Highlight
This study reports that an Arabidopsis mitochondria-localized RRL protein positively regulates ABA signalling through downstream regulatory transcription factor ABI4-mediated mitochondrial retrograde regulation during seed germination and seedling growth.
Key words: ABA signalling, ABI4, AOX1a, mitochondria, mitochondrial retrograde regulation RRL.
Abstract
The molecular mechanisms of abscisic acid (ABA) signalling have been studied for many years; however, how mitochondria-localized proteins play roles in ABA signalling remains unclear. Here an Arabidopsis mitochondria-localized protein RRL (RETARDED ROOT GROWTH-LIKE) was shown to function in ABA signalling. A previous study had revealed that the Arabidopsis mitochondria-localized protein RRG (RETARDED ROOT GROWTH) is required for cell division in the root meristem. RRL shares 54% and 57% identity at the nucleotide and amino acid sequences, respectively, with RRG; nevertheless, RRL shows a different function in Arabidopsis. In this study, disruption of RRL decreased ABA sensitivity whereas overexpression of RRL increased ABA sensitivity during seed germination and seedling growth. High expression levels of RRL were found in germinating seeds and developing seedlings, as revealed by β-glucuronidase (GUS) staining of ProRRL–GUS transgenic lines. The analyses of the structure and function of mitochondria in the knockout rrl mutant showed that the disruption of RRL causes extensively internally vacuolated mitochondria and reduced ABA-stimulated reactive oxygen species (ROS) production. Previous studies have revealed that the expression of alternative oxidase (AOX) in the alternative respiratory pathway is increased by mitochondrial retrograde regulation to regain ROS levels when the mitochondrial electron transport chain is impaired. The APETALA2 (AP2)-type transcription factor ABI4 is a regulator of ALTERNATIVE OXIDASE1a (AOX1a) in mitochondrial retrograde signalling. This study showed that ABA-induced AOX1a and ABI4 expression was inhibited in the rrl mutant, suggesting that RRL is probably involved in ABI4-mediated mitochondrial retrograde signalling. Furthermore, the results revealed that ABI4 is a downstream regulatory factor in RRL-mediated ABA signalling in seed germination and seedling growth.
Introduction
The phytohormone abscisic acid (ABA) regulates many important aspects of plant development, such as promotion of seed maturation and dormancy and inhibition of the transitions from the embryonic to the germinative phase (Rock, 2000; Finkelstein et al., 2002; Hirayama and Shinozaki, 2007). ABA is also a key endogenous signal to help plants adapt to various biotic and abiotic stresses (Rock, 2000; Shinozaki and Yamaguchi-Shinozaki, 2000). Understanding of ABA signalling has progressed greatly, and a recent breakthrough is the discovery of the ABA perception mechanism in the primary ABA signal transduction pathway (Raghavendra et al., 2010). ABA binds to the receptor PYR1/PYLs/RCARs, which together inactivate type 2C protein phosphatases (PP2Cs) such as ABI1 and ABI2. Inactivation of the protein phosphatases enables the release of active OST1 and related SNF1-type kinases (SnRKs) via PP2C inactivation to initiate ABA signal transduction (Fujii et al., 2009; Miyazono et al., 2009; Nishimura et al., 2009). OST1 actives the anion channel SLAC1 (Geiger et al., 2009; Lee et al., 2009) and inhibits the cation channel KAT1 (Sato et al., 2009) by phosphorylation to induce stomatal closure. In ABA signalling to the nucleus, OST1 and related SnRKs target ABA-responsive element-binding factor (ABFs/AREBs) such as the basic leucine zipper (bZIP) transcriptional factor ABI5 (Finkelstein et al., 2005) to regulate ABA-responsive gene expression. ABI3 is a B3 transcriptional factor which binds to ABI5 to facilitate ABI5 binding to ABA-responsive elements (ABREs) (Nakamura et al., 2001). In addition, ABI4, an APETALA2 (AP2)-type transcription factors binds the CE1 cis-element in the promoter of target genes in ABA signalling (Woodson and Chory, 2008).
ABI4 plays essential positive roles in the ABA biogenesis and signal transduction pathway, especially during seed dormancy and germination (Finkelstein et al., 1998; Soderman et al. 2000; Shu et al., 2013). abi4 was identified as an ABA-insensitive mutant during seed germination (Finkelstein et al., 1998), whereas overexpression of ABI4 represses seed germination when exogenous ABA is applied (Holdsworth et al., 2008). The mRNA level of ABI4 is high in developing seed and relatively low in the vegetative tissues, and ABI4 protein is degraded through the 26S proteasomal pathway from the germination and seedling establishment stages (Finkelstein et al., 2011). Moreover, an increasing number of findings have demonstrated that ABI4 is a versatile regulatory factor. ABI4 is found to regulate diverse processes including lipid biosynthesis and breakdown (Penfield et al., 2006), redox regulation (Pontier et al., 2007), cell wall metabolism (Talboys et al., 2011), lateral root development (Shkolnik-Inbar and Bar-Zvi, 2010), salt responses (Quesada et al, 2000), nitrogen deficiency response (Yang et al., 2011), sugar signalling (Arenas-Huertero et al., 2000; Laby et al., 2000), and mitochondrial and chloroplast retrograde signalling (Koussevitzky et al., 2007; Giraud et al., 2009; Sun et al., 2011). High sugar levels lead to delayed seed germination (Dekkers et al., 2004), slow seedling growth, impaired chloroplast development, and pale cotyledons. However, abi4 mutants are insensitive to 6% glucose, lacking all the characteristics of sugar-directed arrested growth (Arenas-Huertero et al., 2000; Huijser et al., 2000). Elevated ABI4 mRNA levels result in glucose oversensitivity (Teng et al., 2008; Cui et al., 2012), which suggests that glucose promotes the sugar-directed growth arrest by the accumulation of ABI4 protein in young seedlings. Furthermore, ABI4 also functions as a point of convergence for anterograde (nuclear to mitochondrial) and retrograde (mitochondrial to nuclear) signalling pathways (Giraud et al., 2009; Ng et al., 2014). ABI4 mediates mitochondrial retrograde signals through regulation of the expression of ALTERNATIVE OXIDASE1a (AOX1a), which is a marker gene of mitochondrial retrograde regulation. ABI4 binds to the CE1 cis-element in the promoter of AOX1a and represses AOX1a gene expression. However, the repression due to the binding of ABI4 can be derepressed by ABA signal (Giraud et al., 2009). The accumulation of AOX1a increases mitochondrial reactive oxygen species (mtROS) levels to keep the balance of the redox status and generate ROS signalling by mitochondrial retrograde regulation when the mitochondrial electron transport chain (mETC) is inhibited in plant cells (Popov, 2003; Ng et al., 2014).
Mitochondria are responsible for both ATP and mtROS production in cells, and the mechanisms of mitochondrial biogenesis and function are relatively well understood. However, there is little knowledge of the regulatory factors involved in the various developmental processes and signalling pathways in mitochondria, which calls for more research on the biological role of mitochondria-localized protein at a molecular level (Ng et al., 2014). A previous study discovered that the Arabidopsis RETARDED ROOT GROWTH (RRG) gene functions in the regulation of cell division in the root meristem (Zhou et al., 2011). RRG is a mitochondria-localized protein containing a conserved DUF155 domain of unknown function. Disruption of RRG causes retarded root growth because of reduced numbers of dividing cells, the rate of cell production, and endoreduplication in the root meristem. Here, the function of another Arabidopsis mitochondria-localized protein with a DUF155 domain named RRL (RETARDED ROOT GROWTH-LIKE) in ABA signalling was characterized. RRL plays a positive role in ABA signal transduction in seed germination and primary root growth with regulation of the expression of the AP2-type transcription factor ABI4 and ABI4-meditated mitochondrial retrograde signalling.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana ecotype Columbia (Col-0) was used as the wild-type control in this study. rrl (SALK_022878) and abi4-1 (CS8104) were from the Arabidopsis Biological Resource Center (ABRC; http://www.arabidopsis.org/abrc/). Seeds were surface sterilized and plated on MS medium (Murashige and Skoog, 1962) containing 0.8% (w/v) agar and 1% (w/v) sucrose. All plates were vernalized at 4 °C for 2 d in the dark followed by incubation under a 16h light/8h dark photoperiod and 70% humidity at 23 °C. For germination and seedling growth assay, ABA (Sigma, USA) was added to the medium.
Plasmid constructions
For complementation assay, a 4.0kb genomic DNA fragment including upstream, coding, and downstream regions of the RRL gene was cloned into the binary vector pCAMBIA3301 at the BamHI/PstI cloning sites to construct the ProRRL-RRL plasmid. In order to generate the RRL-OE plasmid and overexpress RRL in plants, the 1.2kb full-length cDNA of the RRL gene was inserted into the NcoI/PmlI cloning sites of pCAMBIA3301 to replace the β-glucuronidase (GUS) gene driven by the Cauliflower mosaic virus 35S promoter. To make the plasmid ProRRL–GUS for analyses of the expression pattern, a 2kb RRL promoter fragment was cloned into the binary vector pBI101–GUS at the KpnI/PstI cloning sites. To detect subcellular localization of RRL, the plasmid RRL–GFP was constructed by cloning the RRL full-length cDNA sequence in the SalI/PstI cloning sites of vector p35S-SUNGFP, inserting the RRL gene in the N-terminus of the green fluorescent protein (GFP). The sequences of primers used for plasmid construction are listed in Supplementary Table S1 available at JXB online.
Generation of transgenic plants, double mutant abi4-1 rrl, and hybrid abi4-1 OE-4
Agrobacterium tumefaciens strain GV3101 harbouring the plasmid RRL–GFP was used to transform Arabidopsis wild-type (Col-0) plants by floral dip-mediated infiltration (Clough and Bent, 1998) in order to generate RRL–GFP transgenic plants for subcellular localization analysis. For the generation of RRL-overexpression transgenic plants, Agrobacterium GV3101 harbouring the plasmid RRL-OE was used to transform the Col-0 plants. The transgenic lines carrying plasmids ProRRL-RRL and ProRRL–GUS were also generated in the Col-0 background for complementation assay and expression pattern determination, respectively. Homozygous T3 transgenic lines were selected for analyses. Double mutant abi4-1 rrl and hybrid abi4-1 OE-4 lines were generated by crossing the mutant abi4-1 (maternal) with rrl (paternal) and the RRL-overexpression transgenic line OE-4 (paternal), respectively. The abi4-1 mutant contained a G-deletion mutation at base pair 619 that causes early termination of ABI4 translation (Finkelstein et al., 1998). A dCAPS (derived cleaved amplified polymorphic sequence) marker (amplified by primers ABI4dcapsFP and ABI4dcapsRP) was designed to identify homozygous abi4-1 plants from the F2 population derived from the cross between abi4-1 and rrl mutants. PCR products from individual F2 plants were digested by NIaIV. The sequences of ABI4dcapsF and ABI4dcapsR are listed in Supplementary Table S2 at JXB online. Homozygous F2 plants were used for further study.
RT-PCR analyses
Semi-quantitative real-time PCR (RT-PCR) was performed to analyse the expression of RRL. Total RNA was isolated from 5-day-old seedlings using an RNAprep Pure Plant kit (Bio TeKe, China) supplemented with the RNase-free DNase I set, according to the manufacturer’s instructions. RNA samples were reverse transcribed with a ReverAid First Strand cDNA Synthesis Kit (Thermo Scientific, USA). The expression level of the ACTIN2 gene (AT3G18780) was used as a loading control. Quantitative RT-PCR analysis was performed to assay expression levels of mitochondria-associated and ABA-responsive genes with or without ABA treatment. The cDNA was amplified using a SYBR Green master mixture (Bio-Red, USA) with a LightCycler 96 (Roche, USA). Amplification of the β-ACTIN8 (AT1G49240) gene was used as an internal control. The sequences of primers used for semi-quantitative and quantitative RT-PCR analyses are listed in Supplementary Table S2 at JXB online.
Histochemical analysis and subcellular localization analysis
GUS staining was performed to determine the expression pattern of RRL in the ProRRL–GUS transgenic lines. Various tissues of transgenic seedlings were incubated with 5mM K4Fe(CN)6, K3Fe(CN)6, 0.2mM PBS buffer (KH2PO4 and K2HPO4, pH 7.0), 0.1% Triton X-100, 0.5mg ml–1 X- Gluc (Sigma, USA) at 37 °C overnight and washed with 70% ethanol three times (Jefferson et al., 1987). A stereomicroscope (Nikon, smz1000, Japan) was used for examination and photography. To determine the subcellular localization of RRL, leaves of RRL–GFP transgenic Arabidopsis plants were stained with a mitochondrion-selective probe MitoTracker Red (fluorescent dye) (CMTMRos; Invitrogen) following the instruction manual. A transient expression experiment was also carried out to analyse the subcellular localization of RRL as described previously by Voinnet et al. (2003). Agrobacterium GV3101 cells were transformed with the plasmid RRL–GFP. The transformed cells were harvested and resuspended in 10mM MES-KOH, pH 5.6, containing 10mM MgCl2 and 150mM acetosyringone, and then mixed with Agrobacterium expressing the silencing suppressor p19 of Tomato bushy stunt virus (Voinnet et al., 2003) to a final optical density at 600nm (OD600) of 0.8. After incubation for 3h at room temperature, the Agrobacterium suspension was injected into expanded leaves of 4-week-old tobacco plants (Nicotiana benthamiana cv. SR1). Leaves were observed after 3–4 d with a laser scanning confocal imaging system (TCS SP2, Leica).
Transmission electron microscopy
The ultrastructure of mitochondria was observed by transmission electron microscopy. The cut roots from 5-day-old Arabidopsis seedlings were fixed and rinsed in phosphate buffer containing 5% glutaraldehyde and 0.1M cacodylate (pH 7.4), and then were post-fixed in 1% osmium tetroxide for 2.5h at 5 °C. The fixed samples were embedded in catalysed epon (TAAB resin; Energy Beam Sciences), and polymerized at 65 °C for 3 d after dehydration in a graded concentration of acetone. Ultrathin sections were cut with a Reichert Ultracut S ultramicrotome and collected on mesh copper. Uranyl acetate was used to stain the sections, and the stained ultrathin sections were observed with an H-7650 transmission electron microscope (Hitachi).
Detection of ROS
ROS production was detected using the fluorescent dye dichlorofluorescein (DCF) (Pei et al., 2000). In order to quantify ABA-induced ROS levels, 5-day-old seedlings were loaded with 50 μM 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) for 30min and washed with ddH2O before ABA treatment. The loaded seedlings were then treated with 50 μM ABA (Sigma) for 1h. All the pictures were taken by a laser scanning confocal imaging system (TCS SP2, Leica) with excitation at 488nm and emission at 525nm to detect DCF fluorescence. The fluorescent images were analysed with Image J software (National Institutes of Health, USA).
Results
RRL (RETARDED ROOT GROWTH-LIKE) is an Arabidopsis mitochondria-localized protein like RRG (RETARDED ROOT GROWTH)
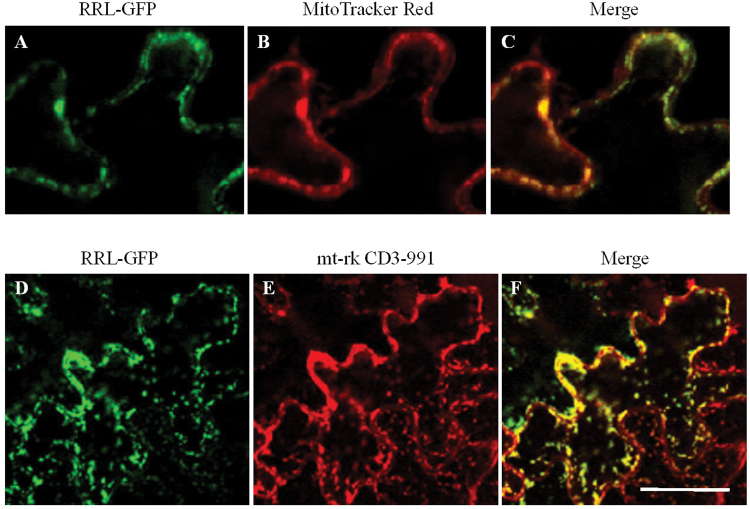
A previous study revealed that an Arabidopsis mitochondria-localized protein RRG (At1G69380) with a domain of unknown function (DUF155) is required for cell division in the root meristem. Disruption of RRG causes the reduction of dividing cells, the rate of cell production, and endoreduplication, which thus decreases the meristem size and root growth rate (Zhou et al., 2011). At5G13610 is a homologous gene of At1G69380 and encodes another protein containing the DUF155 domain. At5G13610 shares 54% and 57% identity in nucleotide and amino acid sequences, respectively, with RRG (At1g69380) (see Supplementary Fig. S1A at JXB online) and was thus named RETARDED ROOT GROWTH-LIKE (RRL). The RRL gene is predicted to encode a novel protein with 402 amino acids (http://www.arabidopsis.org/servlets/TairObject?type=aa_sequence&id=1009134071). The RRL protein contains a conserved domain of unknown function (DUF155) and a putative C-terminal transmembrane domain (http://smart.embl-heidelberg.de/smart/show_motifs.pl) (see Supplementary Fig. S1B). To explore the subcellular localization of RRL protein, transgenic plants were generated with construct RRL–GFP in Arabidopsis. Fluorescence analyses of the expression of RRL–GFP revealed small punctate structures in leaf epidermal cells of the transgenic plants, which appeared to be localized to mitochondria. The mitochondrial localization of RRL–GFP was further confirmed by co-localization with a mitochondrion-selective dye MitoTracker Red (Fig. 1A–C). Additionally, the mitochondrial localization of RRL–GFP was also determined by tobacco leaf infiltration experiments with a well-known mitochondria marker, mt-rk CD3-991 (Nelson et al., 2007), which was generated with a red fluorescent protein, mCherry (Shaner et al., 2004). The RRL–GFP fusion protein co-localized with mt-rk CD3-991 by fluorescence analyses (Fig. 1D–F). These results showed that RRL is localized to mitochondria like RRG (Zhou et al., 2011; Fig. 1).
Fig. 1.
Mitochondrial localization of the RRL–GFP fusion protein. (A–C) Subcellular localization of RRL–GFP fusion protein in leaf epidermal cells of Arabidopsis. (A) RRL–GFP, (B) MitoTracker Red staining, (C) co-localization of RRL–GFP and MitoTracker Red. Scale bar=10 μm. (D–F) Subcellular localization of RRL–GFP fusion protein in leaf epidermal cells of tobacco. (D) RRL–GFP, (E) mt-rk CD3-991 (a mitochondrial marker). (F) The overlay image (merge) shows co-localization of RRL–GFP and the mitochondria marker. Scale bar=30 μm. (This figure is available in colour at JXB online.)
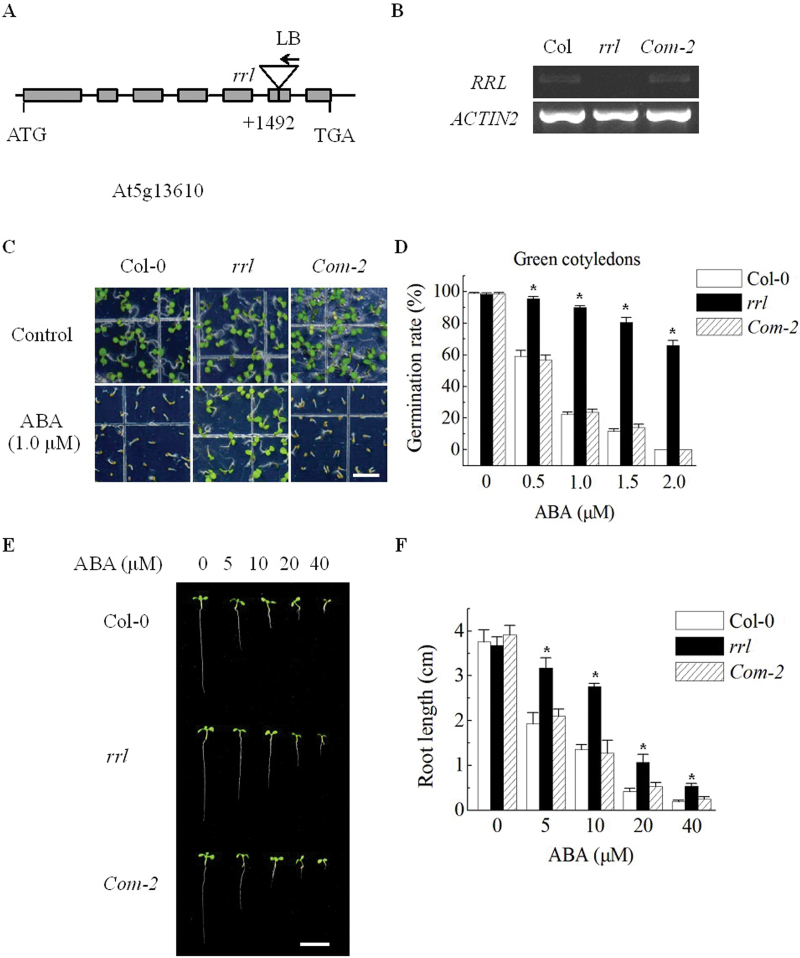
To test if this gene has a similar function to RRG, a null mutant (SALK_022878C) with the T-DNA inserted in the sixth exon of the RRL gene (+1492) was obtained from the ABRC (Fig. 2A). At5G13610 transcripts were detected in the wild type (Col-0) but not in the rrl mutant (Fig. 2B), suggesting that rrl is a null mutant. The rrl mutants showed normal root growth like the wild type (Col-0) plants (see Supplementary Fig. S2 at JXB online), which is different from the retarded root growth of rrg mutants. Moreover, introduction of RRL cDNA driven by the RRG promoter could not recover the retarded root growth of the rrg mutant in complementation assays (see Supplementary Fig. S2). These findings indicated that RRL has different roles in mitochondria in Arabidopsis.
Fig. 2.
ABA-insensitive phenotype of the rrl knockout mutant in seed germination and seedling growth. (A) Schematic drawing (not to scale) of T-DNA insertion sites in the loss-of-function mutant allele of the RRL gene (At5G13610). Grey boxes and lines represent exons and introns, respectively. Arrowheads indicate orientations of T-DNA inserts. (LB, left border primer for T-DNA insertion; ATG, translation start codon; TGA, translation stop codon). (B) Semi-quantitative RT-PCR analysis of RRL gene expression in 5-day-old seedlings of Col-0 (wild type), the rrl mutant, and the Com-2 line (complementation line). ACTIN2 (AT3G18780) was used as the internal control. (C) Seeds were sown on MS medium supplemented or not with 1.0 μM ABA. Photographs were taken at day 5 after stratification. Scale bar=0.5cm. (D) Seeds were sown on MS medium supplemented with 0, 0.5, 1.0, 1.5, or 2.0 μM ABA. Germination rates (%) were analysed at day 5 after stratification. Values are the means ±SE of three independent experiments (n=100). *P<0.05 compared with Col-0 control. (E) Seedlings grew for 10 d after transfer from MS medium to MS medium supplemented with 0, 5, 10, 20, or 40 μM ABA. Seedlings were transferred from ABA-free medium to ABA-containing medium 48h after stratification. Scale bar=1cm. (F) Primary root lengths were assayed at day 10 after transfer. Values are the means ±SE of three independent experiments (n=50). *P<0.05 compared with Col-0 control. (This figure is available in colour at JXB online.)
Disruption of RRL decreases ABA responsiveness in seed germination and primary root growth
In order to predict the function of RRL, the sequence of the RRL promoter was analysed using PLACE (http://www.dna.affrc.go.jp/PLACE/signalup.html). Several ABA-responsive element- (ABRE-) related motifs were found in the promoter region of RRL. Therefore, the relationship between RRL and the ABA signalling pathway was investigated. To test the response of the rrl mutant to ABA, Col-0 and rrl seeds were sown on MS medium supplemented with 1% sucrose and 1.0 μM ABA. More than 90% of rrl seeds are able to germinate, and grow green and open cotyledons, whereas the germination of Col-0 seeds was inhibited at day 5 after stratification (Fig. 2C). Next the germination rates of the rrl mutant on MS medium containing 1% sucrose and a range of ABA concentrations (0, 0.5, 1.0, 1.5, and 2.0 μM ABA) was assayed by counting seedlings with green cotyledons (Fig. 1D). As shown in Fig. 2C and D, the rrl mutant was more insensitive to ABA than Col-0 in terms of seed germination. For example, ~66% (63–69%) of rrl seeds germinated and grew green cotyledons; in contrast, none of the Col-0 seeds germinated at day 5 after stratification when sown on MS medium supplemented with 2.0 μM ABA. In addition, ABA also inhibits primary root elongation during seedling growth as well as seed germination (Finkelstein et al., 2002). Therefore, a primary root growth assay was used to investigate the ABA response of seedling growth. To assess ABA’s effect on seedling growth of the rrl mutant, germinating seed was transferred 48h after stratification from the ABA-free MS medium to MS medium containing 0, 5, 10, 20, or 40 μM ABA for 10 d. In these assays, stronger ABA inhibition of primary root elongation was observed in Col-0 seedlings than in ther rrl mutant when the ABA concentration was >5 μM (Fig. 2E, F).
In order to determine whether the ABA-insensitive phenotypes of the rrl mutant were caused by disruption of the RRL gene, the full-length cDNA was transformed under the control of its native promoter into rrl mutant plants. Thirteen independent transgenic lines were obtained. RT-PCR analysis indicated that the RRL gene was expressed after introducing the RRL gene into the rrl mutant (Fig. 2B) and the expression of RRL could complement the ABA-insensitive phenotype of the rrl mutant in both seed germination (Fig. 2C, D) and root growth assays (Fig. 2E, F). Taken together, these results suggest that disruption of RRL leads to the reduced response to ABA of seed germination and primary root growth.
Overexpression of RRL increases ABA sensitivity of seed germination and primary root growth
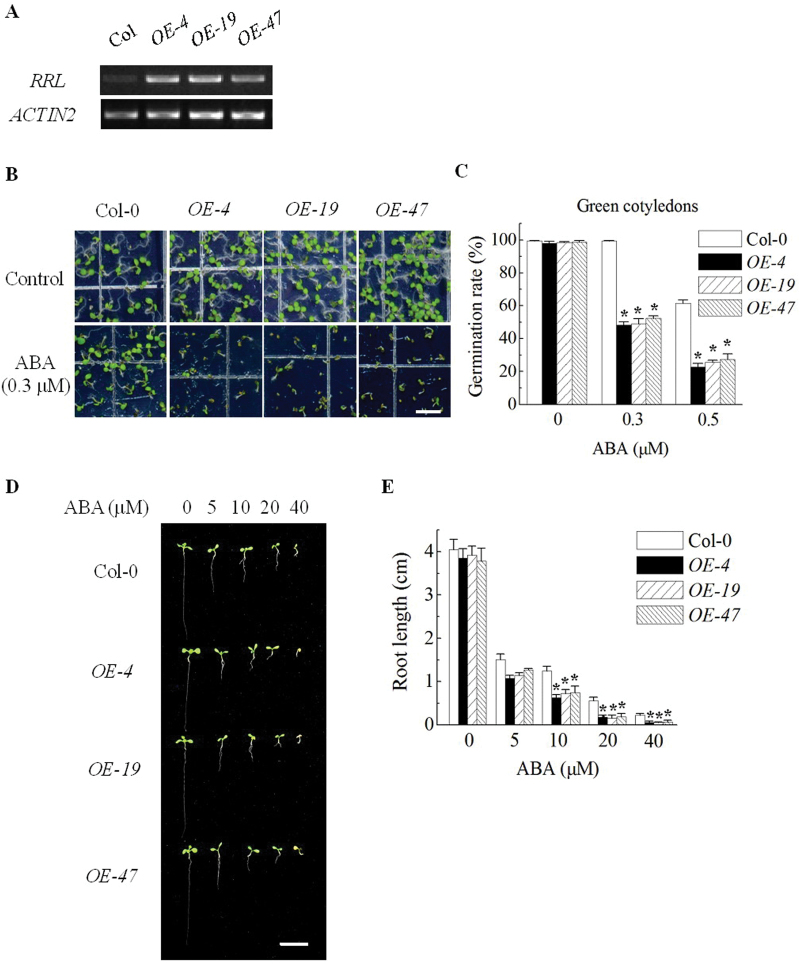
Fifty independent transgenic plants overexpressing RRL in the Col-0 background were generated. Homozygous T3 lines (OE-4, OE-19, and OE-47) with higher levels of RRL transcripts were selected and their responses to ABA were investigated (Fig. 3A). Seed germination and primary root growth assays were carried out in these overexpression lines to examine their responses to ABA. Taking OE-4 as an example, only ~48% (46–50%) and ~23% (20–25%) of OE-4 seeds germinated and grew green cotyledons, whereas the germination rate of Col-0 seeds was ~99% (99–100%) and ~61% (59–64%) when germinated on MS medium supplemented with 1% sucrose and 0.3 μM or 0.5 μM ABA at day 5 after stratification, respectively. (Fig. 3B, C). In root growth assays, the growth of primary roots was significantly inhibited in MS medium containing >10mM ABA compared with that of Col-0 seedlings. These results suggest that overexpression of RRL caused the increased response to ABA of seed germination and primary root growth.
Fig. 3.
Overexpression of RRL cause the ABA-hypersensitive phenotype in seed germination and seedling growth. (A) Semi-quantitative RT-PCR was performed to assay expression levels of the RRL gene in 5-day-old seedlings of Col-0, OE-4, OE-19, and OE-47 overexpression lines. ACTIN2 gene expression represents the loading control. (B) Seeds were sown on MS medium supplemented or not with 0.3 μM ABA. Photographs were taken at day 5 after stratification. Scale bar=0.5cm. (C) Seeds were sown on MS medium supplemented with 0, 0.3, or 0.5 μM ABA. Germination rates (%) were analysed at day 5 after stratification. Values are the means ±SE of three independent experiments (n=100). *P<0.05 compared with Col-0 control. (D) Seedlings grew 10 d after transfer from MS medium to MS medium supplemented with 0, 5, 10, 20, or 40 μM ABA. Seedlings were transferred from ABA-free medium to ABA-containing medium 48h after stratification. Scale bar=1cm. (E) Primary root lengths were assayed at day 10 after transfer. Values are the means ±SE of three independent experiments (n=50). *P<0.05 compared with Col-0 control. (This figure is available in colour at JXB online.)
Because the rrl mutant and the RRL-overexpression lines showed decreased and increased ABA sensitivity, respectively (Figs 2, 3), it was next determined whether disruption or overexpression of RRL would alter the plant response to drought stress. Two-week-old plants were deprived of water for 14 d and then rewatered. The wilting plants of Col-0, the mutant rrl, or the overexpression line OE-4 could not recover by rehydration (see Supplementary Fig. S3A at JXB online). The transpirational water loss of detached rosette leaves was also measured on the 4-week-old Col-0, the mutant rrl, and the overexpression line OE-4. No significantly different water loss was found during 0–6h (see Supplementary Fig. S3B). A stomatal bioassay was also perfomed and it was found that there was no significant difference in ABA-induced stomatal closure of the three lines (see Supplementary Fig. S3C). Moreover, the rrl mutant displayed normal development of guard cells compared with Col-0 (see Supplementary Fig. S4). These results showed that Col-0, rrl, and OE-4 displayed a similar response to drought stress, suggesting that RRL is not involved in ABA-dependent drought signal transduction.
High expression of RRL in germinating seeds and developing seedlings
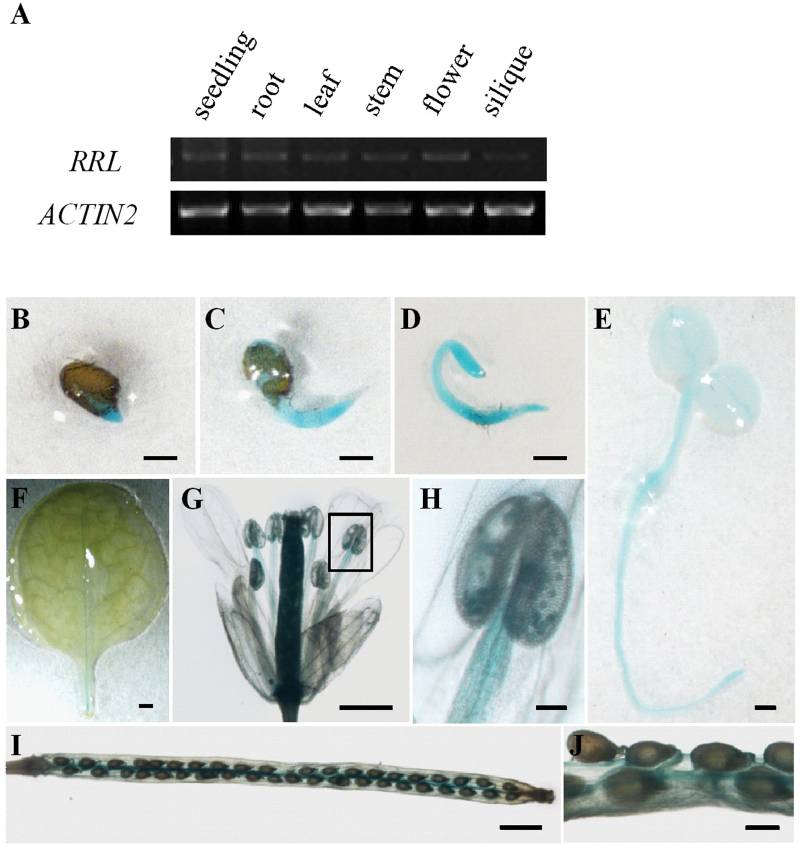
In order to study the specific function of RRL in seed germination and seedling growth, the expression pattern of the RRL gene was next investigated. Semi-quantitative RT-PCR was performed to assess RRL gene expression in various tissues including 7-day-old seedlings, roots, rosette leaves, stem, flowers, and siliques. RRL was expressed in various tissues (Fig. 4A). In the more detailed GUS staining analyses, 10 independent transgenic Arabidopsis lines harbouring the ProRRL–GUS construct were obtained and homozygote T3 lines were used for further analyses. Strong GUS staining was detected in germinating seeds and the early developing seedlings (24–96h after seed germination; Fig. 4B–E). In contrast, very weak GUS signal was observed in vascular tissues of rosette leaves (Fig. 4F). Moreover, GUS staining could also be detected in filaments, pollen in flowers (Fig. 4G, H), and false dissepiments and seed stalks in siliques (Fig. 4I, J). The high expression of RRL in germinating seeds and developing seedlings suggests a role for RRL in these developmental stages, consistent with the finding that RRL mediates ABA signalling in seed germination and seedling growth.
Fig. 4.
The expression pattern of RRL. (A) Semi-quantitative analysis of RRL gene expression in various tissues. ACTIN2 was used as a loading control. (B–J) RRL promoter-driven GUS expression in germinating seeds and developing seedlings (B–D), 4-day-old seedlings (E), rosette leaves (F), flowers (G, H), and siliques (I, J). Scale bars=0.1mm (B, C, D, E, J); 1mm (F, G, I). (H) Magnification of the areas outlined in (G). Scale bar=0.1mm (H). (This figure is available in colour at JXB online.)
The rrl mutant exhibits internal vacuolization of mitochondria and reduced ABA-induced ROS production
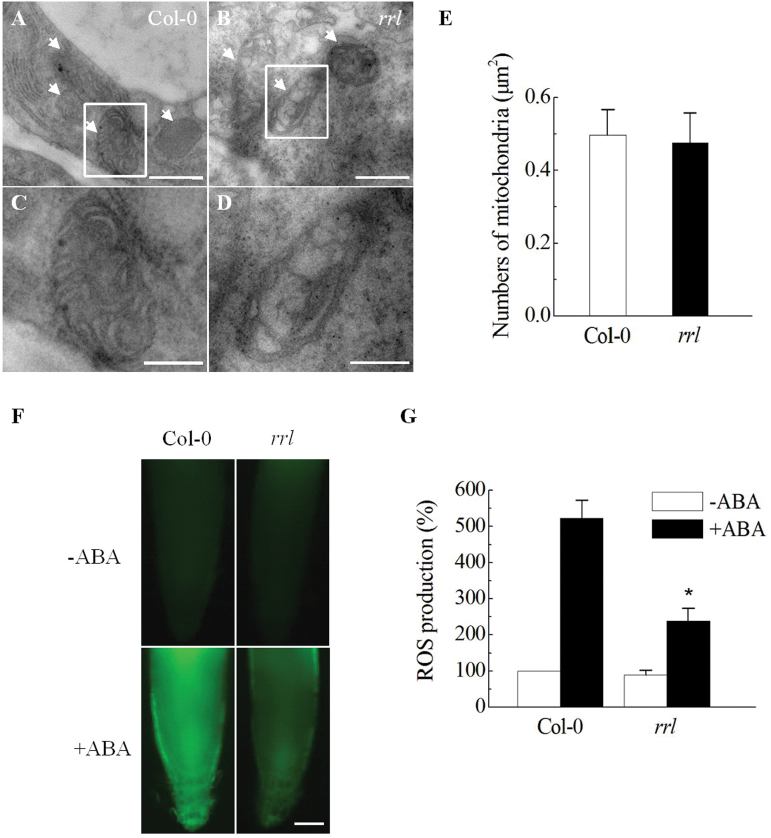
As shown in Fig. 1, RRL is a mitochondria-localized protein in Arabidopsis. Next it was examined whether the disruption of the RRL gene affects the number and structure of mitochondria. Transmission electron microscopy was used to observe the mitochondria in 5-day-old seedling roots of Col-0 and the rrl mutant. A total of 76% of mitochondria in mutant cells exhibited extensive internal vacuolization compared with Col-0 (Fig. 5A–D), but the number of mitochondria per unit of cell area in mutant rrl (0.50mm–2) was found to be the same as that of Col-0 (0.48mm–2) (Fig. 5E). These results suggest that the disruption of the RRL gene results in aberrant mitochondrial structure, but does not affect the number of mitochondria (Fig. 5A–E).
Fig. 5.
Structure and numbers of mitochondria and ABA-induced ROS production in the rrl mutant. (A–D) The mitochondrial structure in Col-0 (A) and the rrl mutant (B) was observed by transmission electron microscopy. (C, D) Magnification of the areas outlined in (A, B). Arrows indicate mitochondria. Scale bar=0.5 μm (A, B); 0.25 μm (C, D). (E) Numbers of mitochondria per square micrometre in 5-day-old seedlings of Col-0 and the rrl mutant were determined. Values are the means ±SD from three independent measurements. (F) ROS production was detected with the fluorescent dye DCF. Five-day-old seedlings were incubated with 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) for 30min. –ABA, ABA-free treatment; +ABA, 50 μM ABA treatment. Scale bar=20 μm. (G) Quantification of ROS levels in Col-0 and the rrl mutant before or after 50 μM ABA treatment. (n=30; ±SD; *P<0.001 compared with ABA-treated Col-0 plants). The fluorescent intensity in Col-0 with ABA treatment was taken as 100%.(This figure is available in colour at JXB online.)
Mitochondria produce ROS continuously in metabolically active cells, and the plant hormone ABA could induce ROS production via the mtETC (Miller et al., 2008). Because of the mitochondrial localization of RRL and the aberrant mitochondrial structure in the rrl mutant cells, it was next determined whether the mutation of RRL led to ABA-induced ROS production. To this end, the ROS levels were compared in 5-day-old seedling roots of Col-0 and the rrl mutant with or without ABA treatment. A reduced ABA-induced ROS level was detected after 50 μM ABA treatment for 1h in the rrl mutant (Fig. 5F). The ROS level increased ~5.2-fold in Col-0, whereas there was only a 2.7-fold increase in ROS levels in the rrl mutant, suggesting that RRL is required for ROS production in response to ABA (Fig. 5G).
Transcriptional alterations of mtETC-associated gene expression and expression of downstream targets in ABA signalling
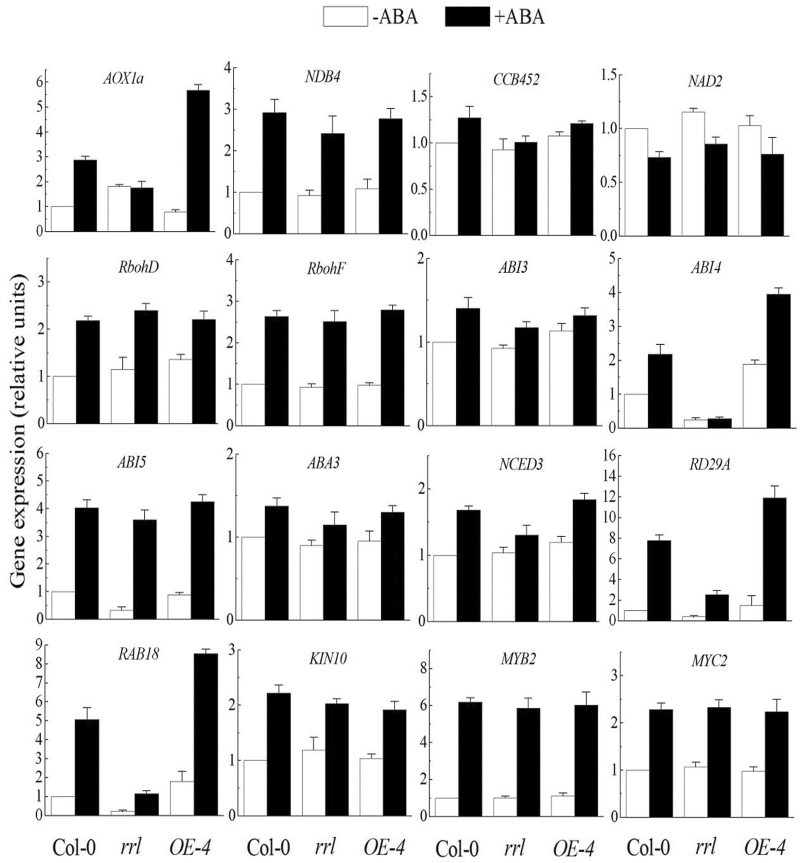
The requirement for RRL for maintaining the structure and function of mitochondria (Fig. 5) prompted an assay of the expression of mtETC-associated genes including AOX1a, NDB4, CCB452, and NAD2 to be carried out. An isoform of alternative oxidase AOX1a functions as a non-phosphorylating bypass of mitochondrial electron transport, and NDB4 is a alternative type II NAD(P)H dehydrogenase without proton translocation activity in the alternative respiratory pathway (Melo et al., 2004). CCB452 is associated with biogenesis of cytochrome c (van Dongen et al., 2011) and NAD2 encodes a subunit of NAD(P)H dehydrogenase in mitochondrial respiratory chain complex I (Heazlewood et al., 2003). Expression levels of genes were quantitatively analysed after the 5-day-old seedlings were subjected to ABA for 6h (Fig. 6). The expression of mtETC-associated genes including CCB45, NDB4, and NAD2 was not altered by disruption or overexpression of RRL in both the absence and presence of the ABA treatments, except for AOX1a (Fig. 6). In this report, ABA-induced expression of AOX1a was found in Col-0 plants as described previouly (Giraud et al., 2009). It should be noted that AOX1a expression was increased in the rrl mutant in the absence of the ABA treatments, but the ABA induction was affected by mutation of RRL. Overexpression of RRL significantly enhanced the expression level of AOX1a in response to ABA (Fig. 6). Because AOX1a induction also serves as a marker of mitochondrial retrograde responses in Arabidopsis (Rhoads and Subbaiah, 2007), this result suggests that RRL is involved in the retrograde signalling between the mitochondria and nucleus in response to ABA.
Fig. 6.
Expression levels of mitochondria-associated genes and ABA-responsive genes in the RRL loss-of-function mutant and overexpression line. The expression levels of some mitochondria-associated and ABA-responsive genes were assayed by quantitative RT-PCR with 5-day-old seedlings of Col-0, rrl, and OE-4. –ABA, ABA-free treatment; +ABA, 50 μM ABA treatment. Gene expression levels were analysed as relative units, with that of ABA-free treated Col-0 plants being taken as 1. Values are the means ±SE of three independent experiments.
To understand whether the ABA-insensitive phenotypes of the rrl knockout mutant and ABA-hypersensitive phenotypes of the transgenic overexpression lines in seed germination and seedling growth (Figs 2, 3) were due to altered ABA signalling, the expression levels of the following ABA-responsive genes were also tested: RbohD and RbohF (Kwak et al., 2008), ABI3 (Giraudat et al., 1992), ABI4 (Finkelstein et al., 1998), ABI5 (Finkelstein and Lynch, 2000), ABA3 (Léon-Kloosterziel et al., 1996), NCED3 (Iuchi et al., 2001), RD29A (Yamaguchi-Shinozaki and Shinozaki, 1994), RAB18 (Lang and Palva, 1992), KIN10 (Baena-González et al, 2007), and MYB2 and MYC2 (Abe et al., 2003). Results from gene expression assays showed that disruption or overexpression of RRL did not affect the expression of RbohD, RbohF, ABI3, ABI5, ABA3, NCED3, KIN10, MYB2, and MYC2 in both the absence and presence of ABA treatment. ABA3 and NCED3 are two key genes which function in ABA biosynthesis. Similar expression levels of ABA3 and NCED3 in Col-0, the rrl mutant, and the overexpression line in response to ABA indicated that RRL gene expression did not alter the ABA content. Disruption of RRL down-regulated expression of ABI4, RD29A, and RAB18 in both the absence and presence of ABA treatments; in contrast, overexpression of RRL amplified the ABA-induced expression effects on these downstream targets in ABA signalling (Fig. 6). The AP2-type transcription factor ABI4 plays a positive role in ABA signal transduction (Finkelstein et al., 1998). It binds to and keeps the AOX1a promoter constitutively repressed, and ABA can lift the repression to stimulate AOX1a gene expression by mediating mitochondrial retrograde signalling (Giraud et al., 2009). The expression of ABI4 was significantly deceased and ABA-induced AOX1a expression was impaired in the rrl mutant in the presence of ABA treatments, suggesting that ABI4 is required for RRL-mediated ABA signalling and mitochondrial retrograde regulation. Taken together, these results on gene expression suggested that the altered ABA responsiveness in germination and seedling growth of the rrl mutant and the OE-4 overexpression line is probably due to ABA signalling modulated by ABI4 expression and ABI4-mediated mitochondrial retrograde regulation.
RRL mediates ABA signal transduction by regulation of ABI4 expression
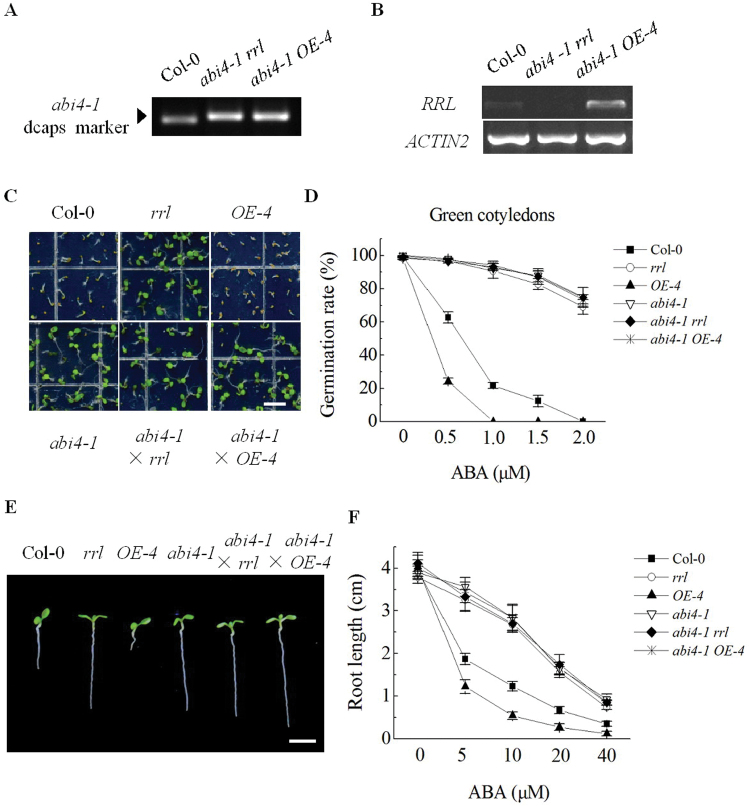
To determine whether ABI4 gene expression is necessary for the RRL-mediated ABA signalling pathway, mutant abi4-1 (maternal) was crossed with rrl (paternal) or the overexpression line OE-4 (paternal). The abi4-1 mutation locus was isolated by a dCAPS marker and RRL expression was analysed by semi-quantitative RT-PCR in the F2 generation seedlings of crossed plants (Fig. 7A, B). In the seed germination assay, the abi4-1 mutant showed an ABA-insensitive phenotype, consistent with a previous study (Finkelstein et al., 1998). The double mutant abi4-1 rrl and hybrid abi4-1 OE-4 showed a similar ABA-insensitive phenotype to abi4 mutants when 1.0 μM ABA was applied (Fig. 7C). Furthermore, germination rates with 0, 0.5, 1.0, 1.5, and 2.0 μM ABA concentrations were assessed by scoring the open green cotyledons at day 5 after stratification. The germination rates of the double mutant abi4-1 rrl and hybrid abi4-1 OE-4 were significantly higher than those of Col-0, which were comparable with those of rrl and abi4-1 mutants. In contrast, the germination rates of the overexpression line OE-4 were drastically reduced compared with Col-0 (Fig. 7D). The length of the primary root of seedling plants grown on MS medium containing the indicated concentration of ABA for 10 d were also measured. The primary root lengths of the double mutant abi4-1 rrl and hybrid abi4-1 OE-4 were also comparable with those of mutant rrl and mutant abi4-1 when the seedlings grew on MS medium containing the indicated concentrations of ABA, whereas the primary root elongation of the overexpression line OE-4 was significantly inhibited by the ABA treatments compared with Col-0 (Fig. 7E, F). Taken together, the data suggest that ABI4 plays a role downstream of RRL in ABA signalling during seed germination and primary root growth.
Fig. 7.
Genetic analysis of the double mutant abi4-1 rrl and the hybrid abi4-1 OE-4. (A, B) F2 generation seedlings of crossed plants were subjected to single nucleotide polymorphism (SNP) identification to isolate abi4 mutation loci (A) and semi-quantitative RT-PCR to determine expression of RRL (B). ACTIN2 was used as the internal control. (C) Seeds were sown on MS medium supplemented with 1.0 μM ABA. Photographs were taken at day 5 after stratification. Scale bar=0.5cm. (D) Seeds were sown on MS medium supplemented with the indicated concentration of ABA. Germination rates (%) were analysed at day 5 after stratification. Values are the means ±SE of three independent experiments (n=100). (E) Seedlings grew for 10 d after transfer from MS medium to MS medium supplemented with 10 μM ABA. Seedlings were transferred from ABA-free medium to ABA-containing medium 48h after stratification. Scale bar=0.5cm. (F) Primary root lengths were assayed after seedlings were transferred to MS medium supplemented with 0, 5, 10, 20, or 40 μM ABA for 10 d. Values are the means ±SE of three independent experiments (n=50). (This figure is available in colour at JXB online.)
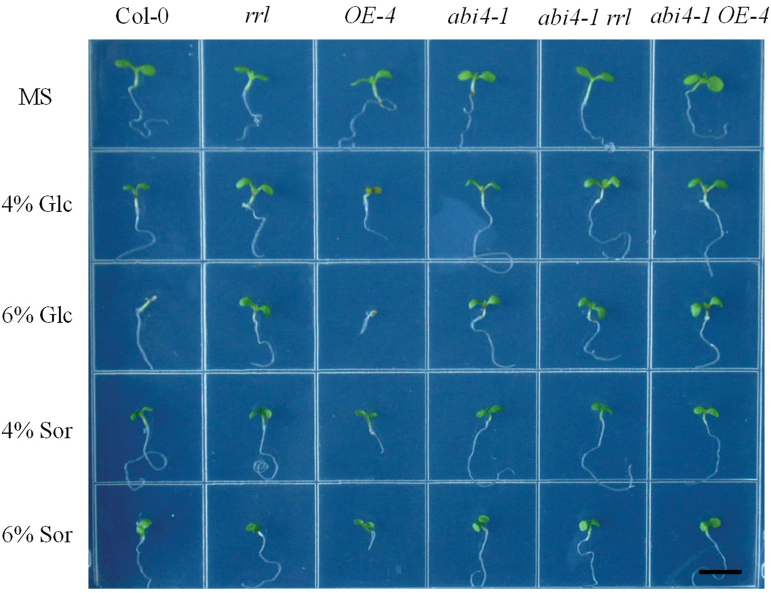
Previous findings have revealed that ABI4 is involved in the ABA-mediated glucose response. Early seedling development is inhibited in the wild type in response to sugars. However, the abi4 mutant displays expanded green cotyledons and true leaves on 6% glucose (Arenas-Huertero et al., 2000; Huijse, et al., 2000). Thus the responses of the rrl mutant, the OE-4 overexpression line, the abi4-1 mutant, the abi4-1 rrl double mutant, and the abi4-1 OE-4 hybrid to glucose were next tested. The results showed that the rrl mutant, the abi4-1 rrl double mutant, and the abi4-1 OE-4 hybrid exhibited the same reduced sensitivity to sugar as the abi4-1 mutant on MS medium supplemented with 4% and 6% glucose. In contrast, the OE-4 overexpression line displayed increased sensitivity to 4% and 6% glucose compared with Col-0 (Fig. 8). Sorbitol at 4% and 6% was used as the osmotic control in this experiment. These results suggest that disruption of RRL renders seedlings sugar insensitive and RRL may participate in ABI4-mediated sugar signalling.
Fig. 8.
Responses to glucose of the rrl knockout mutant, the OE-4 overexpression line, the abi4-1 rrl double mutant, and the abi4-1 OE-4 hybrid. Glucose hypersensitivity of the RRL overexpression line OE-4 (4–6% glucose) and glucose insensitivity of the rrl knockout mutant, abi4-1 mutant, abi4-1 rrl double mutant, and abi4-1 OE-4 hybrid (4–6% glucose) in early seedling development. Sor, sorbitol osmotic control; Glc, glucose. Scale bar=0.5cm. (This figure is available in colour at JXB online.)
Discussion
Mitochondrial proteome studies have discovered that >1000 proteins are present in plant mitochondria. A great number of mitochondria-localized proteins regulated by anterograde (nuclear to mitochondrial) or retrograde (mitochondrial to nuclear) signalling are involved in growth, development, and stress signalling pathways in plant. Therefore, mitochondria are important hubs of the internal and external signals in the cells (Ng et al., 2014). Plants are able to alter the endogenous levels of hormones such as ABA in response to abiotic and biotic stresses, which can target mitochondria directly and lead to mitochondrial dysfunction. Mitochondria in turn play vital roles in transmission and execution of the signals during overall plant stress responses. However, the molecular mechanisms of mitochondrial responses to a variety of stresses are still unclear, although a few components in the signalling pathway have begun to be found (Finkelstein et al., 2002; Rhoads and Subbaiah, 2007; Ng et al., 2014). In this study, the function of an Arabidopsis mitochondria-localized protein RRL in the ABA signalling pathway has been identified. RRL is expressed in germinating seeds and early developing seedlings, and plays a positive role in ABA signal transduction during seed germination and seedling growth. Moreover, RRL-mediated ABA signalling is regulated by mitochondrial retrograde regulation mediated by the AP2 transcription factor ABI4.
RRL positively regulates ABA signalling during seed germination and seedling growth
This study revealed that RRL was a positive regulatory factor in ABA signalling during seed germination and primary root growth (Figs 2, 3). However, the expression of RRL did not alter the response of plants to drought stress (see Supplementary Fig. S3 at JXB online). The expression pattern of RRL was further determined in various tissues. High expression levels of RRL were found in germinating seeds and developing seedlings; however, only very low expression levels of RRL were found in vascular tissues of rosette leaves, showing a close correlation to the functions of RRL in ABA signalling in seed germination and seedling growth (Fig. 4). In order to investigate how RRL regulates ABA signalling, the expression levels of some ABA-responsive genes were next examined. The data from quantitative RT-PCR experiments showed that the expression levels of the marker genes of ABA signalling such as RD29A (Yamaguchi-Shinozaki and Shinozaki, 1994) and RAB18 (Lang and Palva, 1992) were significantly reduced in the rrl mutant but elevated in the OE-4 overexpression line with ABA treatment; in contrast, RbohD, RbohF (Kwak et al., 2008), KIN10 (Baena-González et al, 2007), and MYB2 and MYC2 (Abe et al., 2003) displayed similar expression levels in both the rrl mutant and the OE-4 overexpression line in the presence or absence of ABA treatment (Fig. 6). These results suggest that RRL mediates ABA signalling but not through the regulation of the above components. As shown in Figs 2–4, the high expression levels of RRL and altered plant responses to ABA were all found in germinating seeds and developing seedlings; thus the expression of ABI3 (Giraudat et al., 1992), ABI4 (Finkelstein et al., 1998), and ABI5 (Finkelstein and Lynch, 2000), which are three transcription factors playing essential roles in ABA signalling in seeds and seedlings (Finkelstein et al., 2002), was investigated. The results showed that the expression of ABI4 was significantly down-regulated in the rrl mutant and up-regulated in the OE-4 overexpression line after ABA treatment, whereas the expression of ABI3 and ABI5 was not affected in either the rrl mutant or the OE-4 overexpression line (Fig. 6). These results suggest that it is probable that ABI4 acts downstream of RRL-mediated ABA signalling during seed germination and seedling growth. Genetic analysis was next performed to confirm that RRL and ABI4 function in the same pathway and that ABI4 is a downstream component of RRL-mediated ABA signal transduction (Fig. 7).
Interestingly, in addition to the ABA-insensitive phenotype of the rrl mutant, its insensitive phenotype to high sugar levels was also found. The abi4-1 mutant was insensitive to 6% glucose as described previously, and the rrl mutant showed the same response as the abi4-1 mutant (Fig. 7). In addition, the results from genetic analysis suggest that ABI4 also acts downstream of RRL in sugar signalling (Fig. 7). Previous findings have demonstrated that sugar signalling pathway interacts with the ABA signalling pathway, and common signalling components shared in the both signalling pathways have been reported by intensive studies (Cheng et al., 2002; Rolland et al., 2006). In this report, RRL was found to be such a common component functioning in both the sugar signalling pathway and the ABA signalling pathway (Figs 7, 8).
The function of RRL in mitochondrial retrograde regulation
Anterograde regulation is a top-down signalling pathway from the nucleus to the organelle, whereas retrograde regulation is a bottom-up signalling pathway from the organelle to the nucleus (Leister, 2005; Liu and Butow, 2006). Retrograde regulation takes place when endogenous or external stimuli alter the functioning of organelles and signals originating from the organelles in response control nuclear gene expression. Modulated anterograde regulation can in turn affect the functioning of organelles (Butow and Avadhani, 2004; Pesaresi et al., 2007). Abiotic stress, biotic stress, or mutation results in a dysfunctional mETC in mitochondria, and the expression of AOXs and alternative NAD(P)H dehydrogenases was subsequently induced by mitochondrial retrograde regulation, which can compensate ROS production to regain the redox homeostasis and ROS signalling. AOX1a encodes an isoform of AOX and functions as a marker for mitochondrial retrograde regulation. A previous study has revealed that ABI4 keeps AOX1a expression repressed by binding of a CCAC cis-acting element of the AOX1a promoter, and the repression can be lifted by ABA (Giraud et al., 2009).
In the present study, it was found that RRL was localized to mitochondria (Fig. 1), and disruption of RRL impaired the normal structure of mitochondria (Fig. 5A–D). The aberrant structure has an impact on the functioning of mitochondria such as the reduced production of ABA-stimulated ROS (Fig. 5F, G). MtROS originated from the mETC which is localized at the inner mitochondrial membrane. Two partially overlapping respiratory pathways are reported for mETC, namely the cytochrome respiratory pathway with cytochrome oxidase (COX) as the terminal oxidase and the alternative respiratory pathway with alternative oxidase (AOX) as the terminal oxidase (Finnegan et al., 2004; McDonald and Vanlerberghe, 2004). Interestingly, a putative C-terminal transmembrane domain was predicted in the mitochondria-localized RRL protein sequence (http://smart.embl-heidelberg.de/smart/show_motifs.pl), implying its possible relationship to the respiratory pathway in mitochondria (see Supplementary Fig. S1B at JXB online). Thus the expression of AOX1a and NDB4 associated with the alternative respiratory pathway and CCB452 and NAD2 associated with the cytochrome respiratory pathway was examined in the rrl mutant and OE-4 overexpression line with or without ABA treatment. In contrast to the similar expression levels of NDB4, CCB452, and NAD2 compared with the wild type (Col-0), an enhanced expression level of AOX1a was found in the rrl mutant in the absence of ABA treatment. Because a previous report has revealed that AOX1a expression is repressed by ABI4 under normal conditions (Giraud et al., 2009), the elevated AOX1a expression might be caused by down-regulated expression of ABI4 in the rrl mutant (Fig. 6). It is should be noted, however, that ABA-induced AOX1a expression was affected by the disruption of RRL after ABA treatment, which would explain the reduced ABA-induced ROS production in the rrl mutant (Giraud et al., 2009; Figs 5F, G, 6). Taken together, the present study provides evidence that RRL-mediated mitochondrial retrograde regulation is modulated by the expression of ABI4 and AOX1a. RRL probably plays a positive role in ABA-induced AOX1a accumulation, which consequently leads to increased levels of mtROS; the mtROS, as signalling molecules, are capable of mediating ABA signal transduction in seed germination and seedling growth (El-Maarouf-Bouteau and Bailly, 2008; Gomes and Garcia, 2013). The molecular mechanisms of mitochondrial retrograde regulation have not been elucidated to date, and how ROS-stimulated signalling is transmitted to the nucleus to trigger gene expression in mitochondrial retrograde signalling remains unclear. A recent study has revealed that a group of endoplasmic reticulum (ER)-bound NAC transcription factors such as ANAC017 would help to explain how ROS signals can be transmitted in mitochondrial retrograde signalling (Ng et al., 2013). The ER association with actin filaments facilitates a close interaction between the ER and mitochondria, which would set the stage for ROS diffusion over the ER to activate the release of the membrane-bound NAC transcription factors from the ER. The released NAC transcription factors finally migrate into the nucleus to relay the signals, providing the possibility of transmission of ROS signalling. Additionally, a group of WRKY transcription factors such as WRKY40 and WRKY63 also seem to be the responsive regulators in ROS-mediated mitochondrial retrograde signal transduction (Ng et al., 2014). More studies are required to identify the functional components to better understand the mitochondrial retrograde signalling pathway.
In conclusion, the findings in this study demonstrated the positive function of RRL in ABA signalling in Arabidopsis seed germination and seedling growth. RRL mediated ABA signal transduction through the downstream regulatory transcription factor ABI4 during seed germination and seedling growth. Moreover, mitochondria-localized RRL probably increased ROS levels in response to ABA, and ROS signals were sequentially transmitted to the nucleus to regulate gene expression in mitochondrial retrograde signalling.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Analysis of the RRL protein sequence.
Figure S2. Disruption of RRL does not cause retarded root growth.
Figure S3. Mutation of RRL does not alter the plant response to drought stress.
Figure S4. rrl mutants show normal development of guard cells.
Table S1. Primer sequences used for plasmid constructions in this study.
Table S2. Primer sequences used for semi-quantitative and quantitative RT-PCR experiments in this study.
Acknowledgements
We thank Dr Jian Xu for generously providing the p35S-SUNGFP vectors; Dr Honghong Hu for sharing the stereomicroscope (Nikon, smz1000, Japan); Dr Jianbo Cao for carrying out transmission electron microscopy; and Dr Yan Wu for providing the mitochondrion-selective probe MitoTracker Red. This work was supported by the National Natural Science Foundation of China (31201088) and the Specialized Research Fund for the Doctoral Program of Higher Education of China (20120146120013).
Glossary
Abbreviations:
- ABA
abscisic acid
- ABF/AREB
ABA-responsive element binding factor
- ABRE
ABA-responsive element
- AOX
alternative oxidase
- bZIP
basic leucine zipper
- COX
cytochrome oxidase
- dCAPS
derived cleaved amplified polymorphic sequences
- DCF
dichlorofluorescein
- GUS
β-glucuronidase
- H2DCFDA
2′,7′-dichlorodihydrofluorescein diacetate
- mETC
mitochondrial electron transport chain
- mtROS
mitochondrial ROS
- ROS
reactive oxygen species.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. 2003. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcription activators in abscisic acid signaling. The Plant Cell 15, 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas-Huertero F, Arroyo A, Zhou L, Sheen J, Leon P. 2000. Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes and Devolopment 14, 2085–2096. [PMC free article] [PubMed] [Google Scholar]
- Baena-González1 E, Rolland F, Johan M., Thevelein JM, Sheen J. 2007. A central integrator of transcription networks in plant stress and energy signaling. Nature 448, 938–942. [DOI] [PubMed] [Google Scholar]
- Butow RA, Avadhani NG. 2004. Mitochondrial signaling: the retrograde response. Molecular Cell 14, 1–15. [DOI] [PubMed] [Google Scholar]
- Cheng WH, Endo A, Zhou L, et al. 2002. A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. The Plant Cell 14, 2723–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Hao Y, Kong D. 2012. SCARECROW has a SHORT-ROOT-independent role in modulating the sugar response. Plant Physiology 158, 1769–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers BJ, Schuurmans JA, Smeekens SC. 2004. Glucose delays seed germination in Arabidopsis thaliana . Planta 218, 579–588. [DOI] [PubMed] [Google Scholar]
- El-Maarouf-Bouteau H, Bailly C. 2008. Oxidative signaling in seed germination and dormancy. Plant Signaling and Behavior 3, 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R, Gampala SS, Lynch TJ, Thomas TL, Rock CD. 2005. Redundant and distinct functions of the ABA response loci ABA-INSENSITIVE (ABI)5 and ABRE-BINDING FACTOR (ABF)3 . Plant Molecular Biology 59, 253–267. [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SSL, Rock CD. 2002. Abscisic acid signaling in seeds and seedlings. The Plant Cell 14, S15–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R, Lynch T, Reeves W, Petitfils M, Mostachetti M. 2011. Accumulation of the transcription factor ABA-insensitive (ABI)4 is tightly regulated post-transcriptionally. Journal of Experimental Botany 62, 3971–3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM. 1998. The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA2 domain protein. The Plant Cell 10, 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan PM, Soole KL, Umbach AL. 2004. Alternative mitochondrial electron transport proteins in higher plants. In: Day DA, Millar AH, Whelan J, eds. Plant mitochondria: from genome to function, advances in photosynthesis and respiration . Dordrecht: Kluwer Academic Press, 163–230. [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cuter SR, Sheen J, Rodriguez PL, Zhu JK. 2009. In vitro reconstitution of an abscisic acid signalling pathway. Nature 462, 660–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, et al. 2009. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase–phosphatase pair. Proceedings of the National Academy of Sciences, USA 106, 21425–21430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J, Hauge B, Valon C, Smalle J, Parcy F, Goodman H. 1992. Isolation of the Arabidopsis ABI3 gene by positional cloning. The Plant Cell 4, 1251–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E, Van Aken O, Ho LH, Whelan J. 2009. The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of ALTERNATIVE OXIDASE1a . Plant Physiology 150, 1286–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes MP, Garcia QS. 2013. Reactive oxygen species and seed germination. Biologia 68, 351–357. [Google Scholar]
- Heazlewood JL, Howell KA, Millar AH. 2003. Mitochondrial complex I from Arabidopsis and rice: orthologs of mammalian and fungal components coupled with plant-specific subunits. Biochimica et Biophysica Acta 1604, 159–169. [DOI] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K. 2007. Perception and transduction of abscisic acid signals: key to the function of the versatile plant hormone ABA. Trends in Plant Science 12, 343–351. [DOI] [PubMed] [Google Scholar]
- Holdsworth MJ, Bentsink L, Soppe WJJ. 2008. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytologist 179, 33–54. [DOI] [PubMed] [Google Scholar]
- Huijser C, Kortstee A, Pego J, Weisbeek P, Wisman E, Smeekens S. 2000. The Arabidopsis SUCROSE UNCOUPLED-6 gene is identical to ABSCISIC ACID INSENSITIVE-4: involvement of abscisic acid in sugar responses. The Plant Journal 23, 577–585. [DOI] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K. 2001. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. The Plant Journal 27, 325–333. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: β-glucuronidase as a sensitive and versaltile gene fusion marker in higher plants. EMBO Journal 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J. 2007. Signals from chloroplasts converge to regulate nuclear gene expression. Science 316, 715–719. [PubMed] [Google Scholar]
- Kwak JM, Mäser P, Schroeder JI. 2008. The clickable guard cell, version II: interactive model of guard cell signal transduction mechanisms and pathways. The Arabidopsis Book 6, e0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laby RJ, Kincaid MS, Kim DG, Gibson SI. 2000. The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. The Plant Journal 23, 587–596. [DOI] [PubMed] [Google Scholar]
- Lang V, Palva ET. 1992. The expression of a rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.). Plant Molecular Biology 20, 951–962. [DOI] [PubMed] [Google Scholar]
- Lee SC, Lan W, Buchanan BB, Luan S. 2009. A protein kinase–phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proceedings of the National Academy of Sciences, USA 106, 21419–21424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister D. 2005. Genomics-based dissection of the cross-talk of chloroplasts with the nucleus and mitochondria in Arabidopsis. Gene 354, 110–116. [DOI] [PubMed] [Google Scholar]
- Léon-Kloosterziel KM, Gil MA, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JA, Koornneef M. 1996. Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. The Plant Journal 10, 655–661. [DOI] [PubMed] [Google Scholar]
- Liu Z, Butow RA. 2006. Mitochondrial retrograde signaling. Annual Review of Genetics 40, 159–185. [DOI] [PubMed] [Google Scholar]
- McDonald A, Vanlerberghe G. 2004. Branched mitochondrial electron transport in the Animalia: presence of alternative oxidase in several animal phyla. IUBMB Life 56, 333–341. [DOI] [PubMed] [Google Scholar]
- Melo AM, Bandeiras TM, Teixeira M. 2004. New insights into type II NAD(P)H:quinone oxidoreductases. Microbiology and Molecular Biology Reviews 68, 603–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Shulaev V, Mittler R. 2008. Reactive oxygen signaling and abiotic stress. Physiologia Plantarum 133, 481–489. [DOI] [PubMed] [Google Scholar]
- Miyazono KI, Miyakawa T, Sawano Y, et al. 2009. Structural basis of abscisic acid signalling. Nature 462, 609–614. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. 1962. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum 15, 473–497. [Google Scholar]
- Nakamura S, Lynch T, Finkelstein R. 2001. Physical interactions between ABA response loci of Arabidopsis. The Plant Journal 26, 627–635. [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenfuhr A. 2007. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. The Plant Journal 51, 1126–1136. [DOI] [PubMed] [Google Scholar]
- Ng S, De Clercq I, Van Aken O, Law SR, Ivanova A, Willems P, Giraud E, Van Breusegem F, Whelan J. 2014. Anterograde and retrograde regulation of nuclear genes encoding mitochondrial proteins during growth, development, and stress. Molecular Plant 7, 1075–1093. [DOI] [PubMed] [Google Scholar]
- Ng S, Ivanova A, Duncan O, et al. 2013. A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis . The Plant Cell 25, 3450–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Hitomi K, Arvai AS, Rambo RP, Hitomi C, Cutler SR, Schroeder JI, Getzoff ED. 2009. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science 326, 1373–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei ZM, Murata Y, Benning G, Thomine S, Klüsener B, Allen GJ, Grill E, Schroeder JI. 2000. Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406, 731–734. [DOI] [PubMed] [Google Scholar]
- Penfield S, Li Y, Gilday AD, Graham S, Graham IA. 2006. Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. The Plant Cell 18, 1887–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaresi P, Schneider A, Kleine T, Leister D. 2007. Interorganellar communication. Current Opinion in Plant Biology 10, 600–606. [DOI] [PubMed] [Google Scholar]
- Pontier D, Albrieux C, Joyard J, Lagrange T, Block MA. 2007. Knock-out of the magnesium protoporphyrin IX methyltransferase gene in Arabidopsis. Effects on chloroplast development and on chloroplast-to-nucleus signaling. Journal of Biological Chemistry 282, 2297–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov VN. 2003. Possible role of free oxidation processes in the regulation of reactive oxygen species production in plant mitochondria. Biochemical Society Transactions 31, 1316–1317. [DOI] [PubMed] [Google Scholar]
- Quesada V, Ponce MR, Micol JL. 2000. Genetic analysis of salt-tolerant mutants in Arabidopsis thaliana. Genetics 154, 421–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra AS, Gonugunta VK, Christmann A, Grill E. 2010. ABA perception and signaling. Trends in Plant Science 15, 395–401. [DOI] [PubMed] [Google Scholar]
- Rhoads DM, Subbaiah CC. 2007. Mitochondrial retrograde regulation in plants. Mitochondrion 7, 177–94. [DOI] [PubMed] [Google Scholar]
- Rock C. 2000. Pathways to abscisic acid-regulated gene expression. New Phytologist 148, 357–396. [DOI] [PubMed] [Google Scholar]
- Rolland F, Baena-González E, Sheen J. 2006. Sugar sensing and signaling in plants: conserved and novel mechanisms. Annual Review of Plant Biology 57, 675–709. [DOI] [PubMed] [Google Scholar]
- Sato A, Sato Y, Fukao Y, et al. 2009. Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochemical Journal 424, 439–448. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BNG, Palmer AE, Tsien RY. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nature Biotechnology 22, 1567–1572. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. 2000. Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Current Opinion in Plant Biology 3, 217–223. [PubMed] [Google Scholar]
- Shkolnik-Inbar D, Bar-Zvi D. 2010. ABI4 mediates abscisic acid and cytokinin inhibition of lateral root formation by reducing polar auxin transport in Arabidopsis . The Plant Cell 22, 3560–3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu K, Zhang H, Wang S, Chen M, Wu Y, Tang S, Liu C, Feng Y, Cao X, Xie Q. 2013. ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis. PLoS Genetics 9, e1003577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderman EM, Brocard IM, Lynch TJ, Finkelstein RR. 2000. Regulation and function of the Arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiology 124, 1752–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Feng P, Xu X, Guo H, Ma J, Chi W, Lin R, Lu C, Zhang L. 2011. A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus. Nature Communication 2, 477. [DOI] [PubMed] [Google Scholar]
- Talboys PJ, Zhang HM, Paul Knox J. 2011. ABA signalling modulates the detection of the LM6 arabinan cell wall epitope at the surface of Arabidopsis thaliana seedling root apices. New Phytologist 190, 618–626. [DOI] [PubMed] [Google Scholar]
- Teng S, Rognoni S, Bentsink L, Smeekens S. 2008. The Arabidopsis GSQ5/DOG1 Cvi allele is induced by the ABA-mediated sugar signalling pathway, and enhances sugar sensitivity by stimulating ABI4 expression. The Plant Journal 55, 372–381. [DOI] [PubMed] [Google Scholar]
- van Dongen JT, Gupta KJ, Ramirez-Aguilar SJ, Araujo WL, Nunes-Nesi A, Fernie AR. 2011. Regulation of respiration in plants: a role for alternative metabolic pathways. Journal of Plant Physiology 168, 1434–1443. [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. 2003. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. The Plant Journal 33, 949–956. [DOI] [PubMed] [Google Scholar]
- Woodson JD, Chory J. 2008. Coordination of gene expression between organellar and nuclear genomes. Nature Reviews Genetics 9, 383–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. 1994. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. The Plant Cell 6, 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Yu XC, Song LF, An CC. 2011. ABI4 activates DGAT1 expression in Arabidopsis seedlings during nitrogen deficiency. Plant Physiology 156, 873–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Li Q, Chen X, Liu J, Zhang Q, Liu Y, Liu K, Xu J. 2011. The Arabidopsis RETARDED ROOT GROWTH gene encodes a mitochondria-localized protein that is required for cell division in the root meristem. Plant Physiology 157, 1793–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.