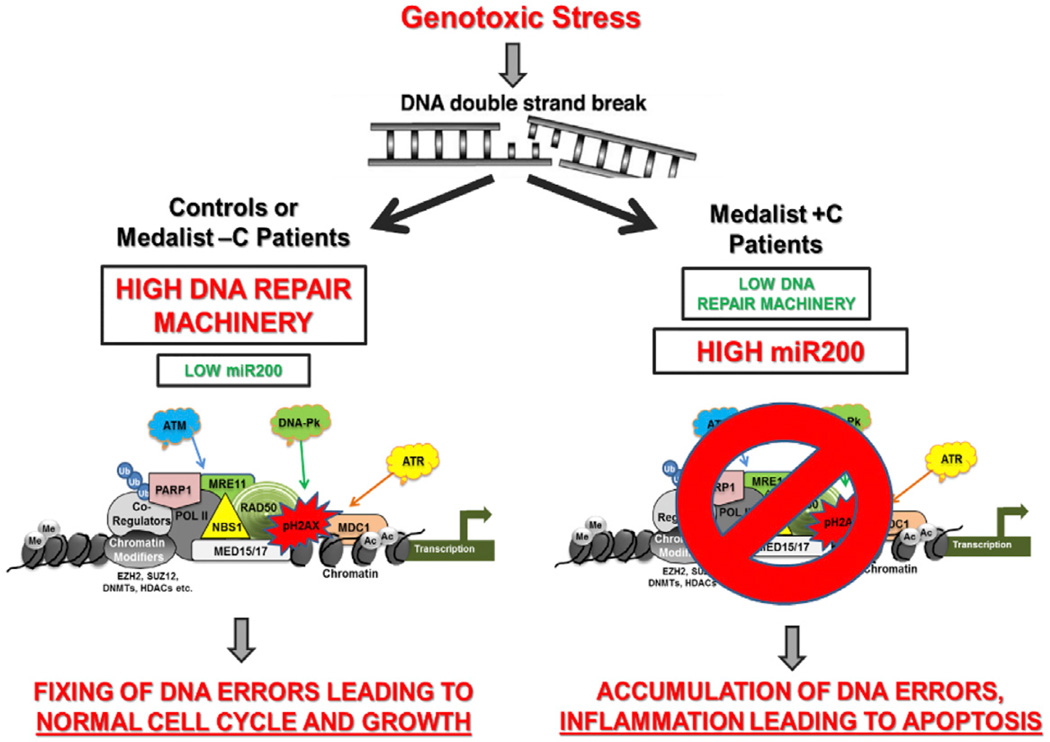
SUMMARY
The mechanisms underlying the development of complications in type 1 diabetes (T1D) are poorly understood. Disease modeling of induced pluripotent stem cells (iPSCs) from patients with longstanding T1D(disease duration ≥ 50 years) with severe (Medalist +C) or absent to mild complications (Medalist −C) revealed impaired growth, reprogramming, and differentiation in Medalist +C. Genomics and proteomics analyses suggested differential regulation of DNA damage checkpoint proteins favoring protection from cellular apoptosis in Medalist −C. In silico analyses showed altered expression patterns of DNA damage checkpoint factors among the Medalist groups to be targets of miR200, whose expression was significantly elevated in Medalist +C serum. Notably, neurons differentiated from Medalist +C iPSCs exhibited enhanced susceptibility to genotoxic stress that worsened upon miR200 overexpression. Furthermore, knockdown of miR200 in Medalist +C fibroblasts and iPSCs rescued checkpoint protein expression and reduced DNA damage. We propose miR200-regulated DNA damage checkpoint pathway as a potential therapeutic target for treating complications of diabetes.
Graphical Abstract
INTRODUCTION
Type 1 diabetes (T1D) is associated with micro- and macrovascular complications (Chang-Chen et al., 2008; Forbes and Cooper, 2013; Rask-Madsen and King, 2013; Fang et al., 2004; Schalkwijk and Stehouwer, 2005), but the mechanisms underlying their development remain elusive, due in part to a lack of suitable cellular models for molecular investigation (Calcutt et al., 2009; Reddy and Natarajan, 2011; Reddy et al., 2012; Lacolley et al., 2009). Diabetic complications affecting the heart and vascular cells (cardiovascular), kidney (nephropathy), eyes (retinopathy), or nerves (neuropathy) arise as a consequence of stress-induced apoptosis, resulting in loss of the functional cellular pool and a concomitant failure of the body’s inherent mechanism to compensate (Brownlee, 2001, 2005). Animal modeling of these phenotypes has met with challenges over the years, in part due to an inability to precisely mimic the human disease phenotype. To overcome such challenges, we derived induced pluripotent stem cells (iPSCs) from individuals with long-standing T1D ( ≥ 50 years), termed Medalist patients (Keenan et al., 2007; Maehr et al., 2009; Park et al., 2008; Tiscornia et al., 2011), and age-matched healthy controls. The Medalists were extensively phenotyped by clinical examination and assessed for the presence of complications and classified as those with severe (Medalist +C) and those with absent to mild (Medalist −C) complications (Keenan et al., 2007, 2010). Mass spectrometry (MS)-based quantitative proteomics analyses of these iPSCs, along with microarray gene expression profiling of the patient fibroblasts used to derive the iPSCs, implicated preserved DNA damage checkpoint pathway function, due to suppressed miR200 expression as a mechanism underlying protection against diabetic complications in the Medalist −C subgroup. Consistently, we observed elevated miR200 levels in sera from Medalist +C patients. Furthermore, overexpression of miR200 in two target cell types, namely primary human neurons and primary endothelial cells, caused downregulation of ATM protein, increased pH2AX, and cellular apoptosis. Corroboratively, neurons differentiated from Medalist +C iPSCs showed increased susceptibility to DNA damage-induced apoptosis, as compared to neurons derived from Medalist −C or control iPSCs, implying clinical significance. The reversal of DNA damage upon knockdown of miR200 in fibroblasts as well as iPSCs from Medalist +C patients points to a direct mechanistic role for miR200. Collectively, our work highlights regulation of the DNA damage checkpoint pathway by miR200 as a mechanism underlying protection against diabetic complications. Targeting of this pathway may lead to more effective interventions to reduce the burden of T1D.
RESULTS
Impaired Growth, Reprogramming, and Self-Renewal in Medalist +C Patient iPSCs
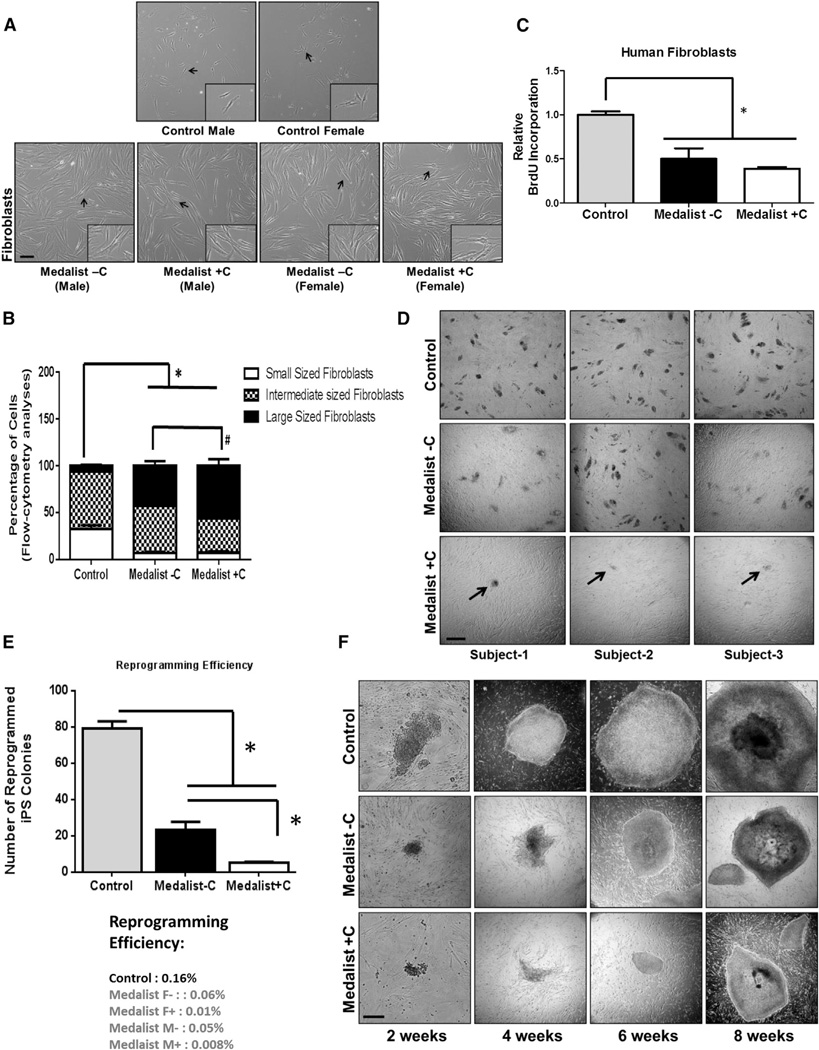
We report, for the first time to our knowledge, derivation of iPSCs from patients with long-standing T1D with severe (Medalist +C) or absent to mild (Medalist −C) complications. Reprogramming of skin fibroblasts from the T1D patients or age-matched controls was performed by cre-excisable lentivirus (Figure S1A). Table 1 summarizes the clinical characteristics of the fibroblast donors. We derived six iPSC lines for each group including three clones per line (a total of 54 lines). Interestingly, microscopic examination and flow cytometry analyses revealed larger cell size (Figures 1A and 1B) and reduced growth potential in fibroblasts from T1D patients (Figure 1C) compared to controls and confirmed this in independent experiments (Figure S1B). We also observed a striking impairment in the reprogramming efficiency of fibroblasts from Medalist +C (Figure 1D), as assessed by quantitative analyses of alkaline phosphatase positive colonies (Figure 1E). The growth potential of emerging iPSC colonies from Medalist +C was also impaired (Figure 1F). Despite these phenotypes, the pluripotency of iPSCs from all clinical groups was confirmed by expression of pluripotency markers including OCT4, NANOG, SOX2, and SSEA4 (Figures S1C–S1F), and staining for early (alkaline phosphatase, Figure S1G) and late (Tra1-60, Figure S1H; Tra1-81, Figure S1I) pluripotent cell-surface markers by live-cell imaging.
Table 1.
Clinical Characteristics of Longstanding T1D Patients Used for iPSC Derivation
| Group | Gender | Age (years) | Duration of Diabetes (years) |
|---|---|---|---|
| Control | M = 4; F = 2 | 71.2 ± 11.1 | n/a |
| Medalist −C | M = 4; F = 2 | 74.5 ± 12.9 | 64.3 ± 11.3 |
| Medalist +C | M = 2; F = 4 | 77.2 ± 5.0 | 67.5 ± 8.5 |
Summarized in the table are clinical characteristics of patients with longstanding T1D with severe (Medalist +C) or absent to mild (Medalist −C) diabetic complications as well as age-matched control or healthy subjects that were used for the derivation of iPSCs. Clinical characteristics of Medalists with absent to mild complications (Medalist −C; % 1 complications: M1, M4, M10, M14, M17, and M22), Medalists with complications (Medalist +C;R2: M3, M6, M9, M11, M12, and M18), and age-matched controls (Control: control 1, control 2, control 3, control 4, control 5, and control 6), used for the derivation of fibroblasts from skin biopsies. Human induced pluripotent stem cells (hiPSCs) reprogrammed from these fibroblasts were used in the study. The number of patients in each group is 6. Data are presented as mean ± SD. Medalist −C: 0/6 with CVD, 5/6 manifest DN Class 0-IIA, and 6/6 manifest no to mild NPDR. Medalist +C: 6/6 with CVD, 5/6 exhibit DN Class IIB or IV, and 4/6 exhibit PDR.
CVD, cardiovascular disease; DN class, diabetic nephropathy class; PDR, proliferative diabetic retinopathy; NPDR, non-proliferative diabetic retinopathy; M, male; F, female; n/a, not applicable.
Figure 1. Reduced Growth and Reprogramming Efficiency of Primary Fibroblasts from Medalist +C Type 1 Diabetes Patients.
(A) Morphological characteristics of primary skin fibroblasts obtained from T1D Medalist −C and +C and age-matched controls, Scale bar: 50 µm. Inset shows magnified image.
(B) Flow cytometry analyses: quantitative size assessment of fibroblasts from the three clinical groups (Controls, Medalist −C, and Medalist +C).
(C) Flow cytometry-based quantitation of BrdU incorporation, a surrogate for cell proliferation, in fibroblasts.
(D) Reprogramming of fibroblasts from the three groups, using lentiviral expression of OCT4, SOX2, KLF4, and c-MYC and assessment of number of emerging iPSC clusters on day 14 of reprogramming. Scale bar: 100 µm.
(E) Quantitation of reprogramming efficiency across n = 6 reprogramming experiments.
(F) Dynamics of growth of Control, Medalist −C, and Medalist +C iPSCs over time, 2, 4, 6, and 8 weeks after reprogramming with a polycistronic lentivirus. Scale bar: 200 µm.
Data represent N = 6 independent subjects per clinical group in each experiment, repeated three times, represented as mean ± SD. # p value < 0.05; *p value < 0.001. See also Figure S1.
Impaired Cellular Differentiation in Medalist +C Patient iPSCs
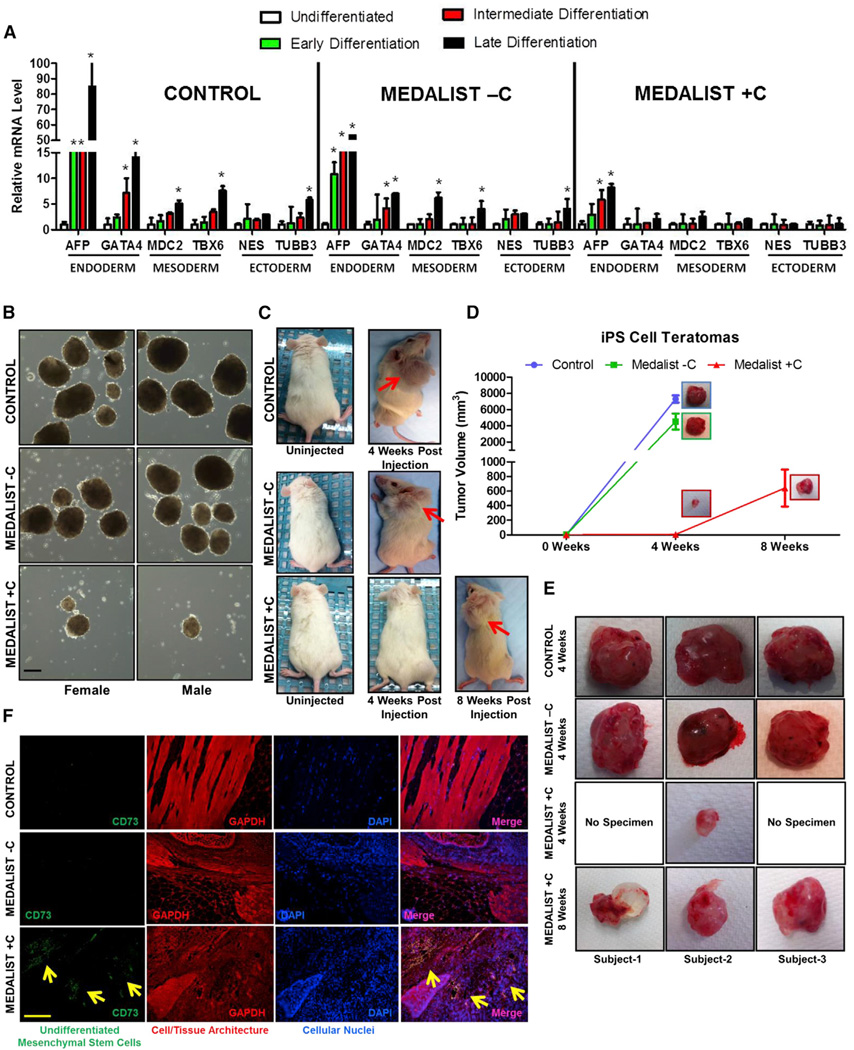
The differentiation potential of iPSCs was assessed by differentiation at different time points over 8 weeks (early [0–2.5 weeks], intermediate [2.5–5 weeks], or late [5–8weeks] periods). Expression of germ layer markers, using real-time qPCR analyses showed impairment in the differentiation of Medalist +C iPSCs into the endoderm lineage and a complete blockade of mesoderm or ectoderm lineages (Figure 2A). In addition, in contrast to the normal differentiation of control iPSCs, Medalist −C iPSCs showed delayed expression of mesodermal and ectodermal differentiation markers and blunted expression of markers of endodermal differentiation (Figure S2A). Consistently, in vitro assays showed smaller and fewer embryoid bodies in Medalist +C (Figure 2B).
Figure 2. Impaired Differentiation Potential of iPSCs from Medalist +C Type 1 Diabetes Patients.
(A) Assessment of markers of the three germ layers, by quantitative real-time qPCR analysis in iPSCs (Control, Medalist −C, and Medalist +C) at various stages in the spontaneous differentiation process.
(B) Embryoid body formation assay: in vitro differentiation of iPSCs. Scale bar: 100 µm.
(C) Teratoma assay: in vivo differentiation of iPSCs, post-injection into NOD-SCID mice.
(D) Quantitative assessment of teratoma size/volume resulting from the in vivo differentiation of iPSCs.
(E) Morphological imaging of differentiated tissue harvested from NOD-SCID mice after injection with iPSCs from Control, Medalist −C, and Medalist +C subjects.
(F) Fluorescence immunostaining analyses for CD73, a marker for undifferentiated mesenchymal stem cells, in the differentiated tissue harvested from NOD-SCID mice after injection with iPSCs from Control, Medalist −C, and Medalist +C subjects. Scale bar: 200 µm.
Data represent N = 4 independent subjects per clinical group and n = 3 clonal iPSC lines per subject in each experiment, repeated three times, represented as mean ± SD. # p value < 0.05; *p value < 0.001. See also Figure S2.
To investigate the physiological significance of these observations, we next assessed iPSC differentiation in vivo in non-obese diabetic mice with severe combined immune deficiency (NODSCID) injected with equal numbers of iPSCs from each experimental group. Histological analyses (Figure S2B) and immunostaining for germ layer markers (Figures S2C–S2E) revealed that iPSCs from all three groups gave rise to tissues of all three germ layers. However, a significantly impaired cellular differentiation in Medalist +C was identified both by a delay in the timescale of differentiation (8 weeks for Medalist +C versus 4 weeks for controls or Medalist −C, Figures 2C–2E) and the presence of undifferentiated mesenchyme in >70% of the transplanted graft, evident upon histo-pathological investigation and staining for the mesenchymal stem cell marker, CD73 (Figure 2F). Consistently, we detected a significant loss of ACINUS protein expression (Figures S2F–S2H) and consequently serine-1004 phosphorylation-dependent activation, in Medalist +C cells (Figures S2F, S2G, and S2I). Failure to activate expression of ACINUS, which is necessary for early differentiation of embryonic stem cells (ESCs) (Rigbolt et al., 2011; Van Hoof et al., 2009) may, in part, underlie the impairment in differentiation observed in Medalist +C patients. It is possible that exposure of fibroblasts to prolonged periods of hyperglycemia in the Medalist patients, and consequent hyperosmotic stress, would have impacted their ability to reprogramming as well as the subsequent potential for proliferation and/or differentiation (Madonna et al., 2013) and requires further investigation.
Preserved DNA Damage Checkpoint Pathway Function in Medalist −C Protects against DNA Damage-Induced Cell Death
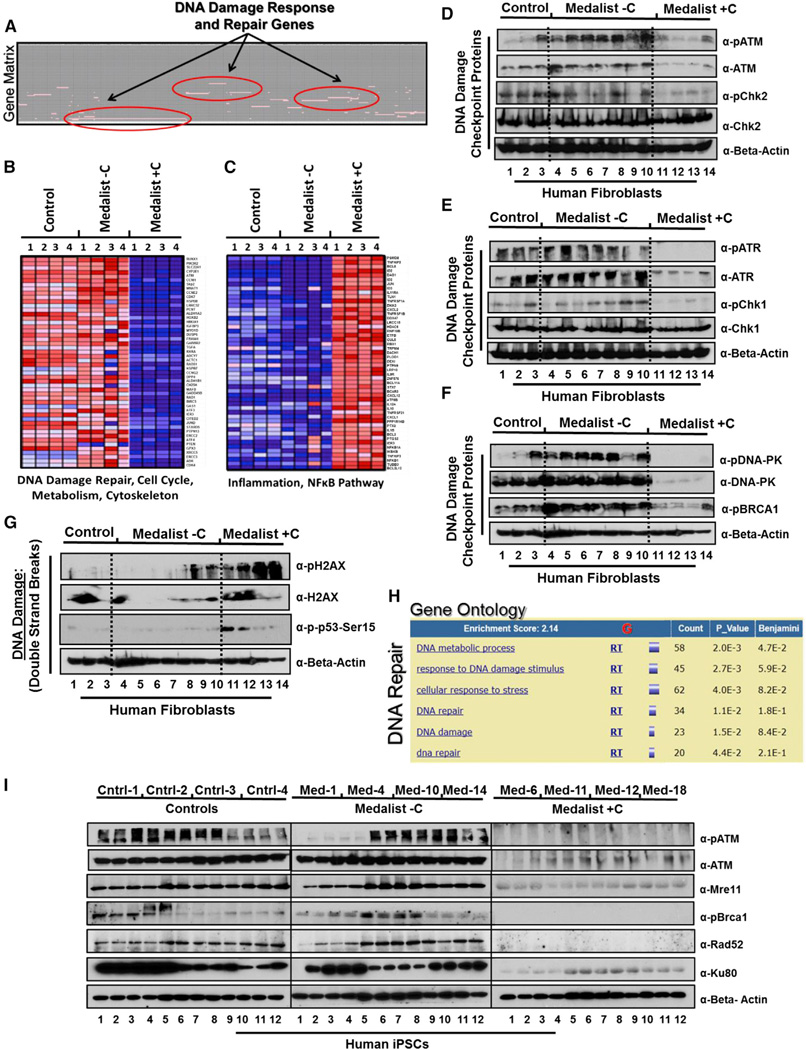
To investigate molecular defects underlying the complications in Medalist +C and/or the protective factors preventing their occurrence in Medalist −C, we subjected donor fibroblasts to microarray gene expression profiling. Comparative marker selection revealed differential expression of genes associated with diabetes between control and T1D patients (Table S1A; A, B, and C), confirming the suitability of these cells for biomarker profiling. Members of the DNA damage checkpoint family were differentially expressed between Medalist −C and +C (Figures 3A and 3B; Figure S3A; Table S1B and Table S2). Further analyses revealed overexpression of genes related to NF-κB and other inflammatory pathways in Medalist +C fibroblasts (Figure 3C). Western analysis showed a protein level impairment of all DNA damage checkpoint proteins, including ATM (Figure 3D), ATR (Figure 3E), and DNA-PK (Figure 3F) regulated in Medalist +- C fibroblasts compared to both Medalist −C and controls. Consistently, Medalist +C cells showed an increased expression of phosho-H2AX and phospho-Ser15-p53 (Figure 3G), suggesting a higher burden of DNA damage due to an impaired repair process. These observations were further supported by a positive enrichment profile of genes in the ATR/BRCA pathway (Figure S3B), UV response cluster (Figure S3C), and proteasome complex (Figure S3D) in fibroblasts from Medalist −C. To investigate the abundance of DNA damage checkpoint proteins in iPSCs, we next performed MS-based quantitative proteomics (Figures S4A and S4B) and confirmed the impairment of the DNA damage checkpoint machinery in Medalist +C iPSCs (Figure 3H; Table S3 and Table S4; see accessory information on Figures 1A–1D in Supplemental Information). High enrichment scores of ATM/ATR/BRCA-DNA repair proteins (Figure S4C), UV response pathway (Figure S4D), and proteasomal DNA repair components (Figure S4E) in Medalist −C iPSCs supported our initial observations. Interestingly, expression of most DNA damage checkpoint proteins was highest in Medalist −C patients (Figure S4F). We confirmed these results by western immunoblot analyses in iPSCs showing significantly reduced expression of key proteins in the DNA damage checkpoint pathway, including but not limited to pATM, Mre11, pBrca1, Rad52, and Ku80 in Medalist +C iPSCs compared to both Medalist −C and control iPSCs (Figure 3I). Furthermore, phospho-proteomics analyses revealed a significant impairment in the activation of MDC1 in Medalist +C iPSCs (see accessory information on Figures 1E–1G in Supplemental Information), a key factor that initiates the DNA damage checkpoint by binding pH2AX in response to cellular DNA damage (Spycher et al., 2008; Stucki et al., 2005) while tightly coordinating the repair with cell cycle and mitosis (Branzei and Foiani, 2008; Dephoure et al., 2008; Goldberg et al., 2003; Stewart et al., 2003). Of clinical relevance, integrated pathway analyses revealed a significant association of proteins differentially expressed between Medalist −C and +C cells to cardiovascular, neurological, ophthalmic, and renal diabetic complications, secondary to defects in DNA damage checkpoint pathway and recombination, chromosome assembly, and cellular response to stress (Table S5).
Figure 3. Impairment of DNA Double Strand Break Repair in Fibroblasts and iPSCs From Medalist +C Type 1 Diabetes Patients.
(A) Gene matrix representing DNA damage repair as one of the most differentially expressed functional gene clusters in microarray analysis of fibroblasts from the three clinical groups (Control, Medalist −C, and Medalist +C) (N = 2; n = 4).
(B) Heatmaps showing downregulation
(blue) of genes involved in DNA repair, cell cycle, metabolism, etc. in Medalist +C fibroblasts compared to Medalist −C and controls.
(C) Heatmaps showing upregulation (red) of genes involved in inflammation and NF-κB pathway in Medalist +C fibroblasts compared to Medalist −C and controls.
(D) Western immunoblot analysis in fibroblasts highlighting impairment in DNA damage checkpoint proteins involving ATM.
(E) DNA damage checkpoint proteins involving ATR.
(F) DNA damage checkpoint proteins involving DNA-PK.
(G) Western immunoblot analysis in fibroblasts, highlighting increased expression of pH2AX and pSer15-p53, surrogate for DNA damage, in Medalist +C compared to Medalist −C or controls.
(H) MS-based quantitative proteomics analysis shows DNA damage repair proteins to be most differentially expressed between Medalist −C and Medalist +C iPSCs (N = 4; n = 2).
(I) Western immunoblot analysis in iPSCs from the three clinical groups, highlighting impairment of DNA damage checkpoint proteins in Medalist +C compared to Medalist −C or controls.
Data represent N = 4 independent subjects per clinical group and n = 3 clonal iPSC lines per subject in each experiment unless indicated otherwise and repeated at least three times. # p value < 0.05; *p value < 0.001. For Figures 3D–3G, additional fibroblast lines were used for Medalist −C samples. See also Figures S3 and S4 and accessory information on Figure 1 in Supplemental Information.
Taken together, these data suggest a preserved DNA damage checkpoint pathway is essential for protection from diabetic complications in the Medalist −C subgroup.
Suppressed miR200 in Medalist –C Preserves DNA Damage Checkpoint Pathway and Protects against Apoptosis
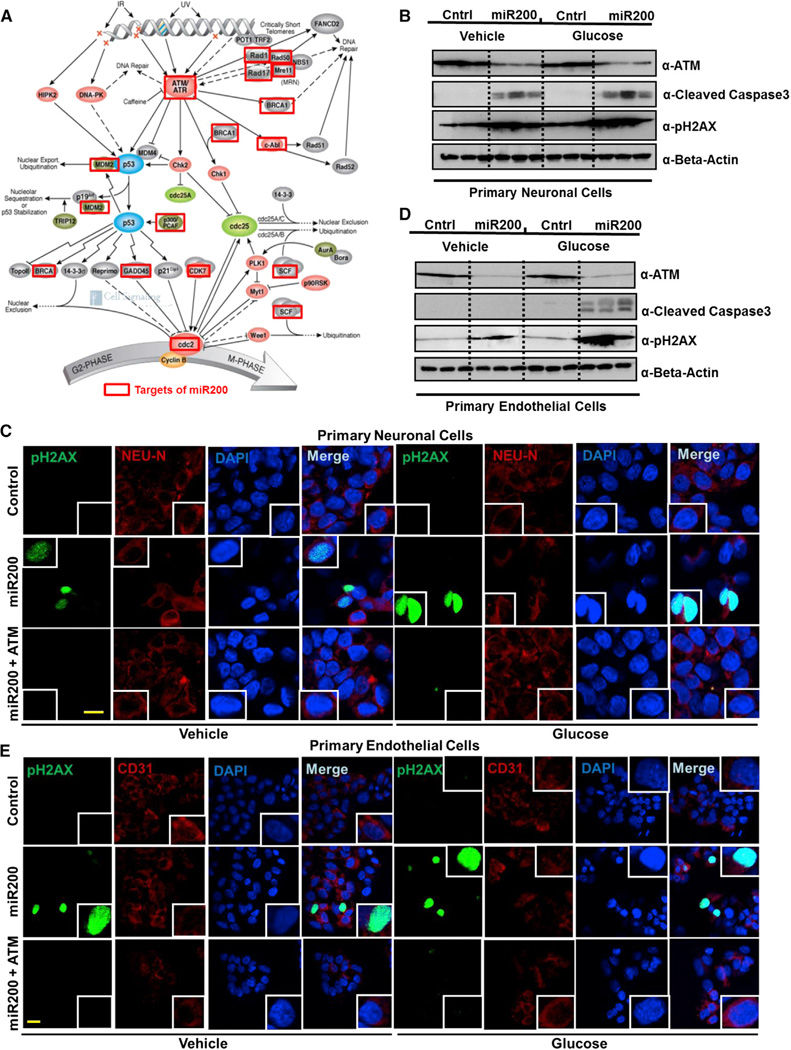
In silico analyses of genomics and proteomics data demonstrated an enrichment of direct miR200 targets within the DNA damage checkpoint pathway repair genes/proteins differentially expressed between Medalist −C and +C (e.g., ATM, ATR, Rad1, 17, 50, Mre11, Brca1) (depicted by red boxes in Figure 4A, Table S6; A and B, and Figure S5A). Other proteins that were non-targets of miR200, such as p53 and Cyclin B, were expressed at comparable levels among the three groups. To directly evaluate this pathway, we used two cell types known to be affected in T1D, neurons and endothelial cells. Indeed, overexpression of miR200 in neuronal cells resulted in a loss of ATM (a key component of the DNA damage checkpoint pathway), with concomitant increases in cleaved caspase-3 and nuclear pH2AX (Figures 4B and 4C; Figures S5B–S5D). These findings were corroborated in primary human endothelial cells, a target cell involved in diabetic complications (Figures 4D and 4E; Figure S5E). Previous reports indicate that cells exposed to stress undergo apoptosis due to nuclear loss of DNA repair proteins, particularly Ku isoforms (Song et al., 2003), downregulated in the Medalist +C (Figure 3I), presumably due to upregulation of miR200. Supporting this finding, we identified the miR200 target sequence in several components of the DNA damage checkpoint pathway (Figure S5A), a majority of which were downregulated in Medalist +C cells (Figures 3D–3F and 3I).
Figure 4. Impaired Expression of DNA Damage Checkpoint Pathway Proteins Secondary to miR200 Overexpression Leads to Apoptosis in Medalist +C Type 1 Diabetes Patients.
(A) Schematic of DNA damage checkpoint pathway machinery, adapted from Cell Signaling, Inc., showing genes predicted to be miR200 targets in red box.
(B) Western immunoblot analyses in primary human neurons subjected to miR200 overexpression (48 hr) followed by 24-hr treatment with either vehicle (5 mM glucose) or high glucose (25 mM glucose).
(C) Immunostaining analyses of pH2AX in primary human neurons subjected to miR200 overexpression (48 hr) followed by rescue of the DNA damage phenotype by re-expression of ATM and 24-hr treatment with either vehicle (5 mM glucose) or high glucose (25 mM glucose) (pH2AX, green; DAPI, blue). Scale bar: 50 µm.
(D) Western immunoblot analyses in primary human endothelial cells subjected to miR200 overexpression (48 hr) followed by 24-hr treatment with either vehicle (5 mM glucose) or high glucose (25 mM glucose).
(E) Immunostaining analyses of pH2AX in primary human endothelial cells subjected to miR200 overexpression (48 hr) followed by rescue of the DNA damage phenotype by re-expression of ATM and 24-hr treatment with either vehicle (5 mM glucose) or high glucose (25 mM glucose) (pH2AX, green, DAPI, blue). Scale bar: 50 µm.
All experiments were repeated three times, represented as mean ± SD. *p value < 0.001. See also Figure S5.
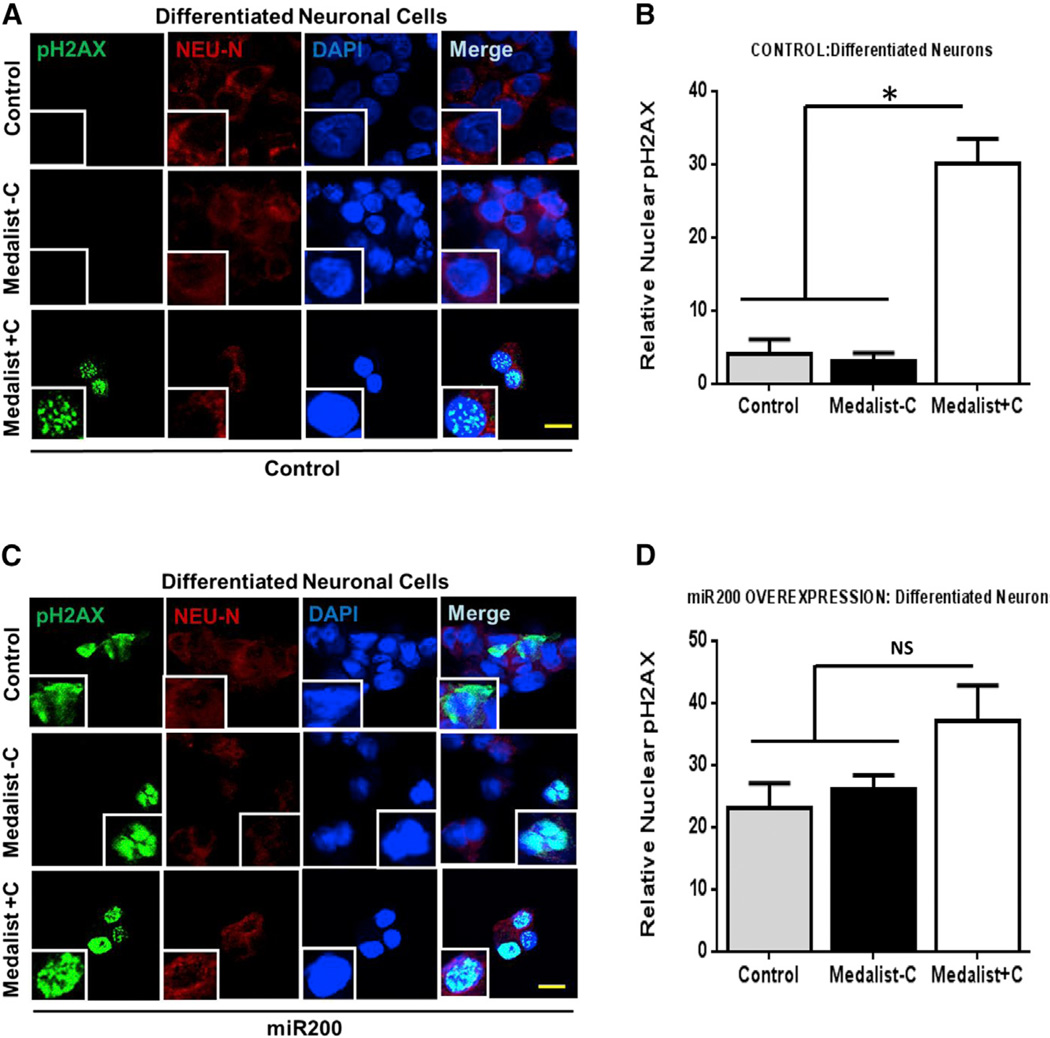
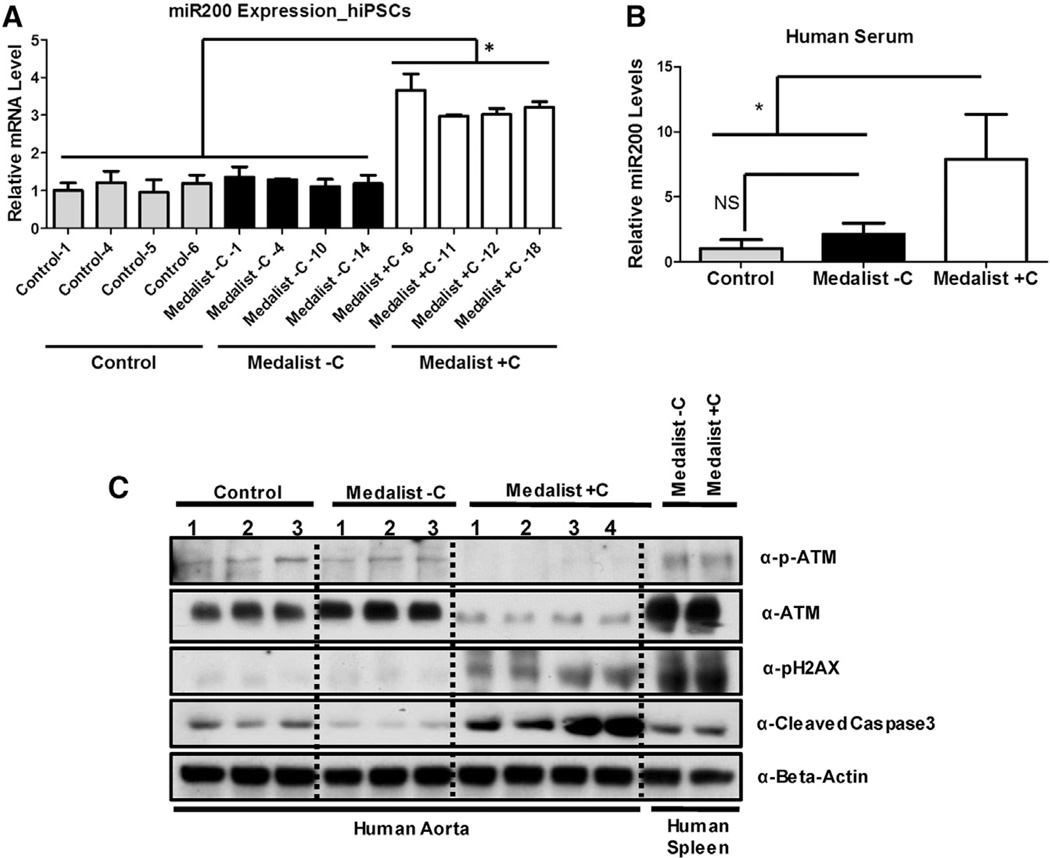
Restoration of ATM function in the presence of elevated miR200 reduced DNA damage to control levels, confirming that miR200-mediated increase in DNA damage/cellular apoptosis occurs, in part, through its actions on ATM protein (Figures 4C and 4E, lower panels). Our findings in both neuronal and endothelial cells provide compelling evidence for broad clinical relevance of these observations. Notably, directed differentiation of these patient-derived iPSCs into the neuronal lineage revealed an increase in DNA damage, observed by a 6-fold upregulation of nuclear pH2AX staining, in Medalist +C-derived neurons, presumably due to an impaired DNA damage checkpoint pathway response (Figures 5A and 5B; Figures S6A and S6B). Subjecting these differentiated neurons to overexpression of miR200 further increased DNA damage while reducing cellular viability (as observed by Trypan blue staining), with more dramatic effects in Medalist +C compared to Medalist −C or controls (Figures 5C and 5D). Of likely clinical relevance, elevated levels of miR200 were observed in iPSCs (Figure 6A) and sera from Medalist +C (Figure 6B). The ability to detect miR200 in blood is consistent with previous reports that miRNAs are present in the circulation (reviewed in Kato et al., 2013; Epstein, 2014). Indeed, circulating miRNAs have been suggested to originate from several sources relevant to our studies including inflammatory foci (Kong et al., 2010), endothelial cells (Chamorro-Jorganes et al., 2013), or mononuclear cells and platelets (Hunter et al., 2008).
Figure 5. Increased DNA Damage Worsened by miR200 Overexpression in Neurons Differentiated from Medalist +C Type 1 Diabetes Patients.
(A) Immunostaining analyses of pH2AX (marker for DNA damage) and Neu-N (marker for neurons) in neuronal-like cells obtained from directed differentiation of human iPSCs from the three clinical groups including Control 2 and Control 3 as healthy controls, M4 and M14 as Medalists without neuropathy, and M6 and M9 as Medalists with neuropathy (pH2AX, green; Neu-N, red; DAPI, blue). Scale bar: 50 µm.
(B) Quantitation of nuclear H2Ax foci across multiple experiments described in (A).
(C) Immunostaining analyses of pH2AX (marker for DNA damage) and Neu-N (marker for neurons) in neuronal-like cells obtained from directed differentiation of human iPSCs from the three clinical groups post-miR200 overexpression (48 hr) (pH2AX, green; Neu-N, red; DAPI, blue).
(D) Quantitation of nuclear H2Ax foci across multiple experiments described in (A). Scale bar: 50 µm.
See also Figure S6. All experiments were repeated three times, represented as mean ± SD. *p value < 0.001.
Figure 6. Increased miR200 Expression in Medalist +C Patients Impairs DNA Damage Checkpoint Pathway Protein Function, Increasing Susceptibility to Cell Death and Apoptosis.
(A and B) Quantitative real-time qPCR analysis in (A) iPSCs (controls: N = 4, Medalist −C: N = 4, and Medalist +C: N = 4) and (B) human sera (controls: N = 6, Medalist −C: N = 2, and Medalist +C: N = 6) to assess relative circulating levels of miR200.
(C) Protein expression analyses in aorta obtained from human subjects from the three groups. All experiments were repeated three times, represented as mean ± SD. *p value < 0.001.
Enhanced presence of miR200 presumably reflects its epigenetic de-repression by potential chromatin modifiers differentially regulated between Medalist −C and +C in proteomics analyses (Table S3 and Table S4), as suggested by previous reports (Vrba et al., 2010). Consistently, we observed reduced expression of DNA damage checkpoint protein(s) such as ATM and increased expression of pH2AX and cleaved caspase-3 in primary aortic tissue obtained from Medalist +C, compared to both Medalist −C and controls (Figure 6C).
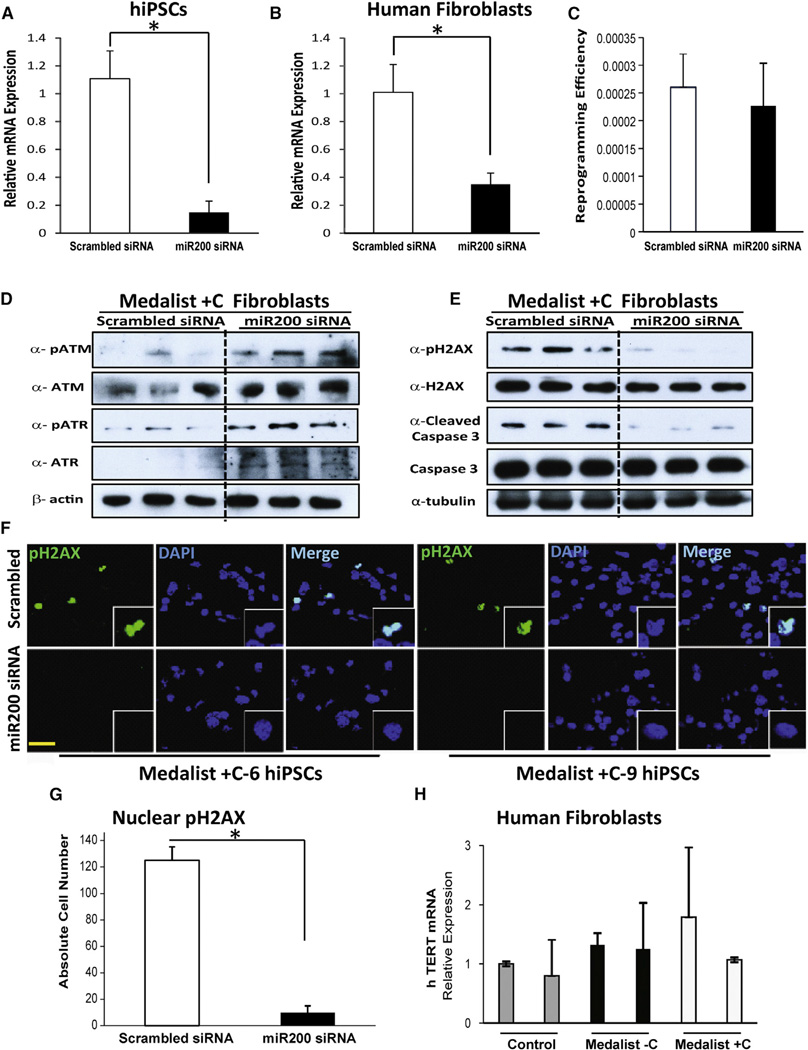
miR200 Knockdown Restores Expression of DNA Damage Checkpoint Proteins in Both Medalist +C Fibroblasts and Medalist +C Human iPSCs
To evaluate whether miR200 is directly involved in reprogramming and in modulating the expression of proteins in the DNA damage checkpoint pathway, we first successfully knocked down (KD) miR200 (A, B, and C together) in Medalist +C iPSCs (Figure 7A) and in Medalist +C fibroblasts (Figure 7B). Examination of reprogramming efficiency of Medalist +C fibroblasts treated with miR200 siRNA revealed no significant differences compared with Medalist +C fibroblasts treated with scrambled siRNA (Figure 7C). However, western analysis showed rescue of DNA damage checkpoint protein expression as evidenced by elevated levels of ATM and ATR in miR200 KD Medalist +C fibroblasts (Figure 7D). Rescue of this pathway was also supported by reduced presence of phospho-H2AX and cleaved caspase-3 in Medalist +C fibroblasts with a KD of miR200, suggesting a direct role for miR200 in the regulation of DNA damage (Figure 7E). Furthermore, the rescue effects were also detected in Medalist +C iPSCs with a KD of miR200, as shown by reduced immunostaining for phospho-H2AX protein in these cells, as compared to scrambled siRNA treated Medalist +C hiPSCs (Figures 7F and 7G). Quantitative-PCR analysis showed a similar level of hTERT mRNA expression between the different experimental groups, indicating that telomere length is not likely to be altered in the age-matched control, Medalist −C and Medalist +C fibroblasts (Figure 7H). Together, these data support a direct role for miR200 in regulating the DNA damage checkpoint pathway machinery by modulating the expression of DNA damage checkpoint proteins. Thus, in a subset of patients with T1D, such as the Medalist −C, the DNA damage checkpoint pathway is preserved secondary to suppressed miR200 expression in order to prevent apoptosis and to protect against the development of diabetic complications.
Figure 7. Rescue of DNA Damage Checkpoint Proteins by miR200 Knockdown in Human Medalist +C Fibroblasts and iPSCs.
(A and B) Quantitative real-time qPCR analysis to assess combined knockdown (KD) of miR200A, miR200B, and miR200C in (A) iPSCs of Medalist +C patients (N = 6) or (B) Medalist +C fibroblasts (M6, M9, and M12; N = 3).
(C) Quantitation of reprogramming efficiency of human Medalist +C fibroblasts (M6, M9, M12) treated with scrambled siRNA or miR200 siRNA (N = 3).
(D) Immunoblot analysis highlighting the restoration of ATM/ATR proteins in Medalist +C fibroblasts (M6, M9, M12) treated with miR200 siRNA compared to scrambled siRNA (N = 3).
(E) Immunoblot analysis showed decreased expression of pH2AX/cleaved caspase-3 in Medalist +C fibroblasts treated with miR200 siRNA (N = 3).
(F) Immunostaining analysis of pH2AX in miR200 siRNA-treated Medalist +C human iPSCs as compared with scrambled siRNA-treated iPSCs. pH2AX, green; DAPI, blue. Scale bar: 50 µm.
(G) Quantitative analyses of immunostaining of pH2AX in human iPSCs (N = 6) derived from Medalist +C patients and subjected to siRNA-mediated knockdown of miR200 (72 hr).
(H) Quantitative real-time PCR analysis showing the expression of hTERT gene in control (cntrl1, cntrl2), Medalist −C (M4, M10), and Medalist +C fibroblasts (M6, M12) (N = 2).
All data are presented as mean ± SD. *p value < 0.001.
DISCUSSION
Lack of Suitable Cellular or Animal Models Mimicking Human Diabetic Complications
Micro- and macro-vascular diabetic complications are a major cause of morbidity and mortality, highlighting the urgent need for therapeutic intervention (Georgescu, 2011; Mabed and Shahin, 2012). Although several molecular defects have been implicated, and modulators of vascular endothelial growth factor (VEGF), advanced glycation end products (AGE), and its receptor (RAGE) have reached human clinical trials (Brownlee et al., 1988; Harja et al., 2008; Nishikawa et al., 2000; Park et al., 1998), therapeutic success has been limited, due in part to the lack of cellular/animal models that fully mimic the human disease phenotype (Calcutt et al., 2009; Onat et al., 2011). iPSCs from our unique medalist group of patients with longstanding T1D with and without complications (Keenan et al., 2007, 2010) provide a excellent opportunity for potential cell-based therapy and cellular disease modeling, while retaining the genetic make-up of the patients. Unbiased microarray and proteomic profiling of primary fibroblasts and iPSCs, respectively, revealed impairment of key regulators of metabolism, energy homeostasis and mitochondrial function (electron transport chain) in T1D Medalist cells, irrespective of complication status. These data suggest the cells manifest effects secondary to the diabetes in the patients and confirm the usefulness of the iPSCs in modeling the disease (Table S1 A, B, and C). The retention of a majority of the differentially regulated genes or proteins in patient fibroblasts and iPSCs indicates conservation of the genetic make-up of the patients and signifies their suitability for disease modeling.
Preserved DNA Damage Checkpoint Pathway Function in Medalist −C, Secondary to Suppressed miR200 Expression, Protects against Diabetic Complications
An unbiased systems biology approach involving genomics and proteomics analyses to compare Medalist −C and Medalist +C revealed alteration(s) in the DNA damage checkpoint pathway machinery. These “protective factors” were significantly elevated in Medalist −C. Further interrogation revealed an elevated basal expression of miR200 in fibroblasts (miR200 transcript was undetectable in fibroblasts from controls and Medalist −C) and iPSCs derived from Medalist +C, compared to Medalist −C and controls. Strikingly, miR200 KD restored DNA damage checkpoint protein expression in both fibroblasts and iPSCs derived from Medalist +C patients. Indeed, consistent with our findings, previous studies have reported silencing of miR200 expression in fibroblasts under normal conditions due to increased promoter methylation (Vrba et al., 2010). Future studies aimed at comparing gene/protein expression in fibroblasts and iPSCs from these clinical groups may aid in identifying epigenetic regulators that underlie the methylation-based silencing of miR200 in fibroblasts while being derepressed in iPSCs and could be therapeutically important in the context of targeting miR200-regulated pathways (Liu et al., 2013; Price and D’Andrea, 2013; Xu et al., 2012b; Zhou et al., 2011). Furthermore, we found a significant impairment in the activation of MDC1 (serine-793), a protein suggested to be key for the recruitment of DNA damage checkpoint pathway machinery to the damaged site to regulate DNA damage checkpoint pathway function and for insuring normal cell cycle progression (Bennetzen et al., 2010; Branzei and Foiani, 2008; Dephoure et al., 2008; Goldberg et al., 2003; Stewart et al., 2003; Lee and Paull, 2005; Falck et al., 2005). We also observed that topoisomerases, PARPs, and proteasomal components that are implicated in early steps of DNA damage checkpoint pathway damage recognition and repair showed reduced expression in Medalist +C (Lin et al., 2009; Xu et al., 2012a; Furuta et al., 2003). Elevated expression of cleaved caspase-3, concomitant with the DNA damage checkpoint pathway impairment, was observed in multiple target cells (neurons and endothelial cells), with elevation in the presence of high glucose, modeling the manifestations of diabetic complications in Medalist +C and protection in Medalist −C (due to elevated DNA damage checkpoint pathway machinery, despite hyperglycemia) or controls (due to optimal DNA damage checkpoint pathway function and normoglycemia). Corroboration of these novel findings in two cell types affected in T1D (neurons and endothelial cells) and the increased susceptibility of neurons differentiated from Medalist +C iPSCs to genotoxic stress (exposure to high glucose, UV irradiation, or Etoposide), compared to Medalist −C or controls, underscore the clinical relevance of these observations. Further, the confirmation of our findings in both in vitro differentiated cells (e.g., neurons derived from differentiation of patient iPSCs) as well as tissues involved in diabetic complications (e.g., aortic tissue obtained from the patients), and the ability to reverse expression of DNA damage checkpoint pathway proteins upon KD of miR200 in fibroblasts and iPSCs, highlights the significance of this pathway for potential therapeutic targeting. Whether the circulating miR200 in our studies originates from damaged endothelial cells, which are known to exist in diabetic patients and/or other cell types, and could serve as a potential biomarker for diabetic complications (Kato et al., 2013) warrants careful investigation.
Impaired Cellular Differentiation Potential in Medalist +C Underscores Manifestation of Diabetic Complications
Under normal conditions, humans have an inherent capacity for differentiation and self-renewal to make up for the loss of functionally mature cells in a tissue, secondary to normal senescence or stress-induced apoptosis, from a pool of progenitor cells that maintain the levels of functional lineage committed and/or mature cells required for normal function (Nakada et al., 2011; Ruiz et al., 2011). However, in disease states, when this endogenous mechanism fails, complications arise requiring pharmacological intervention, cell-replacement therapy (β cells), or organ transplantation (kidney, heart etc.). Consistently, we observed a severe impairment in the differentiation potential of iPSCs derived from Medalist +C patients, both in vitro and in vivo. Further investigation revealed that Medalist +C cells have reduced levels and activation of ACINUS protein, which is implicated in the early differentiation and lineage commitment in embryonic stem cells, through phosphorylation at serine-1004. In the Medalist +C group, our findings reveal increased inflammation due to macrophage infiltration and increased apoptosis due to elevated DNA damage, resulting in cellular loss, which is not compensated by differentiation of progenitor cells, presumably due to impairment in ACINUS protein function.
Collectively, our findings underscore the relevance of miR200 in regulating the DNA damage checkpoint pathway and cellular capacity of self-renewal and differentiation while its impairment leads to cellular inflammation and apoptosis. We therefore propose the design of new interventions targeting miR200 to protect from the onset and progression of diabetes-associated complications in humans.
EXPERIMENTAL PROCEDURES
Primary Human Fibroblast Derivation and Culture
All studies involving human material were approved by the Institutional Review Board, Joslin Diabetes Center and were in accordance with NIH guidelines. Skin biopsies were obtained from human subjects and sustained in DMEM (10-027, Cellgro Inc.) supplemented with 10% heat-inactivated fetal bovine serum, 1% sodium pyruvate (Life Technologies), and 1% penicillin-streptomycin (Mediatech Inc.), over a period of 4 weeks, in a 6-well plate, with media supplementation every other day. Subsequently, as fibroblasts emerged from the primary explant, a brief trypsinization (0.25% Trypsin) was used to separate and further expand the cells in a 10-cm2 plate. For ligand stimulation studies, primary human fibroblasts were cultured in DMEM supplemented with 1 mM glucose followed by 48-hr treatment with either Vehicle (5 mM glucose) or 25 mM glucose.
Human Studies in the Medalist Patients
Individuals who had documented 50 or more years of insulin use for T1D were invited to participate in a baseline visit. Informed consent was obtained from all subjects prior to participation in the study and after approval by the Joslin Diabetes Center Committee on Human Studies.
Assessment of Complications Status
Nephropathy, retinopathy, and neuropathy status have been defined previously (Keenan et al., 2010). Briefly, diabetic nephropathy was defined by an estimated glomerular filtration rate (CKDeGFR) of <45 ml/min/1.73 m2. A dilated eye examination was performed, and retinopathy status was graded using guidelines from the Early Treatment Diabetic Retinopathy Study (ETDRS). Proliferative diabetic retinopathy (PDR) was defined as an ETDRS ≥ 60. The Michigan Neuropathy Screening Instrument was used to assess neuropathy; scores >2 were considered positive. Cardiovascular disease status was based on self-reported history of coronary artery disease, angina, heart attack, prior cardiac or leg angioplasty, or bypass graft surgery. Coronary artery disease required assessment by a clinician for the presence of angina, heart attack, history of cardiac angioplasty, or bypass graft surgery.
Generation of Human iPSCs by Lentiviral Reprogramming
Human skin fibroblasts were transduced with hSTEMMCA Cre-Excisable Polycitronic lentivirus (EMD Millipore) expressing OCT4, SOX2, KLF4, and cMYC (Sommer et al., 2009), driven by a single promoter, in the presence of 5 µg/ml Polybrene (EMD Millipore). Details are in Supplemental Experimental Procedures.
Embryoid Body Formation Assay
hiPSC colonies were manually scored under a microscope. Cell clumps were briefly spun at 200 g for 2 min at room temperature and resuspended into 4 ml embryoid body (EB) media (hiPSC growth media with 2% FBS, without bFGF supplementation) and seeded onto 12-well untreated ultra-low attachment plates (3737, Corning). Plates were shaken every 12–16 hr to prevent attachment of cells onto the plate surface. Culture media was replaced every 3 days until the EB formation could be observed in approximately 10–14 days.
Animal Housing and In Vivo Differentiation Assays
All animal study protocols were approved by the Institutional Animal Care and Use Committee (IACUC), Joslin Diabetes Center and were in accordance with NIH guidelines. NOD SCID male mice (NOD.CB17-Prkdcscid/NCrCrl, Charles River) were obtained when 4 weeks old, housed in pathogen-free facilities, and maintained on a standard rodent chow diet (2020X, Harlan Tekland Global) with a 12 hr light/12 hr dark cycle at the Animal Care Facilities of Joslin Diabetes Center. Mice were anesthetized with Avertin (240 mg/kg intraperitoneally) before intramuscular injection of human iPSCs (approximately 2 × 106 cells harvested as cell clumps, in a 5:1 mixture with Matrigel [BD Biosciences] and final volume of 150 µl) with a 21G needle. Animals were monitored over the next 4–8 weeks while the teratomas started to emerge/grow, and the size measurements were recorded every week using Vernier calipers.
Lentiviral Overexpression of miR200
For miR200 overexpression, genomic fragments encoding cluster-A (miR200b/200a/429) or cluster-B (miR200c/141) cloned into pLenti 4.1Ex backbone vector were obtained from Add gene plasmid repository, plasmids 35533 and 35534, respectively. Lentivirus production involved five plasmid co-transfection of 293FT cells, involving the lentiviral expression cassette containing the gene of interest (GOI), tat, rev, gag/pol, and vsv-g plasmids (in the ratio 20:1:1:1:2) using calcium phosphate transfection method (Clontech Laboratories Inc.) and LentiX (Clontech Laboratories Inc.)-mediated concentration of viral particles and titering before storage at −80°C in aliquots for long-term use. All virus infections used 5 µg/ml Polybrene reagent (EMD Millipore).
miR200 Expression Analyses in Human Sera, Fibroblasts, and iPSCs
For miR200 expression analyses, total RNA was extracted using either fibroblasts, iPSCs, or serum samples obtained from three clinical groups (Control, Medalist −C, and Medalist +C) using standard TRIzol-based RNA extraction protocols in the presence of RNase inhibitors (Life Technologies) and DNase treatment (QIAGEN). RNA measurements used ND-1000 spectrophotometer V3.5 (NanoDrop Technologies, Inc.). Samples with a 260/280 optical densitometry ratios of 2 were used for further analyses that included cDNA conversion using miRCURY LNA Universal RT microRNA PCR cDNA synthesis kit (Exiqon). A total of 5 ng/µl cDNA was thereafter subjected to a nested quantitative real-time PCR analysis using primers for miR200a, miR200b, and miR200c in equimolar ratios (500 nM each) in a standard 10-µl reaction volume and 384-well format, SYBR Green-based assay (Life Technologies, Roche). Primer sequences can be found in Table S7.
miR200 Knockdown in Human Fibroblasts and hiPSCs
Human fibroblasts from Medalist +C patients were plated as a single-cell suspension into 6-well plates and subjected to siRNA-mediated KD of miR200 using 50 nM siRNA inhibitors from Exiqon for 48 hr. Thereafter, cells were collected and lysed for western blot analysis described in Experimental Procedures. Similarly, iPSCs from Medalist +C patients were plated as single-cell suspension into 24-well plates and subjected to siRNA-mediated KD of miR200 using 20 nM siRNA inhibitors from Exiqon for 72 hr. Thereafter, cells were fixed and subjected to immunostaining analyses, described in Experimental Procedures. In both cases, scrambled siRNA was used as a control.
Whole-Genome Microarray Gene Expression Profiling
Primary human fibroblasts from healthy controls, T1D Medalist −C, and T1D Medalist +C (N = 2, n = 4), cultured as described previously, were subjected to RNA extraction using RNeasy Plus kit (QIAGEN) following manufacturer guidelines. In the present study, “N” represents biological replicates and “n” denotes clonal (in case of iPSCs) or technical replicates. RNA integrity and quality assessment was performed using Agilent 2100 Bioanalyzer (Agilent Technologies) at the Microarray and Genomics core facility at Children’s Hospital, Boston. RNA amplification utilized Ambion Illumina Amplification Kit. A total of 850 ng of amplified RNA was used for subsequent labeling, adaptor ligation, and hybridization onto HumanHT-12 v4.0 Expression BeadChip (Illumina), using manufacturer protocols and recommendations. The BeadChip that consists of more than 47,000 probes can run 12 samples simultaneously and has its content derived from NCBI RefSeq Release 38 (November 7, 2009), as well as legacy UniGene content. Beadchips were scanned using the Illumina BeadArray Reader, and expression data were extracted using the BeadStudio software, using background subtraction. Rescaling was used to eliminate negative values, if and wherever applicable, followed by quantile normalization.
LC-MS/MS Proteomics Analyses
Peptide mixtures were analyzed on a high-resolution, reversed-phase capillary LC system coupled with a Thermo Fisher Scientific LTQ-Orbitrap Velos MS. Due to the limitation of four-plex iTRAQ labeling reagents, samples were processed in two batches (Experiment 1 had a total of 12 samples, N = 2, n = 2 for each clinical group and Experiment 2 had a total of 12 samples, N = 2, n = 2 for each clinical group). Sample preparation and other details are in Supplemental Experimental Procedures.
Proteomics Data Analyses
iTRAQ-labeled peptides were identified based on tandem MS/MS spectra using the Sequest search algorithm against a human protein database (UniprotKB, released 2012-04), and the abundance information across fourplex samples was extracted from the report ion intensities within a given spectra. All peptides were identified with < 0.1% False Discovery Rate by using a MS-Generating Function Score (MS-GF) < 1E-10 and a decoy database searching strategy. The reporter ion intensities for each peptide were summed for all identified spectra for each channel in each biological condition. Relative abundances at peptide level were rolled up to the protein level using the software tool DAnTE (Polpitiya et al., 2008) with the abundances being log2 transformed and normalized by the central tendency approach.
Primary Human Neuronal and Endothelial Cell Culture
Primary human neurons obtained from ScienCell Research Laboratories (Cat# 1520) were sustained in growth factor (Cat# 1562) supplemented media (Cat# 1521) with 10% heat-inactivated fetal bovine serum, as per vendor recommendations. Primary human endothelial cells were obtained from Lonza (Cat# CC-7030 AMP) and maintained in basal growth media supplemented with growth factors (CC-3202 KT). Both cell types maintained normal growth and phenotypic characteristics and were used for ten passages. For ligand stimulation studies, cells were cultured in growth media with 1 mM glucose followed by 48-hr treatment with either Vehicle (5 mM glucose) or 25 mM glucose.
Directed Differentiation of Human iPSCs into Neuronal Lineage
Human iPSCs were cultured under sterile conditions in mTeSR media (Stem- Cell Technologies). Standard tissue culture plates were coated with BD Matrigel hESC-qualified Matrix (BD Biosciences) for 1 hr at room temperature. For passaging and/or directed differentiation, iPSCs were dissociated using Collagenase (StemCell Technologies Inc.), washed with Phosphate-Buffered Saline (pH 7.4) (GIBCO), and replated using mTeSR supplemented with 3 µg/ml Y-27632 Rho Kinase inhibitor (Cat# CD0141; Chemdea) and/or frozen using mFreSR media (StemCell Technologies). rtTA3 and Ngn2 lentiviral constructs for inducing neuronal differentiation of human iPSCs were cloned as described in Supplemental Experimental Procedures and viruses were prepared similar to protocols described in the earlier section for miR200 virus generation. Equal number of manually scored iPSCs from all three clinical groups were then infected with rtTA3 and Ngn2 and induced with 0.5 µg/ml doxycycline (Sigma). Two to four days post-induction, cells were selected with puromycin and hygromycin. Cells with neuronal-like projections that stained positive to neuronal markers such as Neu-N appeared within a week and were further assessed for morphological, phenotypic differences between clinical groups.
Human Tissue Extraction and Protein Expression Analyses in Aortic Bio-Specimens
Human aortic tissue biopsies were obtained from patients with long-standing T1D, snap frozen in liquid nitrogen, and stored at −80°C for long-term use. For protein analyses, the tissue was suspended in tissue protein extraction reagent, T-Per, from Thermo Scientific, in the presence of protease and phosphatase inhibitor cocktail from the same manufacturer. The tissue specimen was thereafter lysed through mechanical shearing using a tissue homogenizer from Omni International and incubated at 4°C for 30 min with vortexing at maximum speed for 30 s after every 10 min. Protein estimation and western analyses were performed as described in Supplemental Experimental Procedures.
Supplementary Material
Highlights.
Impaired reprogramming and differentiation of iPSCs in T1D with complications
Low cell apoptosis due to intact DNA checkpoint pathway in T1D without complications
Poor DNA repair rescued by knockdown of miR200 in T1D patients with complications
Differentiated neurons from T1D with complications exhibit elevated DNA damage
ACKNOWLEDGMENTS
The authors acknowledge the microarray facility, Children’s Hospital, Boston; C. Cahill, Advanced Microscopy Facility; iPS Core Facility, DRC, Joslin Diabetes Center (NIH DK036836); and Therese Rw Clauss, Pacific Northwest National Laboratory. The authors thank G. Daley (Children’s Hospital, Boston) for discussions. This research was supported in part by NIH RO1 DK67536 and NIH RO1 103215 (R.N.K.), UC4 DK104167-01 (W.J.Q., R.N.K.), DP2OD006668 (W.J.Q.), NIH DP3 DK094333-01 (G.L.K.), JDRF 17-2013-310 (H.K.), K99DK090210 and R00DK090210 (C.W.L.), 3-APF-2014-182-A-N (A.E.), JDRF 10-2012-240 (G.W.), and P41 GM103493 (R.D.S.). S.B. is the recipient of postdoctoral research fellowship from Mary K. Iacocca Foundation. Proteomics experiments were performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by DOE/BER and located at Pacific Northwest National Laboratory, which is operated by Battelle Memorial Institute for the DOE under Contract DE-AC05-76RL0 1830. A.J.W. is an Early Career Scientist of the Howard Hughes Medical Institute. Content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or other funding agencies.
Footnotes
ACCESSION NUMBERS
All microarray datasets have been deposited into NCBI Gene Expression Omnibus under accession number GEO: GSE70752. The mass spectrometry datasets have been deposited into the MassIVE repository with accession number MSV000079185. The data were also shared with ProteomeXchange under accession number PXD002504.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, six figures, and seven tables and can be found with this article online at http://dx.doi.org/10.1016/j.cmet.2015.07.015.
AUTHOR CONTRIBUTIONS
S.B. conceived of, designed, and executed the study; performed experiments, data analyses, and interpretation; and wrote the manuscript. M.K.G. assisted with characterization of iPSCs and western blotting studies, performed revision experiments, and edited the manuscript. M.K. assisted with analyses of fibroblasts, assisted in revisions, and edited the manuscript. R.M., T.T., A.E., E.D., I.V., and C.W.L. provided technical assistance with experiments. J.S. assisted with the revisions. G.W. assisted with primary neuron studies. B.K.W. performed analysis of the microarray data. M.A.G., T.R.C., R.D.S., and W.J.Q. contributed to the mass spectrometry-based proteomics experiments. P.G., V.B., R.W., and G.C. contributed to neuronal differentiation experiments. G.L.K. and H.A.K. provided access to human skin and aortic tissues from subjects recruited for the Joslin Medalist Study. A.J.W. provided suggestions on iPSC derivation and critiqued the manuscript. R.N.K. conceived of, designed, and supervised the study and wrote the manuscript. All authors read and agreed to the contents of the manuscript.
REFERENCES
- Bennetzen MV, Larsen DH, Bunkenborg J, Bartek J, Lukas J, Andersen JS. Site-specific phosphorylation dynamics of the nuclear proteome during the DNA damage response. Mol. Cell. Proteomics. 2010;9:1314–1323. doi: 10.1074/mcp.M900616-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature. 2001;414:813–820. doi: 10.1038/414813a. [DOI] [PubMed] [Google Scholar]
- Brownlee M. The pathobiology of diabetic complications: a unifying mechanism. Diabetes. 2005;54:1615–1625. doi: 10.2337/diabetes.54.6.1615. [DOI] [PubMed] [Google Scholar]
- Brownlee M, Cerami A, Vlassara H. Advanced glycosylation end products in tissue and the biochemical basis of diabetic complications. N. Engl. J. Med. 1988;318:1315–1321. doi: 10.1056/NEJM198805193182007. [DOI] [PubMed] [Google Scholar]
- Calcutt NA, Cooper ME, Kern TS, Schmidt AM. Therapies for hyperglycaemia-induced diabetic complications: from animal models to clinical trials. Nat. Rev. Drug Discov. 2009;8:417–429. doi: 10.1038/nrd2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamorro-Jorganes A, Araldi E, Suárez Y. MicroRNAs as pharmacological targets in endothelial cell function and dysfunction. Pharmacol. Res. 2013;75:15–27. doi: 10.1016/j.phrs.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang-Chen KJ, Mullur R, Bernal-Mizrachi E. Beta-cell failure as a complication of diabetes. Rev. Endocr. Metab. Disord. 2008;9:329–343. doi: 10.1007/s11154-008-9101-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villén J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. USA. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein DM. Special delivery: microRNA-200-containing extracellular vesicles provide metastatic message to distal tumor cells. J. Clin. Invest. 2014;124:5107–5108. doi: 10.1172/JCI79191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- Fang ZY, Prins JB, Marwick TH. Diabetic cardiomyopathy: evidence, mechanisms, and therapeutic implications. Endocr. Rev. 2004;25:543–567. doi: 10.1210/er.2003-0012. [DOI] [PubMed] [Google Scholar]
- Forbes JM, Cooper ME. Mechanisms of diabetic complications. Physiol. Rev. 2013;93:137–188. doi: 10.1152/physrev.00045.2011. [DOI] [PubMed] [Google Scholar]
- Furuta T, Takemura H, Liao ZY, Aune GJ, Redon C, Sedelnikova OA, Pilch DR, Rogakou EP, Celeste A, Chen HT, et al. Phosphorylation of histone H2AX and activation of Mre11, Rad50, and Nbs1 in response to replication-dependent DNA double-strand breaks induced by mammalian DNA topoisomerase I cleavage complexes. J. Biol. Chem. 2003;278:20303–20312. doi: 10.1074/jbc.M300198200. [DOI] [PubMed] [Google Scholar]
- Georgescu A. Vascular dysfunction in diabetes: The endothelial progenitor cells as new therapeutic strategy. World J. Diabetes. 2011;2:92–97. doi: 10.4239/wjd.v2.i6.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg M, Stucki M, Falck J, D’Amours D, Rahman D, Pappin D, Bartek J, Jackson SP. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature. 2003;421:952–956. doi: 10.1038/nature01445. [DOI] [PubMed] [Google Scholar]
- Harja E, Bu DX, Hudson BI, Chang JS, Shen X, Hallam K, Kalea AZ, Lu Y, Rosario RH, Oruganti S, et al. Vascular and inflammatory stresses mediate atherosclerosis via RAGE and its ligands in apoE−/ − mice. J. Clin. Invest. 2008;118:183–194. doi: 10.1172/JCI32703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, Xiao T, Schafer J, Lee ML, Schmittgen TD, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Castro NE, Natarajan R. MicroRNAs: potential mediators and biomarkers of diabetic complications. Free Radic. Biol. Med. 2013;64:85–94. doi: 10.1016/j.freeradbiomed.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan HA, Costacou T, Sun JK, Doria A, Cavellerano J, Coney J, Orchard TJ, Aiello LP, King GL. Clinical factors associated with resistance to microvascular complications in diabetic patients of extreme disease duration: the 50-year medalist study. Diabetes Care. 2007;30:1995–1997. doi: 10.2337/dc06-2222. [DOI] [PubMed] [Google Scholar]
- Keenan HA, Sun JK, Levine J, Doria A, Aiello LP, Eisenbarth G, Bonner-Weir S, King GL. Residual insulin production and pancreatic β-cell turnover after 50 years of diabetes: Joslin Medalist Study. Diabetes. 2010;59:2846–2853. doi: 10.2337/db10-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong XY, Du YQ, Li L, Liu JQ, Wang GK, Zhu JQ, Man XH, Gong YF, Xiao LN, Zheng YZ, et al. Plasma miR-216a as a potential marker of pancreatic injury in a rat model of acute pancreatitis. World J. Gastroenterol. 2010;16:4599–4604. doi: 10.3748/wjg.v16.i36.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacolley P, Challande P, Osborne-Pellegrin M, Regnault V. Genetics and pathophysiology of arterial stiffness. Cardiovasc. Res. 2009;81:637–648. doi: 10.1093/cvr/cvn353. [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- Lin CP, Ban Y, Lyu YL, Liu LF. Proteasome-dependent processing of topoisomerase I-DNA adducts into DNA double strand breaks at arrested replication forks. J. Biol. Chem. 2009;284:28084–28092. doi: 10.1074/jbc.M109.030601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Kim J, Oberdoerffer P. Metabolic modulation of chromatin: implications for DNA repair and genomic integrity. Front. Genet. 2013;4:182. doi: 10.3389/fgene.2013.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabed M, Shahin M. Mesenchymal stem cell-based therapy for the treatment of type 1 diabetes mellitus. Curr. Stem Cell Res. Ther. 2012;7:179–190. doi: 10.2174/157488812799859829. [DOI] [PubMed] [Google Scholar]
- Madonna R, Görbe A, Ferdinandy P, De Caterina R. Glucose metabolism, hyperosmotic stress, and reprogramming of somatic cells. Mol. Biotechnol. 2013;55:169–178. doi: 10.1007/s12033-013-9668-2. [DOI] [PubMed] [Google Scholar]
- Maehr R, Chen S, Snitow M, Ludwig T, Yagasaki L, Goland R, Leibel RL, Melton DA. Generation of pluripotent stem cells from patients with type 1 diabetes. Proc. Natl. Acad. Sci. USA. 2009;106:15768–15773. doi: 10.1073/pnas.0906894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada D, Levi BP, Morrison SJ. Integrating physiological regulation with stem cell and tissue homeostasis. Neuron. 2011;70:703–718. doi: 10.1016/j.neuron.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa T, Edelstein D, Du XL, Yamagishi S, Matsumura T, Kaneda Y, Yorek MA, Beebe D, Oates PJ, Hammes HP, et al. Normalizing mitochondrial superoxide production blocks three pathways of hyperglycaemic damage. Nature. 2000;404:787–790. doi: 10.1038/35008121. [DOI] [PubMed] [Google Scholar]
- Onat D, Brillon D, Colombo PC, Schmidt AM. Human vascular endothelial cells: a model system for studying vascular inflammation in diabetes and atherosclerosis. Curr. Diab. Rep. 2011;11:193–202. doi: 10.1007/s11892-011-0182-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park L, Raman KG, Lee KJ, Lu Y, Ferran LJ, Jr, Chow WS, Stern D, Schmidt AM. Suppression of accelerated diabetic atherosclerosis by the soluble receptor for advanced glycation end products. Nat. Med. 1998;4:1025–1031. doi: 10.1038/2012. [DOI] [PubMed] [Google Scholar]
- Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polpitiya AD, Qian WJ, Jaitly N, Petyuk VA, Adkins JN, Camp DG, 2nd, Anderson GA, Smith RD. DAnTE: a statistical tool for quantitative analysis of -omics data. Bioinformatics. 2008;24:1556–1558. doi: 10.1093/bioinformatics/btn217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price BD, D’Andrea AD. Chromatin remodeling at DNA double-strand breaks. Cell. 2013;152:1344–1354. doi: 10.1016/j.cell.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rask-Madsen C, King GL. Vascular complications of diabetes: mechanisms of injury and protective factors. Cell Metab. 2013;17:20–33. doi: 10.1016/j.cmet.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy MA, Natarajan R. Epigenetic mechanisms in diabetic vascular complications. Cardiovasc. Res. 2011;90:421–429. doi: 10.1093/cvr/cvr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy MA, Jin W, Villeneuve L, Wang M, Lanting L, Todorov I, Kato M, Natarajan R. Pro-inflammatory role of microrna-200 in vascular smooth muscle cells from diabetic mice. Arterioscler. Thromb. Vasc. Biol. 2012;32:721–729. doi: 10.1161/ATVBAHA.111.241109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigbolt KT, Prokhorova TA, Akimov V, Henningsen J, Johansen PT, Kratchmarova I, Kassem M, Mann M, Olsen JV, Blagoev B. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci. Signal. 2011;4:rs3. doi: 10.1126/scisignal.2001570. [DOI] [PubMed] [Google Scholar]
- Ruiz S, Panopoulos AD, Herrerías A, Bissig KD, Lutz M, Berggren WT, Verma IM, Izpisua Belmonte JC. A high proliferation rate is required for cell reprogramming and maintenance of human embryonic stem cell identity. Curr. Biol. 2011;21:45–52. doi: 10.1016/j.cub.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalkwijk CG, Stehouwer CD. Vascular complications in diabetes mellitus: the role of endothelial dysfunction. Clin. Sci. 2005;109:143–159. doi: 10.1042/CS20050025. [DOI] [PubMed] [Google Scholar]
- Sommer CA, Stadtfeld M, Murphy GJ, Hochedlinger K, Kotton DN, Mostoslavsky G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JY, Lim JW, Kim H, Kim KH. Role of NF-kappaB and DNA repair protein Ku on apoptosis in pancreatic acinar cells. Ann. N Y Acad. Sci. 2003;1010:259–263. doi: 10.1196/annals.1299.044. [DOI] [PubMed] [Google Scholar]
- Spycher C, Miller ES, Townsend K, Pavic L, Morrice NA, Janscak P, Stewart GS, Stucki M. Constitutive phosphorylation of MDC1 physically links the MRE11-RAD50-NBS1 complex to damaged chromatin. J. Cell Biol. 2008;181:227–240. doi: 10.1083/jcb.200709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- Tiscornia G, Vivas EL, Izpisúa Belmonte JC. Diseases in a dish: modeling human genetic disorders using induced pluripotent cells. Nat. Med. 2011;17:1570–1576. doi: 10.1038/nm.2504. [DOI] [PubMed] [Google Scholar]
- Van Hoof D, Muñoz J, Braam SR, Pinkse MW, Linding R, Heck AJ, Mummery CL, Krijgsveld J. Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell. 2009;5:214–226. doi: 10.1016/j.stem.2009.05.021. [DOI] [PubMed] [Google Scholar]
- Vrba L, Jensen TJ, Garbe JC, Heimark RL, Cress AE, Dickinson S, Stampfer MR, Futscher BW. Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PLoS ONE. 2010;5:e8697. doi: 10.1371/journal.pone.0008697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Xu Y, Gursoy-Yuzugullu O, Price BD. The histone variant macroH2A1.1 is recruited to DSBs through a mechanism involving PARP1. FEBS Lett. 2012a;586:3920–3925. doi: 10.1016/j.febslet.2012.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Ayrapetov MK, Xu C, Gursoy-Yuzugullu O, Hu Y, Price BD. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol. Cell. 2012b;48:723–733. doi: 10.1016/j.molcel.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Bellenguez C, Spencer CC, Bennett AJ, Coleman RL, Tavendale R, Hawley SA, Donnelly LA, Schofield C, Groves CJ, et al. GoDARTS and UKPDS Diabetes Pharmacogenetics Study Group; Wellcome Trust Case Control Consortium 2; MAGIC investigators. Common variants near ATM are associated with glycemic response to metformin in type 2 diabetes. Nat. Genet. 2011;43:117–120. doi: 10.1038/ng.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.