVP was then checked in pedicels of NtBPL transgenic plants, and no significant change was found (Supplementary Fig. S5E at JXB online), indicating that NtBPL is indeed located downstream of NtSVP and there was no feedback regulation on NtSVP. To confirm this notion further, NtSVP/NtBPL double RNAi plants were generated by crossing RNAi lines of the two genes. The NtSVP and NtBPL double RNAi plants exhibited shortened pedicels, similar to those of NtBPL-RNAi plants, suggesting a downstream location of NtBPL relative to NtSVP (Fig. 3). In other words, NtSVP and NtBPL may possibly work on the same regulatory pathway for tobacco pedicel development.
Fig. 3.
Genetic interaction of NtSVP and NtBPL by double RNAi analysis. (A) Pedicels of flowers at anthesis from wild-type (WT), NtSVP-RNAi, NtBPL NtSVP double RNAi plants L1 and L2, and NtBPL-RNAi plants. Scale bar=10mm (B) Quantitative measurement of pedicel lengths of the plants described in (A). Values are means ±SE (n=8×5, 40 pedicels from five individual plants per genotype). **P<0.01 by Student’s t-test.
NtSVP acts as a transcription repressor of NtBPL by directly binding to its promoter
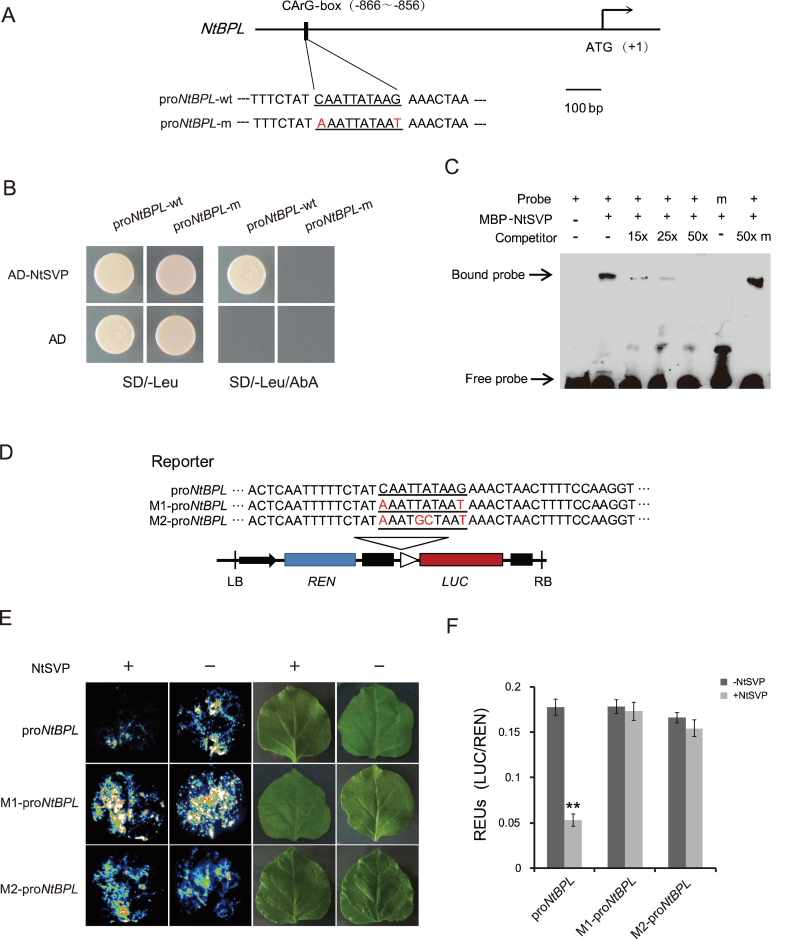
Since genetic analysis indicated that NtBPL acted downstream of NtSVP, experiments were carried out to determine whether NtBPL was a direct target of NtSVP regulation. To test this hypothesis, ~1kb of the promoter sequence of NtBPL was cloned and cis-element prediction was performed using the PLACE program. A CArG-box (5’CAATATTAAG3’), a consensus MADS-box transcription factor binding site, was found on the NtBPL promoter at –866bp to –856bp from the initiation codon (Fig. 4A; Supplementary Fig. S6 at JXB online), providing a condition for direct binding of NtSVP to the promoter of NtBPL. Yeast one-hybrid assay showed that NtSVP could indeed activate the expression of a reporter gene driven by the NtBPL promoter (Fig. 4B). This was further confirmed by EMSA using the MBP–NtSVP fusion protein. As shown in Fig. 4C, the MBP–NtSVP fusion protein was able to bind to DNA probes containing the wild-type CArG-box, but failed to bind to the mutated version. Furthermore, increasing the concentration of unlabelled wild-type probes in the binding reactions resulted in much weaker retarded bands. Meanwhile, the formation of proNtBPL and the MBP–NtSVP complex was not inhibited by an excess amount of mutated probes, indicating that NtSVP may specifically bind to the NtBPL promoter under these conditions.
Fig. 4.
NtSVP as a direct transcription repressor of NtBPL. (A) The design of the wild-type (WT) probe containing the CArG-box (proNtBPL-wt) and the mutated probe (proNtBPL-m) in the NtBPL promoter. Mutated bases are highlighted in red. Numbers indicate sequence locations. (B) Yeast one-hybrid assays showing interactions between the NtSVP protein and the NtBPL promoter. pGADT7 (AD) was used as control. (C) EMSA showing MBP–NtSVP fusion protein binding to the probes of proNtBPL as described in (A). m represents proNtBPL-m as a control. + indicates the presence and – the absence of corresponding components as indicated. (D) The design of reporter constructs for the dual-luciferase assay. The WT and two mutant version of proNtBPL (M1, M2) are indicated. (E) LUC activities in N. benthamiana leaves infiltrated with the Agrobacterium strain harbouring the indicated reporter in the presence (+) or absence (–) of the co-transfected Agrobacterium strain harbouring the plasmid expressing the NtSVP protein. (F) Dual-luciferase assay of relative reporter activity of samples shown in (E). The relative LUC activities were normalized to REN activity and presented as relative expression units (REUs, n=5). **P<0.01.
Next a transient expression assay was performed using the dual-luciferase system in N. benthamiana leaves to study further the mode of action of NtSVP on the transcription of NtBPL. Three reporter constructs were generated, one containing the wild-type CArG-box and the other two containing mutated CArG-boxes (Fig. 4D). An effector plasmid expressing the full-length NtSVP protein was also constructed. As shown in Fig. 4E, when the NtSVP effector was co-expressed with the LUC reporter gene driven by the NtBPL promoter, the expression of LUC reporter genes was significantly repressed. On the other hand, when the reporter construct carried a mutated CArG-box, no significant change in LUC activity was observed whether NtSVP was present or not (Fig. 4E, F). These results support the idea that NtSVP is a repressor of NtBPL and negatively regulates its expression by direct binding to its promoter.
Global gene expression analysis indicates involvement of hormonal regulation in tobacco pedicel development
To investigate further genes that are involved in tobacco pedicel development, the transcriptomes in the pedicels of wild-type and NtBPL-RNAi plants were compared using Agilent tobacco microarrays (http://sbc.biomart.cn). A total of 657 differentially expressed genes were detected between the wild-type and RNAi pedicels [absolute log2 value ≥1; P-value ≤0.001; false discovery rate (FDR) ≤0.01] (Supplementary Dataset 1 at JXB online). Among them, 427 were down-regulated and 230 were up-regulated when NtBPL was knocked down. Four genes were randomly selected for qRT-PCR detection, and the results confirmed the consistency of the microarray data (Supplementary Fig. S7 at JXB online). GO enrichment analysis showed that genes for response to stimulus and various abiotic stresses were significantly enriched among those down-regulated genes (Supplementary Fig. S8A at JXB online). There were fewer enriched classes of genes among up-regulated genes, including ‘response to stimulus’, ‘localization’, ‘transport’, and ‘cellular amino acid and derivative metabolic process’ functions (Supplementary Fig. S8B at JXB online).
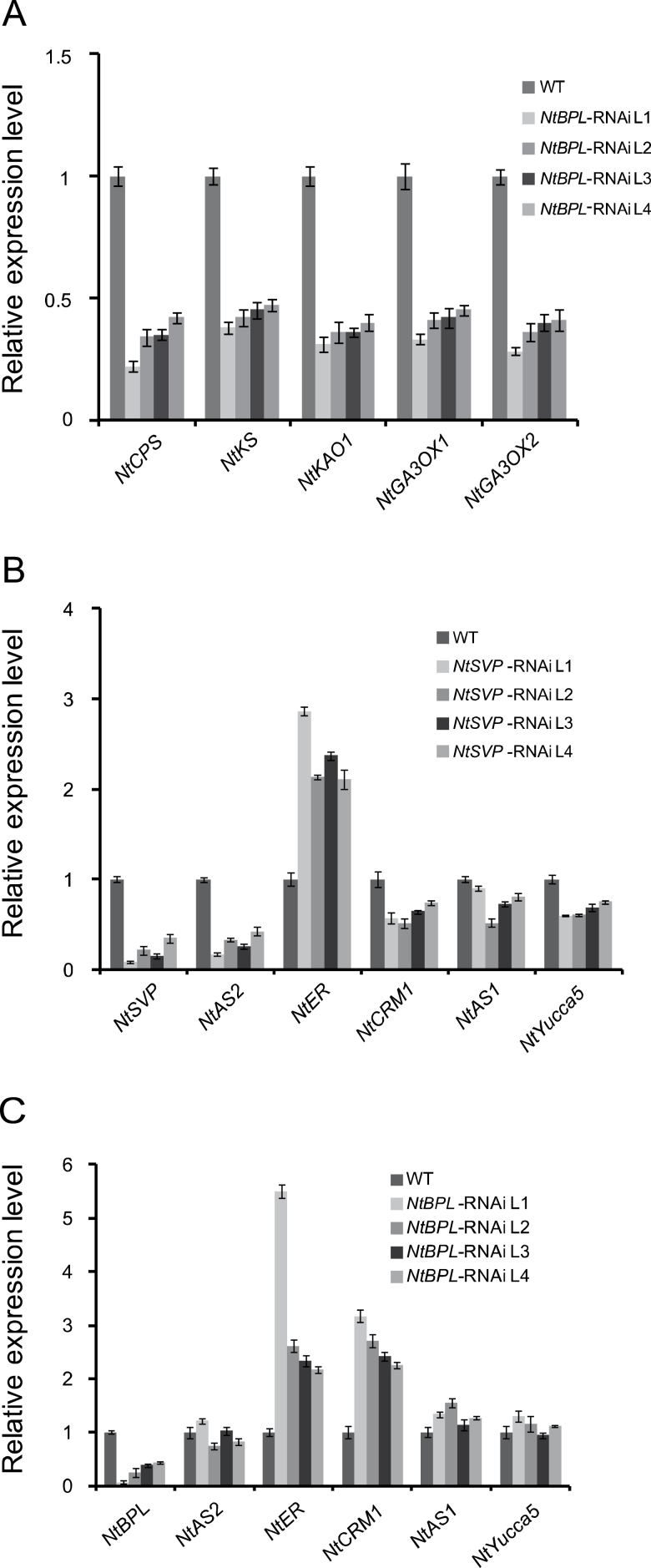
Those differentially expressed genes were then further classified using the MapMan program (http://mapman.gabipd.org), and it was found that the hormone metabolism pathway was notably enriched among down-regulated genes (Supplementary Fig. S8C at JXB online). Hormone biogenesis and signalling genes for auxin, ethylene, gibberellin (GA), and brassinosteriod were found among the differentially expressed genes (Supplementary Table S1 at JXB online). There were seven genes associated with GA functions, five of which were for GA biosynthesis, including homologues of ent-copalyl diphosphate synthase (CPS), ent-kaurene synthase (KS), ent-kaurenoic acid oxidase (KAO), and GA 3-oxidase (GA3OX1/2) (Yamaguchi, 2008). These genes were subject to further characterization by qRT-PCR. As shown in Fig. 5A, NtCPS, NtKS, and NtKAO1 were down-regulated in NtBPL-RNAi lines, together with two homologues, NtGA3OX1 and NtGA3OX2, whose products putatively catalyse the last step of GA biogenesis. These data indicate that disruption of NtBPL may significantly affect GA biogenesis which may play a significant role in tobacco pedicel development. This is consistent with GA as a key hormone for plant cell elongation and division (Shani et al., 2013).
Fig. 5.
Expression analysis of tobacco homologues of Arabidopsis genes for pedicel development. (A) qRT-PCR detection of transcript levels of differently expressed GA biosynthetic genes (Heinrich et al., 2013) in pedicels of NtBPL-RNAi plants compared with the wild type (WT) according to microarray results. NtACTIN9 was used as an internal control. (B) qRT-PCR detection of transcript levels of homologues to Arabidopsis pedicel-regulating genes in pedicels of NtSVP-RNAi and WT plants. Sequence information was obtained from the SOL Genomics Network by Blast using Arabidopsis genes as queries. NtAS1, mRNA_73569; NtAS2, mRNA_104917; NtER, mRNA_128857; NtCRM1, mRNA_59455; NtYucca5, mRNA_8257). (C) qRT-PCR detection of transcript levels of the above genes in pedicels of NtBPL-RNAi and WT plants.
In Arabidopsis, genes that are known to be involved in pedicel development include AS1/2, ER, CRM1, and Yucca5. To test the possible relationships of these genes in the NtSVP–NtBPL pathway, homologues of these genes were isolated from tobacco and their expression patterns were studied in NtSVP and NtBPL-RNAi plants. As shown in Fig. 5B and C, NtAS2 was significantly down-regulated in NtSVP-RNAi plants. NtER, however, was up-regulated when either NtSVP or NtBPL was down-regulated. It is not clear how these two genes were integrated into the NtSVP–NtBPL pathway in tobacco. In addition, the tobacco CRM1 homologue NtCRM1 was significantly up-regulated in NtBPL-RNAi lines. These data suggest that auxin and perhaps additional hormones are involved in tobacco pedicel development as has been shown in Arabidopsis (Yamaguchi and Komeda, 2013). Thus the present work put forward an improved regulatory model for plant pedicel development.
Discussion
Functional divergence of the SVP family genes
MADS-box genes are ubiquitous among flowering plants. Many floral development-associated MADS-box genes are conserved in distantly related species, although subfunctionalization has been reported for genes with duplicated copies (Rijpkema et al., 2006). Despite this, existing data show that genes in the SVP family are divergent in functions even in closely related species. The tomato J and its interacting partners MACROCALXY (MC) and SEPALLATA-like SlMBP21 are essential for AZ formation in tomato (Mao et al., 2000; Nakano et al., 2012; Liu et al., 2014). J is also involved in determining the sympodial meristem identity and hence the tomato inflorescence architecture (Szymkowiak and Irish, 2006). In contrast, the Arabidopsis SVP works as a flowering repressor (Hartmann et al., 2000), while a second SVP-like MADS-box gene AGL24 is a flowering promoter (Michaels et al., 2003). In snapdragon, INCO is responsible for prophyll development as well as floral meristem identity (Masiero et al., 2004). In woody perennial vine kiwifruit (Actinidia spp.), overexpression of an SVP gene affects reproductive development and suppresses anthocyanin biosynthesis in petals, but has no effect on vegetative growth, dormancy, or flowering time (Wu et al., 2014). The involvement of SVP or SVP-like genes in genetic pathways determining inflorescence architecture was recently found to be conserved in Arabidopsis and rice (Liu et al., 2013). Other spike-related phenotypes in grass species may also be related to pedicel development such as those caused by rice Dense and Erect Panicle1 (DEP1), DEP2, and DEP3 (Huang et al., 2009; Li et al., 2010; Qiao et al., 2011; Jiang et al., 2014). Although common tobacco belongs to the Solanaceae family, functional divergence of the SVP family genes may also occur. This is in line with the findings that NtSVP is involved in pedicel development and does not seem to be implicated in AZ development, in contrast toclosest tomato homologue J (Mao et al., 2000). Such an observation may contribute to the possibility of the existence of additional homologues of SVP in tobacco.
Conservation and divergence of regulatory pathways for plant pedicel development
KNOX genes regulate various aspects of development in the plant kingdom and play a key role in controlling the establishment and maintenance of the SAM (Hay and Tsiantis, 2009, 2010). In Arabidopsis, the class I KNOX gene BP is the major player in pedicel development. In tobacco, NtBPL has conserved, but not identical, functions. NtBPL is involved in pedicel elongation; its effect on the pedicel angle seems to be limited. The average pedicel angles are widened, but not as severely as in an Arabidopsis bp mutant where flowers became downward pointing (Venglat et al., 2002). In Arabidopsis, BP is a repressor of KNAT6 and the ARABIDOPSIS THALIANA HOMEOBOX GENE1 (ATH1) gene, which was not observed among differentially expressed genes in the present microarray data. One possibility is that the molecular pathways for pedicel development may be different between the two species. The other possibility is that the corresponding gene is not included in the current version of the tobacco microarray. Also no regulatory relationship between AS1/2 homologues and NtBPL was observed, while in Arabidopsis, the AS1–AS2 complex represses BP and KNAT2 (Phelps-Durr et al., 2005; Luo et al., 2012; Lodha et al., 2013). On the other hand, no MADS-box transcription factor has been shown to be involved in pedicel development. Thus, the discovery of an SVP-like MADS-box gene as a repressor for NtBPL in tobacco may represent a divergent, or as yet uncovered regulatory step for plant pedicel development.
An extended molecular pathway for tobacco pedicel development
In Arabidopsis, a number of genes have been reported to regulate BP in pedicel development. In addition to AS1 and AS2 as repressors of BP and KNAT2 (Lin et al., 2003; Xu et al., 2003; Guo et al., 2008; Rast and Simon, 2012), TCP transcription factors have recently been reported to interact with AS2 to repress class I KNOX genes (Li et al., 2012). Thus, the specification of pedicel development is strictly regulated, with the implication of a variety of transcription factors, mostly playing roles as suppressors. Such a mode of action seems to be conserved in tobacco. Here, NtSVP, a MADS-box transcription factor gene, is found to be associated with tobacco pedicel development as a transcription suppressor of NtBPL. The data suggest that NtSVP may exert its function on pedicel development by direct binding to the promoter of NtBPL. Interestingly, NtAS2 was found to be severely suppressed in NtSVP-RNAi pedicels, putting NtAS2 in a position in between NtSVP and NtBPL. The molecular mechanism of how NtSVP interacts with other NtBPL regulators such as AS1/2 and TCP genes as shown in Arabidopsis needs further investigation.
Hormonal regulation of tobacco pedicel development
In Arabidopsis, auxin is considered to be involved in pedicel development, as shown by the mutation of the auxin transport-related gene BIG/CRM1 that results in shortened pedicels and internodes (Douglas and Riggs, 2005; Yamaguchi and Komeda, 2013). The function of CRM1 is positively correlated with the activity of the auxin transporter PIN1. However, BP affects several aspects of pedicel development, including cell division, cell differentiation, and cell elongation, with no polarity in either internode or pedicel, indicating that as far as BP is concerned, auxin may not be the major downstream hormonal effector (Venglat et al., 2002).
Transcriptome analysis showed that when NtBPL was disrupted, several classes of hormones were affected; however, genes involved in GA biosynthesis and signalling were more likely to be responsible for pedicel development in tobacco. The functions of KNOX proteins in regulating GA-related genes (GA20OX and GA2OX) have been reported in Arabidopsis stems and shoot meristem, where KNOX proteins induce cytokinin biosynthesis, directly suppress GA synthesis via GA 20-oxidase repression, and promote GA deactivation via GA 2-oxidase activation (Sakamoto et al., 2001; Hay et al., 2002). Furthermore, in the triple mutants of the gibberellin receptor genes GID1a-1, GID1b-1, and GID1c-1, the elongation of the pedicel is strikingly reduced when compared with the single mutant (Griffiths et al., 2006). Transcriptome analysis revealed seven GA-related genes that were differentially expressed when NtBPL was disrupted. Such an observation supports the idea that GA may be the major player for tobacco pedicel development. Such a hypothesis should be studied in more detail.
Tobacco has been a research model for a long time despite its tetraploid nature. The high similarity between the homoeologues makes it feasible to carry out functional characterization by manipulating the copies simultaneously. Nevertheless, a number of questions related to NtSVP and NtBPL genes remain to be answered. Would additional copies of the NtSVP gene be responsible for AZ development as its closest homologue is in tomato? Since AS2 seems to be down-regulated in NtSVP-RNAi lines and AS2 is known to suppress BP in Arabidopsis, how does NtSVP work together with AS2 in regulating NtBPL? What kind of genes are the direct targets of NtBPL? Future study using tobacco as a model should provide interesting insight into the molecular mechanisms to fine-tune plant pedicel development.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Protein sequence analysis and tissue-specific expression of NtSVP.
Figure S2. Phenotypes of the NtSVP transgenic plants.
Figure S3. Protein sequence analysis and tissue-specific expression of NtBPL.
Figure S4. Developmental-specific expression of NtSVP and NtBPL in pedicels of wild-type plants.
Figure S5. Phenotypes of the NtBPL transgenic plants.
Figure S6. The promoter sequence of NtBPL.
Figure S7. Confirmation of microarray data.
Figure S8. GO enrichment analysis of differentially expressed genes in NtBPL-RNAi pedicels and their MapMan classification.
Table S1. Hormone-related genes that are differentially expressed due to down-regulation of NtBPL.
Table S2. Primers used in this work.
Dataset 1. List of differentially expressed genes between the wild-type and NtBPL-RNAi pedicels.
Acknowledgements
We thank Drs Chentao Lin and Bin Liu for help with the luciferase assay experiment. This work is financially supported by the National Science Foundation of China (#31070260) and the CAAS Innovation Program.
References
- Douglas SJ, Riggs CD. 2005. Pedicel development in Arabidopsis thaliana: contribution of vascular positioning and the role of the BREVIPEDICELLUS and ERECTA genes. Developmental Biology 284, 451–463. [DOI] [PubMed] [Google Scholar]
- Evers JB, van der Krol AR, Vos J, Struik PC. 2011. Understanding shoot branching by modelling form and function. Trends in Plant Science 16, 464–467. [DOI] [PubMed] [Google Scholar]
- Gleave AP. 1992. A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Molecular Biology 20, 1203–1207. [DOI] [PubMed] [Google Scholar]
- Gregis V, Sessa A, Colombo L, Kater MM. 2006. AGL24, SHORT VEGETATIVE PHASE, and APETALA1 redundantly control AGAMOUS during early stages of flower development in Arabidopsis. The Plant Cell 18, 1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregis V, Sessa A, Colombo L, Kater MM. 2008. AGAMOUS-LIKE24 and SHORT VEGETATIVE PHASE determine floral meristem identity in Arabidopsis. The Plant Journal 56, 891–902. [DOI] [PubMed] [Google Scholar]
- Gregis V, Sessa A, Dorca-Fornell C, Kater MM. 2009. The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes. The Plant Journal 60, 626–637. [DOI] [PubMed] [Google Scholar]
- Griffiths J, Murase K, Rieu I, et al. 2006. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. The Plant Cell 18, 3399–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Thomas J, Collins G, Timmermans MC. 2008. Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. The Plant Cell 20, 48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. 1994. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Molecular Biology 25, 989–994. [DOI] [PubMed] [Google Scholar]
- Hamant O, Pautot V. 2010. Plant development: a TALE story. Current Opinion in Plant Biology 333, 371–381. [DOI] [PubMed] [Google Scholar]
- Hartmann U, Hohmann S, Nettesheim K, Wisman E, Saedler H, Huijser P. 2000. Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. The Plant Journal 21, 351–360. [DOI] [PubMed] [Google Scholar]
- Hay A, Kaur H, Phillips A, Hedden P, Hake S, Tsiantis M. 2002. The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Current Biology 12, 1557–1565. [DOI] [PubMed] [Google Scholar]
- Hay A, Tsiantis M. 2009. A KNOX family TALE. Current Opinion in Plant Biology 12, 593–598. [DOI] [PubMed] [Google Scholar]
- Hay A, Tsiantis M. 2010. KNOX genes: versatile regulators of plant development and diversity. Development 137, 3153–3165. [DOI] [PubMed] [Google Scholar]
- Heinrich M, Hettenhausen C, Lange T, Wunsche H, Fang J, Baldwin IT, Wu J. 2013. High levels of jasmonic acid antagonize the biosynthesis of gibberellins and inhibit the growth of Nicotiana attenuata stems. Plant Journal 73, 591–606. [DOI] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA. 2005. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch RB, Rogers SG, Fraley RT. 1985. Transgenic plants. Cold Spring Harbor Symposia on Quantitative Biology 50, 433–437. [DOI] [PubMed] [Google Scholar]
- Huang X, Qian Q, Liu Z, Sun H, He S, Luo D, Xia G, Chu C, Li J, Fu X. 2009. Natural variation at the DEP1 locus enhances grain yield in rice. Nature Genetics 41, 494–497. [DOI] [PubMed] [Google Scholar]
- Iwata T, Nagasaki O, Ishii HS, Ushimaru A. 2012. Inflorescence architecture affects pollinator behaviour and mating success in Spiranthes sinensis (Orchidaceae). New Phytologist 193, 196–203. [DOI] [PubMed] [Google Scholar]
- Jiang G, Xiang Y, Zhao J, Yin D, Zhao X, Zhu L, Zhai W. 2014. Regulation of inflorescence branch development in rice through a novel pathway involving the pentatricopeptide repeat protein sped1-D. Genetics 197, 1395–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Li W, Tang J, Chen J, Tong H, Hu B, Li C, Fang J, Chen M, Chu C. 2010. Rice DENSE AND ERECT PANICLE 2 is essential for determining panicle outgrowth and elongation. Cell Research 20, 838–849. [DOI] [PubMed] [Google Scholar]
- Li Z, Li B, Shen WH, Huang H, Dong A. 2012. TCP transcription factors interact with AS2 in the repression of class-I KNOX genes in Arabidopsis thaliana. The Plant Journal 71, 99–107. [DOI] [PubMed] [Google Scholar]
- Lin WC, Shuai B, Springer PS. 2003. The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial–abaxial patterning. The Plant Cell 15, 2241–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Teo ZW, Bi Y, Song S, Xi W, Yang X, Yin Z, Yu H. 2013. A conserved genetic pathway determines inflorescence architecture in Arabidopsis and rice. Developmental Cell 24, 612–622. [DOI] [PubMed] [Google Scholar]
- Liu D, Wang D, Qin Z, Zhang D, Yin L, Wu L, Colasanti J, Li A, Mao L. 2014. The SEPALLATA MADS-box protein SLMBP21 forms protein complexes with JOINTLESS and MACROCALYX as a transcription activator for development of the tomato flower abscission zone. The Plant Journal 77, 284–296. [DOI] [PubMed] [Google Scholar]
- Liu YG, Whittier RF. 1995. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25, 674–681. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lodha M, Marco CF, Timmermans MC. 2013. The ASYMMETRIC LEAVES complex maintains repression of KNOX homeobox genes via direct recruitment of Polycomb-repressive complex2. Genes and Development 27, 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Yu CW, Chen FF, Zhao L, Tian G, Liu X, Cui Y, Yang JY, Wu K. 2012. Histone deacetylase HDA6 is functionally associated with AS1 in repression of KNOX genes in arabidopsis. PLoS Genetics 8, e1003114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L, Begum D, Chuang HW, Budiman MA, Szymkowiak EJ, Irish EE, Wing RA. 2000. JOINTLESS is a MADS-box gene controlling tomato flower abscission zone development. Nature 406, 910–913. [DOI] [PubMed] [Google Scholar]
- Masiero S, Li MA, Will I, Hartmann U, Saedler H, Huijser P, Schwarz-Sommer Z, Sommer H. 2004. INCOMPOSITA: a MADS-box gene controlling prophyll development and floral meristem identity in Antirrhinum. Development 131, 5981–5990. [DOI] [PubMed] [Google Scholar]
- Michaels SD, Ditta G, Gustafson-Brown C, Pelaz S, Yanofsky M, Amasino RM. 2003. AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. The Plant Journal 33, 867–874. [DOI] [PubMed] [Google Scholar]
- Meng Y, Li H, Wang Q, Liu B, Lin C. 2013. Blue light-dependent interaction between cryptochrome2 and CIB1 regulates transcription and leaf senescence in soybean. The Plant Cell 25, 4405–4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Kimbara J, Fujisawa M, Kitagawa M, Ihashi N, Maeda H, Kasumi T, Ito Y. 2012. MACROCALYX and JOINTLESS interact in the transcriptional regulation of tomato fruit abscission zone development. Plant Physiology 158, 439–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N, Eshed Y, Chuck G, Bowman JL, Hake S. 2000. Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127, 5523–5532. [DOI] [PubMed] [Google Scholar]
- Phelps-Durr TL, Thomas J, Vahab P, Timmermans MC. 2005. Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. The Plant Cell 17, 2886–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y, Piao R, Shi J, Lee S.I, Jiang W, Kim BK, Lee J, Han L, Ma W, Koh HJ. 2011. Fine mapping and candidate gene analysis of dense and erect panicle 3, DEP3, which confers high grain yield in rice (Oryza sativa L.). Theoretical and Applied Genetics 122, 1439–1449. [DOI] [PubMed] [Google Scholar]
- Ragni L, Belles-Boix E, Gunl M, Pautot V. 2008. Interaction of KNAT6 and KNAT2 with BREVIPEDICELLUS and PENNYWISE in Arabidopsis inflorescences. The Plant Cell 20, 888–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rast MI, Simon R. 2012. Arabidopsis JAGGED LATERAL ORGANS acts with ASYMMETRIC LEAVES2 to coordinate KNOX and PIN expression in shoot and root meristems. The Plant Cell 24, 2917–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Kuhlemeier C. 2002. Plant architecture. EMBO Reports 3, 846–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijpkema AS, Royaert S, Zethof J, van der Weerden G, Gerats T, Vandenbussche M. 2006. Analysis of the Petunia TM6 MADS box gene reveals functional divergence within the DEF/AP3 lineage. The Plant Cell 18, 1819–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Kamiya N, Ueguchi-Tanaka M, Iwahori S, Matsuoka M. 2001. KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes and Development 15, 581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shani E, Weinstain R, Zhang Y, Castillejo C, Kaiserli E, Chory J, Tsien RY, Estelle M. 2013. Gibberellins accumulate in the elongating endodermal cells of Arabidopsis root. Proceedings of the National Academy of Sciences, USA 110, 4834–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymkowiak EJ, Irish EE. 2006. JOINTLESS suppresses sympodial identity in inflorescence meristems of tomato. Planta 223, 646–658. [DOI] [PubMed] [Google Scholar]
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruge P, Selbig J, Muller LA, Rhee SY, Stitt M. 2004. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. The Plant Journal 37, 914–939. [DOI] [PubMed] [Google Scholar]
- van Zanten M, Snoek LB, Proveniers MC, Peeters AJ. 2009. The many functions of ERECTA . Trends in Plant Science 14, 214–218. [DOI] [PubMed] [Google Scholar]
- Venglat SP, Dumonceaux T, Rozwadowski K, Parnell L, Babic V, Keller W, Martienssen R, Selvaraj G, Datla R. 2002. The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proceedings of the National Academy of Sciences, USA 99, 4730–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, et al. 2001. Construct design for efficient, effective and high-throughput gene silencing in plants. The Plant Journal 27, 581–590. [DOI] [PubMed] [Google Scholar]
- Wu R, Wang T, McGie T, Voogd C, Allan AC, Hellens RP, Varkonyi-Gasic E. 2014. Overexpression of the kiwifruit SVP3 gene affects reproductive development and suppresses anthocyanin biosynthesis in petals, but has no effect on vegetative growth, dormancy, or flowering time. Journal of Experimental Botany 65, 4985–4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Li Z, Zhu Y, Wang H, Ma H, Dong A, Huang H. 2008. Arabidopsis genes AS1, AS2, and JAG negatively regulate boundary-specifying genes to promote sepal and petal development. Plant Physiology 146, 566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Xu Y, Dong A, Sun Y, Pi L, Huang H. 2003. Novel as1 and as2 defects in leaf adaxial–abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development 130, 4097–4107. [DOI] [PubMed] [Google Scholar]
- Yamaguchi N, Komeda Y. 2013. The role of CORYMBOSA1/BIG and auxin in the growth of Arabidopsis pedicel and internode. Plant Science 209, 64–74. [DOI] [PubMed] [Google Scholar]
- Yamaguchi N, Suzuki M, Fukaki H, Morita-Terao M, Tasaka M, Komeda Y. 2007. CRM1/BIG-mediated auxin action regulates Arabidopsis inflorescence development. Plant and Cell Physiology 48, 1275–1290. [DOI] [PubMed] [Google Scholar]
- Yamaguchi N, Yamaguchi A, Abe M, Wagner D, Komeda Y. 2012. LEAFY controls Arabidopsis pedicel length and orientation by affecting adaxial–abaxial cell fate. The Plant Journal 69, 844–856. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S. 2008. Gibberellin metabolism and its regulation. Annual Review of Plant Biology 59, 225–251. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Li B, Mu S, Han B, Cui R, Xu M, You Z, Dong H. 2013. TTG2-regulated development is related to expression of putative AUXIN RESPONSE FACTOR genes in tobacco. BMC Genomics 14, 806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.