Abstract
The trillions of bacteria that naturally reside in the human gut collectively constitute the complex system known the gut microbiome, a vital player for the host’s homeostasis and health. However, there is mounting evidence that dysbiosis, a state of pathological imbalance in the gut microbiome is present in many disease states. In this review, we present recent insights concerning the gut microbiome’s contribution to the development of colorectal adenomas and the subsequent progression to colorectal cancer (CRC). In the United States alone, CRC is the second leading cause of cancer deaths. As a result, there is a high interest in identifying risk factors for adenomas, which are intermediate precursors to CRC. Recent research on CRC and the microbiome suggest that modulation of the gut bacterial composition and structure may be useful in preventing adenomas and CRC. We highlight the known risk factors for colorectal adenomas and the potential mechanisms by which microbial dysbiosis may contribute to the etiology of CRC. We also underscore novel findings from recent studies on the gut microbiota and colorectal adenomas along with current knowledge gaps. Understanding the microbiome may provide promising new directions towards novel diagnostic tools, biomarkers, and therapeutic interventions for CRC.
Keywords: colorectal adenoma, colorectal cancer (CRC), gut microbiota, bacterial dysbiosis, inflammation, microbiome
Introduction
Globally in 2012 alone, colorectal cancer (CRC) accounted for approximately 694,000 deaths (approx. 8.5% of total cancer deaths) and 1.36 million new cases [1]. In the United States, CRC is the third most commonly diagnosed cancer and the second leading cause of cancer deaths and will account for about 136,830 new cases and 50,310 deaths in 2014 [2]. The annual economic burden of CRC in 2010 was approx. 14.1 billion dollars and this is expected to increase to about 17.4 billion dollars in 2020 [3]. Mortality from CRC is more broadly associated with metastatic disease; therefore, early detection and screening are vital.
CRC occurs in a stepwise fashion beginning with abnormal cell proliferation, and aberrant crypt foci leading to the development of adenomatous polyps, which are widely considered to be CRC precursors [4]. Colonic polyps are mostly classified on the basis of their properties to progress to malignancy (hyperplastic or adenomatous) as well as their structure including types (sessile, pedunculated, and flat), shape (tubular, villous, serrated) and size (small 1–5 mm, medium 5–10 mm, and large ≥ 10mm) [5]. Hyperplastic polyps are usually small, located in the rectum and sigmoid colon, and are generally thought to have no malignant potential. However, subsets of serrated hyperplastic polyps are associated with a risk of CRC [6]. Adenomatous polyps or adenomas account for approximately 70% of colon polyps and have the potential to progress to CRC over time if not screened and removed by colonoscopy or sigmoidoscopy [7].
In clinical settings, the number, and structure (shape, size, and type) of adenomatous polyps are vital indicators when predicting which patients are more prone to develop CRC based on polyp morphology. Thus, adenomas are important intermediates in colorectal carcinogenesis and identifying adenoma risk factors is important in preventing CRC. Although the specific etiologic agents responsible for adenomas and CRC are unknown, several genetic and environmental risk factors have been implicated.
Risk factors for colorectal adenomas and CRC
The role of genetic alterations in the progression of adenomas to CRC was initially described by Fearon and Vogelstein [2]. Genetic mutations in oncogenes (KRAS), tumor suppressor genes such as adenomatous polyposis coli (APC), CTNNB1 and p53, [2, 8–13], and alterations in pathways that revolve around chromosomal and microsatellite instability (MSI), mismatch repair (MMR) [14, 15], and CpG island methylation (CIMP) [16, 17] are key players in colorectal adenomas and CRC [18].
In addition, findings from genome-wide association studies (GWAS) support a polygenic model of CRC in which several common low penetrance susceptibility genes such genetic variants in vitamin D [19], cyclin D1, and Smad7 [20] contribute to increased risk of adenoma and CRC [21, 22]. Family history and age are also considered to be important CRC predictors as they have been associated with higher risk of adenomas and CRC. Studies suggest that genetic predisposition and somatic alterations in combination with environmental factors are responsible for CRC as a complex disease [20].
The most common environmental factors implicated in association with colorectal adenomas and CRC are lifestyle and diet. Several studies demonstrate that unhealthy diets such as those high in fat, alcohol, red meat, and low in fiber are associated with increased risk of adenomas and CRC [23]. Moreover, smoking, obesity, low physical activity [24, 25], sex (increased risk for males), ethnicity (predominantly in Non-Hispanic Black population [19, 26], and lifestyle (lack of physical exercise) all contribute to the development of CRC. Adopting a healthy lifestyle, incorporating regular exercise and a diet high in fruits, vegetables, and high-fiber foods could potentially reduce the risk of CRC. However, not all the results from dietary studies are consistent. A pooled study of fiber and CRC reported inconsistent findings in which about half of the studies showed a protective effect of fiber while the others did not [27]. These discrepancies could relate to the influence of the gut microbiota on fiber. The gut microbiota was not assessed in these studies.
Gut microbiota
The human colon hosts a very diverse and complex microbial community comprising an estimated 100 trillion bacteria of more than 1,000 heterogeneous species (harboring approx. 4 million genes) along with viruses, archaea, and fungi. The collective bacterial genome referred to as the gut microbiome, harbors 150-fold more genes than the human genome [28, 29]. Bacterial cells of the gut exceed the total number of host cells in the human body by 10-fold [30]. These bacteria play key roles in modulating host metabolism such as absorption of indigestible carbohydrates, production of vitamins B and K, and promotion, maturation and development of innate and cell-mediated immunity and also help to maintain intestinal barrier function and appropriate immune response against pathogens [31, 32]. Under normal physiological conditions, the gut bacteria and the host co-exist in a state of homeostasis. However, the gut microbiota is increasingly associated with a variety of diseases including obesity, inflammatory bowel diseases, adenomas, and CRC [12, 33, 34].
Gut microbiota, adenomas, and CRC
Several studies implicate microbial dysbiosis, a pathological imbalance in the microbial community, in the etiology of colorectal adenomas and CRC. This is summarized in Fig. 1A. Shen et al. used molecular fingerprinting and clone sequencing methods to characterize the adherent bacterial composition in normal rectal mucosal biopsies and observed that the gut bacterial composition of subjects with adenomas differed significantly from that of control subjects without adenomas [35]. They reported a higher proportion of Proteobacteria and lower abundance of Bacteroidetes in cases than in controls. These initial findings were confirmed in a follow-up study that used 16S rRNA gene amplicon 454 pyrosequencing methods to characterize the gut bacteria. Sanapareddy et al. [36] found an overabundance of potential pathogens, Pseudomonas, Helicobacter, Acinetobacter and other genera belonging to the phylum Proteobacteria in rectal mucosal biopsies of adenoma cases compared to non-adenoma controls [36]. Brim et al. compared the fecal microbiota from a small sample group of African American patients with or without colorectal adenomas and noted a trend of altered microbial changes between adenoma patients and healthy controls [37]. In experimental models of CRC, Wei et al. observed dysbiosis associated with an increased abundance of Ruminococcus obeum, and Allobaculum spp. in precancerous lesions [38]. These findings suggest that changes in the gut adherent microbial community composition may play a role in the development of adenomas.
Figure 1.

A. Schematic diagram of colonic microbiota and adenomas progression to CRC.
Shifts in the balance of host-microbial symbiotic relationships derail the state of homeostasis (normal physiology) in the human gut. Dysbiosis, an imbalance of microbial population dynamics, is characterized by decreased beneficial commensals/symbionts, overexpression of pathogenic microbiota such as genotoxic bacteria, invasive and inflammation triggering microbiota, procarcinogenic bacteria and cancer enhancing bacterial antigens and metabolites. Consequences of the microbial dysbiosis lead to the chronic inflammation after damaging the host defenses (natural barrier) can further drive to the enhancement of small adenomas to adenocarcinoma by multistep processes.
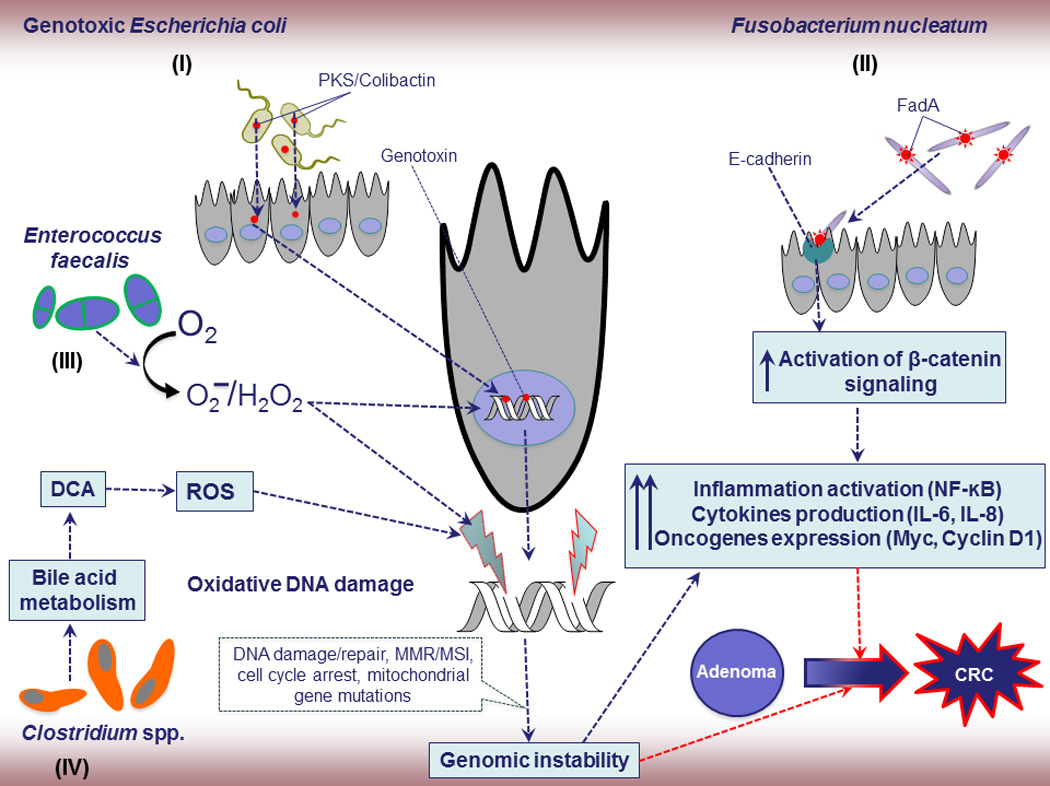
B. Proposed mechanisms of specific bacteria and CRC.
- E. coli, Gram-negative facultative anaerobic bacterium, considered as one of the potential etiological agents of CRC due to its genotoxins such as Colibactin, and cytolethal distending toxin (CDT). These products could induce DNA damage and influence the progression of CRC due to genomic instability from MSI, MMR, and mutations.
- F. nucleatum, Gram-negative anaerobic bacterium, has been linked to CRC progression but the exact underlying mechanisms are still unknown. A potential F. nucleatum-driven CRC mechanism is its invasion into epithelial cells and activation of oncogenic and inflammatory responses through its unique FadA adhesin. Active FadA binds to E-cadherin, mediating Fusobacterium attachment and invasion into the epithelial cells. This activates β-catenin signaling, leading to increased activation of inflammatory genes (NF-κB) and secretion of cytokines interleukin-6 (IL-6), IL-8, and IL-18, and oncogenes and drives to adenoma to adenocarcinoma.
- E. faecalis, has been shown to produce extracellular superoxide and hydrogen peroxide, which damage DNA and also further enhances chromosomal instability in colonic epithelial cells. Chromosomal instability, a common cause of genomic instability in tumors, is characterized by nucleotide additions or deletions, inversions, translocations, and complex rearrangements, and ultimately contributes to the dramatic and unstable alteration in genomic state critical for tumor initiation in the colorectum.
- Gram-positive, spore forming bacteria in cluster IX of the genus Clostridium spp. convert primary bile acids into a secondary bile acid such as deoxycholic acid (DCA). DCA is widely considered as a carcinogen that is associated with DNA damage via the production of free radicals or reactive oxygen species (ROS) and implicated to adenoma-inflammation-CRC through enhancing genomic instability and inflammation.
Other studies have also examined the microbiota in relation to CRC (Table 1). Marchesi et al. assessed the microbiota in colon tumors and matching normal tissue and observed bacterial dysbiosis in the tumors [39]. In particular, they noted an overabundance of Fusobacterium in tumors compared to matching normal tissue. Their initial findings for Fusobacterium and CRC have been confirmed by others [40–45]. Furthermore, some studies characterized the microbiota in fecal samples from CRC subjects and healthy controls. Sobhani et al. examined fecal samples from CRC patients and controls and found that bacterial dysbiosis was associated with CRC and was characterized by an increased abundance of Prevotella [46]. Bacterial dysbiosis associated with CRC has been reported to have relative decreased abundance of obligate anaerobes, increased potential pathogenic bacteria, and reduction in proportions of beneficial butyrate-producing bacteria [45, 47–49]. Zackular et al. demonstrated that changes in the gut microbiota associated with inflammation and tumorigenesis directly contribute to colorectal cancer [50]. In experimental models, they transferred the fecal microbiota of tumor bearing mice to germ free mice and showed that the microbiota from the tumor bearing mice (donor) promoted tumorigenesis in recipient animals with twice as many colon tumors than mice given healthy microbiota. Similar to the donor microbiota, the microbiota of recipient mice was characterized by elevated abundance of Akkermansia, Odoribacter, and Bacteroides. Their observations suggest that the gut microbiota may be amenable to manipulation with antibiotics or probiotics to prevent the development of adenomas and CRC.
Table 1.
Human studies of gut bacteria and colorectal cancer
| Study | Sampling site | Disease | Findings | Reference |
|---|---|---|---|---|
| Geng et al. 2013 | Tumor/matching normal tissue of Chinese CRC patients | CRC | Overabundance of Fusobacterium spp., Roseburia in tumor tissues and over-representation of Microbacterium, Anoxybacillus bacteria away from tumor site | [45] |
| McCoy et al. 2013 | Rectal mucosa | Adenoma | Fusobacterium spp., higher abundance in adenoma subjects. | [43] |
| Castellarin et al. 2012 | Tumor/matching normal tissue | CRC | Overabundance of Fusobacterium nucleatum sequences | [41] |
| Chen et al. 2012 | Intestinal lumen, mucosa (rectal swabs), fecal samples, tumor/matching normal tissue | CRC | Lower bacterial diversity in tumor, altered microbial structures in CRC lumen compared to mucosa. CRC might be due to cometabolism by lumen microflora and direct interaction between host and mucosa-associated microbiota. |
[81] |
| Kostic et al. 2012 | Tumor/matching normal tissue | CRC | Altered microbiota, high abundance of Fusobacterium sequences and low Bacteroides and Firmicutes sequences in tumors | [42] |
| Sanapareddy et al. 2012 | Rectal mucosa | Adenoma | Bacterial dysbiosis, altered diversity and increased richness | [37] |
| Marchesi et al. 2011 | Tumor/matching normal tissue | CRC | Bacterial dysbiosis, high abundance of Fusobacterium in tumors | [40] |
| Shen et al. 2010 | Colonic mucosa of adenoma/non- adenoma | Adenoma | Bacterial dysbiosis, altered diversity, higher abundance of Proteobacteria and lower abundance of Bacteroides in adenoma cases | [36] |
| Ahn et al. 2013 | Fecal sample | CRC | Reduced bacterial diversity in CRC cases | [83] |
| Brim et al. 2013 | Fecal sample | Adenoma | Microbiota changes at the sub-genus level but not genome/functions level in colon polyps. | [38] |
| Ohigashi et al. 2013 | Fecal samples from CRC/adenoma/non-adenoma | CRC& Adenoma | Significant differences in the intestinal environment; altered microbiota (decreased particularly obligate anaerobes), decreased SCFAs, and elevated pH in CRC. | [48] |
| Ohigashi et al. 2013 | Fecal sample before/after surgery | CRC | Marked decreased of obligate anaerobes, increased pathogenic bacteria, and reduction of short chain fatty acids detected after surgery for CRC | [49] |
| Weir et al. 2013 | Fecal sample | CRC | Decrease butyrate producing bacteria | [50] |
| Wu et al. 2013 | Fecal sample | CRC | Bacterial dysbiosis, altered diversity, enriched Bacteroides, more abundant of Fusobacteriumand Campylobacter sps. Decreased butyrate producing bacteria | [46] |
| Sobhani et al. 2011 | Fecal sample | CRC | Bacterial dysbiosis linked with elevated IL-17 in CRC patients | [47] |
The overall consensus from these studies is that a combination of the expansion of procarcinogenic bacteria concomitant with the reduction of tolerogenic commensals such as Faecalibacterium prausnitzii [51] or spore-forming Clostridium clusters IV and XIV [52] may link bacterial dysbiosis to the risk of adenomas and CRC. However, it is difficult to discern from human studies whether gut bacterial dysbiosis is a cause or consequence of adenomas and CRC.
Specific gut bacteria, adenoma, and CRC
Overall, the mechanisms by which the gut microbiota influences adenoma and CRC development remain to be fully established. Moreover, the contribution of specific bacterial signatures and potential mechanisms are not yet elucidated. Potential mechanisms include promotion of chronic inflammation, DNA damage, and production of bioactive carcinogenic metabolites. We describe current reports on some specific bacteria.
Fusobacterium nucleatum: Various studies suggest that overabundance of Fusobacterium spp. is a common feature of CRC that may contribute to disease progression from adenoma to cancer. However, it is not clear whether Fusobacterium spp. is a cause or consequence of adenomas and CRC [53]. Two recent experimental studies provide further mechanistic insights into the relationship between F. nucleatum and colorectal neoplasia. Rubinstein et al. [54] observed that binding of F. nucleatum via its FadA adhesion molecule to E-cadherin leads to activation of β-catenin signaling to induce pro-oncogenic and inflammatory pathways (Fig. 1B. I). The second study by Kostic et al. showed that Fusobacterium modulates the tumor immune microenvironment to promote inflammation and tumorigenesis [43]. In the APC min mouse model of CRC, they showed that Fusobacterium increases infiltration of myeloid cells such as CD11b positive T cells, macrophages, and dendritic cells to induce an NF-κB-driven pro-inflammatory response to promote CRC. In a companion human study, increased FadA expression (> 10–100 times) correlated with elevated expression of oncogenic and inflammatory genes in CRC subjects. While these findings support a role for Fusobacterium spp. and FadA in colorectal carcinogenesis, it is too early to determine their potential as a CRC biomarker or their utility as potential diagnostic and therapeutic targets. Thus, additional studies are needed.
Streptococcus gallolyticus (formerly S. bovis): DNA from S. gallolyticus is present in about 20–50% of colon tumors compared to less than 5% in the normal colon [55]. It has also been associated with increased colonization of collagen-rich surfaces of colorectal adenomas and tumors [56]. It is thought that S. gallolyticus may contribute to neoplastic transformation in the colon via invasion through a breach in the epithelial barrier or virulence factors, which ultimately enhance inflammation and tumorigenesis [55, 56].
Enterotoxigenic Bacteroides fragilis (ETBF): Other bacteria possessing virulence traits such as ETBF are pro-oncogenic and may remodel the microbiota as a whole to promote mucosal immune responses and epithelial changes, which promote colorectal adenomas and cancer. ETBF produces a toxin known as fragilysin (B. fragilis toxin; BFT) which activates the Wnt/β-catenin signaling pathway to increase cell proliferation [57]. BFT also activates NF-kB to induce production of inflammatory mediators. This leads to mucosal inflammation and, ultimately, colorectal carcinogenesis [58, 59]. ETBF was shown to promote tumorigenesis in a study by Wu et al. in which they colonized the APC min model of intestinal neoplasia with a pig isolate of ETBF. They observed a marked increase in colon adenoma and tumor formation in mice colonized with ETBF compared to control mice [60]. The enhanced tumorigenesis by ETBF could occur via activation of Stat3, induction of IL-17 [61] and DNA damage [62]. These observations support a link between bacterial antigens, virulence factors and colon adenomas and CRC.
Enterococcus faecalis: In experimental models, certain strains of E. faecalis have been associated with CRC and colitis- associated CRC. Some strains promote release of extracellular superoxide in host cells. The superoxide is converted by hydrogen peroxide could induce DNA damage [63], chromosome instability [64], and cancer in germfree Interleukin-10 (IL-10−/−) mice (Fig. 1B. III) [65, 66].
Escherichia coli: DNA damage induced by genotoxic E. coli strains could result in CRC-initiating lesions. E. coli possessing the polyketide synthase (pks) Genotoxic Island, which encodes the enzymatic machinery to make Colibactin may also promote CRC via induction of DNA double strand breaks (Fig. 1B. I) [67]. Arthur et al. recently showed that deletion of pks from a strain of E. coli results in reduced DNA damage, tumor numbers, and tumor invasion, but not inflammation in mono-associated IL10−/− mice treated with azoxymethane (AOM) [68]. A few human studies suggest that E. coli harboring the pks is more common in CRC and inflammatory bowel disease patients [68, 69]. Thus, these findings lend strong support to the contribution of genotoxic E. coli in colorectal cancer.
Acidovorax: Acidovorax spp., an acid degrading member of the phylum Proteobacteria is also associated with increased risk of adenomas [36]. Acidovorax may promote colon neoplasia through increased metabolism of nitro-aromatic compounds [70] in the gut as well as induction of local inflammation by its flagellar proteins [71, 72].
In addition to DNA damage and superoxide release, activation of inflammation is a common theme across these studies. Further research is needed to identify additional mechanisms by which bacteria and their virulence factors promote colorectal carcinogenesis. While monoassociation studies involving individual bacteria provide useful mechanistic insights, they may not fully represent the complex interactions between gut bacterial communities and adenomas and CRC.
Bacteria metabolites, adenomas, and CRC
The colonic microbiota influences a wide range of metabolic processes and functions that may lead to beneficial or detrimental effects within the human colon. Metabolites produced by colonic microbiota might play a critical role in the progression of adenomas to CRC, though limited information about the function of most of the gut bacteria and their metabolites is known to date. Certain gut bacteria produce short chain fatty acids (SCFAs) such as butyrate, which can serve as an energy source for colonic epithelial cells. Wang and colleagues observed a reduction in butyrate-producing bacteria in feces of CRC patients suggesting that microbial metabolites may contribute to the etiology of CRC [73]. A few members of the Clostridium cluster IX, XI, and XVIa are capable of metabolizing primary bile acids into secondary bile acids [74]. Secondary bile acids such as deoxycholic acid (DCA) might contribute to CRC progression (Fig. 1B. IV) by interacting with host metabolism and immunity [75–78].
Few human studies have evaluated the metabolome and microbiota in relation to adenomas or CRC. Findings from a recent study suggests that there is a correlation between bacterial dysbiosis, the metabolome, and colorectal adenomas [79]. More studies are needed to fully explore the relationship between the microbiota, metabolome, adenomas, and CRC.
Summary and conclusions
Although gut bacterial dysbiosis is increasingly recognized as a phenomenon in colorectal carcinogenesis, host-bacterial interactions still remain to be fully elucidated. In studying the gut microbiota and adenomas or CRC, it is unclear whether sampling the mucosa or the luminal content is the most appropriate. Bacteria in the lumen are transient and may be more influenced by diet while the adherent mucosal bacteria are considered residents and may be more relevant to CRC because of their close contact with the host mucosa and immune cells. To date, there is no clear consensus. Studies suggest that bacteria communities in the feces differ from the mucosa [80, 81]. Findings from two studies that compared the microbiota in mucosa, rectal swabs, and feces of the same patients were inconclusive [82, 83]. Additional studies are needed to further define the best sampling location so as to enhance uniformity and reproducibility among studies.
The role of the gut microbiota in the progression from adenomas to CRC is undoubtedly multifactorial and can affect the various stages of the neoplastic process. Microbial dysbiosis, induction of mucosal inflammation, and production of reactive metabolites are all processes that might act in concert to set the colonic mucosa on the initial stage of the adenoma-carcinoma process. Further research in experimental animal models is necessary to better understand the mechanisms that underlay the association between the gut microorganisms and CRC. The intestinal microbiota represents an enormous reservoir for the discovery of novel signatures that could be potentially useful as biomarkers and predictors for adenomas and CRC. Manipulation of the gut microbiota to restore normal physiologic balance might be beneficial in preventing colon adenomas and CRC. Furthermore, beneficial or “friendly” bacteria that have been specifically engineered to provide desired inflammatory responses and epigenetic expression could have the potential to be useful therapeutically in CRC.
In conclusion, the advances in microbiome research provides an opportunity to elucidate the exact connections between the host gut microbiome and the onset of CRC, which will hopefully lead to safer and more efficacious treatments in the near future.
ACKNOWLEDGMENTS
Special thanks to Ms. Amber McCoy, Dr. Andrea Azcarate-Peril and Dr. Janelle Arthur for reviewing drafts of the manuscript.
Funding Sources
NIH NCI R01 CA136887, NIH NCI R01 CA044684, NIH P30 DK 034987
Footnotes
Author Contributions
All authors contributed to the writing of this manuscript and have given approval to the final version.
The authors declare no competing interests.
References
- 1.GLOBOCON. IARC, WHO; 2012. Estimated cancer incidence, mortality and prevalence in 2012. [Google Scholar]
- 2.Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. CA: a cancer journal for clinicians. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 3.Mariotto AB, Yabroff KR, Shao Y, et al. Projections of the cost of cancer care in the United States: 2010–2020. J Natl Cancer Inst. 2011;103:117–128. doi: 10.1093/jnci/djq495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muto T, Bussey HJ, Morson BC. The evolution of cancer of the colon and rectum. Cancer. 1975;36:2251–2270. doi: 10.1002/cncr.2820360944. [DOI] [PubMed] [Google Scholar]
- 5.Parente F, Bargiggia S, Boemo C, et al. Anatomic distribution of cancers and colorectal adenomas according to age and sex and relationship between proximal and distal neoplasms in an i-FOBT-positive average-risk Italian screening cohort. International journal of colorectal disease. 2014;29:57–64. doi: 10.1007/s00384-013-1759-9. [DOI] [PubMed] [Google Scholar]
- 6.Sugumar A, Sinicrope FA. Serrated polyps of the colon. F1000 medicine reports. 2010;2:89. doi: 10.3410/M2-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sillars-Hardebol AH, Carvalho B, de Wit M, et al. Identification of key genes for carcinogenic pathways associated with colorectal adenoma-to-carcinoma progression. Tumour Biol. 2010;31:89–96. doi: 10.1007/s13277-009-0012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fearon ER. Molecular genetics of colorectal cancer. Annual review of pathology. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 9.Laken SJ, Petersen GM, Gruber SB, et al. Familial colorectal cancer in Ashkenazim due to a hypermutable tract in APC. Nat Genet. 1997;17:79–83. doi: 10.1038/ng0997-79. [DOI] [PubMed] [Google Scholar]
- 10.Hamilton SR. Molecular genetic alterations as potential prognostic indicators in colorectal carcinoma. Cancer. 1992;69:1589–1591. doi: 10.1002/1097-0142(19920315)69:6+<1589::aid-cncr2820691314>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 11.Fearon ER, Cho KR, Nigro JM, et al. Identification of a chromosome 18q gene that is altered in colorectal cancers. Science. 1990;247:49–56. doi: 10.1126/science.2294591. [DOI] [PubMed] [Google Scholar]
- 12.Hold GL, Smith M, Grange C, et al. Role of the gut microbiota in inflammatory bowel disease pathogenesis: What have we learnt in the past 10 years? World journal of gastroenterology : WJG. 2014;20:1192–1210. doi: 10.3748/wjg.v20.i5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends in genetics : TIG. 1993;9:138–141. doi: 10.1016/0168-9525(93)90209-z. [DOI] [PubMed] [Google Scholar]
- 14.Carethers JM. Differentiating lynch-like from lynch syndrome. Gastroenterology. 2014;146:602–604. doi: 10.1053/j.gastro.2014.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang CL, Marra G, Chauhan DP, et al. Oxidative stress inactivates the human DNA mismatch repair system. Am J Physiol Cell Physiol. 2002;283:C148–C154. doi: 10.1152/ajpcell.00422.2001. [DOI] [PubMed] [Google Scholar]
- 16.Arends MJ. Pathways of colorectal carcinogenesis. Applied immunohistochemistry & molecular morphology : AIMM / official publication of the Society for Applied Immunohistochemistry. 2013;21:97–102. doi: 10.1097/PAI.0b013e31827ea79e. [DOI] [PubMed] [Google Scholar]
- 17.Sakai E, Nakajima A, Kaneda A. Accumulation of aberrant DNA methylation during colorectal cancer development. World J Gastroenterol. 2014;20:978–987. doi: 10.3748/wjg.v20.i4.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Al-Sohaily S, Biankin A, Leong R, et al. Molecular pathways in colorectal cancer. Journal of gastroenterology and hepatology. 2012;27:1423–1431. doi: 10.1111/j.1440-1746.2012.07200.x. [DOI] [PubMed] [Google Scholar]
- 19.Pibiri F, Kittles RA, Sandler RS, et al. Genetic variation in vitamin D-related genes and risk of colorectal cancer in African Americans. Cancer causes & control : CCC. 2014 doi: 10.1007/s10552-014-0361-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esteban-Jurado C, Garre P, Vila M, et al. New genes emerging for colorectal cancer predisposition. World journal of gastroenterology : WJG. 2014;20:1961–1971. doi: 10.3748/wjg.v20.i8.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tenesa A, Farrington SM, Prendergast JG, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet. 2008;40:631–637. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomlinson IP, Webb E, Carvajal-Carmona L, et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet. 2008;40:623–630. doi: 10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- 23.Willett WC, Stampfer MJ, Colditz GA, et al. Relation of meat, fat, and fiber intake to the risk of colon cancer in a prospective study among women. N Engl J Med. 1990;323:1664–1672. doi: 10.1056/NEJM199012133232404. [DOI] [PubMed] [Google Scholar]
- 24.Blum T, Moore MA. Failure of hydrodynamics within the vortex-liquid phase. Physical review B, Condensed matter. 1995;51:15359–15362. doi: 10.1103/physrevb.51.15359. [DOI] [PubMed] [Google Scholar]
- 25.Huxley RR, Ansary-Moghaddam A, Clifton P, et al. The impact of dietary and lifestyle risk factors on risk of colorectal cancer: a quantitative overview of the epidemiological evidence. Int J Cancer. 2009;125:171–180. doi: 10.1002/ijc.24343. [DOI] [PubMed] [Google Scholar]
- 26.O'Keefe SJ, Chung D, Mahmoud N, et al. Why do African Americans get more colon cancer than Native Africans? J Nutr. 2007;137:175S–182S. doi: 10.1093/jn/137.1.175S. [DOI] [PubMed] [Google Scholar]
- 27.Park Y, Hunter DJ, Spiegelman D, et al. Dietary fiber intake and risk of colorectal cancer: a pooled analysis of prospective cohort studies. JAMA : the journal of the American Medical Association. 2005;294:2849–2857. doi: 10.1001/jama.294.22.2849. [DOI] [PubMed] [Google Scholar]
- 28.Proctor LM. The Human Microbiome Project in 2011 and beyond. Cell Host Microbe. 2011;10:287–291. doi: 10.1016/j.chom.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Wu GD, Lewis JD. Analysis of the human gut microbiome and association with disease. Clin Gastroenterol Hepatol. 2013;11:774–777. doi: 10.1016/j.cgh.2013.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gill SR, Pop M, Deboy RT, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hullar MA, Burnett-Hartman AN, Lampe JW. Gut microbes, diet, and cancer. Cancer treatment and research. 2014;159:377–399. doi: 10.1007/978-3-642-38007-5_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones MLMC, Ganopolsky JG, Labbé A, Prakash S. The human microbiome and bile acid metabolism: dysbiosis, dysmetabolism, disease and intervention. Expert Opin Biol Ther. 2014;14:467–482. doi: 10.1517/14712598.2014.880420. [DOI] [PubMed] [Google Scholar]
- 33.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Candela M, Turroni S, Biagi E, et al. Inflammation and colorectal cancer, when microbiota-host mutualism breaks. World journal of gastroenterology : WJG. 2014;20:908–922. doi: 10.3748/wjg.v20.i4.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen XJ, Rawls JF, Randall T, et al. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes. 2010;1:138–147. doi: 10.4161/gmic.1.3.12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanapareddy N, Legge RM, Jovov B, et al. Increased rectal microbial richness is associated with the presence of colorectal adenomas in humans. The ISME journal. 2012;6:1858–1868. doi: 10.1038/ismej.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brim H, Yooseph S, Zoetendal EG, et al. Microbiome analysis of stool samples from african americans with colon polyps. PloS one. 2013;8:e81352. doi: 10.1371/journal.pone.0081352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei H, Dong L, Wang T, et al. Structural shifts of gut microbiota as surrogate endpoints for monitoring host health changes induced by carcinogen exposure. FEMS microbiology ecology. 2010;73:577–586. doi: 10.1111/j.1574-6941.2010.00924.x. [DOI] [PubMed] [Google Scholar]
- 39.Marchesi JR, Dutilh BE, Hall N, et al. Towards the human colorectal cancer microbiome. PloS one. 2011;6:e20447. doi: 10.1371/journal.pone.0020447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome research. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome research. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCoy AN, Araujo-Perez F, Azcarate-Peril A, et al. Fusobacterium is associated with colorectal adenomas. PloS one. 2013;8:e53653. doi: 10.1371/journal.pone.0053653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell host & microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geng J, Fan H, Tang X, et al. Diversified pattern of the human colorectal cancer microbiome. Gut pathogens. 2013;5:2. doi: 10.1186/1757-4749-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu N, Yang X, Zhang R, et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microbial ecology. 2013;66:462–470. doi: 10.1007/s00248-013-0245-9. [DOI] [PubMed] [Google Scholar]
- 46.Sobhani I, Tap J, Roudot-Thoraval F, et al. Microbial dysbiosis in colorectal cancer (CRC) patients. PloS one. 2011;6:e16393. doi: 10.1371/journal.pone.0016393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohigashi S, Sudo K, Kobayashi D, et al. Changes of the intestinal microbiota, short chain fatty acids, and fecal pH in patients with colorectal cancer. Digestive diseases and sciences. 2013;58:1717–1726. doi: 10.1007/s10620-012-2526-4. [DOI] [PubMed] [Google Scholar]
- 48.Ohigashi S, Sudo K, Kobayashi D, et al. Significant changes in the intestinal environment after surgery in patients with colorectal cancer. Journal of gastrointestinal surgery : official journal of the Society for Surgery of the Alimentary Tract. 2013;17:1657–1664. doi: 10.1007/s11605-013-2270-x. [DOI] [PubMed] [Google Scholar]
- 49.Weir TL, Manter DK, Sheflin AM, et al. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One. 2013;8:e70803. doi: 10.1371/journal.pone.0070803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zackular JP, Baxter NT, Iverson KD, et al. The gut microbiome modulates colon tumorigenesis. mBio. 2013;4:e00692–e00613. doi: 10.1128/mBio.00692-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Atarashi K, Tanoue T, Shima T, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keku TO, McCoy AN, Azcarate-Peril AM. Fusobacterium spp. and colorectal cancer: cause or consequence? Trends in microbiology. 2013;21:506–508. doi: 10.1016/j.tim.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rubinstein MR, Wang X, Liu W, et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell host & microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abdulamir AS, Hafidh RR, Bakar FA. Molecular detection, quantification, and isolation of Streptococcus gallolyticus bacteria colonizing colorectal tumors: inflammation-driven potential of carcinogenesis via IL-1, COX-2, and IL-8. Molecular cancer. 2010;9:249. doi: 10.1186/1476-4598-9-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boleij A, van Gelder MM, Swinkels DW, et al. Clinical Importance of Streptococcus gallolyticus infection among colorectal cancer patients: systematic review and meta-analysis. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2011;53:870–878. doi: 10.1093/cid/cir609. [DOI] [PubMed] [Google Scholar]
- 57.Sokol SY. Wnt signaling and dorso-ventral axis specification in vertebrates. Current opinion in genetics & development. 1999;9:405–410. doi: 10.1016/S0959-437X(99)80061-6. [DOI] [PubMed] [Google Scholar]
- 58.Sears CL. Enterotoxigenic Bacteroides fragilis: a rogue among symbiotes. Clinical microbiology reviews. 2009;22:349–369. doi: 10.1128/CMR.00053-08. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shiryaev SA, Remacle AG, Chernov AV, et al. Substrate cleavage profiling suggests a distinct function of Bacteroides fragilis metalloproteinases (fragilysin and metalloproteinase II) at the microbiome-inflammation-cancer interface. The Journal of biological chemistry. 2013;288:34956–34967. doi: 10.1074/jbc.M113.516153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu S, Rhee KJ, Albesiano E, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nature medicine. 2009;15:1016–1022. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tosolini M, Kirilovsky A, Mlecnik B, et al. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer research. 2011;71:1263–1271. doi: 10.1158/0008-5472.CAN-10-2907. [DOI] [PubMed] [Google Scholar]
- 62.Goodwin AC, Destefano Shields CE, Wu S, et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:15354–15359. doi: 10.1073/pnas.1010203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huycke MM, Abrams V, Moore DR. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis. 2002;23:529–536. doi: 10.1093/carcin/23.3.529. [DOI] [PubMed] [Google Scholar]
- 64.Wang X, Huycke MM. Extracellular superoxide production by Enterococcus faecalis promotes chromosomal instability in mammalian cells. Gastroenterology. 2007;132:551–561. doi: 10.1053/j.gastro.2006.11.040. [DOI] [PubMed] [Google Scholar]
- 65.Wang X, Yang Y, Moore DR, et al. 4-hydroxy-2-nonenal mediates genotoxicity and bystander effects caused by Enterococcus faecalis-infected macrophages. Gastroenterology. 2012;142:543–551. doi: 10.1053/j.gastro.2011.11.020. e547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang Y, Wang X, Huycke T, et al. Colon Macrophages Polarized by Commensal Bacteria Cause Colitis and Cancer through the Bystander Effect. Translational oncology. 2013;6:596–606. doi: 10.1593/tlo.13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cuevas-Ramos G, Petit CR, Marcq I, et al. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11537–11542. doi: 10.1073/pnas.1001261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arthur JC, Perez-Chanona E, Muhlbauer M, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Buc E, Dubois D, Sauvanet P, et al. High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PloS one. 2013;8:e56964. doi: 10.1371/journal.pone.0056964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Malkan AD, Strollo W, Scholand SJ, et al. Implanted-port-catheter-related sepsis caused by Acidovorax avenae and methicillin-sensitive Staphylococcus aureus. Journal of clinical microbiology. 2009;47:3358–3361. doi: 10.1128/JCM.01093-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tanaka N, Che FS, Watanabe N, et al. Flagellin from an incompatible strain of Acidovorax avenae mediates H2O2 generation accompanying hypersensitive cell death and expression of PAL, Cht-1, and PBZ1, but not of Lox in rice. Molecular plant-microbe interactions : MPMI. 2003;16:422–428. doi: 10.1094/MPMI.2003.16.5.422. [DOI] [PubMed] [Google Scholar]
- 72.Takakura Y, Che FS, Ishida Y, et al. Expression of a bacterial flagellin gene triggers plant immune responses and confers disease resistance in transgenic rice plants. Molecular plant pathology. 2008;9:525–529. doi: 10.1111/j.1364-3703.2008.00477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang T, Cai G, Qiu Y, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. The ISME journal. 2012;6:320–329. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ridlon JM, Kang DJ, Hylemon PB, et al. Bile acids and the gut microbiome. Current opinion in gastroenterology. 2014 doi: 10.1097/MOG.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Barrasa JI, Olmo N, Lizarbe MA, et al. Bile acids in the colon, from healthy to cytotoxic molecules. Toxicology in vitro : an international journal published in association with BIBRA. 2013;27:964–977. doi: 10.1016/j.tiv.2012.12.020. [DOI] [PubMed] [Google Scholar]
- 76.Da Silva MGKJ, Verstraeten SV, Erlejman AG, Fraga CG, Oteiza PI. Large procyanidins prevent bile-acid-induced oxidant production and membrane-initiated ERK1/2, p38, and Akt activation in Caco-2 cells. Free Radic Biol Med. 2012;52:151–159. doi: 10.1016/j.freeradbiomed.2011.10.436. [DOI] [PubMed] [Google Scholar]
- 77.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bernstein C, Holubec H, Bhattacharyya AK, et al. Carcinogenicity of deoxycholate, a secondary bile acid. Arch Toxicol. 2011;85:863–871. doi: 10.1007/s00204-011-0648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nugent JL, McCoy AN, Addamo CJ, et al. Altered Tissue Metabolites Correlate with Microbial Dysbiosis in Colorectal Adenomas. Journal of proteome research. 2014 doi: 10.1021/pr4009783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zoetendal EG, von Wright A, Vilpponen-Salmela T, et al. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Applied and environmental microbiology. 2002;68:3401–3407. doi: 10.1128/AEM.68.7.3401-3407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen W, Liu F, Ling Z, et al. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PloS one. 2012;7:e39743. doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kang M, Edmundson P, Araujo-Perez F, et al. Association of plasma endotoxin, inflammatory cytokines and risk of colorectal adenomas. BMC cancer. 2013;13:91. doi: 10.1186/1471-2407-13-91. [DOI] [PMC free article] [PubMed] [Google Scholar]


