The Musashi-2 (MSI2) gene, at chromosome band 17q22, is recurrently associated with regulating normal hematopoiesis and promoting leukemia progression [1-2]. MSI2 is overexpressed in human myeloid leukemia cell lines and its knockdown leads to decreased proliferation and increased apoptosis [2]. High expression of MSI2 protein may predict unfavorable outcome in acute myeloid leukemia (AML) and adult B-cell acute lymphoblastic leukemia [2-4]. MSI2 rearrangement was first reported in two patients who presented with chronic myelogenous leukemia and cryptic balanced translocations involving chromosomes 7 and 17 were discovered during disease progression [5]. The translocation involved identical breakpoints in chromosome 17q22 within the MSI2 gene in both cases [5]. One of these translocations resulted in a MSI2-HOXA9 chimeric fusion gene [5]. MSI2 rearrangement has also been reported in patients with myeloid leukemia and a 3;17 translocation leading to EVI1 gene overexpression [6]. Here, we present a case of de novo AML exhibiting an unbalanced 10;17 translocation leading to a TTC40-MSI2 fusion gene. To our knowledge this is the first report of this specific translocation.
A previously healthy 66-year-old woman presented with fatigue and was found to have abnormal blood counts: hemoglobin 11.1 g/dL, white blood cells 58 × 10k/ul with 80% blasts, and platelets 87 × 10k/ul. Her peripheral smear showed marked leukocytosis with increased blast forms that were intermediate in size and had round nuclei, dispersed chromatin, indistinct nucleoli, and scant cytoplasm with rare azurophilic granules. No Auer rods were identified. Red blood cells and platelets were unremarkable. The bone marrow was hypercellular (95% cellularity), with a predominance (90-95% of cellularity) of intermediate-sized blasts, similar to those seen in the blood. Maturing myeloid and erythroid cells and megakaryocytes were rare. Immunophenotyping by flow cytometry identified a population of abnormal myeloid blasts (96.9% of total cells) expressing CD117 (partial), CD13 (increased), CD15 (partial), CD33 (increased), CD38 (decreased), CD45, CD56 (partial), CD64 (dim/equivocal), and CD71, but not CD34, HLA-DR, or CD14. Fluorescence in situ hybridization (FISH) using probe sets for the LSI PML-RARA and RARA break-apart (Abbott Molecular) did not show a 15;17 translocation or RARA gene rearrangement. A diagnosis of AML without maturation (or with minimal differentiation) was made. The patient underwent three cycles of induction chemotherapy with two chemo drugs and achieved complete remission. Then, she had one cycle of consolidation chemotherapy with high dose cytarabine (1 g/m2, 6 days). 9 months after complete remission, she had relapsed. She was treated with 5 days of intravenous mitoxantrone (8 mg/m2), etoposide (100 mg/m2) and cytarabine (1 g/m2) along with 4 doses of MDX-1338 (1000 mg). She expired shortly, 14 months after the initial diagnosis.
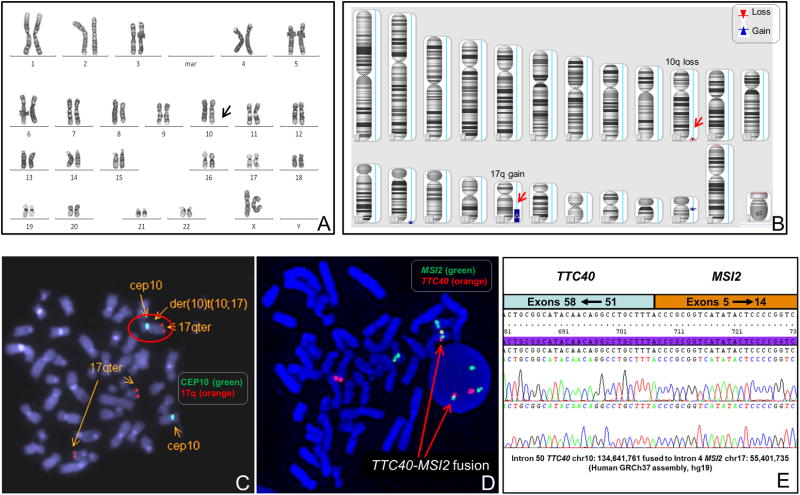
Cytogenetic analysis was performed on bone marrow cells at the time of diagnosis. All 20 metaphase cells analyzed had an abnormal chromosome 10 with additional material of unknown origin on the distal long arm of chromosome 10, 46,XX,add(10)(q26)[20] (Figure 1A). Whole genome CNV/SNP chromosomal microarray assay (CMA) with a total of 2.6 million copy number variant and SNP markers (Affymetrix CytoscanHD array) was performed on the DNA extracted from bone marrow cells and revealed a 25.6 Mb gain of the distal long arm of chromosome 17 (17q22-qter) and a 783 Kb loss of the distal long arm of chromosome 10 (10q26.3), arr[hg19]10q26.3(134644013-135427143)×1,17q22q25.3(55401945-81041938)×3 (Figure 1B). Based on the CMA findings the breakpoint of the translocation on chromosome 10 was mapped within the tetratricopeptide repeat domain 40 (TTC40) gene at band 10q26.3 and the breakpoint on chromosome 17 was within the MSI2 gene at band 17q22. The TTC40 gene contains 58 exons spanning 134 Kb, and is a novel gene with high GC content.
Figure 1.
Cytogenetic and molecular analyses of unbalanced 10;17 translocation with TTC40-MSI2 fusion gene. (A) Representative metaphase showing a derivative chromosome 10 with additional material of unknown origin on 10q. (B) Chromosomal microarray assay revealing a small deletion of the 10q26.3 region (783 Kb) and a large gain of the 17q22-q25.3 region (25.6 Mb). (C) Metaphase FISH assay confirming the presence of chromosome 17 material on the derivative chromosome 10 product of an unbalanced 10;17 translocation. (D) Dual-color FISH using RP11-166N4 covering the MSI2 gene (labeled in spectrum green) and RP11-384O10/RP11-702C24 covering the TTC40 gene (labeled in spectrum orange) showing the fusion of the MSI2 gene and the TTC40 gene (yellow color) on the derivative chromosome 10 in a metaphase and an interphase cell. (E) Partial chimeric gene sequence. TTC40 intron 50 sequence fused to within MSI2 intron 4 with an “A” base that is derived from either the TTC40 intron 50 or the MSI2 intron 4 sequence.
FISH analysis was performed using BAC clones covering the MSI2 gene on 17q22 and the TTC40 gene on 10q26.3, as well as chromosome 10 centromere probe and the 17q subtelomere probe D17S928 mapping to 17q25 (Abbott Molecular). Analysis of abnormal metaphase cells confirmed the presence of chromosome 17 material on the derivative chromosome 10 (Figure 1C) and revealed the presence of the TTC40-MSI2 gene fusion on interphase and metaphase cells (Figure 1D).Sequencing analysis following long-range polymerase chain reaction using the primer pair TTC40-1F:5′-GACGGGACTGGACGCTGCAC-3′ and MSI2-1R: 5′-GGTGCATGTGGCCCAGTCAGT-3′ revealed that the MSI2 gene was fused to the TTC40 gene with breaks in introns 4 and 50, respectively (Figure 1E). These findings confirm that the derivative chromosome 10 is the product of an unbalanced translocation between chromosome bands 10q26.3 and 17q22, der(10)t(10;17)(q26.3;q22), leading to a TTC40-MSI2 fusion gene.
The unbalanced 10;17 translocation also resulted in a small terminal deletion of the long arm of chromosome 10 distal to 10q26.3 and a large duplication of the long arm of chromosome 17 including the 17q22-q25.3 region. The deleted 10q26.3 region contains 29 genes (10 OMIM genes) (http://genome.ucsc.edu). This 10q26.3 deletion has also been reported in normal individuals without phenotype, suggesting that the region might be a population variant /polymorphic and does not contain haploinsufficient genes (http://dgv.tcag.ca). However, the duplicated 17q22-q25.3 region contains ∼ 480 genes (235 OMIM genes and known disease genes) (http://genome.ucsc.edu). Among them, at least 20 genes are involved in solid tumors, including ASPSCR1, AXIN2, BCAS3, BRIP1, CLTC, DDX5, ENPP7, MIR21, PPMID,PRKAR1A, RAC3, SLC9A3R1, SLC16A3, SOCS3, SPSF1, ST6GALNAC1, TBX2, TIMP2, USP32, VMP1 (http://atlasgeneticsoncology.org). A number of other genes are thought to play a role in the pathogenesis of hematologic malignancies. The RNF213 and CLTC genes are engaged in anaplastic large cell lymphoma [7], the SEPT9 gene (a fusion partner gene of MLL) is involved in de novo and treatment related leukemia [8], and the GRB2 gene is implicated in the pathogenesis of Philadelphia chromosome positive leukemia [9].
This case illustrates the importance of comprehensive morphologic, cytogenetic, chromosomal microarray and sequence analysis in characterizing a de novo AML with unbalanced 10;17 translocation, leading to discovery of a novel TTC40-MSI2 fusion gene. This unbalanced translocation der(10)t(10;17)(q26.3;q22) in our case is the first to be reported, and further studies are warranted to elucidate the roles of the involved genes in leukemogenesis. Collecting and reporting data on rare chromosomal abnormalities in AML will add important information regarding disease pathogenesis and prognosis, and may eventually translate to targeted therapies.
Footnotes
Conflict of interest statement: The authors declare no conflict of interest.
References
- 1.Ito T, Kwon H, Zimdahl B, et al. Regulation of myeloid leukemia by the cell fate determinant Musashi. Nat. 2010;466:765–768. doi: 10.1038/nature09171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kharas M, Lengner C, Al-Shahrour F, et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat Med. 2010;16:903–908. doi: 10.1038/nm.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mu Q, Wang Y, Chen B, et al. High expression of Musashi-2 indicates poor prognosis in adult B-cell acute lymphoblastic leukemia. Leuk Res. 2013;37:922–927. doi: 10.1016/j.leukres.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 4.Byers R, Currie T, Tholouli E, et al. MSI2 protein expression predicts unfavorable outcome in acute myeloid leukemia. Blood. 2011;118:2857–2867. doi: 10.1182/blood-2011-04-346767. [DOI] [PubMed] [Google Scholar]
- 5.Barbouti A, Hoglund M, Johansson B, et al. A novel gene, MSI2, encoding a putative RNA-binding protein is recurrently rearranged at disease progression of chronic myeloid leukemia and forms a fusion gene with HOXA9 as a result of the cryptic t(7;17)(p15;q23) Cancer Res. 2003;63:1202–1206. [PubMed] [Google Scholar]
- 6.Weer A, Speleman F, Cauwelier B, et al. EVI1 overexpression in t(3;17) positive myeloid malignancies results from juxtaposition of EVI1 to the MSI2 locus at 17q22. Haematologica. 2008;93:1903–1907. doi: 10.3324/haematol.13192. [DOI] [PubMed] [Google Scholar]
- 7.Cools J, Wlodarska I, Somers R, et al. Identification of novel fusion partners of ALK, the anaplastic lymphoma kinase, in anaplastic large-cell lymphoma and inflammatory myofibroblastic tumor. Genes, chromosomes & cancer. 2002;34:354–362. doi: 10.1002/gcc.10033. [DOI] [PubMed] [Google Scholar]
- 8.Hall PA, Russell SE. The pathobiology of the septin gene family. J Pathol. 2004;204:489–505. doi: 10.1002/path.1654. [DOI] [PubMed] [Google Scholar]
- 9.Adams S, Aydin I, Celebi J. GAB2 - a scaffolding protein in cancer. Mol Cancer Res. 2012;10:1265–1270. doi: 10.1158/1541-7786.MCR-12-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]