Highlight
An integrated transcriptomics and metabolomics analysis of the nitrogen-deprivation response in the diatom Phaeodactylum tricornutum uncovered remobilization of internal nitrogen-containing resources, and remodelling of carbon and lipid structures.
Key words: Carbon metabolism, diatom, metabolomics, nitrogen deprivation, Phaeodactylum tricornutum, transcriptome, triacylglycerol.
Abstract
Algal growth is strongly affected by nitrogen (N) availability. Diatoms, an ecologically important group of unicellular algae, have evolved several acclimation mechanisms to cope with N deprivation. In this study, we integrated physiological data with transcriptional and metabolite data to reveal molecular and metabolic modifications in N-deprived conditions in the marine diatom Phaeodactylum tricornutum. Physiological and metabolite measurements indicated that the photosynthetic capacity and chlorophyll content of the cells decreased, while neutral lipids increased in N-deprived cultures. Global gene expression analysis showed that P. tricornutum responded to N deprivation through an increase in N transport, assimilation, and utilization of organic N resources. Following N deprivation, reduced biosynthesis and increased recycling of N compounds like amino acids, proteins, and nucleic acids was observed at the transcript level. The majority of the genes associated with photosynthesis and chlorophyll biosynthesis were also repressed. Carbon metabolism was restructured through downregulation of the Calvin cycle and chrysolaminarin biosynthesis, and co-ordinated upregulation of glycolysis, the tricarboxylic acid cycle, and pyruvate metabolism, leading to funnelling of carbon sources to lipid metabolism. Finally, reallocation of membrane lipids and induction of de novo triacylglycerol biosynthesis directed cells to accumulation of neutral lipids.
Introduction
Phytoplankton blooms vary temporally and spatially in accordance with nutrient availability (Brandes et al., 2007; Falkowski and Raven, 2007). Under upwelling conditions, high levels of available nitrate and iron lead to an increase in phytoplankton biomass, which is generally dominated by diatoms (Kudela and Dugdale, 2000; Capone and Hutchins, 2013). Inorganic nitrogen (N) in the form of ammonia or nitrate is utilized by several phytoplankton (Dham et al., 2005); some phytoplankton are also able to use organic forms of nitrogen such as amino acids, nucleic acids, and urea (Baker et al., 2009; Solomon et al., 2010).
Diatoms are a group of unicellular heterokont microalgae believed to include some 200 000 species (Armbrust, 2009). It is estimated that marine diatoms are responsible for about 32% of global phytoplankton primary production (Uitz et al., 2010). Unlike plants and green algae, diatoms and other heterokonts originate from a serial secondary endosymbiosis event, in which a green alga and subsequently a red alga were engulfed by a heterotrophic eukaryote (Moustafa et al., 2009; Bowler et al., 2010). In addition, a large number of horizontal gene transfer events have further increased the gene repertoire. As a result, diatom genomes contain unique combinations of nutrient assimilation and metabolic pathways that have contributed to their ecological success in the ocean (Prihoda et al., 2012).
The whole-genome sequences of the centric diatom Thalassiosira pseudonana (Armbrust et al., 2004) and the pennate diatom Phaeodactylum tricornutum (Bowler et al., 2008) have provided valuable information on the regulatory and metabolic inventory of these diatoms. The genomes of T. pseudonana and P. tricornutum contain several transporter proteins for uptake of inorganic and organic nitrogen (Rees and Syrett, 1979; Armbrust et al., 2004; Allen, 2005; Hildebrand, 2005; Solomon et al., 2010). Nitrate entering the cell is first reduced to nitrite and ammonium (Allen et al., 2005; Bowler et al., 2010). Ammonium is then assimilated by glutamate synthase/glutamine synthetase to amino acids (Zehr and Falkowski, 1988; Takabayashi et al., 2005). Diatoms possess plastidial glutamine synthetase (GSII) and glutamate synthase (Fd-GOGAT) as well as mitochondrial NAD(P)H-GOGAT and GSIII (Bowler et al., 2010; Allen et al., 2011). Mitochondrial GSIII may catalyse the assimilation of glutamine from ammonium derived from cytosolic catabolic reactions, e.g. deamination and hydrolysis of organic N (Hockin et al., 2012; Kissen et al., 2010; Parker and Armbrust, 2005).
N and carbon metabolism are closely connected to each other. N assimilation and amino acid biosynthesis require reducing equivalents from photosynthesis and carbon skeletons from the tricarboxylic acid (TCA) cycle (Hockin et al., 2012). Moreover, in the photosynthetic apparatus, assimilated N is used for example in ribulose-1,5-bisphosphate carboxylase (Rubisco) and the light-harvesting complex (LHC) (Orellana and Perry, 1995; Foyer et al., 2003; Nunes-Nesi et al., 2010). In response to N deprivation, diatoms reprogram several metabolic pathways. The impact of N deprivation on pigments, photosynthesis, carbon fixation, and N assimilation has been studied in diatoms (Syrett et al., 1986; Kolber et al., 1988; Geider et al., 1993; Granum et al., 2009; Bender et al., 2014). Diatoms store carbon in the form of 1,3-β-d-glucan (chrysolaminarin) or lipids (Kroth et al., 2008). Under optimal conditions, chrysolaminarin is the major sink of carbon storage in the vacuoles (Granum and Myklestad, 2002). Under several stress conditions, in particular N starvation, diatoms change their carbon storage patterns in favour of neutral lipid accumulation (Eizadora et al., 2009; Norici et al., 2011; Sharma et al., 2012; Valenzuela et al., 2012). Neutral lipids produced from microalgae have been proposed as a sustainable substitute biofuel for fossil fuels (Wijffels and Barbosa, 2010). Other N-containing compounds, such as proteins and nucleic acids, are also affected by a decrease in cellular N content (Olson et al., 1986; Mock and Kroon, 2002; Bertozzini et al., 2013; Mus et al., 2013).
To understand how the oleaginous marine diatom P. tricornutum responds to N deprivation, cells were grown in f/2 medium and in N-free medium, and samplings were conducted at 48 and 72h after N deprivation. We combined transcriptional and metabolite analyses to monitor the effect of N deprivation at different molecular levels in order to get a better insight into the acclimation strategies employed by P. tricornutum under N deprivation. These data were further complemented by physiological data such as measurements of cell growth, neutral lipids, and other cell chemistry measurements. We use this data to predict metabolic changes in N-deprived cells leading to remodelling of lipid metabolism and triacylglycerol (TAG) accumulation.
Materials and methods
Growth conditions and treatments
Axenic cultures of P. tricornutum clone Pt1 8.6 (CCMP632) were grown in f/2 medium and kept in exponential growth at 15 °C under continuous white fluorescent light (60 µmol photons m–2 s–1) for 3 weeks. Bacterial contamination was checked regularly by inoculation in peptone-enriched f/2 medium (Andersen et al., 1997). Growth medium (f/2) was made from 0.2 µm-filtered seawater, autoclaved, and enriched with macro- and micronutrients (Guillard, 1975). Three or four replicates of the start culture (6–7ml) were transferred to 220ml of medium supplemented with complete f/2 nutrients (replete) or f/2 without added nitrate (deprived). The nitrate concentration in the seawater used for the experiments was measured to 10 μM, which is 1.1% of the f/2 nitrate concentration. Cells were incubated in batch cultures with a starting cell density of 5×104 ml–1 in sterile culture flasks with a 75cm2 growth area. Cell counting and maximum quantum yield of photosystem II (PSII) (Fv/Fm) was measured daily using a Bürker–Türk counting chamber and AquaPen-C AP-C 100 (Photon Systems Instruments), respectively. For the other experiments, samples were harvested 48 and 72h after the beginning of the treatment. Samples for RNA and metabolite analysis were stored at –80 °C, while samples for nutrient and pigment analysis were stored at –23 °C until analysis.
Nutrient analysis
Triplicate cultures for particulate N, carbon, and phosphorus analysis were collected on pre-combusted GF/F filters (particulate C and N analysis) or 0.2 µm GF/F filters (particulate phosphorus), and the flowthrough was used for detection of medium phosphate and nitrate concentration. Triplicate samples for particulate N and carbon analysis along with blank filters were treated with HCl vapour (37%), packed in tin capsules, dried for 2 days at 60 °C, and analysed by an ECS 4010 element analyser (Costech Instruments). All these processes were performed according to Chauton et al. (2013). Inorganic nutrients were measured in the filtrate. + and were analysed in parallel according to I.O. Analytical cartridge Part A002603 and A002604, respectively, as described by Hansen and Koroleff (1999). Particulate phosphate was first oxidized to according to Norwegian standard NS4725, and then analysed as inorganic .
Pigment analysis
Pigment analysis (fg per cell) was performed based on the protocol by Rodríguez et al. (2006). Briefly, 60ml (N replete) or 100ml (N deprived) of cultures was collected on GF/F filters. The cells were extracted with 6ml of 100% ethanol, and extracts were filtered through Millipore 0.45 μm filters. A volume of 73 µl of the final extracts was mixed with 23 µl of water and injected into a Hewlett-Packard HPLC 1100 Series system. Pigments were separated on a Waters Symmetry C8 column using the high-performance liquid chromatography (HPLC) method of Zapata et al. (2000). Chlorophyll a and fucoxanthin were detected by absorbance at 440nm and identified by a diode array detector (λ=350–370nm, 1.2nm spectral resolution). Standard curves were made by isolating pigments separated by HPLC, verifying their identity and quantifying on a spectrophotometer, and running a dilution series on the HPLC instrument. The specific extinction coefficients (α: 1g–1 cm–1) provided by Egeland et al. (2011) were used for pigment quantification.
Neutral lipid measurement
A volume of 1ml of culture was stained with 1 µl of 0.1 µg ml–1 of BODIPY 505/515 (4,4-difluoro-1,3,5,7-tetramethyl-4-bora-3a-4a-diaza-s-indancene; Life Technologies) dissolved in 2% (w/v) dimethyl sulfoxide and shaken carefully by hand (Govender et al., 2012). After 5min, 30 µl of culture was transferred to a microscope slide, and a coverslip was placed on top of the culture and sealed using dental wax. At least 20 cells from two replicates were analysed for BODIPY 505/515 fluorescence on a Leica TCS SP5 confocal laser scanning microscope using a ×63 water objective. Z-Sectional images were made using argon laser excitation at 488nm (17% of maximal intensity), and emission was detected with a spectral detector set from 495 to 550nm. Non-confocal bright-field images were made simultaneously. A z-stack consisting of 10 scans was made for each cell, encompassing the complete fluorescent part of the cell. The length of the z-stack varied between 4.00 and 5.78 μm; consequently, the z-slice step size varied between 0.44 and 0.64 μm. Laser power, PMT gain, and offset were kept constant for all scans. Image stacks containing the fluorescence channel were imported into ImageJ (Abràmoff et al., 2004). To determine the total fluorescence detected in the z-stack, a region was drawn around each cell to be measured, and three regions next to the selected cell that had no fluorescence were used for background subtraction. The corrected total cell fluorescence for each cell was calculated using the following formula (Gavet and Pines, 2010; Potapova et al., 2011):
Background fluorescence, as measured from five z-sectional images of unstained cells, was negligible (<1% of stained N-replete cells). A total of 20–30 cells were analysed for each treatment.
Harvesting and extraction for metabolite profiling
Depending on cell density, 60–100ml of culture was collected on 0.65 µm Durapore membrane filters, washed off the filter using 1ml of f/2 medium (N-deprived cells were washed with f/2 without nitrate supplement), and centrifuged at 13 000rpm for 1min at 4 °C. Care was taken to minimize the harvesting time, which was less than 3min. The supernatant was removed and pellets were flash frozen in liquid N2 and stored at–80 °C. Metabolites were extracted by adding 1ml of a pre-cooled water:methanol:chloroform (1:2.5:1) mixture containing ribitol as an internal standard (100 µg ml–1). Samples were treated for 60min at 60 °C in an ultrasonic bath, centrifuged for 10min at 13 000rpm, and 600 µl aliquots of supernatant were transferred to 2ml Eppendorf tubes.
Sample derivatization and gas chromatography/mass spectrometry (GC-MS) analysis
Samples were dried in a SavantTM SpeedVac plus SC210A (Thermo Scientific) overnight and stored at –80 °C before derivatization. Dried samples were redissolved in 80 µl of methoxyamine hydrochloride in pyridine (20mg ml–1), derivatized for 90min at 30 °C, further treated with 80 µl of N-methyl-N-(trimethylsilyl)trifluoroacetamide for 30min at 37 °C, and finally transferred to 1.5ml autosampler vials with glass inserts prior to GC-MS. Separations were performed on an Agilent 6890/5975 GC-MS (Agilent Technologies) equipped with a HP-5MS capillary column (30 m×0.25mm internal diameter, film thickness 0.25 µm) (Agilent Technologies). Sample volumes of 3 µl were injected with a split ratio of 15:1. Injection and interface temperature were set to 230 and 250 °C, respectively. The GC temperature program was held isothermically at 70 °C for 5min, ramped from 70 to 310 °C at 5 °C min–1, and finally held at 310 °C for 7min (run time: 60min). The MS source was adjusted to 230 °C and a mass range of m/z 70–600 was recorded (EI mode).
Metabolite data analysis
Chromatogram visualization and peak identification was carried out using Agilent ChemStation software (Agilent Technologies), AMDIS software (version 2.71; National Institute of Standards and Technology), and OpenChrom Community Edition Synge (version 0.6.0) (Peter Wenig; http://www.openchrom.net). NIST05 spectral library (National Institute of Standards and Technology) in combination with a metabolite target library (Hummel et al., 2010) were used for tentative compound identification. GC-MS data integration, normalization (total signal), and alignment were carried out using the MetAlign software (PRI-Rikilt). Based on distinct quantifier ions, detected metabolites were quantified using the internal standard ribitol (normalized response) and finally expressed in ng per 106 cells. Statistical analysis was carried out using one-way analysis of variance across all time points and N conditions.
A total of 119 metabolites and metabolite tags were detected, 110 of which were tentatively identified based on the MS library. Based on the extraction properties of the solvent mixture, a broad range of metabolites were simultaneously extracted (Nappo et al., 2009), including lipophilic alkanes, fatty acids, and glycerides, and polar compounds such as amino acids, organic acids, sugars, and polyols. A total of 94 metabolites are presented in Supplementary Table S1 (available at JXB online), showing the ratio of compound levels in N-deprived cultures to replete conditions after 48 and 72h.
RNA isolation
Depending on cell density, 60–100ml of cultures was collected on 0.65 µm Durapore membrane filters, washed off the filter using 1ml of f/2 medium (N-deprived cells were washed with f/2 without nitrate supplement), and centrifuged at 13 000rpm for 1min at 4 °C. The supernatant was removed and pellets were flash frozen in liquid N2 and stored at –80 °C. Frozen samples were homogenized using a TissueLyser system (Qiagen) for 2×2min at 25 Hz. The samples were placed in a pre-cooled (–80 °C) adapter set for the first shaking step. Before the second shaking step, the samples were transferred to a room temperate adapter set, and 0.5ml of lysis buffer (SpectrumTM Plant Total RNA kit; Sigma-Aldrich) was added to each tube. Total RNA was isolated with a SpectrumTM Plant Total RNA kit (Sigma-Aldrich). To eliminate genomic DNA, an on-column digestion was performed using an RNAase-free DNase I set (Qiagen). Total RNA was quantified using a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies). The RNA quality was verified using formaldehyde gel electrophoresis. In addition, RNA integrity was checked on a 2100 Bioanalyzer (Agilent). All samples had RNA integrity numbers above 7.
cDNA microarray experiments
Total RNA (200ng) was reverse transcribed, amplified, and labelled according to a Low Input Quick Amp Labeling Kit, One-Color (Agilent Technologies). A total of 1650ng of cRNA from each sample was fragmented and hybridized with a Gene Expression Hybridization Kit (Agilent Technologies) on 4×44K P. tricornutum whole-genome 60-mer oligonucleotide microarrays (Agilent Technologies) in an Agilent G2545A Hybridization Rotary Oven at 10rpm, 65 °C for 17.5h. Slides were washed with washing buffer 1 and 2 using a Gene Expression Wash Buffer Kit (Agilent Technologies) and directly scanned using a laser scanner (G2505 B; Agilent Technologies) based on the ‘dynamic range expander’ option in the scanner software. Images were processed by Agilent Feature Extraction software version 9.5.
Statistical analysis
The Limma package (version 3.20.1) (Smyth, 2005) and R version 3.0.3 were used for statistical analysis and identification of significant differentially expressed genes. Single-colour feature expression files from the Agilent microarray scans were imported, and spots identified as feature outliers were excluded from the analysis. Weak or undetected spots were given reduced weight. The data were normalized using the quantile method, and no background subtraction was performed. A design matrix was created and pair-wise comparisons between the samples, DN48 (nitrogen-deprived 48h) and R48 (replete 48h) and DN72 (nitrogen-deprived 72h) and R72 (replete 72h) were performed. The method of Benjamini and Hochberg (1995) was used to estimate the false discovery rate. Genes with an adjusted P value of <0.05 were regarded as significantly differentially expressed and were included in the analysis if all oligonucleotides for each gene had a mean adjusted P value of <0.05. The study is MIAME compliant. Raw data has been deposited in GEO (accession no. GSE58946).
The Gene Ontology (GO) dataset for biological process was downloaded from the P. tricornutum database at Joint Genome Institute (http://genome.jgi-psf.org/Phatr2/Phatr2.home.html). GO terms assigned to significantly regulated genes at each time point were listed separately for up- and downregulated genes. Metabolic pathways were analysed using the DiatomCyc database (Fabris et al., 2012).
cDNA synthesis and quantitative real-time PCR
cDNA synthesis was performed using 1 µg of total RNA with a QuantiTect Reverse Transcription Kit (Qiagen) following the manufacturer’s instructions. cDNA samples were diluted five times in ddH2O before use for quantitative real-time PCR (qRT-PCR) analysis.
Three biological replicates from all treatments were used to perform qRT-PCR on a LightCycler 480 using a LightCycler 480 SYBR Green I Master kit (Roche Applied Science), with a program comprising pre-incubation for 5min at 95 °C, followed by 50 cycles of amplification consisting of 10 s at 95 °C, 10 s at 55 °C, and 10 s at 72 °C. Primer sequences used in the qRT-PCR experiment are given in Table 1. The microarray dataset was screened for genes that were non-responsive to N deprivation at both time points. Based on this screen, Exportin1 (Phatr2_24186) and Aureochrome1 (Phatr2_8113) were selected as reference genes for the qRT-PCR analysis. PCR efficiencies and C t values were calculated by linear regression using the LinRegPCR software (Ramakers et al., 2003; Ruijter et al., 2009), and the mean PCR efficiency was calculated for each primer pair. PCR efficiencies and C t values were used in the REST 2009 software (Pfaffl et al., 2002) to calculate the statistical significance of difference in expression levels in various treatments. The target genes were normalized to the reference genes in the REST 2009 software.
Table 1.
Genes analysed by real-time qPCR and their respective primers
| Phatr2 ID | Accession | Description | Orientation | Sequence (5′→3′) | Amplicon size (bp) |
|---|---|---|---|---|---|
| 54101 | XP_002177983 | Nitrate transporter | Forward | GGAATACTTGCTGTTCCTATGC | 58 |
| Reverse | AGGAGACTCTAGGCTTCGATCT | ||||
| 34373 | XP_002178768 | Molybdopterin biosynthesis-like protein CNX5 | Forward | ATGCTCAAGGACCGATGCAAAC | 130 |
| Reverse | CAGCTTGGTTCTTACGTCAACA | ||||
| 17344 | XP_002176623 | Adenine/guanine permease | Forward | AACTTTACCAGCGATCTTTCGG | 87 |
| Reverse | CTAGAAGAGTACCGCTTGTATC | ||||
| 18049 | XP_002177871 | LHC protein LHCF1 | Forward | GCAACAACTACCTCGACTTTGG | 82 |
| Reverse | TCCCTGGTTGAGTTCGATAGCA | ||||
| 49339 | XP_002183906 | Pyruvate carboxylase PYC2 | Forward | GTGGAACTCGTTTCTATCCAAG | 116 |
| Reverse | CGAATCTCCTAACAAGTTCTGG | ||||
| 24186 | XP_002185483 | Exportin 1-like protein XPO1 | Forward | TCTATTGTTTGGGCGATGAAGC | 89 |
| Reverse | CTTACCGACATTAACCAGCAGT | ||||
| 8113 | XP_002183783 | Aureochrome AUREO1a | Forward | GGCTTTCTCAACTTGACGGGAT | 116 |
| Reverse | TTCAATGGCCTTACGGATACGC |
Results
Effect of deprived levels of N on physiological responses in P. tricornutum
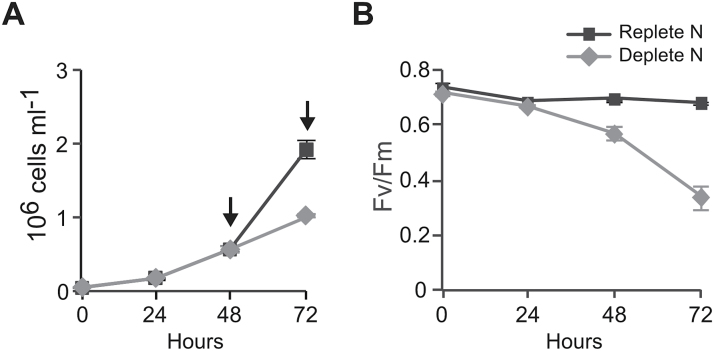
P. tricornutum cell growth was monitored daily in both cultures. All N-replete cultures remained in the exponential phase throughout the time course of the experiment. During the experiment period, cell density increased from 5×104 to 1.92×106 cells ml–1 in replete cultures (Fig. 1A). N-free cultures showed similar cell growth compared with N-replete cultures until 48h, but significantly lower growth at 72h, with a cell density of 1.02×106 cells ml–1. In order to compare physiological and transcriptional responses before and after the N deprivation started to affect cell growth, the time points of 48 and 72h were chosen for further physiological and molecular experiments.
Fig. 1.
Physiological responses of P. tricornutum to nitrate deprivation. Growth curves (A) and changes in maximum quantum yield (Fv/Fm) (B) of P. tricornutum in N-replete (f/2 medium) and N-deprived (f/2 medium minus nitrate) cultures. Arrows indicate sampling time points. Values are means±standard deviation of four biological replicates.
Nutrient assays of dissolved inorganic nitrate and phosphate demonstrated that none of the replete cultures encountered any deprivation in dissolved inorganic nitrate or phosphate during the whole experiment (Table 2). An increase in the C:N ratio was observed at both time points in N-deprived cells. In N-deprived cells, C:N deviated from the Redfield ratio (Redfield, 1934). The N:P ratio in replete cells was well below the Redfield ratio, but the cultures were still in exponential phase. Reduction of N:P in deprived cells coincided with nitrate loss in the medium.
Table 2.
Changes in chemical composition, medium nutrient concentration, and pigment concentration of nitrogen-replete (N+) and nitrogen-deprived (N–) cultures 48 and 72h after N deprivation (n=4)
| N+ 48 h | N– 48 h | N+ 72 h | N– 72 h | |
|---|---|---|---|---|
| Cellular nutrient content | ||||
| µg C: µg N | 6.03±0.32 | 9.46±0.73 | 5.44±0.19 | 14.65±0.77 |
| µg N: µg P | 4.24±0.26 | 2.37±0.31 | 4.89±0.18 | 1.67±0.1 |
| Medium concentration | ||||
| µg PO4 3- /l | 612,77±2,39 | 618±2,24 | 316,99±26,73 | 576,72±31,38 |
| µg (NO3 -+NO2 -) /l | 8797,38±49,23 | 3,98±1,07 | 7550,87±108,79 | 3,8±0,7 |
| Pigment concentration | ||||
| fg Chl a cell-1 | 271.4±25.89 | 153.2±5.96 | 271.6±26.29 | 76.9±9.42 |
| fg fucoxanthin cell-1 | 100.7±3.21 | 56.8±2.25 | 96.2±7.73 | 32.4±3.98 |
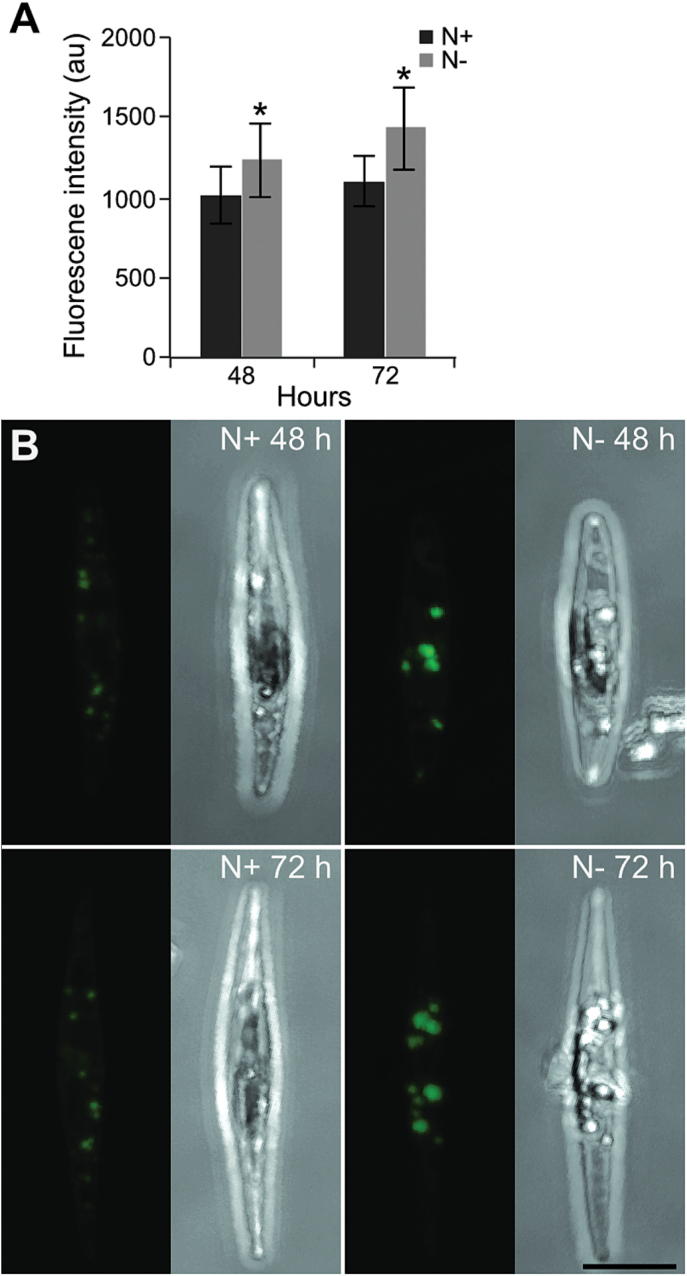
Measurements of chlorophyll a and fucoxanthin (the major carotenoid in diatoms) levels per cell showed that the content of these pigments declined progressively in N-deprived cells at both time points, while both pigments were stable in control cells (Table 2). In contrast, the ratio between chlorophyll a and fucoxanthin did not change. We also monitored the effect of N deprivation on the activity of PSII. Maximum quantum yield of PSII (Fv/Fm), applied as a proxy measure of photosynthesis, was similar in N-replete and N-deprived cultures after 24h. A clear drop in Fv/Fm was observed in N-deprived cells (Fv/Fm=0.33) after 72h, while the ratio remained unchanged (Fv/Fm=0.68) in the control cultures (Fig. 1B). Quantification of neutral lipids by confocal laser scanning microscopy and BODIPY 505/515 showed that the neutral lipid content increased significantly at 72h (t-test: P=1.87 E–06) and was 29.3% higher in N-deprived cells compared with N-replete cells (Fig. 2A). Representative images from the analysis showed that the lipid droplets were larger and more strongly stained by the BODIPY marker in N-deprived cells than in replete cells, implying higher lipid accumulation (Fig. 2B).
Fig. 2.
Accumulation of neutral lipids during nitrate deprivation. (A) Fluorescence intensity in P. tricornutum cells stained with BODIPY 505/515 at 48 and 72h after N deprivation. The level of lipid fluorescence was measured in 20–30 randomly selected cells using confocal microscopy. Statistical differences (*P<0.01) between nitrate-replete (N+) and nitrate deprived (N–) cultures are indicated. au, Arbitrary units. (B) Z-stack projections of P. tricornutum in N+ and N– cultures at 48 and 72h after N deprivation. Bar, 5 μm. (This figure is available in colour at JXB online.)
Metabolite profiling of the responses to N deprivation was performed using GC-MS. Strong effects on the central metabolism were revealed, with a significant decrease in most of the N-containing metabolites and major fatty acids (Supplementary Table S1). The regulation of biosynthesis and significance of distinct metabolites is further discussed in subsequent sections.
Gene expression
Transcriptome responses at 48 and 72h after N deprivation were analysed using whole-genome oligonucleotide microarrays. The treatment led to strong transcriptome responses: 5279 genes were significantly regulated (P<0.05) in N-deprived cultures compared with N-replete cultures 48h after N deprivation. As expected, the stronger N deprivation at 72h affected even more genes (6629). Comparison of the N-replete cultures at 48 and 72h resulted in only 22 significantly regulated genes, probably reflecting higher cell densities (results not shown).
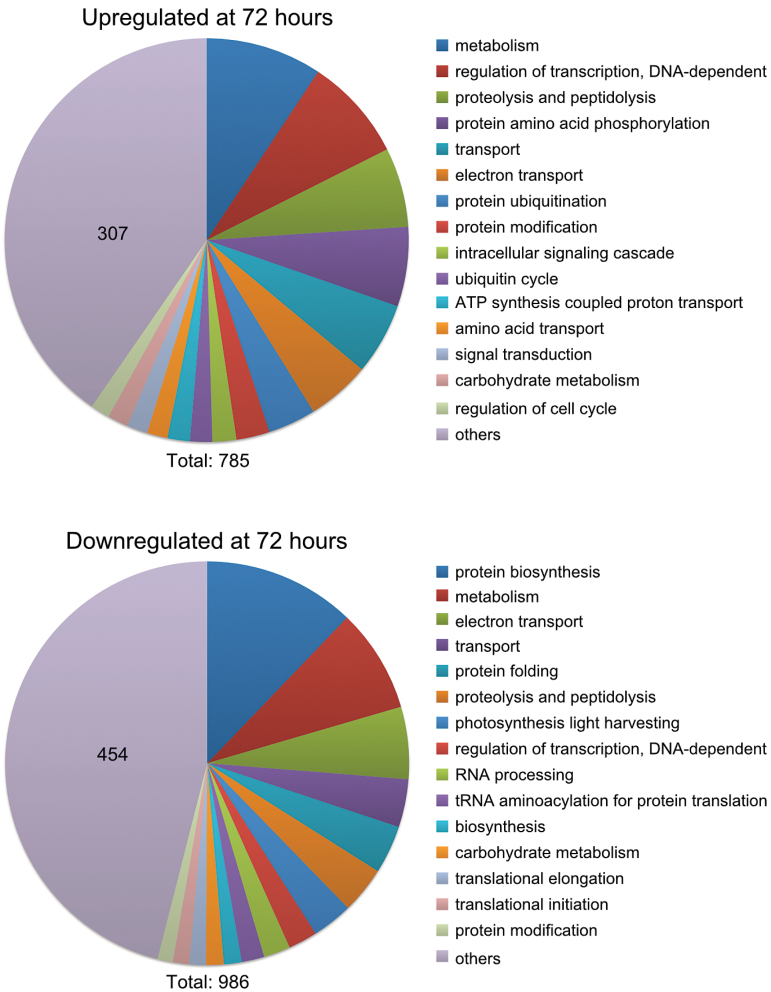
GO analysis was performed on the dataset. As the P. tricornutum genome is still poorly annotated, GO terms are assigned to a limited number of genes. The GO analysis still provided an overview of the processes most affected by N deprivation. The most enriched GO terms were similar at 48h (Supplementary Fig. S1, available at JXB online) and 72h (Fig. 3); however, there were large differences between GO terms enriched in up- and downregulated genes. The most frequent GO term among the downregulated genes was protein biosynthesis. Other GO terms related to ribosomal assembly and translation were also enriched among downregulated genes, indicating reduced protein biosynthesis. Furthermore, photosynthesis light harvesting was the fourth most used GO term among the downregulated genes, indicating downsizing of the light-harvesting apparatus. In contrast, the upregulated genes were enriched in GO terms related to signal transduction and ubiquitination.
Fig. 3.
GO analysis of significantly regulated genes after 72h of nitrate deprivation. The dataset was divided into up- and downregulated genes and analysed for process GO terms. The 15 most frequent GO terms are listed, and the rest were combined into ‘others’. The number in the ‘others’ section indicates the number of hits within this category. The total number of GO term hits is listed below the graphs.
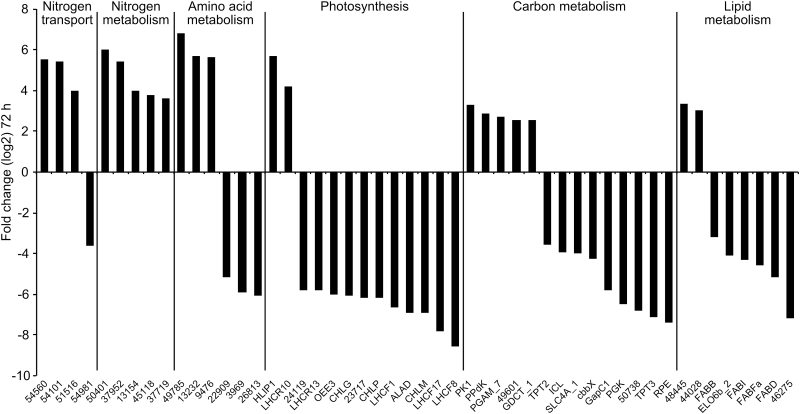
Strong transcriptional responses were observed for genes encoding proteins involved in processes such as photosynthesis, central carbon metabolism, lipid metabolism, nitrogen metabolism and transport, and amino acid metabolism, as discussed below. The responses of the strongest regulated genes within these categories 72h after N deprivation are shown in Fig. 4. In order to verify the results of microarray analysis, qRT-PCR was performed on five selected genes involved in photosynthesis, and N and carbon metabolism, respectively, that were differentially regulated at 48 and 72h in the microarray analysis. The qRT-PCR results correlated well with the microarray analysis (Supplementary Fig. S2, available at JXB online).
Fig. 4.
Genes strongly regulated by N deprivation. The genes most up- or downregulated after 72h of nitrate deprivation are shown for the processes listed at the top of the graph. The ratios were log2 transformed. Numbers indicate Phatr2 gene IDs.
Effect of N deprivation on N metabolism
N uptake and assimilation
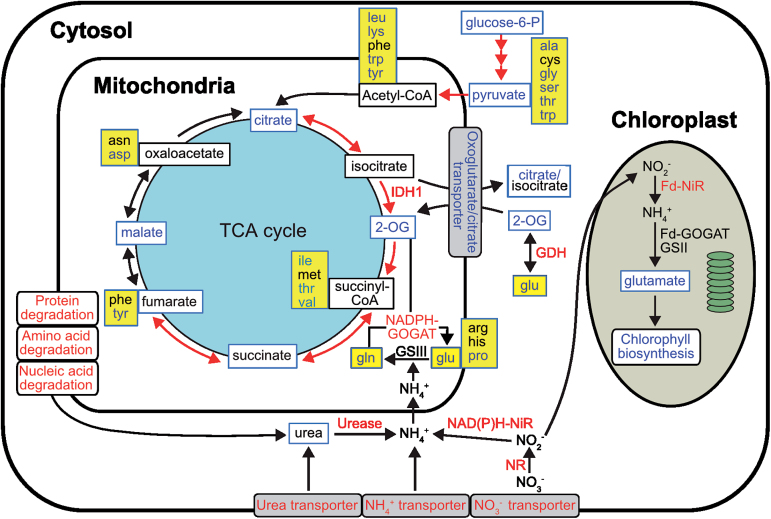
Transcriptional responses to N deprivation of P. tricornutum showed that uptake, assimilation, and scavenging mechanisms were activated (Fig. 5). In our experiment, transcript levels of genes involved in , , and urea transport were upregulated (Fig. 4 and Supplementary Dataset S1, available at JXB online). Of four ammonium transporters detected in our microarray data, three were upregulated. The induction of a nitrate transporter (Phatr2_54101) was confirmed by qRT-PCR (Supplementary Fig. S2). Increased transcription of genes encoding nitrate reductase (NR) and both NAD(P)H- and Fd-dependent nitrite reductase was observed at 72h after deprivation (Figs 4 and 5). Interestingly, two genes encoding molybdopterin biosynthesis proteins were induced (Supplementary Dataset S1). These enzymes might be orthologues of the Arabidopsis thaliana cofactor of NR and xanthine dehydrogenase CNX5 and CNX2, respectively (Schwarz and Mendel, 2006). The biosynthesis of molybdenum cofactor (Moco), which forms the active site of molybdenum (Mo) enzymes in eukaryotes, involves six enzymes. The qRT-PCR result also confirmed upregulation of the CNX5 orthologue (Phatr2_34373; Supplementary Fig. S2). None of the genes encoding plastidial GSII/Fd-GOGAT and mitochondrial GSIII (GLNA), which are required for ammonium assimilation, were regulated (P<0.05). However, increased transcript levels of two different isoforms of NAD(P)H-dependent glutamate synthase (NADPH-GOGAT, GltD and GltX) were observed in N-deprived cells (Fig. 5). Glutamate dehydrogenase (GDH) is another enzyme that catalyses the reversible conversion of 2-oxoglutarate (2-OG) to glutamate. We observed increased expression of an NADP-GDH (Phatr2_13951; Fig. 5 and Supplementary Dataset S1).
Fig. 5.
Cellular pathways and processes related to N metabolism under N deprivation in P. tricornutum. Metabolites detected are indicated by a blue box frame. Red, blue, and black text indicate up-, down-, and no regulation of pathways, genes, or metabolites by N deprivation, respectively. Amino acids are indicated by a yellow background. Red arrows depict gene transcripts found to be upregulated. Fd-GOGAT, ferredoxin-dependent glutamate synthase; GSII, ferredoxin-dependent glutamine synthetase; Fd-NiR, ferredoxin-dependent nitrite reductase; GDH, glutamate dehydrogenase; GSIII, bacterial-origin glutamine synthetase; IDH, isocitrate dehydrogenase; NADPH-GOGAT, NAD(P)H-dependent glutamate synthase; NAD(P)H-NiR, NAD(P)H-dependent nitrite reductase; NR, nitrate reductase.
N scavenging from various organic compounds
We observed induction of genes encoding two glutamyl-tRNA(Gln) amidotransferase-like proteins (Phatr2_50401 and Phatr2_45118) and three acetamidase/formamidases (Phatr2_54476, Phatr2_37952, and Phatr2_37719) at both time points in N-deprived cells (Fig. 4). Phylogenetic analyses indicated that Phatr2_54476 is related to FmdA-type formamidases (Supplementary Fig. S3, available at JXB online); the main substrate of Methylophilus methylotropus FmdA (Wyborn et al., 1994), as well as lupin LaFmd (Rath et al., 2010), is formamide. Phatr2_37952 and Phatr2_37719, which were strongly induced at both time points, encode amidohydrolases belonging to a poorly characterized clade with low similarity to FmdA-type formamidases (Supplementary Fig. S3).
Purine and pyrimidine biosynthesis and degradation
Most of the transcripts involved in biosynthesis of purine and pyrimidine were downregulated (Supplementary Dataset S1). Simultaneously, we observed upregulation of several transcripts involved in their catabolic processes, such as purine and pyrimidine deaminases. Furthermore, uracil-xanthine permease (Phatr2_16991) and adenine/guanine permease (Phatr2_17344) transcripts were upregulated in N-deprived cultures; the response of the latter was confirmed by qRT-PCR analysis (Supplementary Fig. S2). Urease (Phatr2_29702), which catalyses the hydrolysis of urea into CO2 and , was also transcriptionally induced following N deprivation (Supplementary Dataset S1).
Protein biosynthesis, folding, and degradation
N deprivation influenced both biosynthesis and degradation of proteins and amino acids. Most amino acid biosynthesis pathways were transcriptionally repressed; the strongest downregulation was found for transcripts encoding homoserine dehydrogenase (Phatr2_26813) and N-acetylglutamate kinase (Phatr2_3969) (Fig. 4). Reduction of protein biosynthesis could be observed as a decrease in mRNA levels of many genes encoding aminoacyl-tRNA synthetases, as well as ribosomal subunits and translation elongation factors. The transcription levels of genes encoding 18 peptidylprolyl isomerases and two protein disulfide isomerases that catalyse protein folding were also reduced. In contrast, the transcript levels of genes encoding amino acid degradation enzymes, such as those related to catabolism of branched-chain amino acids, were upregulated (Fig. 4). Five identified autophagy-related genes were upregulated at one or both time points in N-deprived cells (Supplementary Dataset S1). The mRNA levels of several genes involved in ubiquitination were upregulated, but most of the proteasome subcomponents were downregulated.
Effect of N deprivation on photosynthesis and pigment biosynthesis
Glutamate, the main precursor of chlorophyll biosynthesis, declined under N-deprived conditions (Supplementary Table S1). In line with the reduced chlorophyll a and fucoxanthin levels in N-deprived cells, the expression levels of most of the genes encoding enzymes involved in the chlorophyll a and carotenoid biosynthetic pathways were also repressed (Figs 4 and 5, and Supplementary Dataset S1). Of 39 differentially regulated genes encoding LHC proteins, only red algal-like LHCR10 and two high-light-induced proteins (HLIP1 and HLIP1b) were significantly upregulated at both time points, whereas the LI818-like LHCX4 and LHCR7 showed moderate upregulation 72h after N deprivation (Fig. 4). Among the downregulated LHCs, repression of LHCF1 was confirmed by qRT-PCR (Supplementary Fig. S2). Similarly, a majority of the transcripts involved in photosynthesis were downregulated (Figs 4 and 5, and Supplementary Dataset S1). Furthermore, a chloroplastic ferredoxin-NADP reductase (Phatr2_23717) was strongly downregulated, indicating that NAD(P)H production through photosynthesis was reduced (Fig. 4).
Effect of N deprivation on carbon and lipid metabolism
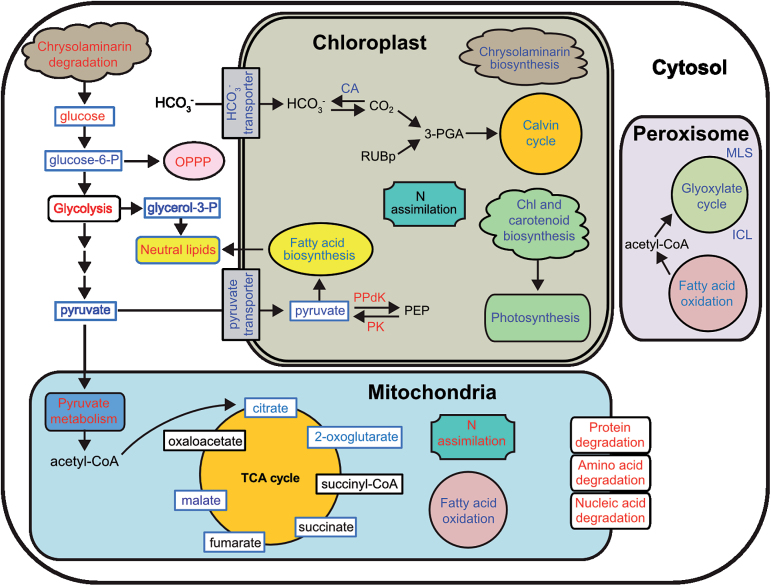
Carbon fixation
Downregulation of several genes connected to the biophysical carbon-concentrating mechanism was observed in N-deprived cells. Of five carbonic anhydrase (CA) genes related to the biophysical carbon-concentrating mechanism that were significantly regulated 72h after N deprivation (Supplementary Dataset S1), transcript levels of CA-III and two β-CAs (PtCa1 and PtCa2) decreased. A chloroplast bicarbonate transporter (SLC4A_1) was also repressed. In accordance with the β-CA and SLC4A_1 downregulation, a majority of the genes encoding enzymes of the Calvin cycle were downregulated (Figs 4 and 6). Upregulation of several transcripts encoding enzymes involved in the mitochondrial decarboxylation under N-deprived conditions was observed in N-deprived cells (Supplementary Dataset S1). Downregulation of plastid-localized pyruvate carboxylase 2 (PYC2) was confirmed by qRT-PCR analysis (Supplementary Fig. S2).
Fig. 6.
Cellular pathways and processes affected under N deprivation in P. tricornutum. Metabolites detected are indicated by a blue box frame. Red, blue,and black text indicates up-, down-, and no regulation of pathways, genes, or metabolites by N deprivation, respectively. 3-PGA, 3-phosphoglycerate; CA, carbonic anhydrase; ICL, isocitrate lyase; MLS, malate synthase; OPPP, oxidative pentose phosphate pathway; PEP, phosphoenolpyruvate; PK, pyruvate kinase; PPdK, pyruvate orthophosphate dikinase; RuBP, ribulose-1,5-bisphosphate.
TCA cycle
Consistent with upregulation of the mitochondrial decarboxylation, TCA cycle transcripts were induced (Figs 5 and 6, and Supplementary Dataset S1). However, genes encoding enzymes towards the end of the TCA cycle were not regulated. Aconitate hydratase (Phatr2_26290) and isocitrate dehydrogenase (Phatr2_14762) transcripts showed the highest level of upregulation. In contrast to the upregulation of TCA transcripts, we observed a decrease in the levels of most of the metabolite intermediates of the TCA cycle (Supplementary Table S1).
Chrysolaminarin biosynthesis and degradation
We observed repression of several genes encoding enzymes potentially involved in gluconeogenesis, as well as chrysolaminarin biosynthesis (Kroth et al., 2008; Chauton et al., 2013), especially 72h after N deprivation (Fig. 6). Inversely, transcript levels of genes encoding enzymes for chrysolaminarin degradation, such as exo-1,3-β-glucosidases, increased. Chrysolaminarin degradation produces glucose; indeed, glucose levels were higher in N-deprived cells (Fig. 6 and Supplementary Table S1). Consistent with the increased glucose level, cytosolic glucokinase was also induced in our experiment.
Oxidative pentose phosphate pathway (OPPP), glycolysis, and pyruvate metabolism
Surprisingly, all OPPP transcripts were induced (Fig. 6). Transcripts of most of the putatively cytosolic glycolytic enzymes, such as phosphoglycerate mutase (PGAM_7), increased in our experiment, while transcript levels of several plastidial enzymes, such as glyceraldehyde-3-phosphate dehydrogenase (GAPC1), showed the opposite regulation (Fig. 4). In contrast to the transcriptional induction of glycolytic enzymes, metabolite levels of glucose-6-phosphate, fructose-6-phosphate, and pyruvate declined (Supplementary Table S1), which might be the result of their quick conversion to other metabolites. Consistent with the decrease in pyruvate, transcript levels of several genes responsible for pyruvate metabolism were upregulated.
Fatty acid biosynthesis and degradation
Most transcripts related to the chloroplast fatty acid biosynthetic pathway were strongly downregulated (Fig. 4 and Supplementary Dataset S1, available at JXB online); the only upregulated transcript was 3-oxoacyl-[acyl-carrier-protein] synthase (FABFb) (Phatr2_18940). We also observed lower levels of total free fatty acids in N-deprived cells (Supplementary Table S1), which might be a consequence of their incorporation into TAG.
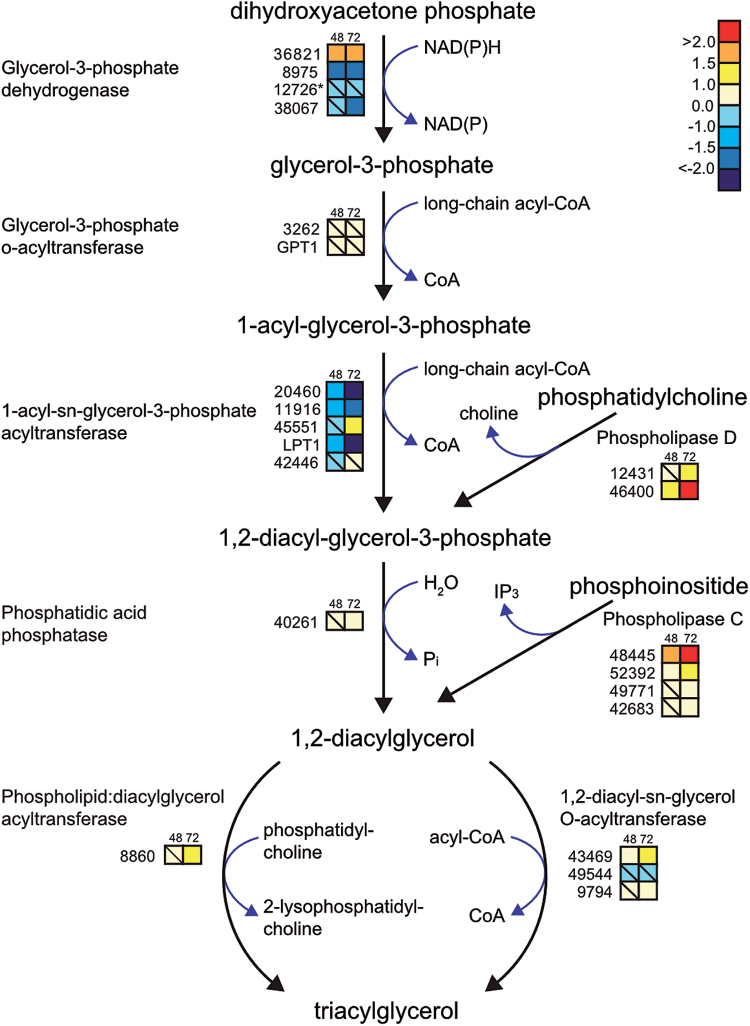
Membrane lipid remodelling and TAG biosynthesis
TAG biosynthetic pathways are illustrated in Fig. 7. Of three differentially regulated glycerol-3-phosphate dehydrogenases, transcript levels of Phatr2_36821 increased in N-deprived cells (Fig. 7). This enzyme consumes NAD(P)H to convert dihydroxyacetone phosphate, an intermediate in glycolysis, to glycerol-3-phosphate. Subsequent transfer of two acyl-CoAs to glycerol-3-phosphate by glycerol-3-phosphate acyltransferase and acyl-glycerol-3-phosphate acyltransferase (AGPAT) result in the formation of phosphatidic acid. Of the five detected isoforms of AGPAT, only one putative isoform (Phatr2_45551) displayed increased transcript levels 72h after deprivation, whereas three others (Phatr2_20460, Phatr2_11916, and LPT1) were suppressed. Phosphatidic acid is dephosphorylated to diacylglycerol, the main precursor of TAG. This process is catalysed by phosphatidic acid phosphatase; a putative PAP (Phatr2_40261) was weakly upregulated. Incorporation of the third fatty acyl-CoA into glycerol-3-phosphate backbone by diacylglycerol O-acyltransferase (DGAT) completes TAG formation. The mRNA levels of two isoforms of DGAT (Phatr2_43469 and Phatr2_9794) were induced in our experiment. Although we could detect TAG accumulation at both the molecular and physiological level, the transcript abundance of several TAG lipases was induced in N-deprived cells.
Fig. 7.
Transcriptional changes in genes related to TAG biosynthesis in response to N deprivation. Coloured squares indicate the regulation pattern of genes encoding putative enzymes functioning in the TAG biosynthetic pathway after 48 and 72h of N deprivation, compared with N-replete cultures. Squares with a diagonal line inside indicate non-significant regulation (P>0.05). The scale on the right represents gene expression ratio values, which were log2 transformed. Numbers indicate Phatr2 gene IDs. Gene ID 12726 (marked with an asterisk) belongs to the Phatr1 database (http://genome.jgi-psf.org/Phatr1/Phatr1.home.html).
Upregulation of four isoforms of phospholipase C and two isoforms of phospholipase D indicated that membrane phospholipids are degraded under N deprivation to provide the TAG precursors phosphatidic acid and diacylglycerol (Fig. 7). The highest induction was seen for phospholipase C (Phatr2_48445), with an approximately 4- and 10-fold increase 48 and 72h after deprivation, respectively. Transcript levels of a putative phospholipid:diacylglycerol acyltransferase enzyme (PDAT, Phatr2_8860) were also upregulated at 72h in N-deprived cells (Fig. 7).
Discussion
Previous studies in diatoms and other microalgae have demonstrated that these organisms undergo dramatic metabolic changes in response to N starvation (Hockin et al., 2012; Valenzuela et al., 2012; Yang et al., 2013). Performing an integrated analysis of the response to N deprivation in P. tricornutum, we confirmed these modifications at the physiological, metabolite, and transcriptome levels.
Reprogramming of N metabolism
We observed a higher C:N ratio than that suggested by the Redfield ratio (Redfield, 1934) in N-replete and N-deprived cultures, indicating that the C:N:P composition of phytoplankton and marine particulate matter is flexible, especially in nutrient-deprived cells (Geider and La Roche, 2002). Furthermore, an increase in the C:N ratio of N-deprived cultures is probably a result of biomass increase after N exhaustion.
Although transcriptional regulation is indicative and not necessarily directly linked to changes at protein level, some trends are evident. Due to a decrease in the N content of N-deprived cells, many processes connected to N metabolism were affected. A major response to N deprivation was to increase the cellular capacity for N uptake and nitrate reduction to ammonium, as observed at the transcript level. This phenomenon was reported previously in N-deprived P. tricornutum and other microalgae at the transcriptome level (Mock et al., 2008; Miller et al., 2010; Valenzuela et al., 2012) (Fig. 7). However, the downregulation of one ammonium transporter (Phatr2_54981) was in agreement with previous reports (Haimovich-Dayan et al., 2013; Krell et al., 2007; Yang et al., 2013). The induction of two molybdopterin biosynthesis genes could be related to an increased need of Moco for NR and xanthine dehydrogenase in P. tricornutum. In contrast to our results, a NR gene was repressed under N stress in Emiliania huxleyi; the authors postulated that expression of the NR gene is stimulated under high concentrations (Bruhn et al., 2010). In addition, they observed no co-ordination between the regulation of nitrate reduction at the transcript and protein levels. Despite the increase in the expression level of genes encoding nitrate-reducing enzymes in N-deficient T. pseudonana (Mock et al., 2008), their protein levels were found to decrease in a proteomic study (Hockin et al., 2012); the authors postulated that the levels of these enzymes might be controlled by post-transcriptional modifications. In photosynthetic eukaryotes, ammonium assimilation primarily occurs inside the chloroplast (Bowler et al., 2010). Although we could not detect any regulation of GSII/Fd-GOGAT or mitochondrial GLNA, increased transcript levels of two different NADPH-GOGAT isoforms (GltD and GltX) indicated that cells increased their N scavenging mechanisms to assimilate more ammonium from other pathways such as protein and amino acid degradation (Fig. 5). In addition, the upregulated levels in N-deprived cells of another ammonium assimilating enzyme, NADP-GDH, might also contribute to ammonium assimilation. GDH is generally believed to act as a catabolic enzyme, catalysing the oxidative deamination of glutamate to 2-OG, and previous studies have implied that GDH is a minor contributor to assimilation (Zehr and Falkowski, 1988; Guerra et al., 2013). However, the observed decrease in 2-OG levels might indicate that GDH also can perform the anabolic reaction to scavenge under N deprivation. Previous studies have demonstrated that diatoms are able to use other sources of N, such as amides, amines, urea, and amino acids (Shah and Syrett, 1982; Baker et al., 2009). Besides the increase in N-scavenging mechanisms, upregulation of amidases and acetamidase/formamidases indicates that P. tricornutum can degrade organic N sources such as amides and formamide from intracellular or possible extracellular sources to produce ammonium when faced with N deprivation, as observed in N-deficient Aureococcus anophagefferens (Wurch et al., 2011). In support of our results, a formamidase transcript was induced under N stress in E. huxleyi (Bruhn et al., 2010).
Biosynthesis of several amino acids relies on the availability of glutamate, which declined after N deprivation as a consequence of a reduced N pool (Supplementary Table S1). The lower glutamate level compared with the control resulted in a strong drop in levels of other amino acids and repression of amino acid biosynthetic pathways. A decline in the cellular amino acid pool as a result of N deprivation is consistent with results from other diatoms (Granum et al., 2002). Simultaneous with the suppression of amino acid biosynthesis, increased degradation of amino acids through various catabolic pathways was observed at the transcript level, which produced several carbon-containing intermediates that can enter the TCA cycle. Consistent with increased transcripts associated with catabolism of branched-chain amino acids in our experiment, Ge et al. (2014) suggested that branched-chain amino acid degradation directs carbon and energy towards TAG accumulation in N-deprived P. tricornutum.
All purine and pyrimidine nitrogen compounds originate from amino acids (glutamine, aspartate, and glycine) (Zrenner et al., 2006). Reduced biosynthesis of purine and pyrimidines is probably a result of decreases in the amino acid pools (Supplementary Table S1) and cell growth (Fig. 1A). At the same time, recycling of purines and pyrimidines provides the cells with an important N source during N deprivation. Upregulation of the urease gene could also be related to purine degradation. produced from the hydrolysis of urea is hypothesized to be redirected to mitochondria for amino acid biosynthesis via mitochondrial GS/GOGAT (Allen et al., 2011). The strong induction of purine/pyrimidine permeases might explain the ability of diatoms to import purine and pyrimidine from the environment under limited N conditions, as reported by Allison and Syrett (1987). However, Berg et al. (2008) showed that the purine permease AaURA is expressed during growth of A. anophagefferens on a number of N sources, indicating its role as an important nitrogen source for proliferation of this organism. The high expression of AaURA might be related to the growth habitat of A. anophagefferens in shallow coastal waters, which are in close contact with sediments rich in dissolved organic N.
Eukaryotes utilize autophagy and the ubiquitin–proteasome system for protein degradation (Onodera and Ohsumi, 2004). The ubiquitin–proteasome system is used for rapid degradation of proteins and acts mainly to degrade short-lived proteins such as transcription factors, while the turnover of long-lived proteins, which constitute 99% of cellular proteins, is processed by autophagy (Onodera and Ohsumi, 2004). While N-limited A. anophagefferens showed moderate downregulation of two autophagy-related genes (Berg et al., 2008), the transcript levels of several autophagy-related genes in our experiment and during N limitation of the green microalga Neochloris oleoabundans (Rismani-Yazdi et al., 2012) were induced. Although autophagy generally is not a selective protein degradation process, selective autophagy was stimulated under conditions of nutritional stress, especially N deficiency, in yeast and plants (Onodera and Ohsumi, 2004; Yoshimoto et al., 2004). Therefore, the induction of autophagy-related genes can be explained as a response to N deprivation by selectively degrading excessive proteins into amino acids that are recycled to protein biosynthetic pathways in order to maintain cellular homeostasis. Autophagy is highly regulated at the protein level (Klionsky and Emr, 2000); therefore, it would be worthwhile looking at regulation of autophagy components at the protein level to better understand their role under N deprivation.
Chlorosis is one of the main responses of diatoms to N deprivation. The reduced chlorophyll a level in N-deprived cells (Table 2) is probably caused at least partly by repression of its biosynthesis. In addition, the co-ordinated decrease in chlorophyll a and fucoxanthin content under N deprivation suggests that the biosynthesis of these two pigments is co-ordinated under N deprivation; a similar result was also observed in a previous study in P. tricornutum (Geider et al., 1993). In contrast, N limitation in Chaetoceros gracilis led to changes in the chlorophyll a:fucoxanthin ratio (Cleveland and Perry, 1987). The decrease in pigment biosynthesis and LHC proteins corresponds to a reduced photosynthetic apparatus of N-deprived cells and a lower requirement for pigments. These results clearly indicate that N deprivation reduces the photosynthetic efficiency, in agreement with the observed reduction in maximum quantum yield of PSII (Fv/Fm) (Fig. 1B). Despite the downregulation of LHCF proteins, induction of LHCXs was reported under several stresses in diatoms (Zhu and Green, 2008). Induction of LHCR10 expression by N deprivation was also observed by Yang et al. (2013); furthermore, both LHCX and LHCR-II genes, to which LHCR7 and LHCR10 belong, are induced by high light (Nymark et al., 2009, 2013). The increased transcript levels of LHCX4, along with two LHCRs and two HLIPs, may be related to a photoprotective role during acclimation to low N levels. In summary, N-deprived P. tricornutum modified N metabolism in order to reduce synthesis of nitrogenous compounds and catabolize excessive N-containing compounds in favour of essential N compounds.
Remodelling of carbon metabolism
The major pathway used by diatoms for carbon fixation is the Calvin cycle. Since β-CAs and transporters are required to concentrate inorganic carbon in the vicinity of Rubisco, their downregulation repressed a majority of the genes encoding enzymes of the Calvin cycle (Fig. 6). Although the Calvin cycle was downregulated, cells might employ other mechanisms such as pyruvate orthophosphate dikinase to dissipate excess energy around the photosystems to reduce the production of reactive oxygen species under N deficiency, as reported by Haimovich-Dayan et al. (2013).
Upregulation of several transcripts encoding enzymes involved in the mitochondrial decarboxylation under N-deprived conditions leads to production of oxaloacetate and pyruvate (Supplementary Dataset S1). Oxaloacetate can replenish C4 acids of the TCA cycle, whereas pyruvate can enter the TCA cycle or fatty acid biosynthesis. The TCA cycle could also be upregulated in response to high levels of protein and amino acid degradation, which generates TCA cycle intermediates and provides precursors for resynthesis of certain amino acids, as observed by Hockin et al. (2012). Furthermore, strong upregulation of the genes encoding aconitate hydratase and isocitrate dehydrogenase leads to production of 2-OG, which acts as a precursor in ammonium assimilation. Malate from the TCA cycle could also be directed to the fatty acid biosynthetic pathway through NADP-dependent malic enzyme (Supplementary Dataset S1). Similar regulation of the TCA cycle was reported for other diatoms under N deprivation (Bender et al., 2014). Thus, a co-ordinated upregulation of the TCA cycle and mitochondrial decarboxylation might shift the flow of carbon skeletons towards fatty acid biosynthesis.
Degradation of chrysolaminarin releases glucose. Glucose cannot enter the metabolic pathway directly and must be converted to glucose-6-phosphate by cytosolic glucokinase. Phosphorylated glucose could further enter the glycolytic pathway and/or OPPP (Fig. 6). Upregulation of OPPP produces NAD(P)H supplying NAD(P)H-dependent pathways like lipid synthesis and nitrogen assimilation. Utilization of glucose-6-phosphate through glycolysis produces energy in the form of ATP and NAD(P)H, as well as the glycolysis end product, pyruvate. Further metabolism of pyruvate produces acetyl-CoA, which can enter the TCA cycle, or de novo fatty acid biosynthesis in the chloroplast. Thus, increased degradation of chrysolaminarin, along with induction of OPPP and glycolysis, could provide N-deprived cells with reducing equivalents to balance reduced NAD(P)H production from photosynthesis, as well as carbon fluxes for the TCA cycle and fatty acid biosynthesis.
The microarray data showed downregulation of de novo fatty acid biosynthesis genes (Fig. 4 and Supplementary Data S1), supporting the observed decrease in free fatty acids (Supplementary Table S1). In contrast, Yang et al. (2013) reported increased total fatty acid levels in N-deprived cells, while RNA-sequencing data from their experiment showed that most of the transcripts involved in chloroplast de novo synthesis of fatty acids were downregulated. We conclude that, under exponential growth, the high cell division rate leads to a high demand for membrane lipids in newly synthesized cells. In contrast, cell division slows or halts in cells faced with N limitation, and there is less need for membrane lipids and fatty acids. Even though de novo synthesis of fatty acids is downregulated, fatty acids are still produced and accumulate in the form of neutral lipids. Moreover, repression of the fatty acid β-oxidation pathway might direct fatty acids from degradation of membrane lipids to TAG production.
Under favourable conditions, fatty acids are incorporated into membrane lipids. The main classes of lipids in diatom chloroplasts are sulphoquinovosyldiacylglycerol, monogalactosyldiacylglycerol, digalactosyldiacylglycerol, phosphatidylglycerol, and phosphatidylcholine; however, phosphoglyceride levels are higher in the endoplasmic and plasma membranes (Hu et al., 2008; Goss and Wilhelm, 2010). Therefore, the downregulation in membrane lipid biosynthesis could be related to a reduced demand for membrane lipids due to the reduced cell growth rate. A major effect of N starvation in many microalgae is the switch in lipid production towards neutral lipid accumulation, mainly TAGs, which act primarily as energy reserves inside the cells (Hu et al., 2008). Increased levels of neutral lipids were also observed in our study (Fig. 2). TAG can be synthesized through different pathways, one of which uses glycerol-3-phosphate as a precursor (Fig. 7) (Hu et al., 2008). The accumulation of TAG was consistent with a decrease in the amount of free glycerol-3-phosphate inside the cells (Supplementary Table S1), implying that this compound might be used as a precursor for TAG biosynthesis. Furthermore, TAG can also accumulate via degradation of glycerophospholipids. The increased expression of phospholipase C and phospholipase D suggests that these enzymes contribute substantially to TAG accumulation through processing of phospholipids. An acyl-CoA-independent mechanism for biosynthesis of TAG from diacylglycerol using phospholipids as acyl donors is catalysed by a putative gene encoding phospholipid:diacylglycerol acyltransferase (PDAT, Phatr2_8860). This could also contribute to TAG accumulation upon N deprivation. The upregulation of several TAG lipases could be related to saturated lipid bodies and could act to recycle previously synthesized TAGs with new TAGs, or they might have been induced to make modifications in TAG structure. Overall, our results suggest that both de novo TAG biosynthesis and remodelling of membrane lipids play important roles in TAG accumulation under N deprivation.
Conclusions
A combined analysis of transcriptional and non-targeted metabolite profiling, along with physiological and biochemical experiments, revealed transcriptional, metabolic, and physiological acclimation in the diatom P. tricornutum under conditions of N deprivation. The global expression data suggested that P. tricornutum is able to remobilize N through catabolism of internal N-containing resources such as amino acids and proteins. N deprivation was also accompanied by a reduction of pigment pools and photosynthetic capacity. We also showed large changes in genes related to carbon and lipid metabolism. Decreased levels of carbon skeletons due to suppression of the Calvin cycle were compensated by breakdown of chrysolaminarin, leading to upregulation of OPPP, cytosolic glycolysis, pyruvate metabolism, and the TCA cycle. These pathways provide precursors for fatty acid biosynthesis. In addition, remodelling of membrane lipids and upregulation of the de novo TAG biosynthetic pathway was further supported by increased levels of neutral lipids, indicating TAG accumulation under N deprivation. Our study provides a detailed picture of P. tricornutum acclimation to N deprivation, and can be used as a guide for future metabolic manipulations to increase TAG production.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. GO analysis of significantly regulated genes after 48h of nitrate deprivation.
Supplementary Fig. S2. qRT-PCR analysis of selected genes.
Supplementary Fig. S3. Phylogenetic analysis of FmdA-type amidases/formamidases in P. tricornutum.
Supplementary Table S1. Tentatively identified algal metabolites based on GC-MS profiling.
Supplementary Table S2. Genes analysed by qRT-PCR and their respective primers.
Supplementary Dataset S1. Representative mRNA transcripts grouped by cellular pathway.
Acknowledgements
We thank Torfinn Sparstad for excellent technical assistance with transcriptome analyses, Matilde S. Chauton for access to instrumentation and technical advice in particulate nutrient analysis, Kjersti Andresen for help in pigment and nutrient experiments, and Bjørnar Sporsheim for kind guidance in lipid analysis. This work was supported by the Research Council of Norway through grants 184146 and 207794.
Glossary
Abbreviations:
- CA
carbonic anhydrase
- CCM
carbon-concentrating mechanism
- GC-MS
gas chromatography2-OG, 2-oxoglutarate; /mass spectrometry
- GDH
glutamate dehydrogenase
- GO
Gene Ontology
- HPLC
high-performance liquid chromatography
- LHC
light-harvesting complex
- NR
nitrate reductase
- N
nitrogen
- OPPP
oxidative pentose phosphate pathway
- PSII
photosystem II
- qRT-PCR
quantitative real-time PCR
- TAG
triacylglycerol
- TCA
tricarboxylic acid.
References
- Abràmoff MD, Magalhães PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics International 11, 36–43. [Google Scholar]
- Allen AE. 2005. Beyond sequence homology: redundant ammonium transporters in a marine diatom are not functionally equivalent. Journal of Phycology 41, 4–6. [Google Scholar]
- Allen AE, Dupont CL, Obornik M, et al. 2011. Evolution and metabolic significance of the urea cycle in photosynthetic diatoms. Nature 473, 203–207. [DOI] [PubMed] [Google Scholar]
- Allen AE, Ward BB, Song B. 2005. Characterization of diatom (Bacillariophyceae) nitrate reductase genes and their determination in marine phytoplankton communities. Journal of Phycology 41, 95–104. [Google Scholar]
- Allison G, Syrett PJ. 1987. The metabolism of guanine by the diatom Phaeodactylum tricornutum Bohlin. Journal of Phycology 23, 666–668. [Google Scholar]
- Andersen RA, Morton SL, Sexton JP. 1997. Provasoli-Guillard National Center for culture of marine phytoplankton 1997 list of strains. Journal of Phycology 33, 1–75. [Google Scholar]
- Armbrust EV. 2009. The life of diatoms in the world’s oceans. Nature 459, 185–192. [DOI] [PubMed] [Google Scholar]
- Armbrust EV, Berges JA, Bowler C, et al. 2004. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306, 79–86. [DOI] [PubMed] [Google Scholar]
- Baker KM, Gobler CJ, Collier JL. 2009. Urease gene sequences from algae and heterotrophic bacteria in axenic and nonaxenic phytoplankton cultures. Journal of Phycology 45, 625–634. [DOI] [PubMed] [Google Scholar]
- Bender SJ, Durkin CA, Berthiaume CT, Morales RL, Armbrust E. 2014. Transcriptional responses of three model diatoms to nitrate limitation of growth. Aquatic Microbiology 1, 3. [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B 57, 289–300. [Google Scholar]
- Berg GM, Shrager J, Glöckner G, Arrigo KR, Grossman AR. 2008. Understanding nitrogen limitation in Aureococcus anophagefferens (Pelagophyceae) through cDNA and qRT-PCR analysis. Journal of Phycology 44, 1235–1249. [DOI] [PubMed] [Google Scholar]
- Bertozzini E, Galluzzi L, Ricci F, Penna A, Magnani M. 2013. Neutral lipid content and biomass production in Skeletonema marinoi (Bacillariophyceae) culture in response to nitrate limitation. Applied Biochemistry and Biotechnology 170, 1624–1636. [DOI] [PubMed] [Google Scholar]
- Bowler C, Allen AE, Badger JH, et al. 2008. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239–244. [DOI] [PubMed] [Google Scholar]
- Bowler C, Vardi A, Allen AE. 2010. Oceanographic and biogeochemical insights from diatom genomes. Annual Review of Marine Science 2, 333–365. [DOI] [PubMed] [Google Scholar]
- Brandes JA, Devol AH, Deutsch C. 2007. New developments in the marine nitrogen cycle. Chemical Reviews 107, 577–589. [DOI] [PubMed] [Google Scholar]
- Bruhn A, LaRoche J, Richardson K. 2010. Emiliana huxleyi (Prymnesiophyceae): nitrogen-metabolism genes and their expression in response to external nitrogen sources. Journal of Phycology 46, 266–277. [Google Scholar]
- Capone DG, Hutchins DA. 2013. Microbial biogeochemistry of coastal upwelling regimes in a changing ocean. Nature Geoscience 6, 711–717. [Google Scholar]
- Chauton MS, Winge P, Brembu T, Vadstein O, Bones AM. 2013. Gene regulation of carbon fixation, storage, and utilization in the diatom Phaeodactylum tricornutum acclimated to light/dark cycles. Plant Physiology 161, 1034–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland J, Perry M. 1987. Quantum yield, relative specific absorption and fluorescence in nitrogen-limited Chaetoceros gracilis . Marine Biology 94, 489–497. [Google Scholar]
- Dham VV, Wafar MVM, Heredia AM. 2005. Nitrogen uptake by size-fractionated phytoplankton in mangrove waters. Aquatic Microbial Ecology 41, 281–291. [Google Scholar]
- Egeland G, Garrido J, Clementson L, et al. 2011. Part VI: Data sheets aiding identification of phytoplankton carotenoids and chlorophylls. In: Roy S, Llewellyn CA, Egeland ES, Johnson G, eds. Phytoplankton pigments: characterization, chemotaxonomy and applications in oceanography . Cambridge, UK: Cambridge University Press, 665–822. [Google Scholar]
- Eizadora TY, Zendejas FJ, Lane PD, Gaucher S, Simmons BA, Lane TW. 2009. Triacylglycerol accumulation and profiling in the model diatoms Thalassiosira pseudonana and Phaeodactylum tricornutum (Baccilariophyceae) during starvation. Journal of Applied Phycology 21, 669–681. [Google Scholar]
- Fabris M, Matthijs M, Rombauts S, Vyverman W, Goossens A, Baart GJ. 2012. The metabolic blueprint of Phaeodactylum tricornutum reveals a eukaryotic Entner–Doudoroff glycolytic pathway. The Plant Journal 70, 1004–1014. [DOI] [PubMed] [Google Scholar]
- Falkowski PG, Raven JA. 2007. Aquatic photosynthesis in biogeochemical cycles. In: Falkowski PG, Raven JA. (eds) Aquatic photosynthesis . Princeton, NJ: Princeton University Press, 364–409. [Google Scholar]
- Foyer CH, Parry M, Noctor G. 2003. Markers and signals associated with nitrogen assimilation in higher plants. Journal of Experimental Botany 54, 585–593. [DOI] [PubMed] [Google Scholar]
- Gavet O, Pines J. 2010. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Developmental Cell 18, 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge F, Huang W, Chen Z, Zhang C, Xiong Q, Bowler C, Yang J, Xu J, Hu H. 2014. Methylcrotonyl-CoA carboxylase regulates triacylglycerol accumulation in the model diatom Phaeodactylum tricornutum . The Plant Cell 26, 1681–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geider R, La Roche J. 2002. Redfield revisited: variability of C:N:P in marine microalgae and its biochemical basis. European Journal of Phycology 37, 1–17. [Google Scholar]
- Geider RJ, La Roche J, Greene RM, Olaizola M. 1993. Response of the photosynthetic apparatus of Phaeodactylum tricornutum (Bacillariophyceae) to nitrate, phosphate, or iron starvation. Journal of Phycology 29, 755–766. [Google Scholar]
- Goss R, Wilhelm C. 2010. Lipids in algae, lichens and mosses. In: Wada H, Murata N, eds. Lipids in photosynthesis , vol. 30 Dordrecht, the Netherlands: Springer, 117–137. [Google Scholar]
- Govender T, Ramanna L, Rawat I, Bux F. 2012. BODIPY staining, an alternative to the Nile Red fluorescence method for the evaluation of intracellular lipids in microalgae. Bioresource Technology 114, 507–511. [DOI] [PubMed] [Google Scholar]
- Granum E, Kirkvold S, Myklestad SM. 2002. Cellular and extracellular production of carbohydrates and amino acids by the marine diatom Skeletonema costatum: diel variations and effects of N depletion. Marine Ecology Progress Series 242, 83–94. [Google Scholar]
- Granum E, Myklestad S. 2002. A simple combined method for determination of β-1,3-glucan and cell wall polysaccharides in diatoms. Hydrobiologia 477, 155–161. [Google Scholar]
- Granum E, Roberts K, Raven JA, Leegood RC. 2009. Primary carbon and nitrogen metabolic gene expression in the diatom Thalassiosira pseudonana (Bacillariophyceae): diel periodicity and effects of inorganic carbon and nitrogen. Journal of Phycology 45, 1083–1092. [DOI] [PubMed] [Google Scholar]
- Guerra LT, Levitan O, Frada MJ, Sun JS, Falkowski PG, Dismukes GC. 2013. Regulatory branch points affecting protein and lipid biosynthesis in the diatom Phaeodactylum tricornutum . Biomass and Bioenergy 59, 306–315. [Google Scholar]
- Guillard RL. 1975. Culture of phytoplankton for feeding marine invertebrates. In: Smith W, Chanley M, eds. Culture of marine invertebrate animals . New York, NY: Springer, 29–60. [Google Scholar]
- Haimovich-Dayan M, Garfinkel N, Ewe D, Marcus Y, Gruber A, Wagner H, Kroth PG, Kaplan A. 2013. The role of C4 metabolism in the marine diatom Phaeodactylum tricornutum . New Phytologist 197, 177–185. [DOI] [PubMed] [Google Scholar]
- Hansen HP, Koroleff F. 1999. Determination of nutrients. In: Grasshoff K, Kremling K, Ehrhardt M, eds. Methods of seawater analysis , 3rd edn Weinheim, Germany: Wiley-VCH, 159–228. [Google Scholar]
- Hildebrand M. 2005. Cloning and functional characterization of ammonium transporters from the marine diatom Cylindrotheca fusiformis (Bacillariophyceae). Journal of Phycology 41, 105–113. [Google Scholar]
- Hockin NL, Mock T, Mulholland F, Kopriva S, Malin G. 2012. The response of diatom central carbon metabolism to nitrogen starvation is different from that of green algae and higher plants. Plant Physiology 158, 299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Sommerfeld M, Jarvis E, Ghirardi M, Posewitz M, Seibert M, Darzins A. 2008. Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. The Plant Journal 54, 621–639. [DOI] [PubMed] [Google Scholar]
- Hummel J, Strehmel N, Selbig J, Walther D, Kopka J. 2010. Decision tree supported substructure prediction of metabolites from GC-MS profiles. Metabolomics 6, 322–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissen R, Winge P, Tran DH, Jørstad TS, Størseth TR, Christensen T, Bones AM. 2010. Transcriptional profiling of an Fd-GOGAT1/GLU1 mutant in Arabidopsis thaliana reveals a multiple stress response and extensive reprogramming of the transcriptome. BMC Genomics 11, 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. 2000. Autophagy as a regulated pathway of cellular degradation. Science 290, 1717–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolber Z, Zehr J, Falkowski P. 1988. Effects of growth irradiance and nitrogen limitation on photosynthetic energy conversion in photosystem II. Plant Physiology 88, 923–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krell A, Funck D, Plettner I, John U, Dieckmann G. 2007. Regulation of proline metabolism under salt stress in the psychrophilic diatom Fragilariopsis cylindrus (Bacillariophyceae). Journal of Phycology 43, 753–762. [Google Scholar]
- Kroth PG, Chiovitti A, Gruber A, et al. 2008. A model for carbohydrate metabolism in the diatom Phaeodactylum tricornutum deduced from comparative whole genome analysis. PLoS One 3, e1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudela RM, Dugdale RC. 2000. Nutrient regulation of phytoplankton productivity in Monterey Bay, California. Deep Sea Research Part II: Topical Studies in Oceanography 47, 1023–1053. [Google Scholar]
- Miller R, Wu G, Deshpande RR, et al. 2010. Changes in transcript abundance in Chlamydomonas reinhardtii following nitrogen deprivation predict diversion of metabolism. Plant Physiology 154, 1737–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mock T, Kroon BMA. 2002. Photosynthetic energy conversion under extreme conditions—I: important role of lipids as structural modulators and energy sink under N-limited growth in Antarctic sea ice diatoms. Phytochemistry 61, 41–51. [DOI] [PubMed] [Google Scholar]
- Mock T, Samanta MP, Iverson V, et al. 2008. Whole-genome expression profiling of the marine diatom Thalassiosira pseudonana identifies genes involved in silicon bioprocesses. Proceedings of the National Academy of Sciences, USA 105, 1579–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustafa A, Beszteri B, Maier UG, Bowler C, Valentin K, Bhattacharya D. 2009. Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science 324, 1724–1726. [DOI] [PubMed] [Google Scholar]
- Mus F, Toussaint J-P, Cooksey K, Fields M, Gerlach R, Peyton B, Carlson R. 2013. Physiological and molecular analysis of carbon source supplementation and pH stress-induced lipid accumulation in the marine diatom Phaeodactylum tricornutum . Applied Microbiology and Biotechnology 97, 3625–3642. [DOI] [PubMed] [Google Scholar]
- Nappo M, Berkov S, Codina C, Avila C, Messina P, Zupo V, Bastida J. 2009. Metabolite profiling of the benthic diatom Cocconeis scutellum by GC-MS. Journal of Applied Phycology 21, 295–306. [Google Scholar]
- Norici A, Bazzoni AM, Pugnetti A, Raven JA, Giordano M. 2011. Impact of irradiance on the C allocation in the coastal marine diatom Skeletonema marinoi Sarno and Zingone. Plant, Cell & Environment 34, 1666–1677. [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Fernie AR, Stitt M. 2010. Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Molecular Plant 3, 973–996. [DOI] [PubMed] [Google Scholar]
- Nymark M, Valle KC, Brembu T, Hancke K, Winge P, Andresen K, Johnsen G, Bones AM. 2009. An integrated analysis of molecular acclimation to high light in the marine diatom Phaeodactylum tricornutum . PLoS One 4, e7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nymark M, Valle KC, Hancke K, Winge P, Andresen K, Johnsen G, Bones AM, Brembu T. 2013. Molecular and photosynthetic responses to prolonged darkness and subsequent acclimation to re-illumination in the diatom Phaeodactylum tricornutum . PLoS One 8, e58722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson RJ, Vaulot D, Chisholm SW. 1986. Effects of environmental stresses on the cell cycle of two marine phytoplankton species. Plant Physiology 80, 918–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera J, Ohsumi Y. 2004. Ald6p is a preferred target for autophagy in yeast, Saccharomyces cerevisiae . Journal of Biological Chemistry 279, 16071–16076. [DOI] [PubMed] [Google Scholar]
- Orellana MV, Perry MJ. 1995. Optimization of an immunofluorescent assay of the internal enzyme ribulose-1,5-bisphosphate carboxylase (RUBISCO) in single phytoplankton cells. Journal of Phycology 31, 785–794. [Google Scholar]
- Parker MS, Armbrust EV. 2005. Synergistic effects of light, temperature, and nitrogen source on transcription of genes for carbon, and nitrogen metabolism in the centric diatom Thalassiosira pseudonana (Bacillariophyceae). Journal of Phycology 41, 1142–1153. [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. 2002. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Research 30, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potapova TA, Sivakumar S, Flynn JN, Li R, Gorbsky GJ. 2011. Mitotic progression becomes irreversible in prometaphase and collapses when Wee1 and Cdc25 are inhibited. Molecular Biology of the Cell 22, 1191–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prihoda J, Tanaka A, de Paula WBM, Allen JF, Tirichine L, Bowler C. 2012. Chloroplast-mitochondria cross-talk in diatoms. Journal of Experimental Botany 63, 1543–1557. [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RHL, Moorman AF. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neuroscience Letters 339, 62–66. [DOI] [PubMed] [Google Scholar]
- Rath M, Salas J, Parhy B, et al. 2010. Identification of genes induced in proteoid roots of white lupin under nitrogen and phosphorus deprivation, with functional characterization of a formamidase. Plant and Soil 334, 137–150. [Google Scholar]
- Redfield AC. 1934. On the proportions of organic derivations in sea water and their relation to the composition of plankton. In: Daniel RJ, ed. James Johnstone Memorial Volume . Liverpool: University Press of Liverpool, 177–192. [Google Scholar]
- Rees TAV, Syrett PJ. 1979. The uptake of urea by the diatom, Phaeodactylum . New Phytologist 82, 169–178. [Google Scholar]
- Rismani-Yazdi H, Haznedaroglu BZ, Hsin C, Peccia J. 2012. Transcriptomic analysis of the oleaginous microalga Neochloris oleoabundans reveals metabolic insights into triacylglyceride accumulation. Biotechnology for Biofuels 5, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez F, Chauton M, Johnsen G, Andresen K, Olsen LM, Zapata M. 2006. Photoacclimation in phytoplankton: implications for biomass estimates, pigment functionality and chemotaxonomy. Marine Biology 148, 963–971. [Google Scholar]
- Ruijter J, Ramakers C, Hoogaars W, Karlen Y, Bakker O, Van den Hoff M, Moorman A. 2009. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Research 37, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz G, Mendel RR. 2006. Molybdenum cofactor biosynthesis and molybdenum enzymes. Annual Review of Plant Biology 57, 623–647. [DOI] [PubMed] [Google Scholar]
- Shah N, Syrett PJ. 1982. Uptake of guanine by the diatom, Phaeodactylum tricornutum . Journal of Phycology 18, 579–587. [Google Scholar]
- Sharma KK, Schuhmann H, Schenk PM. 2012. High lipid induction in microalgae for biodiesel production. Energies 5, 1532–1553. [Google Scholar]
- Smyth GK. 2005. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, eds. Bioinformatics and computational biology solutions using R and Bioconductor . New York, NY: Springer, 397–420. [Google Scholar]
- Solomon CM, Collier JL, Berg GM, Glibert PM. 2010. Role of urea in microbial metabolism in aquatic systems: a biochemical and molecular review. Aquatic Microbial Ecology 59, 67–88. [Google Scholar]
- Syrett PJ, Flynn KJ, Molloy CJ, Dixon GK, Peplinska AM, Cresswell RC. 1986. Effects of nitrogen deprivation on rates of uptake of nitrogenous compounds by the diatom, Phaeodactylum tricornutum Bohlin. New Phytologist 102, 39–44. [DOI] [PubMed] [Google Scholar]
- Takabayashi M, Wilkerson FP, Robertson D. 2005. Response of glutamine synthetase gene transcription and enzyme activity to external nitrogen sources in the diatom Skeletonema costatum (Bacillariophyceae). Journal of Phycology 41, 84–94. [Google Scholar]
- Uitz J, Claustre H, Gentili B, Stramski D. 2010. Phytoplankton class-specific primary production in the world’s oceans: seasonal and interannual variability from satellite observations. Global Biogeochemical Cycles 24, GB3016. [Google Scholar]
- Valenzuela J, Mazurie A, Carlson R, Gerlach R, Cooksey K, Peyton B, Fields M. 2012. Potential role of multiple carbon fixation pathways during lipid accumulation in Phaeodactylum tricornutum . Biotechnology for Biofuels 5, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijffels RH, Barbosa MJ. 2010. An outlook on microalgal biofuels. Science 329, 796–799. [DOI] [PubMed] [Google Scholar]
- Wurch LL, Haley ST, Orchard ED, Gobler CJ, Dyhrman ST. 2011. Nutrient-regulated transcriptional responses in the brown tide-forming alga Aureococcus anophagefferens . Environmental Microbiology 13, 468–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyborn NR, Scherr DJ, Jones CW. 1994. Purification, properties and heterologous expression of formamidase from Methylophilus methylotrophus . Microbiology 140, 191–195. [Google Scholar]
- Yang Z-K, Niu Y-F, Ma Y-H, Xue J, Zhang M-H, Yang W-D, Liu J-S, Lu S-H, Guan Y, Li H-Y. 2013. Molecular and cellular mechanisms of neutral lipid accumulation in diatom following nitrogen deprivation. Biotechnology for Biofuels 6, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y. 2004. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. The Plant Cell 16, 2967–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata M, Rodriguez F, Garrido JL. 2000. Separation of chlorophylls and carotenoids from marine phytoplankton: a new HPLC method using a reverse C8 column and pyridine-containing mobile phases. Marine Ecology Progress Series 195, 29–45. [Google Scholar]
- Zehr JP, Falkowski PG. 1988. Pathway of ammonium assimilation in a marine diatom determined with radiotracer 13N1. Journal of Phycology 24, 588–591. [Google Scholar]
- Zhu S-H, Green BR. 2008. Light-harvesting and photoprotection in diatoms: identification and expression of L818-like proteins. In: Allen JF, Gantt E, Golbeck JH, Osmond B, eds. Photosynthesis. Energy from the sun . Dordrecht, Netherlands: Springer, 261–264. [Google Scholar]
- Zrenner R, Stitt M, Sonnewald U, Boldt R. 2006. Pyrimidine and purine biosynthesis and degradation in plants. Annual Review of Plant Biology 57, 805–836. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.