Abstract
An important task in pharmacogenomics (PGx) studies is to identify genetic variants that may impact drug response. The success of many systematic and integrative computational approaches for PGx studies depends on the availability of accurate, comprehensive and machine understandable drug-gene relationship knowledge bases. Scientific literature is one of the most comprehensive knowledge sources for PGx-specific drug-gene relationships. However, the major barrier in accessing this information is that the knowledge is buried in a large amount of free text with limited machine understandability. Therefore there is a need to develop automatic approaches to extract structured PGx-specific drug-gene relationships from unstructured free text literature. In this study, we have developed a conditional relationship extraction approach to extract PGx-specific drug-gene pairs from 20 million MEDLINE abstracts using known drug- gene pairs as prior knowledge. We have demonstrated that the conditional drug- gene relationship extraction approach significantly improves the precision and F1 measure compared to the unconditioned approach (precision: 0.345 vs. 0.11; recall: 0.481 vs. 1.00; F1: 0.402 vs. 0.201). In this study, a method based on co-occurrence is used as the underlying relationship extraction method for its simplicity. It can be replaced by or combined with more advanced methods such as machine learning or natural language processing approaches to further improve the performance of the drug- gene relationship extraction from free text. Our method is not limited to extracting a drug-gene relationship; it can be generalized to extract other types of relationships when related background knowledge bases exist.
Keywords: pharmacogenomics, text mining, information extraction
1. Introduction
Automatically extracting pharmacogenomics (PGx) specific drug-gene pairs from free text is a challenging task. First, gene symbols are short and sometimes ambiguous. Ambiguous gene symbols can introduce false positives during the relationship extraction process. For instance, the symbol “CAD” can represent the gene symbol for “carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase”. “CAD” is also the symbol for a metabolizing gene for the pharmacological substance l-glutamine. In addition, , “CAD” is the abbreviation for “coronary artery disease” in MEDLINE, and co-occurs with many drugs indicated for cardiovascular diseases. When “CAD” co-occurs with a drug, the relationship can be a PGx-specific drug-gene relationship (e.g., l-glutamine-CAD and aspartate-CAD), or drug disease relationship (e.g., cyclosporine-CAD, azathioprine-CAD, and prednisone-CAD). Secondarily, the exact semantic relationships between a drug and its co-occurred genes can be a drug-gene target relationship (e.g., enalapril-ACE, and testosterone-AR), metabolizing (PGx) relationship (e.g., warfarin-CYP2C9, and warfarin-VKORC1) or others. For accurate PGx-specific drug-gene relationship extraction from free text, it is important to disambiguate gene symbols or semantically classify sentences before extracting the drug-gene relationship from sentences.
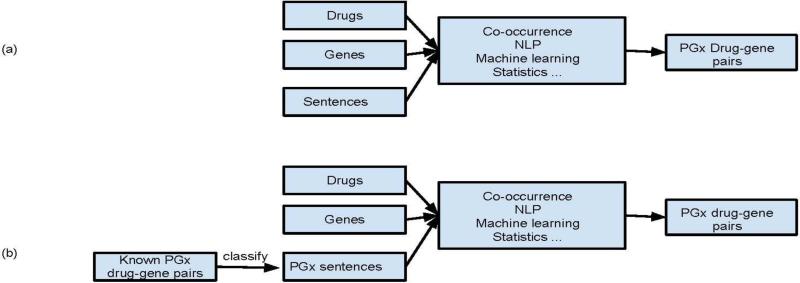
Standard drug- gene relationship extraction algorithms often use natural language processing (NLP), machine learning, co-occurrence statistics, or a combination of these methods to extract drug-gene pairs from sentences (Figure 1a). Unlike standard methods, our conditional methods only extract drug-gene pairs from sentences classified as PGx-related (Figure 1b). More specifically, we first automatically classify sentences as PGx-related or not based on the occurrences of known PGx-specific drug-gene pairs or PGx-specific genes in the sentences. Then, we extract additional drug-gene pairs from the sentences classified as PGx-related using standard relationship extraction approaches (co-occurrence methods used in this study). For example, the sentence “Substrates for CYP2C9 include fluoxetine, losartan, phenytoin, tolbutamide, torsemide, S-warfarin, and numerous NSAIDs” (PMID 09663807) contains a known PGx-specific drug-gene pair warfarin-CYP2C9 and is classified as PGx-related. Additional drug-gene pairs, such as fluoxetine-CYP2C9, losartan-CYP2C9, phenytoin-CYP2C9, tolbutamide-CYP2C9, and torsemide-CYP2C9, will be extracted from this sentence and determined to be PGx-specific.
Figure 1.
(a) Standard and (b) conditional PGx-specific drug-gene relationship extraction methods
2. Background
2.1 Importance of PGx-specific drug-gene relationship extraction from free text
Different patients respond differently to the same drug. Both genetic and non-genetic factors are involved in an individual's drug response, with genetics accounting for 20 to 95 percent of variability [1]. Pharmacogenomics (PGx) is the study of how human genetic variations affect an individual's response to drugs, with focuses on drug metabolism, absorption, distribution and excretion. The assumption underlying personalized medicine is that an individual's genotype profile can be used to predict effects (both efficacy and side effects) of drug treatment [2]. An understanding of the genetic variants associated with various drug responses is an essential step of personalized medicine [3, 4].
New PGx discovery depends on knowledge generated by previous research. PGx research is a knowledge-intensive field whose goal is to discover new drug-gene relationship knowledge and put it to clinical use for disease treatment. In this field, the research focus is rapidly shifting from studying an individual entity (e.g., one disease, drug, or gene) to entire networks of many different biological entities. Computational analysis of the knowledge represented in biomedical networks can uncover important new relationships, generate new testable hypotheses and provide new insight into biological systems [5, 6]. Recent investigations use systems biology methods to examine drug responses, by utilizing a network-based view of the genes involved in complex drug responses [7, 8].
The success of PGx studies largely depends on the availability of accurate, comprehensive and machine understandable drug-gene relationship knowledge. Adequate drug-gene relationship acquisition and integration are therefore becoming fundamentally important for these studies. The number of biomedical research publications, and therefore the underlying biomedical knowledge base, is rapidly expanding. The MEDLINE 2010 database contains over 20 million records (http://www.ncbi.nlm.nih.gov/pubmed). Scientific literature is the ultimate knowledge source for PGx studies. Clearly, with the current rate of growth in published biomedical research, it becomes increasingly likely that important knowledge connecting drugs, genes and diseases is being missed.
There is a need to develop new ways torsemide acquire structured drug-gene relationship knowledge from literature. Biocuration is the activity of transforming the information buried in human natural language into machine understandable knowledge by human curators reading scientific reports and extracting knowledge from published literature [9]. Biocuration has become an essential part of biological discovery and biomedical research. Substantial manual curation efforts have been used to extract PGx knowledge from literature. For example, The Pharmacogenomics Knowledge Base (PharmGKB) is an integrated resource about how variation in human genetics leads to variation in response to drugs [10]. Each of the curation projects involves a large number of curators, but their knowledge base is still limited by their ability to review all current related medical literature in a timely manner. To extract biomedical information, including drug-gene relationships, from published literature manually and to transform it into machine understandable knowledge is a difficult task, since biomedical terminologies and knowledge are huge, dynamic, diversified and complex. In addition, human curators are liable to error and subjective bias. Therefore, any manually curated terminology and knowledge base is deemed to be incomplete [11, 12]. Automated information extraction of structured knowledge from natural language text is crucial to biomedical researchers in their search for complete and up-to-date knowledge from published scientific reports. Compared to potential biocurator errors generated by heavy workload and/or bias, automated extraction will improve the quality and timeliness of the knowledge base.
2.2 Methods for biomedical relationship extraction from free text
Currently there are two major types of approaches for extracting biomedical relationships, including drug-gene relationships, from free text. The simplest and also the most widely used approaches are based on co-occurrence and use frequency-based statistics to rank extracted relationships. Li et al. used the co-occurrence of drug and disease names in MEDLINE abstracts to derive drug–disease relationships and to build a disease specific drug–protein network [13]. Yen et al. developed a co-occurrence approach based on an information retrieval principle to extract gene-disease relationships from text [14]. Blaschke et al. and Rosario et al. extracted semantic relationships among entities based on co-occurrence of two named entities and one semantic type from text [15, 16]. The assumption of co-occurrence methods is that, if two entities appear together, they may be related. Cooccurrence methods often have high recall. However, it is often true that two entities are mentioned together without being semantically related [17]. Therefore, an important shortcoming of these methods is that they introduce many false positives and suffer low precision. In addition, no semantics are provided for the associations. The second type of relationship extraction algorithm is based on NLP techniques to recognize entities and relationships using domain-specific lexicons and syntactic grammars. Syntactic templates and shallow parsing are often used in these NLP-based approaches. NLP methods have the advantage of being able to learn semantic types between entities. However, NLP methods sometimes suffer from low recall [18]. Rindflesch et al. extracted protein-binding relationships from text using NLP methods [19]. Leroy et al. have developed a shallow parser based on closed-class words to capture a variety of relationships from text [20]. Friedman et al. developed an NLP system called GENIE to extract molecular pathways from journal articles [21]. Rindflesh et al. developed a rule-based symbolic natural language processing system called SemRep to extract semantic predications from free text using the Unified Medical Language System (UMLS)] as the underlying knowledge base [22, 23]. However, due to the complexity of natural language, these NLP-based relationship extraction methods often target only specific semantic relations.
2.3 Prior studies of PGx-specific drug-gene relationship extraction from free text
Developing automatic approaches for extraction of PGx-specific drug-gene relationships from free text is a highly active research area. Both co-occurrence and NLP methods have been used. Chang et al. extracted drug-gene pairs from literature using the co-occurrence method and then used supervised machine learning algorithms to classify the extracted relationships into five subcategories such as genotype, clinical outcome, or pharmacokinetics [24]. The co-occurrence algorithm was able to achieve a recall of 78% when evaluated using one review article from the literature. However, the precision was not reported in Chang's study. Garten et al. developed Pharmspresso, a text-mining tool for extracting PGx concepts and relationships from full text [25]. The evaluation in Garten's study was done using manually curated PGx related articles, and the performance of extracting drug-gene relationships from other types of text (e.g., general MEDLINE articles) was not evaluated. Guided by the drug-gene relationships available in PharmGKB, Theobald et al. constructed n-way Bayesian networks based on conditional probability tables extracted from co-occurrence statistics over the entire MEDLINE corpus, and produced a broad-coverage analysis of the relationships between these biological entities [26]. The focus of Theobald's study was on building a Bayesian network. No evaluation was done in terms of the precision and recall of the extracted drug-gene relationships. Hansen et al. recently described an algorithm that uses existing knowledge to rank 12,460 genes in the human genome on the basis of their potential relevance to specific drugs [7]. Strictly speaking, this work did not focus on developing automatic methods for drug-gene relationship extraction from free text; it used existing biomedical knowledge about drug structures and indications in order to improve the precision of ranking PGx-specific genes for a given drug. Garten et al. extended Hansen's work by replacing the drug-gene relationships in PharmGKB with the drug-gene co-occurrence relationships extracted from manually curated PGx-specific full-text articles [27] and showed that a knowledge base derived from cooccurrence relationships mined from PGx specific literature performs as well as the curated knowledge base. Although the focus of these two studies was not on automatic drug-gene relationship extraction from free text, they demonstrate that prior knowledge is important for PGx-specific drug-gene relationship determination and that drug-gene co-occurrences based on highly relevant PGx literature have quality comparable to those curated by humans. Ahlers et al. developed an NLP system (Enhanced SemRep) to extract semantic relationships on pharmacogenomics in Medline citations [28]. The development of Enhanced SemRep depends on domain knowledge in the UMLS. Coulet et al. have developed NLP techniques to build a PGx ontology from 17 million MEDLINE abstracts by using the syntactic dependency structure of MEDLINE sentences to systematically extract common relationships and to map them to a common schema [29]. This method, based on detailed syntactic dependency analysis, achieved high precision. Recall was not reported.
2.4 Special challenges in PGx-specific drug-gene relationship extraction from free text and our approach
As we discussed in the Introduction section, extracting PGx-specific drug-gene relationships from free text is a challenging task. First, gene symbols are sometimes ambiguous. For instance, the symbol “CAD” represents the metabolizing gene “carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase”. “CAD” is also the abbreviation used for “coronary artery disease”. When a drug name co-occurs with the symbol “CAD”, the relationship can be a drug-gene relationship as shown in sentence (1), or a drug-disease relationship as shown in sentence (2).
-
(1)
“Mammalian DHOase (S-dihydroorotate amidohydrolase, EC 3.5.2.3) is part of a large multifunctional protein called CAD, which also has a carbamoyl-phosphate synthetase [carbon-dioxide: L-glutamine amido-ligase (ADP-forming, carbamate-phosphorylating), EC 6.3.5.5] and aspartate transcarbamoylase (carbamoyl-phosphate: L-aspartate carbamoyltransferase, EC 2.1.3.2) activities” (PMID 01967494).
-
(2)
“The possible role of viral infection in the genesis of CAD stimulated the review of 102 patients transplanted since the introduction of triple drug immunosuppression (cyclosporine, azathioprine and prednisone) to assess the importance of posttransplant cytomegalovirus infection in the development of CAD in the cardiac graft” (PMID 02547298)
For accurate drug- gene relationship extraction, it is necessary to disambiguate gene symbols or classify sentences based on the features or prior knowledge inherent in the sentences. In this study, we first automatically classify sentences as PGx-related or non-PGX-related, based on the occurrences of known PGx-specific drug-gene pairs in the sentences. Then, we extract additional drug-gene pairs from the sentences classified as PGx-related. For example, sentence (1) contains one known PGx-specific drug-gene pair CAD-L-aspartate, and is classified as PGx-related. An additional drug-gene pair CAD- L-glutamine is extracted from sentence (1) if this pair is unknown. On the other hand, sentence (2) does not contain any known PGx-specific drug-gene pairs and is classified as non-PGX-related. Thus, no drug- gene relationship extraction is performed for sentence (2).
Secondarily, even though a gene is drug-related, the exact semantic relationships between the gene and co-occurred drugs can be complicated. The semantic relationships between a drug and a gene can be a drug- gene target relationship as shown in sentences (3) and (4), or a drug-gene metabolizing relationship as shown in sentences (5) and (6).
-
(3)
“AR agonists and inhibitors included dihydrotestosterone (DHT), testosterone (T), and flutamide (Flu)” (PMID 17559882).
-
(4)
“ACE inhibitors: enalapril and captopril compared” (PMID 02998719).
-
(5)
“Pharmacogenetic testing of CYP2C9 and VKORC1 alleles for warfarin” (PMID 18281922).
-
(6)
“Substrates for CYP2C9 include fluoxetine, losartan, phenytoin, tolbutamide, torsemide, S-warfarin, and numerous NSAIDs” ” (PMID 09663807).
In this study, we first used known PGx-specific drug-gene relationship knowledge to classify sentences, and then extracted drug-gene pairs from PGx-related sentences. For example, neither sentence (3) nor (4) contains known PGx-specific drug-gene pairs, and our algorithm will classify these two sentences as non-PGx-related. Sentence (5) contains a known PGx-specific drug-gene pair warfarin-CYP2C9 and will be classified as PGx-related. An additional drug-gene pair warfarin-VKORC1 will be extracted from sentence (5) if this pair is unknown. Similarly, sentence (6) will be classified as PGx-related based on the occurrence of a known drug-gene pair warfarin-CYP2C9. Additional PGx-specific drug-gene pairs such as fluoxetine-CYP2C9, losartan-CYP2C9, phenytoin-CYP2C9, tolbutamide-CYP2C9, and torsemide-CYP2C9 will be extracted from this sentence.
The focus of our study is not to develop an independent drug-gene relationship extraction algorithm. Instead, our goal is to demonstrate that drug-gene relationship extraction algorithms can benefit from the addition of existing prior knowledge. The main contribution of this study is that we use prior knowledge, i.e., the known drug-gene pairs available in PharmGKB, to classify sentences as PGx-related or non-PGx-related before applying any drug-gene relationship extraction algorithms. Our assumption is that if a sentence contains one known PGx-specific drug-gene pair, it is very likely that this sentence is a PGx-related sentence. Then additional drug-gene pairs extracted from PGx-related sentences are likely to be PGx-specific drug-gene pairs. In this study we use co-occurrence based on a relationship extraction algorithm for its simplicity. However, the co-occurrence based method can be easily replaced by more advanced methods such as machine learning, NLP or rule-based approaches.
3. Data and methods
We have used 20 million MEDLINE abstracts (roughly 100 million sentences) published from 1965 to 2010 as the text corpus for our task of PGx-specific drug-gene relationship extraction. The drug-gene pairs available in PharmGKB were used as prior knowledge and as an evaluation gold standard. The PharmGKB database, downloaded in January of 2011, contained 10,898 drug-gene pairs, 918 drugs, and 2,388 genes. Annotation of the 20 million MEDLINE abstracts and 100 million sentences with drugs and gene terms was done using ThinTek's high performance tagger (http://www.thintek.com/). ThinTek's tagger is a cloud-based fast general-purpose biomedical named entity recognizer running on multiple processors in parallel. The tagger is based on simple exact string matching; no syntactic parsing was used. The tagger takes as input either user-provided biomedical dictionaries or the biomedical dictionaries that ThinTek provides. In this study, we provided the tagger the lists of drug and gene terms from PharmGKB. For each MEDLINE sentence or abstract, we collected the co-occurring drug-gene pairs.
There are total 918 drugs, 2,388 genes and 10,898 drug-gene pairs in PharmGKB. Only 2,943 (27%) out of the 10,898 pairs appear in MEDLINE sentences, 3,957 (36%) pairs appear in MEDLINE abstracts, and 1,014 pairs appear in MEDLINE abstracts, but not in sentences since a drug and gene can occur in the same abstract but be in different sentences (Table 1). This overall low percentage is partly due to the fact that the drug terms in PharmGKB drug-gene relationships include drug class names such as anticholinesterases, antihypertensives, antimalarials and beta blocking agents, and non-natural language drug names such as “antivirals for treatment of HIV infections, combinations”, “sulfonamides, urea derivatives”, “antiinflammatory and antirheumatic products, non-steroids”, “multivitamins, plain”, and “interferon alfa-2a, recombinant”. These terms are not commonly used in MEDLINE research articles. To evaluate MEDLINE-based PGx-specific drug-gene relationship extraction algorithms, we used the PharmGKB drug-gene pairs that appear in MEDLINE as the gold standard. We used the 2,943 drug-gene pairs that occur in MEDLINE sentences as the gold standard to evaluate drug-gene relationship extraction from sentences, and the 3,957 pairs that appear in MEDLINE abstracts were used as the gold standard for evaluating relationship extraction from MEDLINE abstracts.
Table 1.
PharmGKB drug-gene pair occurrence in MEDLINE
| Drugs | Genes | Drug-Gene Pairs | |
|---|---|---|---|
| PharmGKB | 918 | 2,388 | 10,898 |
| PharmGKB in MEDLINE Sentences | 585 | 718 | 2,943 |
| PharmGKB in MEDLINE Abstracts | 643 | 965 | 3,957 |
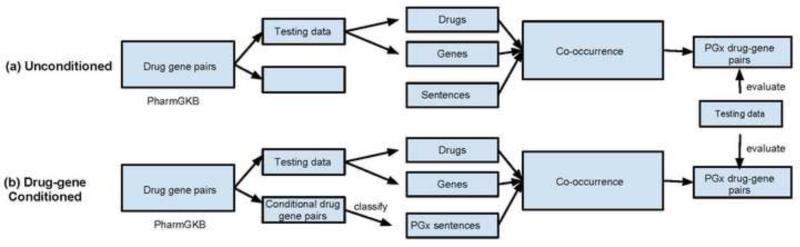
We developed two methods to extract PGx-specific drug-gene pairs from MEDLINE sentences. The algorithm “Unconditioned” is a simple co-occurrence based method in which drug-gene pairs are extracted from unclassified sentences (Figure 2a). The algorithm “Drug-Gene Conditioned” first classifies sentences based on the occurrence of known drug-gene pairs from PharmGKB before relationship extraction (Figure 2b). The drug-gene pairs in PharmGKB are split into two parts: one part is used as training data set in the algorithm “Drug-Gene Conditioned”, but not used in the algorithm “Unconditioned”), and the other part is used as testing data. The same testing data was used for both methods. The Student's t-test was performed for significance evaluation. A comparison evaluation was determined as significant when p value is less than 10E-7.
Figure 2.
(a) Unconditional co-occurrence method for drug-gene relationship extraction from unclassified sentences; (b) Conditional co-occurrence (“Drug-Gene Conditioned”) method for PGx-specific drug-gene relationship extraction from classified sentences containing known PGx-specific drug-gene pairs.
4. Results
4.1 Performance comparison of the unconditional and conditional methods for PGx-specific drug-gene relationship extraction from MEDLINE
We compared the precision, recall and F1 measure of the two co-occurrence based approaches (“Unconditioned” vs. “Drug-Gene Conditioned”) for PGx-specific drug-gene pair extraction from both MEDLINE sentences and abstracts. The “Unconditioned” method is a simple co-occurrence based method for drug-gene extraction from unclassified MEDLINE sentences or abstracts. The “Drug-Gene Conditioned” method uses the occurrence of known drug-gene pairs to classify sentences as PGx-related or not before relationship extraction. The drug-gene pairs from PharmGKB were randomly split into training data set and testing data set at five different training/testing ratios: 10%-90%, 20%-80%, 30%-70%, 40%-60%, and 50%-50%. One part (training set) was used as the prior knowledge for classifying sentences in the method “Drug-Gene Conditioned”, but not used in the method “Unconditioned”. The other part (testing set) was used for evaluation for both methods.
As shown in Table 2, the method “Drug-Gene Conditioned” consistently has significantly better precision and F1 values than the “Unconditioned” method. These improvements were significant for both sentence-based and abstract-based drug-gene pair extraction at all five different training/test ratios. For example, when 10% of PharmGKB drug-gene pairs (294 out of the 2,943 pairs that appear in MEDLINE sentences) were used as prior knowledge for sentence classification, and the remaining 90% were used as testing data, the method “Drug-Gene Conditioned” achieved a precision value of 38.8%, more than a 200% improvement over the Unconditioned method (precision: 11.7%). The recall is lower for the method “Drug-Gene Conditioned”, but the F1 score, which is the balanced measure of precision and recall, is significantly higher (37.7% vs. 20.9%). The precision and F1 improvements are consistent across document types (sentences or abstracts) and not sensitive to the amount of prior knowledge and testing data used. For the “Unconditioned” method, the precision slightly decreased from 11.7% to 9.1% when testing data was decreased from 90% to 50%. For the “Drug-Gene Conditioned” method, the precision also significantly decreased from 38.8% to 24.9% when less test data was used, while recall significantly increased from 36.6% to 65.1% and F1 did not change (37.1% to 36.1%). However, the “Drug-Gene Conditioned” method has significantly better precision and F1 than the “Unconditioned” method, even when the amount of testing data was decreased. In addition, both the “Unconditioned” and “Drug-Gene Conditioned” methods for drug-gene relationship extraction from sentences consistently has significantly better precision and F1 scores than from abstracts (p-values less than 10E-7).
Table 2.
Performance comparison of “Unconditioned” and “Drug-Gene Conditioned” PGx-specified drug-gene relationship extractions from MEDLINE sentences and abstracts
| PharmGKB | Unconditioned | Drug-Gene Conditioned | ||||||
|---|---|---|---|---|---|---|---|---|
| Sentence | Prior | Testing | Precision | Recall | F1 | Precision | Recall | F1 |
| 10% | 90% | 11.7 | 100.0 | 20.9 | 38.8 | 36.6 | 37.7 | |
| 20% | 80% | 11.1 | 100.0 | 20.1 | 34.5 | 48.1 | 40.2 | |
| 30% | 70% | 10.5 | 100.0 | 18.9 | 30.8 | 55.5 | 39.7 | |
| 40% | 60% | 10.0 | 100.0 | 18.1 | 28.4 | 60.2 | 38.5 | |
| 50% | 50% | 9.1 | 100.0 | 16.7 | 24.9 | 65.1 | 36.1 | |
| Abstract | 10% | 90% | 8.1 | 100.0 | 14.9 | 26.8 | 50.5 | 35.0 |
| 20% | 80% | 7.6 | 100.0 | 14.1 | 21.7 | 64.3 | 32.4 | |
| 30% | 70% | 7.2 | 100.0 | 13.4 | 18.4 | 70.8 | 29.2 | |
| 40% | 60% | 6.8 | 100.0 | 12.7 | 16.4 | 74.7 | 26.9 | |
| 50% | 50% | 6.4 | 100.0 | 12.0 | 14.8 | 78.8 | 24.9 | |
These results show that using known drug-gene relationship knowledge to guide drug-gene relationship extraction from free text can significantly reduce false positives while keeping high recall. The occurrence of a known PGx-specific drug-gene pair in a sentence or abstract can implicitly classify the sentence or abstract as PGx-related. Drug-gene relationship extraction from sentences or abstracts classified as PGx-related has significantly better precision and F1 scores than from unclassified sentences. In this study, we used the simple co-occurrence-based relationship extraction method, which can be easily replaced by more advanced methods such as NLP and machine learning methods. We expect that these advanced methods will benefit from extraction from PGx-related sentences.
3.3 Comparison of different conditional methods for PGx-specific drug-gene extraction
We have shown that the occurrence of known PGx-specific drug-gene pairs in a sentence or abstract can implicitly classify the sentence or abstract as PGx-related and therefore improve the overall precision and F1 of subsequent drug-gene relationship extraction (Table 2). We then tested whether or not the appearance of additional drug terms, gene terms or both (not necessarily known PGx-specific drug-gene pairs) can also implicitly classify a sentence or abstract as PGx-related. We have developed five different PGx-specific drug-gene extraction methods: (1) The “Unconditioned” method extracts drug-gene pairs from unclassified sentences, (2) The “Drug-Gene Conditioned” method extracts drug-gene pairs only from sentences containing known PGx-specific drug-gene pairs, (3) The “Drug Conditioned” method performs relationship extraction only from sentences that contain at least one additional drug term, (4) The “Gene Conditioned” method performs relationship extraction only from sentences that contain at least one additional gene, and (5) The “Drug × Gene Conditioned” method extracts drug-gene pairs only from sentences containing at least one additional drug term and one gene term. The method “Drug-Gene Conditioned” is conditioned on the appearance of actual drug-gene pairs from PharmGKB while the method “Drug × Gene Conditioned” is conditioned on the appearance of the cross product of genes and drugs from PharmGKB. We split the drug-gene pairs in PharmGKB into training data set and testing data set. We use the drug terms, gene terms, or drug-gene pairs in the training data set to implicitly classify sentences before relationship extraction for conditioned methods. For example, when we use a randomly selected 10% of PharmGKB drug-gene pairs as training data set, we obtain 294 known drug-gene pairs, 189 drugs, 158 genes, and total of 29,862 (189 × 158) drug-gene pairs. In the “Drug-Gene Conditioned” method, a sentence is classified as positive if it contains at least one of the 294 known drug-gene pairs. In the “Gene Conditioned” method, a sentence is classified as positive when it contains at least one of the 158 genes. In the “Drug × Gene Conditioned” method, a sentence is determined as positive only if contains at least one of the 29,862 (189 × 158) drug-gene pairs; the drug-gene pairs do not necessarily all appear in MEDLINE and are not necessarily valid PGx-specific drug-gene pairs. The 29,862 drug-gene pairs comprise a superset of all valid PGx-specific drug-gene pairs, including the 294 pairs derived from PharmGKB.
As shown in Table 3, the F1 scores for the “Drug-Gene Conditioned”, “Gene Conditioned” and “Drug × Gene Conditioned” methods are similar (37.1% vs. 30.3% vs. 32.4%) when 10% of PharmGKB data was used as training data set. When 50% of PharmGKB data was used as training data set, the F1 score for the “Drug-Gene Conditioned” method is significantly higher than those for the “Gene Conditioned” and “Drug × Gene Conditioned” methods (36.1% vs. 17.3% vs. 19.5%). The same trend holds true for drug-gene extraction from abstracts. All three methods (“Drug-Gene Conditioned”, “Gene Conditioned” and “Drug × Gene Conditioned”) have significantly better precision and F1 values than the “Unconditioned” and “Drug Conditioned” methods. The “Unconditioned” and “Drug Conditioned” methods have similar F1 scores for drug-gene relationship extraction from MEDLINE sentences (20.9% vs. 23.4% and 16.7 vs. 17.0) when 10% or 50% of PharmGKB data is used as training data set. In summary, drug-gene relationship extractions from text documents (sentences or abstracts) containing known PGx specific drug-gene pairs (“Drug-Gene Conditioned”), gene terms (“Gene Conditioned”) or gene terms plus drug terms (“Drug × Gene Conditioned”) have significantly better precision and F1 scores than methods based on unclassified documents (“Unconditioned”) or documents containing additional drug names from training dataset. The “Gene Conditioned” method performs as well as both the “Drug-Gene Conditioned” and “Drug × Gene Conditioned” methods, meaning that the appearance of a PGx-specific gene symbol in a sentence is sufficient to classify it as PGx-related. For example, if a sentence contains a known PGx-specific gene symbol “CYP2C9”, it is likely that any drug-gene pairs extracted from this sentence are PGx-specific. In addition, the occurrence of a PGx-specific symbol such as “CYP2C9” in a sentence can implicitly disambiguate other gene symbols in the same sentence. For example, if the symbol “CAD” appears in a sentence containing the symbol “CYP2C9”, it is highly possible that “CAD” represents the metabolizing gene “carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase”, instead of the short name for “coronary artery disease”. On the other hand, occurrence of additional drug names from known drug-gene pairs cannot classify a sentence as PGx-related or not. Drug terms can appear together with ambiguous gene symbols such as “CAD” in sentences specifying drug-disease relationships, or with gene symbols representing drug-gene target relationships instead of drug-gene metabolizing relationships.
Table 3.
Comparison of five different methods: “Unconditioned”, “Drug-Gene Conditioned”, “Drug Conditioned”, “Gene Conditioned”, and “Drug × Gene Conditioned”.
| Condition | Training | Testing | Precision | Recall | F1 | |
|---|---|---|---|---|---|---|
| Sentence | Unconditioned | 10% | 90% | 11.7 | 100.0 | 20.9 |
| 50% | 50% | 9.1 | 100.0 | 16.7 | ||
| Drug-Gene Conditioned | 10% | 90% | 38.8 | 36.6 | 37.7 | |
| 50% | 50% | 24.9 | 65.1 | 36.1 | ||
| Drug Conditioned | 10% | 90% | 13.6 | 83.5 | 23.4 | |
| 50% | 50% | 9.4 | 95.1 | 17.0 | ||
| Gene Conditioned | 10% | 90% | 19.0 | 75.3 | 30.3 | |
| 50% | 50% | 9.5 | 90.1 | 17.3 | ||
| Drug × Gene Conditioned | 10% | 90% | 21.6 | 64.9 | 32.4 | |
| 50% | 50% | 11.0 | 85.6 | 19.5 | ||
| Abstract | Unconditioned | 10% | 90% | 8.1 | 100.0 | 14.9 |
| 50% | 50% | 6.4 | 100.0 | 12.0 | ||
| Drug-Gene Conditioned | 10% | 90% | 26.8 | 50.5 | 35.0 | |
| 50% | 50% | 14.8 | 78.8 | 24.9 | ||
| Drug Conditioned | 10% | 90% | 9.0 | 94.4 | 16.5 | |
| 50% | 50% | 66.5 | 98.9 | 12.2 | ||
| Gene Conditioned | 10% | 90% | 12.3 | 81.0 | 21.4 | |
| 50% | 50% | 7.2 | 94.7 | 13.3 | ||
| Drug × Gene Conditioned | 10% | 90% | 13.6 | 77.5 | 23.2 | |
| 50% | 50% | 7.3 | 94.0 | 13.5 |
4.3 Detailed analysis of the mechanisms underlying the performance improvements
We use specific examples to show how the “Drug-Gene Conditioned” method can implicitly disambiguate gene symbols. We picked six known ambiguous gene symbols (“PC”, “GC”, “CP”, “BID”, “NP”, and “CAD”) based on our experience. For each of these ambiguous gene symbols, we extracted the full gene names from PharmGKB and non-gene-related names by searching MEDLINE. As shown in the Table 4, the “Drug-Gene Conditioned” method (column 5) significantly reduces false positive rates compared to the “Unconditioned” method (column 4). For example, total 506 different drug names co-occur with the symbol “PC” in MEDLINE. However, more than 99% of the cooccurrences are not PGx-specific gene-drug pairs based on manual examination. The “Drug-Gene Conditioned” method reduces the false positives from 506 to 33 for the symbol “PC’. Similar reductions are obtained for the other five ambiguous gene symbols.
Table 4.
Comparison of “Unconditioned” and “Drug-Gene Conditioned” methods in gene disambiguation
| Gene Symbol | Gene Name | Alternative Name (Non-gene Name) | False Positives (Unconditioned) | False Positives (Drug-Gene Conditioned) |
|---|---|---|---|---|
| PC | Pyruvate carboxylase | Phosphatidylcholine | 506 | 33 |
| GC | Group-specific component | Glucose consumption | 482 | 30 |
| CP | Ceruloplasmin (ferroxidase) | Cyclophosphamide | 562 | 134 |
| BID | BH3 interacting domain death agonist | Twice a day | 315 | 18 |
| NP | Nucleoside phosphorylase | Non-Preferring | 330 | 52 |
| CAD | Carbamoyl-phosphate synthetase | Coronary artery disease | 247 | 25 |
Next we show how the conditional method reduces false positive rates by examining some specific drug-gene pairs involving ambiguous gene symbols. For example, the drug dipyridamole co-occurred with the symbol “CAD” 334 times in MEDLINE; this pair never appears together with known drug-gene pairs from PharmGKB such as “warfarin-CYP2C9” (Table 5). The same is true for the ambiguous symbol “GC”. The symbol “GC” is highly ambiguous; it refers to “glucose consumption”, “glucose clearance”, “glucose cycling”, “glucose clamp”, and “glucocorticoids”. For example, “GC” represents glucocorticoids in the sentence, “In the liver, glucocorticoids (GC) normally regulates the glucose synthesis by acting on PEPCK” (pmid17182006). The drug choline and symbol “GC” appear together in MEDLINE sentences 1,126 times. However, the pair choline-GC has not appeared together with any of the known PGx drug-gene pairs. Since the “Drug-Gene Conditioned” method only extracts drug-gene relationships from sentences containing known PGx-specific drug-gene pairs, the pair dipyridamol-CAD or choline-GC will be extracted.
Table 5.
Comparison of “Unconditioned” and “Drug-Gene Conditioned” methods in drug-gene relationship extraction for ambiguous gene symbols “CAD” and “GC”.
| Symbol | False Positives (Unconditioned) | False Positives (Drug-Gene Conditioned) | |
|---|---|---|---|
| Dipyridamole | CAD | 334 | 0 |
| Aspirin | CAD | 213 | 0 |
| Atorvastatin | CAD | 87 | 0 |
| Choline | GC | 1126 | 0 |
| Ethanol | GC | 232 | 0 |
| Paclitaxel | GC | 156 | 0 |
5. Discussion
We have developed four conditional methods (“Drug-Gene Conditioned”, “Drug Conditioned”, “Gene Conditioned” and “Drug × Gene Conditioned”) for PGx-specific drug-gene relationship extraction from MEDLINE. We have compared conditional methods to an unconditioned method, which extracts drug-gene pairs from unclassified sentences. We have shown that the “Drug-Gene Conditioned”, “Gene Conditioned” and “Drug × Gene Conditioned” methods significantly improve precision and F1 measures, compared to both “Unconditioned” and “Drug Conditioned” methods in extracting PGx-specific drug-gene relationships from MEDLINE. In this study, we used a co-occurrence based method for drug-gene relationship extraction for its simplicity. However, this co-occurrence based method can be replaced by more advanced relationship extraction methods such as machine learning or NLP approaches.
Our approaches have a number of limitations. First, the precision of conditional methods largely depends on the precision of the underlying prior knowledge. If there are incorrect or ambiguous drug-gene pairs in the underlying knowledge base, the errors and ambiguities will propagate into the extracted drug-gene pairs. For example, in the sentence “At rest patients with CAD showed an increased myocardial extraction of glutamate, glucose and lactate and an augmented glutamine and alaninerelease compared with controls” (PMID 02707269), CAD-glutamate is a disease-drug pair. However, the same pair is also a PGx-specific drug-gene pair in PharmGKB. Because of this ambiguous pair, our algorithm will classify the above sentence as PGx-related. Three additional drug-CAD pairs, namely CAD-glucose, CAD-lactate and CAD-alanine, will be extracted as PGx-specific drug-gene pairs. In this situation, the “Gene Conditioned” method will be able to classify this sentence as non-PGx-related since it does not contain additional PGx-specific genes. The “Gene Conditioned” method depends on highly specific PGx-specific gene symbols. For example, the PGx-specific gene symbol “CYP2C9” or “VKORC1” can disambiguate a sentence as PGx-related or not. On the other hand, ambiguous gene symbols such as “PC” or “GC” cannot. Therefore, to further improve precision of conditional methods, we need to develop methods to identify PGx-specific gene symbols or drug-gene pairs and only use non-ambiguous gene symbols or pairs as the prior knowledge. An alternative approach would be to develop text classification methods to classify a sentence or abstract as PGx-related or non-PGx-related based on text features such as the text patterns that researchers use to describe PGx-specific drug-gene relationship.
The recall of the conditional methods depends on the coverage of the underlying prior knowledge. Consider the sentence, “Oxatomide was metabolized by CYP2D6-Val and CYP3A4, but not by CYP1A2, CYP2C9-Arg, CYP2C9-Cys or CYP2C19” (PMID 15133245). The drug oxatomide does not associate with any genes in PharmGKB. The “Drug-Gene Conditioned” algorithm will classify this sentence as non-PGx-related since it does not contain any known PGx-specific drug-gene pairs and will not extract the valid pairs (CYP2D6-oxatomide and CYP3A4-oxatomide) from the sentence. The “Unconditioned” method will extract these pairs, including false positives (CYP1A2-oxatomide, CYP2C9-oxatomide, and CYP2C19-oxatomide).
In this study, we used a co-occurrence based method as the underlying drug-gene relationship extraction for conditional methods. The limitation of any co-occurrence approaches is that they cannot differentiate pure co-occurrences from real semantic relationships. For example, in the sentence “In the present study, the possible role of genetic polymorphism of three drug-metabolizing enzymes, debrisoquine/sparteine hydroxylase (CYP2D6), glutathione S-transferase mu (GSTM1), and N-acetyltransferase (NAT2), as a putative genetic component of human longevity, was explored” (PMID 9654200), there are three drugs (debrisoquine, sparteine, and glutathione) and three drug metabolizing genes (CYP2D6, NAT2 and GSTM1). The gene symbols “CYP2D6”, “NAT2” and “GSTM1” are highly specific drug metabolizing genes. In addition, the drug-gene pairs “debrisoquine-CYP2D6” and“sparteine-CYP2D6” pairs are in PharmGKB. Our conditional methods (“Drug-Gene Conditioned”, “Gene Conditioned” and “Drug × Gene Conditioned”) will correctly classify this sentence as PGx-related. The co-occurrence based relationship extraction methods will extract nine (3 ×3) drug-gene relationships (the same as the “Unconditioned”), method instead of four valid PGx-specific drug-gene pairs (debrisoquine-CYP2D6, sparteine-CYP2D6, glutathione-GSTM1, and glutathione-NAT2). In this situation, more advanced methods such as machine learning, NLP approaches, or human curation will be still needed.
In summary, the performance of the conditional methods for PGx-specific drug-gene relationship extraction depends on the quality of the prior knowledge and the underlying relationship extraction methods. Imprecision and ambiguity of prior knowledge decrease the overall precision of conditional methods. The coverage of the prior knowledge can also affect the recall. In addition, the underlying relationship extraction algorithms (co-occurrence, machine learning, NLP, or rule-based approaches) will affect the performance of the corresponding conditional methods. It will be interesting to investigate how conditional prior knowledge can affect other relationship extraction methods in PGx-specific drug-gene relationship extraction. It may be possible that the conditional methods only have a big impact on methods with low precision such as the co-occurrence method, not on those with high precision such as NLP or rule-based approaches.
6. Conclusions
We have developed knowledge-driven conditional relationship extraction approaches to extract PGx-specific drug-gene pairs from 20 million MEDLINE abstracts. We have used the drug-gene pairs available in PharmGKB as prior knowledge to implicitly classify sentences before applying relationship extraction methods. The conditional methods significantly improve both the precision and F1 measures compared to the traditional (unconditioned) method (precision: 0.345 vs. 0.11; recall: 0.481 vs. 1.00; F1: 0.402 vs. 0.201). Our method is not limited to PGx-specific drug-gene relationship extraction, and it can be generalized to extract other types of biomedical relationships from free text, provided that high quality prior background knowledge exists for a give task. In the future, we will develop automatic approaches to identify ambiguous gene and drug-gene pairs from PharmGKB, to further improve the precision of prior knowledge and of the conditional methods. In addition, we will develop conditional PGx-specific drug-gene extraction methods based on more advanced relationship extraction methods such as NLP approaches.
➢ Knowledge of drug-gene relationships is important for pharmacogenomics (PGx) studies.
➢ Automatic PGx-specific drug-gene relationship extraction from free text is difficult.
➢ We develop a conditional relationship extraction method using prior knowledge.
➢ We compare the conditional method to method using no prior knowledge.
➢ Conditional method has significant better precision and F1 measure than unconditional method.
Acknowledgements
Both Rong Xu and QuanQiu Wang have conceived the idea, designed and implemented the algorithms. Xu has written the paper.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Evans WE, McLeod HL. Pharmacogenomics - drug disposition, drug targets, and side effects. N Engl. J. Med. 2003;348:538. doi: 10.1056/NEJMra020526. [DOI] [PubMed] [Google Scholar]
- 2.Weiss ST, McLeod HL, Flockhart DA, Dolan ME, Benowitz NL, Johnson RA, Ratain MJ, Giacomini KM. Creating and evaluating genetic tests predictive of drug response. Nat. Rev. Drug Discov. 2008;7:568–74. doi: 10.1038/nrd2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swen JJ, Huizinga TW, Gelderblom H, deVries SG, Assendelft WJ, Kirchheinen J, Guchalaar HJ. Translating Pharmacogenomics: Challenges on the road to the clinic. PLoS Med. 2007;4:e209. doi: 10.1371/journal.pmed.0040209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis JC, Furstenthal L, Desai AA, Norris T, Sutaria S, Fleming E, Ma P. The microeconomics of personalized medicine: today's challenge and tomorrow's promise. Nat. Rev. Drug Discov. 2009;8:279–286. doi: 10.1038/nrd2825. [DOI] [PubMed] [Google Scholar]
- 5.Ma'ayan A, Jenkins SL, Goldfarb J, Iyengar R. Network analysis of FDA approved drugs and their targets. Mt. Sinai J. Med. 2007;74:27–32. doi: 10.1002/msj.20002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yildirim MA, Goh KI, Cusick ME, Barabasi AL, Vidal M. Drug-target network. Nat. Biotechnol. 2007;25:1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 7.Hansen NT, Brunak S, Altman RB. Generating genome-scale candidate gene lists for Pharmacogenomics. Clin. Pharmacol. Ther. 2009;86:183–189. doi: 10.1038/clpt.2009.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tatonetti NP, Liu T, Altman RB. Predicting drug side-effects by chemical systems biology. Genome Biol. 2009;10:238. doi: 10.1186/gb-2009-10-9-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howe D, Costanzo M, Fey P, Gojobori T, Hannick L, Hide W, Hill DP, Kania R, Schaffer M, St. Pierre S, Twigger S, White O, Rhee SY. Big data: The future of biocuration. Nature. 2008;455:47–50. doi: 10.1038/455047a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein TE, Chang JT, Cho MK, Easton KL, Fergerson R, Hewett M, Lin Z, Liu Y, Liu S, Oliver DE, Rubin DL, Shafa F, Stuart JM, Altman RB. Integrating genotype and phenotype information: an overview of the PharmGKB project, Pharmacogenetics Research Network and Knowledge Base. Pharmacogenomics J. 2001;1:167–170. doi: 10.1038/sj.tpj.6500035. [DOI] [PubMed] [Google Scholar]
- 11.Hahn U, Wermter J, Blasczyk R, Horn PA. Text mining: powering the database revolution. Nature. 2007;448:130. doi: 10.1038/448130b. [DOI] [PubMed] [Google Scholar]
- 12.Baumgartner WA, Jr., Cohen KB, Fox L, Acquaah-Mensah G, Hunter L. Manual annotation is not sufficient for curating genomic databases. Bioinformatics. 2007;23:i41–i48. doi: 10.1093/bioinformatics/btm229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, Zhu X, Chen JY. Building disease-specific drug–protein connectivity maps from molecular interaction networks and PubMed abstracts. PLoS Comput. Biol. 2009;5:e1000450. doi: 10.1371/journal.pcbi.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yen YT, Chen B, Chiu HW, Lee YC, Li YC, Hsu CY. Developing an NLP and IR-based algorithm for analyzing gene-disease relationships. Methods Inf. Med. 2006;45:321–329. [PubMed] [Google Scholar]
- 15.Blaschke C, Andrade MA, Ouzounis C, Valencia A. Automatic extraction of biological information from scientific text: protein–protein interactions. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1999:60–67. [PubMed] [Google Scholar]
- 16.Rosario B, Hearst MA. Classifying semantic relations in bioscience texts. ACL. 2004:430–437. [Google Scholar]
- 17.Ramani AK, Bunescu RC, Mooney RJ, Marcotte EM. Consolidating the set of known human protein-protein interactions in preparation for large-scale mapping of the human interactome. Genome Biol. 2005;6:R40. doi: 10.1186/gb-2005-6-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fundel K, Kuffner R, Zimmer R. RelEx – relation extraction using dependency parse trees. Bioinformatics. 2007;23:365–371. doi: 10.1093/bioinformatics/btl616. [DOI] [PubMed] [Google Scholar]
- 19.Rindflesch TC, Rajan JV, Hunter L. Extracting molecular binding relationships from biomedical text. Proceedings of the ANLP-NAACL. 2000:188–95. [Google Scholar]
- 20.Leroy G, Chen H, Martinez JD. A shallow parser based on closed-class words to capture relations in biomedical text. J Biomed. Inform. 2003;36:145–158. doi: 10.1016/s1532-0464(03)00039-x. [DOI] [PubMed] [Google Scholar]
- 21.Friedman C, Kra P, Yu H, Krauthammer M, Rzhetsky A. GENIES: a natural-language processing system for the extraction of molecular pathways from journal articles. Bioinformatics. 2001;17(Suppl. 1):S74–S82. doi: 10.1093/bioinformatics/17.suppl_1.s74. [DOI] [PubMed] [Google Scholar]
- 22.Rindflesch TC, Fiszman M. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. J Biomed. Inform. 2003;36:462–477. doi: 10.1016/j.jbi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 23.Rindflesch TC, Fiszman M, Libbus B. Semantic interpretation for the biomedical research literature. In: Chen, Fuller, Hersh, Friedman, editors. Medical informatics: Knowledge management and data mining in biomedicine. Springer; 2005. pp. 399–422. [Google Scholar]
- 24.Chang JT, Altman RB. Extracting and characterizing gene-drug relationships from the literature. Pharmacogenetics. 2004;14:577–586. doi: 10.1097/00008571-200409000-00002. [DOI] [PubMed] [Google Scholar]
- 25.Garten Y, Altman RB. Pharmspresso: a text mining tool for extraction of pharmacogenomic concepts and relationships from full text. BMC Bioinformatics. 2009;10(Suppl. 2):S6. doi: 10.1186/1471-2105-10-S2-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Theobald M, Shah NH, Shrager J. Extraction of conditional probabilities of the relationships between drugs, diseases, and genes from PubMed guided by relationships in PharmGKB, Summit on Translat. Bioinforma. 2009 Mar;1:124–128. 2009. [PMC free article] [PubMed] [Google Scholar]
- 27.Garten Y, Tatonetti NP, Altman RB. Improving the prediction of pharmacogenes using text-derived drug-gene relationships. Pac. Symp. Biocomput. 2010:305–314. doi: 10.1142/9789814295291_0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahlers CB, Fiszman M, Demner-Fushman D, Lang FM, Findflesch TC. Extracting semantic predications from MEDLINE citations for pharmacogenomics. Pac. Symp. Biocomput. 2007:209–20. [PubMed] [Google Scholar]
- 29.Coulet A, Shah NH, Garten Y, Musen M, Altman RB. Using text to build semantic networks for pharmacogenomics. J. Biomed. Inform. 2010;43:1009–1019. doi: 10.1016/j.jbi.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]