Abstract
Unexpected activation of mTOR signaling, measured by ribosomal S6 phosphorylation or ribosomal S6 kinase (p70S6K) activity, has been reported in aging-related settings. Evidence of elevated mTOR activity has been reported in heart and muscle tissue in aged mice and humans, mouse models of progeria, and senescent human fibroblasts. We explore these reports and the possibility that activation of the mTOR/p70S6K kinase pathway may represent a ROS-mediated response to mitochondrial stress leading to the activation of senescence. This activation is a hallmark of both aged tissue and senescent human cells.
Introduction
Target of rapamycin complexes
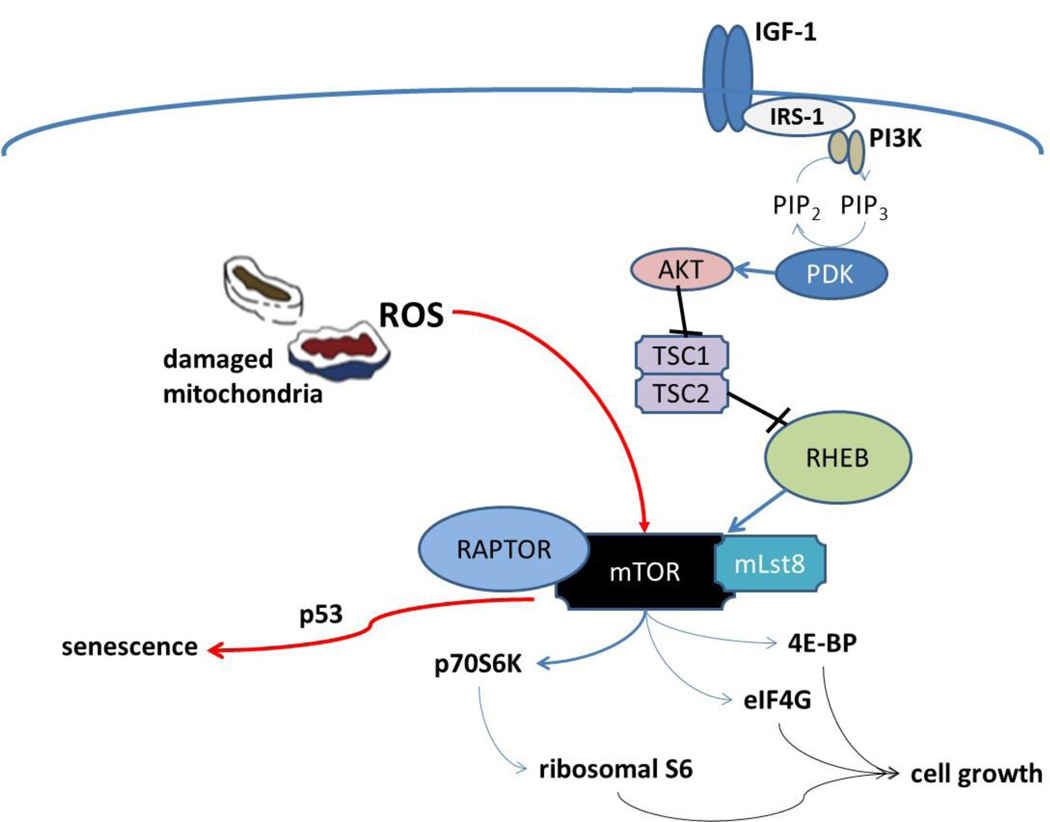
The mechanistic target of rapamycin (mTOR) pathway is a key intracellular signaling pathway that integrates proliferative signals, nutrient availability, and stress (Sengupta et al., 2010). In this review, we will discuss the pathway, it’s known cellular functions, and the possibility that mitochondrial dysfunction serves as an additional regulator of the mTOR pathway. A model of the mTOR pathway including input from the mitochondria is presented in Figure 1.
Figure 1. Possible links between mitochondrial ROS and mTOR.
A model is presented invoking a mitochondrial ROS mediated impact on mTOR signaling during aging and senescence. The mTOR complex is impacted by mitochondrial dysfunction to drive senescence rather than proliferation. This impact would be alleviated by rapamycin producing a reduction in stress signaling in addition to changes which enhance mitochondrial homeostasis such as an increase in autophagy.
The mTOR signaling pathway is highly conserved in eukaryotic organisms and consists of a protein complex that includes the mTOR kinase itself and several accessory proteins and downstream mediators including the ribosomal S6 kinase (p70S6K). The pathway has been functionally defined as the target of the highly-specific antifungal, rapamycin. Rapamycin is produced by the bacterial strain Streptomyces hygroscopicus, originally isolated from the island of Rapa Nui. Rapamycin binds to a12 kDa FK506-binding protein (FKBP12), which then acts as an allosteric inhibitor of the mTOR kinase complex (Laplante and Sabatini 2012; Loewith and Hall 2011). Consequently, rapamycin is also a potent inhibitor of the p70S6K. The p70S6K is a serine/threonine kinase that is a key downstream target of mTOR (Chung et al., 1992; Kuo et al., 1992; Price et al., 1992).
The proteins that comprise the core mTOR complex are the Ser/Thr kinase mTOR, also known as the FKBP12-rapamycin associated protein (FRAP1), and mammalian lethal with SEC13 protein 8 (mLST8). These core components have the capability of forming either of two complexes, mTORC1 or mTORC2, which are distinguishable by their sensitivity to rapamycin. The rapamycin-sensitive mTORC1 contains the scaffolding protein regulatory-associated protein of mTOR (RAPTOR), whereas the rapamycin-insensitive complex mTORC2 contains the scaffolding protein rapamycin-insensitive companion of mTOR (RICTOR). These scaffolding proteins function to direct mTORC1 and mTORC2 to their respective targets (Alessi et al., 2009). Additional components are unique to each complex. For example, Deptor and PRAS40 are inhibitory proteins associated with mTORC1 (Sancak et al., 2007; Vander Haar et al., 2007), whereas the Sin1 and proctor proteins, which are potential mediators of subcellular localization, are associated with mTORC2 (Frias et al., 2006; Pearce et al., 2007; Yang et al., 2006).
The best characterized mTORC1 targets are p70S6K and the translational regulator eukaryotic initiation factor 4E (eIF4E)-binding protein (4E-BP). Phosphorylation of either target by mTORC1 promotes translation. Phosphorylation of eIF4E prevents binding to eIF4E mRNA cap binding protein, enabling assembly of the cap-binding complex and translation initiation (Sonenberg 2008), whereas the activated p70S6K phosphorylates multiple substrates, most notably the ribosomal protein S6, to increase translation initiation and elongation, promoting ribosome biogenesis as well as the translation of proteins regulating cell growth and division (Alessi et al., 2009). Targets of mTORC2 include AKT, serum glucocorticoid-induced kinase (SGK), and some members of the PKC family, which all contribute to cytoskeleton organization and cell survival (Zoncu et al., 2011).
Regulation of mTOR signaling
Growth factors stimulate mTORC1 activity via activation of the lipid kinase PI3K. Activation of PI3K occurs via direct binding to the growth factor receptors or through interaction with the docking proteins insulin receptor substrate (IRS) or GRB2-associated binder (GAB). Upon activation, PI3K generates phosphatidylinositol 3,4,5 triphosphate (PIP3) to produce a membrane-binding site for the protein kinase AKT and results in conformational changes that allow access to critical serine and threonine sites within AKT. Subsequent phosphorylation of AKT by 3-phosphoinositide-dependent kinase 1 (PDK1) and mTORC2 serves to fully activate AKT (Vanhaesebroeck and Alessi 2000). Subsequently, AKT phosphorylates substrates that enhance cell growth and survival. One Akt target is the tuberous sclerosis complex 2 (TSC2) GTPase activating protein (GAP). The phosphorylation of TSC2 blocks TSC2-mediated inhibition of the GTPase Ras homolog enriched in brain (RHEB). Consequently, active RHEB-GTP directly activates mTORC1 (Alessi et al., 2009; Inoki et al., 2002; Manning et al., 2002).
mTORC1 modulates cell growth and cell size through the regulation of translation, nucleotide biosynthesis, lipogenesis, glycolysis, and autophagy (Ben-Sahra et al., 2013; Ganley et al., 2009; Hosokawa et al., 2009; Ma and Blenis 2009; Peterson et al., 2011; Robitaille et al., 2013). Input on the metabolic state of the cell required to optimally regulate translation and proliferation is provided by the AMP kinase, which is activated at elevated AMP levels. Active AMP kinase inhibits mTORC1 through phosphorylation of both RAPTOR and TSC1, thereby reducing mTOR activity when the energy state is low (Gwinn et al., 2008; Inoki et al., 2012; Inoki et al., 2003).
Lifespan extension through reduced mTOR signaling
A reduction in mTOR signaling has been found to extend lifespan in multiple organisms. In yeast, a screening study for long-lived mutants identified the SCH9 gene, which was originally classified as an ortholog of the mammalian AKT (Sobko 2006). SCH9 was later identified as the functional homolog of p70S6K (Urban et al., 2007). Deletion of this gene conferred a 2-to-3 fold increase in lifespan and increased resistance to oxidative stress (Fabrizio et al., 2001). Subsequent studies in Caenorhabditis elegans using RNAi approaches demonstrated that targeting either mTOR (let-363) or RAPTOR (daf-15) extended lifespan (Jia et al., 2004; Vellai et al., 2003). Further, genetic studies in Drosophila melanogaster demonstrated that overproduction of TSC1 or TSC2, or the expression of dominant negative forms of dTOR and dS6K extend lifespan (Kapahi et al., 2004). In mice, deletion of the p70S6K gene resulted in reduced body size and increased lifespan (Selman and Withers 2011). Landmark studies involving mouse colonies at three independent sites demonstrated that oral delivery of rapamycin extends the lifespan of mice, even when administered late in life (Harrison et al., 2009; Miller et al., 2011).
Despite extensive literature, the mechanisms by which rapamycin extends mammalian lifespan are poorly understood. A reduction in neoplasia may contribute to lifespan extension in response to rapamycin. For example, rapamycin has emerged as a promising anti-tumor therapy in some malignancies (Dibble and Manning 2013). Moreover, a reduction in tumor formation has been noted in rapamycin-fed mice (Anisimov et al., 2010; Komarova et al., 2012; Popovich et al., 2014), which suggests that reduced tumor formation may underlie lifespan extension seen in normal mice (Sharp and Richardson 2011). However, rapamycin also has ameliorative effects on degenerative disease states that are clearly not associated with enhanced proliferation. For example, treatment of mice with rapamycin, either through intraperitoneal injection or via oral delivery through compounding in rodent diet, can ameliorate age-related pathologies in models of Alzheimer’s Disease, Parkinson’s Disease, and cardiomyopathy (Harrison et al., 2009; Johnson et al., 2013a; Powers et al., 2006; Robida-Stubbs et al., 2012).
At the cellular level, inhibition of mTORC1 has multiple consequences that may be important to lifespan extension. Two critical cellular processes affected by mTORC1 are translation and autophagy. As previously noted, regulation of translation occurs at the initiation step, through phosphorylation of 4E-BP to prevent association with eIF4E (Thoreen 2013). However, the influence of mTORC1 on translation appears to be greater on some subsets of mRNA. For example, mRNAs that contain a 5’ polypyrimidine sequence are highly sensitive to mTORC1 inhibition (Thoreen 2013). Additionally, it has been suggested that the structure of the 5’ untranslated region of the mRNA may contribute to differential expression under conditions of mTORC1 inhibition. (Kapahi 2010). In yeast, the Gcn4 mRNA is such a target. Gcn4 regulates several genes involved in enhanced stress resistance, and translation of Gcn4 is more effective under conditions of TOR inhibition (Steffen et al., 2008; Thoreen 2013). In C. elegans and D. melanogaster, TOR inhibition leads to preferential expression of a subset of mRNAs related to stress response and mitochondrial activity (Rogers et al., 2011; Zid et al., 2009). This enhanced stress resistance may contribute to lifespan extension.
Autophagy is a critical intracellular pathway for the removal of misfolded proteins and dysfunctional organelles such as mitochondria, and for the recycling of amino acids during starvation (Mizushima et al., 2008). Autophagy decreases with age, potentially contributing to the accumulation of protein aggregates and mitochondrial dysfunction (Cuervo 2008). Rapamycin is known to increase autophagy by inhibiting mTOR-mediated phosphorylation of Ulk-1, a key regulator of autophagosome formation (Egan et al., 2011; Kim et al., 2011).
The induction of autophagy is required for lifespan extension in response to dietary restriction in yeast and C. elegans (Alvers et al., 2009; Hansen et al., 2008). In simple terms, increasing autophagy should serve to enhance mitochondrial clearance and rapamycin provides protection against mitochondrial dysfunction in both a mouse model of Leigh Syndrome (Johnson et al., 2013b) and in response to direct mitochondrial DNA depletion in human cells (Nacarelli et al., 2014). However, the effects of rapamycin on mitochondrial homeostasis appear to be more complex than a simple increase in clearance. For example, transcriptional profiling of mice fed rapamycin revealed significant changes in genes associated with mitochondrial function (Fok et al., 2014a; Fok et al., 2014b; Fok et al., 2014c). Additionally, acute treatment with rapamycin induces a rapid shift in metabolism, reducing metabolic intermediates of mitochondrial function (Ramanathan and Schreiber 2009) and acute treatment of myoblast cells with rapamycin inhibits mitochondrial biogenesis (Cunningham et al., 2007). These acute changes in mTORC1 and mTORC2 signaling have been found to influence the balance between mitochondrial and non-mitochondrial ATP generation (Schieke et al., 2006). In contrast to the acute effects, chronic treatment of human cells with nanomolar concentrations of rapamycin enhances mitochondrial biogenesis, perhaps as a compensatory response (Lerner et al., 2013). Similarly, results in Drosophila indicate that increased expression of 4E-BP increases expression of mitochondrial electron transport chain proteins (Zid et al., 2009). Rapamycin also increases the lifespan of dendritic cells by preserving mitochondrial function (Amiel et al., 2014; Haidinger et al., 2010).
Aberrant activation of mTOR during aging
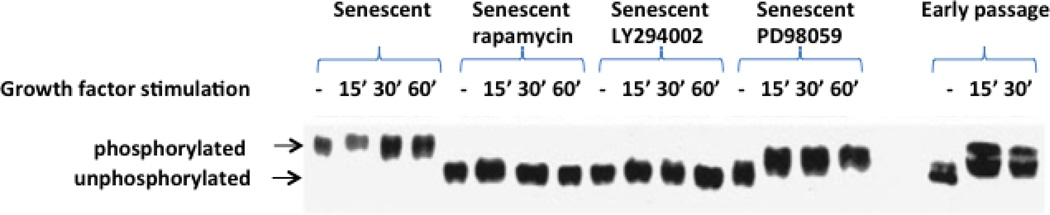
A number of years ago, our laboratory reported that p70S6K was activated during cellular senescence in the absence of growth factor stimulation (Zhang et al., 2000). These studies were performed by placing cells under serum-free conditions in the presence of nutrients and amino acids while using a medium specifically designed for the maintenance of human fibroblasts (McKeehan et al., 1977). In normal (ie. untransformed) cells these conditions abrogate the multiple mTOR inputs described above and lead to dephosphorylation of p70S6K as cells enter a stable G0 state. Stimulation with growth factors rapidly induces p70S6K activation through phosphorylation by mTORC1, which is activated via the PI3K pathway (Ming et al., 1994). Surprisingly, senescent cells maintain p70S6K phosphorylation in the G0 state independent of growth factors. Members of the Ras/mitogen activated protein kinase (MAPK) pathway appear to be responsible for the continued activation of p70S6K in the absence of growth factors, as inhibitors of MEK1/2 block the phosphorylation of p70S6K (Zhang et al., 2000). However, growth factor signaling in the senescent cells was blocked by specific inhibitors of PI3K and mTORC1, but not the MEK1/2 inhibitor. This confirms that the Ras/MAPK contribution to p70S6K phosphorylation in senescent cells is independent of the canonical growth-factor-triggered signaling pathways (see Figure 2). Importantly for this discussion, the PI3K pathway was defined as a pathway distinct from the Ras/MAPK pathway, and these 2 pathways appear to bifurcate at the level of growth factor receptor activation (Ming et al., 1994).
Figure 2. Activation of the p70 S6 kinase in senescent human fibroblasts.
WI-38 normal human fibroblasts were serum deprived for 72 h (SF) and treated for 40 min with the mTOR inhibitor, rapamycin (1 ng/ml), the PI 3-kinase inhibitor, LY294002 (50 mM), or the MEK1/2 inhibitor, PD98059 (50 mM). Cultures were then stimulated with 10% fetal bovine serum for 15 min, 30 min, or 1 h and total cellular proteins isolated. The p70 S6K1 protein was visualized by Western blot analysis using a polyclonal antibody (Santa CruzBiotechnology, Santa Cruz, CA). The mobility of p70 S6K indicates that the phosphorylation status of the enzyme. In early passage cells, the addition of serum to serum-deprived cultures decreased the mobility of p70 S6K indicative of phosphorylation. For illustration, a Western blot of the p70 S6K in serum-deprived (SF) and serum-stimulated early passage cells is included on the right side. Note that the MEK 1/2 inhibitor induces dephosphorylation of p70 S6K (evidences by a lower band) and that growth factor stimulation induces phosphorylation in the presence of this inhibitor. In contrast, growth factors are unable to phosphorylate the enzyme in the presence or either rapamycin or LY294002, which inhibit known growth factor signaling pathways linked to p70 S6K activation. Figure recreated from Zhang et al. (Zhang et al., 2000).
More recently, there have been a growing number of reports of aberrant activation of mTOR signaling in senescence and aging. For example, a multi-center study originally intended to examine components of the IGF-1 axis during aging, identified increased ribosomal S6 phosphorylation as the most consistent finding in both mouse and human muscle (Sandri et al., 2013). Similarly, in a mouse model of progeria, mTORC1 activity based on ribosomal S6 and 4E-BP phosphorylation increased relative to control animals (Ramos et al., 2012). Interestingly, an increase in mTOR, RAPTOR, and ribosomal S6 phosphorylation has been reported in brain tissue of patients with severe Alzheimer’s Disease relative to those with mild or moderate disease (Sun et al., 2014).
Increased mTOR activity has recently been linked to senescence. Work by two groups demonstrated that rapamycin treatment can delay replicative senescence (Kolesnichenko et al., 2012; Lerner et al., 2013) and that rapamycin reduces senescence induced by DNA damage (Demidenko and Blagosklonny 2008). Rapamycin also delays or reduces senescence in several cancer cell lines exposed to chemotherapeutics (Xu et al., 2014) and it has been proposed that mTORC1 signaling dictates whether cells enter quiescence or senescence, a geroconversion (Leontieva et al., 2014).
Rapamycin: protection from mitochondrial stress?
In an effort to understand the negative consequences of insulin-like growth factor 1 (IGF-1) signaling on non-transformed cells, our laboratory has demonstrated that exposure of cells to IGF-1 in the absence of co-stimulatory molecules such as epidermal growth factor leads to mitochondrial dysfunction and the induction of senescence (Bitto et al., 2010). Rapamycin was able to both reduce mitochondrial dysfunction and delay replicative senescence in normal human fibroblasts (Bitto et al., 2010; Lerner et al., 2013). Interestingly, we observed a progressive increase in the percentage of cells harboring dysfunctional mitochondria during replicative senescence (Lerner et al., 2013). It has been suggested that mitochondrial dysfunction may account for the stochastic nature of senescence in cultured cells (Passos et al., 2007). This raises the possibility that aberrant activation of mTOR, that we and others have noted, may be due to mitochondrial stress. As outlined in Figure 1, we put forward the hypothesis that mTORC1 activation by mitochondrial-generated ROS contributes to senescence. Although it is clear that high levels of stress or DNA damage can serve to inhibit mTOR (Sengupta et al., 2010), redox regulation of mTOR has been observed in HEK293 cells following direct application of cellular oxidizing agents (Sarbassov and Sabatini 2005). In this case, activation was independent of nutrient signaling, and although the stimulus was a known oxidant, the concept that mTOR activates p70S6K in response to oxidation (oxidative stress) is consistent with our model.
Subcellular localization of mTOR
The mTOR complex has been localized to several intracellular organelles, including the lysosome, endoplasmic reticulum, nucleus, and mitochondria (Betz and Hall 2013). Mitochondrial localization of mTOR has been described using subcellular fraction and immunoprecipitation techniques (Desai et al., 2002; Ramanathan and Schreiber 2009; Schieke et al., 2006). In one specific experimental model of osmotic stress, mTOR was linked to mitochondrial function. mTOR activity was suppressed by mitochondrial dysfunction in response to acute osmotic stress induced by high levels of sorbitol, (Desai et al., 2002). The results suggested that a complex including mTOR formed at the permeability transition pore (PTP) to modulate mTOR activity in response to shifts in mitochondrial function (Desai et al., 2002). It is highly likely that such a complex would alter mTOR activity in a positive or negative manner depending on the cellular context.
Concluding remarks
It is clear that the activation of mTOR/p70S6K occurs during aging and in senescent cells. The source of this activation is likely to provide insight into the aging process and shed light on additional aspects of the senescent arrest. While metabolic inputs certainly can impact this pathway, we suggest that stress signaling from mitochondrial ROS activates mTOR/p70S6K during aging. Our laboratory is working toward an understanding of the molecular events involved in this activation.
Highlights.
Markers of enhanced mTORC1 activity have been detected in aged tissue.
mTORC1 activation occurs in senescent human cells independent of growth factor signaling.
Mitochondrial dysfunction increases in senescent cells.
Mitochondrial dysfunction may be linked to mTORC1 activation.
Rapamycin alleviates mitochondrial stress and preserves mitochondrial integrity.
Acknowledgements
This work was supported by grants AG22443 and AG39733 to CS and a fellowship from the Drexel Aging Initiative to TN.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Alessi DR, Pearce LR, Garcia-Martinez JM. New insights into mTOR signaling: mTORC2 and beyond. Science signaling. 2009;2:pe27. doi: 10.1126/scisignal.267pe27. [DOI] [PubMed] [Google Scholar]
- Alvers AL, Wood MS, Hu D, Kaywell AC, Dunn WA, Jr, Aris JP. Autophagy is required for extension of yeast chronological life span by rapamycin. Autophagy. 2009;5:847–849. doi: 10.4161/auto.8824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiel E, Everts B, Fritz D, Beauchamp S, Ge B, Pearce EL, Pearce EJ. Mechanistic Target of Rapamycin Inhibition Extends Cellular Lifespan in Dendritic Cells by Preserving Mitochondrial Function. J Immunol. 2014 doi: 10.4049/jimmunol.1302498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimov VN, Zabezhinski MA, Popovich IG, Piskunova TS, Semenchenko AV, Tyndyk ML, Yurova MN, Antoch MP, Blagosklonny MV. Rapamycin extends maximal lifespan in cancer-prone mice. Am J Pathol. 2010;176:2092–2097. doi: 10.2353/ajpath.2010.091050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Sahra I, Howell JJ, Asara JM, Manning BD. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science. 2013;339:1323–1328. doi: 10.1126/science.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz C, Hall MN. Where is mTOR and what is it doing there? J Cell Biol. 2013;203:563–574. doi: 10.1083/jcb.201306041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitto A, Lerner C, Torres C, Roell M, Malaguti M, Perez V, Lorenzini A, Hrelia S, Ikeno Y, Matzko ME, McCarter R, Sell C. Long-term IGF-I exposure decreases autophagy and cell viability. PLoS One. 2010;5:e12592. doi: 10.1371/journal.pone.0012592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J, Kuo CJ, Crabtree GR, Blenis J. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell. 1992;69:1227–1236. doi: 10.1016/0092-8674(92)90643-q. [DOI] [PubMed] [Google Scholar]
- Cuervo AM. Autophagy and aging: keeping that old broom working. Trends Genet. 2008;24:604–612. doi: 10.1016/j.tig.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- Demidenko ZN, Blagosklonny MV. Growth stimulation leads to cellular senescence when the cell cycle is blocked. Cell cycle. 2008;7:3355–3361. doi: 10.4161/cc.7.21.6919. [DOI] [PubMed] [Google Scholar]
- Desai BN, Myers BR, Schreiber SL. FKBP12-rapamycin-associated protein associates with mitochondria and senses osmotic stress via mitochondrial dysfunction. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:4319–4324. doi: 10.1073/pnas.261702698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibble CC, Manning BD. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nature cell biology. 2013;15:555–564. doi: 10.1038/ncb2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan D, Kim J, Shaw RJ, Guan KL. The autophagy initiating kinase ULK1 is regulated via opposing phosphorylation by AMPK and mTOR. Autophagy. 2011;7:643–644. doi: 10.4161/auto.7.6.15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 2001;292:288–290. doi: 10.1126/science.1059497. [DOI] [PubMed] [Google Scholar]
- Fok WC, Bokov A, Gelfond J, Yu Z, Zhang Y, Doderer M, Chen Y, Javors M, Wood WH, 3rd, Zhang Y, Becker KG, Richardson A, Perez VI. Combined treatment of rapamycin and dietary restriction has a larger effect on the transcriptome and metabolome of liver. Aging cell. 2014a;13:311–319. doi: 10.1111/acel.12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fok WC, Chen Y, Bokov A, Zhang Y, Salmon AB, Diaz V, Javors M, Wood WH, 3rd, Zhang Y, Becker KG, Perez VI, Richardson A. Mice fed rapamycin have an increase in lifespan associated with major changes in the liver transcriptome. PloS one. 2014b;9:e83988. doi: 10.1371/journal.pone.0083988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fok WC, Livi C, Bokov A, Yu Z, Chen Y, Richardson A, Perez VI. Short-term rapamycin treatment in mice has few effects on the transcriptome of white adipose tissue compared to dietary restriction. Mech Ageing Dev. 2014c;140C:23–29. doi: 10.1016/j.mad.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias MA, Thoreen CC, Jaffe JD, Schroder W, Sculley T, Carr SA, Sabatini DM. mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr Biol. 2006;16:1865–1870. doi: 10.1016/j.cub.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Molecular cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haidinger M, Poglitsch M, Geyeregger R, Kasturi S, Zeyda M, Zlabinger GJ, Pulendran B, Horl WH, Saemann MD, Weichhart T. A versatile role of mammalian target of rapamycin in human dendritic cell function and differentiation. J Immunol. 2010;185:3919–3931. doi: 10.4049/jimmunol.1000296. [DOI] [PubMed] [Google Scholar]
- Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, Pahor M, Javors MA, Fernandez E, Miller RA. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan JL, Oshiro N, Mizushima N. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Kim J, Guan KL. AMPK and mTOR in cellular energy homeostasis and drug targets. Annual review of pharmacology and toxicology. 2012;52:381–400. doi: 10.1146/annurev-pharmtox-010611-134537. [DOI] [PubMed] [Google Scholar]
- Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nature cell biology. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- Jia K, Chen D, Riddle DL. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 2004;131:3897–3906. doi: 10.1242/dev.01255. [DOI] [PubMed] [Google Scholar]
- Johnson SC, Martin GM, Rabinovitch PS, Kaeberlein M. Preserving youth: does rapamycin deliver? Science translational medicine. 2013a;5:211fs240. doi: 10.1126/scitranslmed.3007316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SC, Yanos ME, Kayser EB, Quintana A, Sangesland M, Castanza A, Uhde L, Hui J, Wall VZ, Gagnidze A, Oh K, Wasko BM, Ramos FJ, Palmiter RD, Rabinovitch PS, Morgan PG, Sedensky MM, Kaeberlein M. mTOR inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science. 2013b;342:1524–1528. doi: 10.1126/science.1244360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahi P. Protein synthesis and the antagonistic pleiotropy hypothesis of aging. Adv Exp Med Biol. 2010;694:30–37. doi: 10.1007/978-1-4419-7002-2_3. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature cell biology. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnichenko M, Hong L, Liao R, Vogt PK, Sun P. Attenuation of TORC1 signaling delays replicative and oncogenic RAS-induced senescence. Cell cycle. 2012;11:2391–2401. doi: 10.4161/cc.20683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova EA, Antoch MP, Novototskaya LR, Chernova OB, Paszkiewicz G, Leontieva OV, Blagosklonny MV, Gudkov AV. Rapamycin extends lifespan and delays tumorigenesis in heterozygous p53+/− mice. Aging (Albany NY) 2012;4:709–714. doi: 10.18632/aging.100498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CJ, Chung J, Fiorentino DF, Flanagan WM, Blenis J, Crabtree GR. Rapamycin selectively inhibits interleukin-2 activation of p70 S6 kinase. Nature. 1992;358:70–73. doi: 10.1038/358070a0. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontieva OV, Demidenko ZN, Blagosklonny MV. Contact inhibition and high cell density deactivate the mammalian target of rapamycin pathway, thus suppressing the senescence program. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:8832–8837. doi: 10.1073/pnas.1405723111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner C, Bitto A, Pulliam D, Nacarelli T, Konigsberg M, Van Remmen H, Torres C, Sell C. Reduced mammalian target of rapamycin activity facilitates mitochondrial retrograde signaling and increases life span in normal human fibroblasts. Aging cell. 2013;12:966–977. doi: 10.1111/acel.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R, Hall MN. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics. 2011;189:1177–1201. doi: 10.1534/genetics.111.133363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nature reviews Molecular cell biology. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Molecular cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- McKeehan WL, McKeehan KA, Hammond SL, Ham RG. Improved medium for clonal growth of human diploid fibroblasts at low concentrations of serum protein. In Vitro. 1977;13:399–416. doi: 10.1007/BF02615100. [DOI] [PubMed] [Google Scholar]
- Miller RA, Harrison DE, Astle CM, Baur JA, Boyd AR, de Cabo R, Fernandez E, Flurkey K, Javors MA, Nelson JF, Orihuela CJ, Pletcher S, Sharp ZD, Sinclair D, Starnes JW, Wilkinson JE, Nadon NL, Strong R. Rapamycin, but not resveratrol or simvastatin, extends life span of genetically heterogeneous mice. The journals of gerontology Series A, Biological sciences and medical sciences. 2011;66:191–201. doi: 10.1093/gerona/glq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming XF, Burgering BM, Wennstrom S, Claesson-Welsh L, Heldin CH, Bos JL, Kozma SC, Thomas G. Activation of p70/p85 S6 kinase by a pathway independent of p21ras. Nature. 1994;371:426–429. doi: 10.1038/371426a0. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacarelli T, Azar A, Sell C. Inhibition of mTOR Prevents ROS Production Initiated by Ethidium Bromide-Induced Mitochondrial DNA Depletion. Frontiers in endocrinology. 2014;5:122. doi: 10.3389/fendo.2014.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passos JF, Saretzki G, Ahmed S, Nelson G, Richter T, Peters H, Wappler I, Birket MJ, Harold G, Schaeuble K, Birch-Machin MA, Kirkwood TB, von Zglinicki T. Mitochondrial dysfunction accounts for the stochastic heterogeneity in telomere-dependent senescence. PLoS biology. 2007;5:e110. doi: 10.1371/journal.pbio.0050110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce LR, Huang X, Boudeau J, Pawlowski R, Wullschleger S, Deak M, Ibrahim AF, Gourlay R, Magnuson MA, Alessi DR. Identification of Protor as a novel Rictor-binding component of mTOR complex-2. The Biochemical journal. 2007;405:513–522. doi: 10.1042/BJ20070540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, Sabatini DM. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovich IG, Anisimov VN, Zabezhinski MA, Semenchenko AV, Tyndyk ML, Yurova MN, Blagosklonny MV. Lifespan extension and cancer prevention in HER-2/neu transgenic mice treated with low intermittent doses of rapamycin. Cancer Biol Ther. 2014;15:586–592. doi: 10.4161/cbt.28164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers RW, 3rd, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes & development. 2006;20:174–184. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DJ, Grove JR, Calvo V, Avruch J, Bierer BE. Rapamycin-induced inhibition of the 70-kilodalton S6 protein kinase. Science. 1992;257:973–977. doi: 10.1126/science.1380182. [DOI] [PubMed] [Google Scholar]
- Ramanathan A, Schreiber SL. Direct control of mitochondrial function by mTOR. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:22229–22232. doi: 10.1073/pnas.0912074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos FJ, Chen SC, Garelick MG, Dai DF, Liao CY, Schreiber KH, MacKay VL, An EH, Strong R, Ladiges WC, Rabinovitch PS, Kaeberlein M, Kennedy BK. Rapamycin reverses elevated mTORC1 signaling in lamin A/C-deficient mice, rescues cardiac and skeletal muscle function, and extends survival. Science translational medicine. 2012;4:144ra103. doi: 10.1126/scitranslmed.3003802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robida-Stubbs S, Glover-Cutter K, Lamming DW, Mizunuma M, Narasimhan SD, Neumann-Haefelin E, Sabatini DM, Blackwell TK. TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell metabolism. 2012;15:713–724. doi: 10.1016/j.cmet.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robitaille AM, Christen S, Shimobayashi M, Cornu M, Fava LL, Moes S, Prescianotto-Baschong C, Sauer U, Jenoe P, Hall MN. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science. 2013;339:1320–1323. doi: 10.1126/science.1228771. [DOI] [PubMed] [Google Scholar]
- Rogers AN, Chen D, McColl G, Czerwieniec G, Felkey K, Gibson BW, Hubbard A, Melov S, Lithgow GJ, Kapahi P. Life span extension via eIF4G inhibition is mediated by posttranscriptional remodeling of stress response gene expression in C. elegans. Cell metabolism. 2011;14:55–66. doi: 10.1016/j.cmet.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Molecular cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Sandri M, Barberi L, Bijlsma AY, Blaauw B, Dyar KA, Milan G, Mammucari C, Meskers CG, Pallafacchina G, Paoli A, Pion D, Roceri M, Romanello V, Serrano AL, Toniolo L, Larsson L, Maier AB, Munoz-Canoves P, Musaro A, Pende M, Reggiani C, Rizzuto R, Schiaffino S. Signalling pathways regulating muscle mass in ageing skeletal muscle: the role of the IGF1-Akt-mTOR-FoxO pathway. Biogerontology. 2013;14:303–323. doi: 10.1007/s10522-013-9432-9. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Sabatini DM. Redox regulation of the nutrient-sensitive raptor-mTOR pathway and complex. J Biol Chem. 2005;280:39505–39509. doi: 10.1074/jbc.M506096200. [DOI] [PubMed] [Google Scholar]
- Schieke SM, Phillips D, McCoy JP, Jr, Aponte AM, Shen RF, Balaban RS, Finkel T. The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J Biol Chem. 2006;281:27643–27652. doi: 10.1074/jbc.M603536200. [DOI] [PubMed] [Google Scholar]
- Selman C, Withers DJ. Mammalian models of extended healthy lifespan. Philos Trans R Soc Lond B Biol Sci. 2011;366:99–107. doi: 10.1098/rstb.2010.0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Molecular cell. 2010;40:310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp ZD, Richardson A. Aging and cancer: can mTOR inhibitors kill two birds with one drug? Target Oncol. 2011;6:41–51. doi: 10.1007/s11523-011-0168-7. [DOI] [PubMed] [Google Scholar]
- Sobko A. Systems biology of AGC kinases in fungi. Sci STKE. 2006;2006:re9. doi: 10.1126/stke.3522006re9. [DOI] [PubMed] [Google Scholar]
- Sonenberg N. eIF4E, the mRNA cap-binding protein: from basic discovery to translational research. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2008;86:178–183. doi: 10.1139/O08-034. [DOI] [PubMed] [Google Scholar]
- Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, Dang N, Johnston ED, Oakes JA, Tchao BN, Pak DN, Fields S, Kennedy BK, Kaeberlein M. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YX, Ji X, Mao X, Xie L, Jia J, Galvan V, Greenberg DA, Jin K. Differential activation of mTOR complex 1 signaling in human brain with mild to severe Alzheimer's disease. Journal of Alzheimer's disease : JAD. 2014;38:437–444. doi: 10.3233/JAD-131124. [DOI] [PubMed] [Google Scholar]
- Thoreen CC. Many roads from mTOR to eIF4F. Biochem Soc Trans. 2013;41:913–916. doi: 10.1042/BST20130082. [DOI] [PubMed] [Google Scholar]
- Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G, Riezman H, Broach JR, De Virgilio C, Hall MN, Loewith R. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Molecular cell. 2007;26:663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nature cell biology. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Alessi DR. The PI3K-PDK1 connection: more than just a road to PKB. The Biochemical journal. 2000;346(Pt 3):561–576. [PMC free article] [PubMed] [Google Scholar]
- Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- Xu S, Cai Y, Wei Y. mTOR Signaling from Cellular Senescence to Organismal Aging. Aging and disease. 2014;5:263–273. doi: 10.14336/AD.2014.0500263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Inoki K, Ikenoue T, Guan KL. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes & development. 2006;20:2820–2832. doi: 10.1101/gad.1461206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Hoff H, Marinucci T, Cristofalo VJ, Sell C. Mitogen-independent phosphorylation of S6K1 and decreased ribosomal S6 phosphorylation in senescent human fibroblasts. Exp Cell Res. 2000;259:284–292. doi: 10.1006/excr.2000.4965. [DOI] [PubMed] [Google Scholar]
- Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, Lu TA, Benzer S, Kapahi P. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell. 2009;139:149–160. doi: 10.1016/j.cell.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nature reviews Molecular cell biology. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]